Significance
Most studies of proteolysis by the ubiquitin-proteasome pathway have focused on the regulation by ubiquitination. However, we showed that pharmacological agents that raise cAMP and activate protein kinase A by phosphorylating a proteasome subunit enhance proteasome activity and the cell’s capacity to selectively degrade misfolded and regulatory proteins. We investigated whether similar adaptations occur in physiological conditions where cAMP rises. Proteasome activity increases by this mechanism in human muscles following intense exercise, in mouse muscles and liver after a brief fast, in hepatocytes after epinephrine or glucagon, and renal collecting duct cells within 5 minutes of antidiuretic hormone. Thus, hormones and conditions that raise cAMP rapidly enhance proteasome activity and the cells’ capacity to eliminate damaged and preexistent regulatory proteins.
Keywords: proteasome phosphorylation, hormones, ubiquitin proteasome system, protein degradation, cAMP
Abstract
Pharmacological agents that raise cAMP and activate protein kinase A (PKA) stimulate 26S proteasome activity, phosphorylation of subunit Rpn6, and intracellular degradation of misfolded proteins. We investigated whether a similar proteasome activation occurs in response to hormones and under various physiological conditions that raise cAMP. Treatment of mouse hepatocytes with glucagon, epinephrine, or forskolin stimulated Rpn6 phosphorylation and the 26S proteasomes’ capacity to degrade ubiquitinated proteins and peptides. These agents promoted the selective degradation of short-lived proteins, which are misfolded and regulatory proteins, but not the bulk of cell proteins or lysosomal proteolysis. Proteasome activities and Rpn6 phosphorylation increased similarly in working hearts upon epinephrine treatment, in skeletal muscles of exercising humans, and in electrically stimulated rat muscles. In WT mouse kidney cells, but not in cells lacking PKA, treatment with antidiuretic hormone (vasopressin) stimulated within 5-minutes proteasomal activity, Rpn6 phosphorylation, and the selective degradation of short-lived cell proteins. In livers and muscles of mice fasted for 12–48 hours cAMP levels, Rpn6 phosphorylation, and proteasomal activities increased without any change in proteasomal content. Thus, in vivo cAMP-PKA–mediated proteasome activation is a common cellular response to diverse endocrine stimuli and rapidly enhances the capacity of target tissues to degrade regulatory and misfolded proteins (e.g., proteins damaged upon exercise). The increased destruction of preexistent regulatory proteins may help cells adapt their protein composition to new physiological conditions.
It is widely assumed that rates of protein degradation by the ubiquitin proteasome system (UPS) are determined only by their rates of ubiquitination, and that 26S proteasomes efficiently degrade ubiquitinated proteins. However, there is growing evidence that protein half-lives can also be altered through changes in proteasome activity under different physiological and pathological conditions (1, 2). Recent studies have demonstrated that proteasome activity in mammalian cells can be increased upon subunit phosphorylation by protein kinases (2, 3), including protein kinase A (PKA) (4, 5), DYRK2 (6, 7), CaMKII (8), and PKG (9, 10). Pharmacological treatments that raise cAMP by stimulating adenylate cyclases (e.g., forskolin) or by inhibiting phosphodiesterase 4 (PDE4) (e.g., rolipram) enhance the 26S proteasome’s ability to hydrolyze ubiquitinated proteins, ATP, and short peptide substrates (4). This activation results from phosphorylation by PKA of the 19S subunit Rpn6 on Serine 14 and leads to a rapid increase in the cell’s ability to hydrolyze short-lived cell proteins, which includes misfolded and regulatory proteins, but not the bulk of cell proteins, which are long-lived components (4). These effects of cAMP and PKA have clear therapeutic promise because pharmacologic agents that raise cAMP levels enhance the capacity of cultured cells (4) and mouse brains (5) to degrade various misfolded, aggregation-prone proteins that cause major neurodegenerative diseases, including mutant forms of tau, which cause Alzheimer’s disease or frontotemporal dementia, and mutant forms of SOD1, FUS, and TDP43, which cause amyotrophic lateral sclerosis. The enhanced degradation of such proteins can reduce their accumulation in cells and prevent their toxic effects (4, 5).
Because cAMP and PKA mediate the actions of many hormones and neurotransmitters and regulate diverse cellular responses, we have examined whether proteasome activation and enhanced degradation by the UPS also accompany hormonal responses and physiological conditions that raise cAMP, such as exercise and fasting. These studies demonstrate that a variety of hormones that activate adenylate cyclases, but have diverse physiological roles, ranging from energy mobilization (e.g., glucagon and epinephrine in liver), to increasing cardiac output (epinephrine), to water reabsorption in kidney (antidiuretic hormone), all stimulate proteasome activity and the selective degradation of short-lived cell proteins. Thus, rapid increases and decreases in the proteasomes’ degradative capacity through subunit phosphorylation must be occurring frequently in different tissues in vivo, and presumably the enhanced degradation of misfolded and damaged proteins helps cells maintain proteolysis during stress (e.g., with exercise), while the accelerated breakdown of preexistent regulatory proteins facilitates changes in cell protein composition upon transitions to new physiological conditions (e.g., with fasting).
Results
Glucagon and Epinephrine Enhance Proteasome Activities, Rpn6 Phosphorylation, and Proteolysis in Hepatocytes.
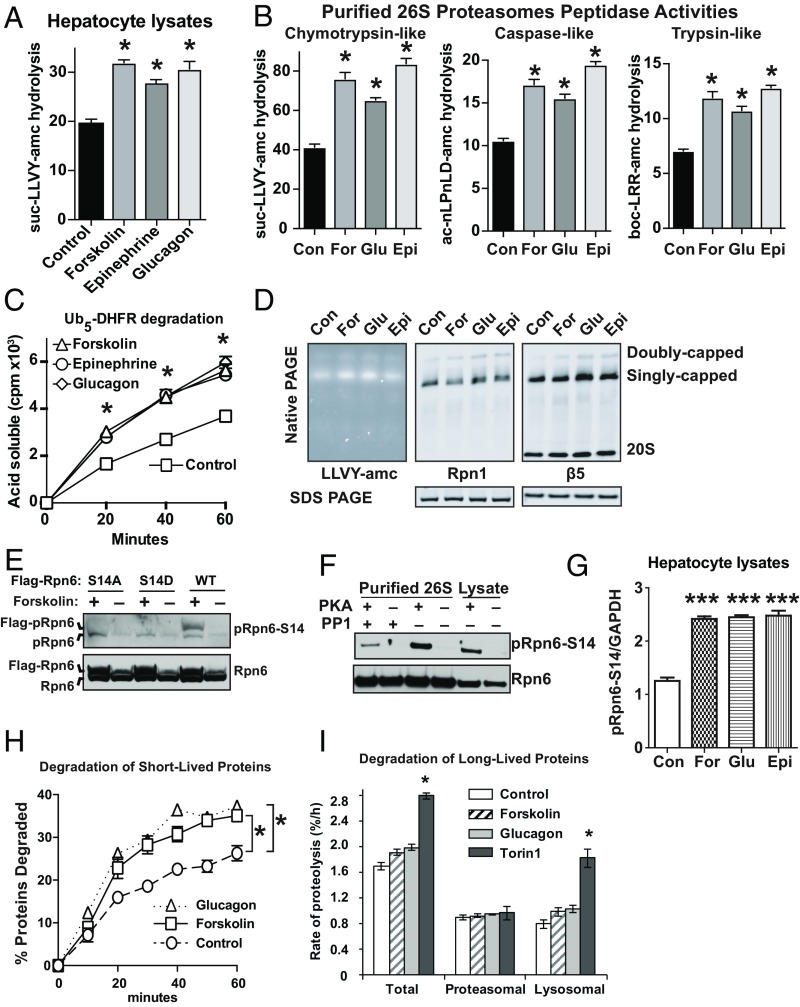
Much of our knowledge of cAMP’s role in signal transduction was first discovered in studies to understand how epinephrine stimulates glycogen breakdown in liver and muscle, which led to the discovery that PKA regulates glycogen phosphorylase (11). cAMP and PKA also mediate the stimulation of hepatic glycogenolysis by glucagon when blood glucose falls (e.g., in fasting). Therefore, we tested in hepatocytes whether epinephrine and glucagon, both of which bind to distinct receptors on hepatocytes and activate adenylate cyclases, also cause proteasome phosphorylation and activation. Mouse primary hepatocytes were incubated for 1 h with these hormones or forskolin, a natural product which directly stimulates adenylate cyclases. The 26S proteasomes’ chymotrypsin-like peptidase activity was then measured in the cell lysates using standard fluorogenic substrates (12). Treatment with forskolin, epinephrine, or glucagon all caused a similar increase in proteasomal peptidase activity (Fig. 1A).
Fig. 1.
Raising cAMP levels with glucagon, epinephrine, or forskolin, enhances 26S proteasome activities and the degradation of short-lived proteins in mouse primary hepatocytes. (A) Mouse primary hepatocytes were incubated with the vehicle (DMSO) control, forskolin (10 μM), epinephrine (1 μg/mL), or glucagon (1 µg/mL) for 1 h and the 26S proteasome’s chymotrypsin-like peptidase activity was measured in cell extracts by following the hydrolysis of suc-LLVY-amc. n = 4, *P < 0.05. In this and subsequent figures, error bars represent the means ± SEM. (B) Raising cAMP levels enhances the peptidase activities of 26S proteasomes purified from mouse primary hepatocytes treated as in A. Proteasomes were affinity-purified by the Ubl-method. The chymotrypsin-like activity was assayed with suc-LLVY-amc, the caspase-like with ac-nLPnLD-amc, and the trypsin-like with boc-LRR-amc. n = 4, *P < 0.05. (C) Degradation of Ub5-DHFR by 26S proteasomes, purified by the Ubl-method from mouse hepatocytes treated as in A. The rates of degradation were measured by following the conversion of radiolabeled protein to TCA-soluble labeled material. n = 4, *P < 0.05. (D) Glucagon and epinephrine increase slightly the amount of assembled 26S proteasomes, both doubly capped and singly capped, in lysates of the mouse primary hepatocytes without increasing proteasome subunit levels. Native PAGE of cell extracts followed by suc-LLVY-amc overlay assay or Western analysis for a 19S subunit (Rpn1) or a 20S subunit (β5). The experiment was performed twice with three samples per condition. (E) Validation of the specificity of the phospho-specific Rpn6-S14 antibody. Plasmids encoding Rpn6-WT, -S14D, and -S14A were transfected into HEK293 cells and after 48 h the cells were treated with forskolin (10 μM) for 5 h and Western blot analysis was performed. There was greater expression of the plasmid-encoded Rpn6 variants, as detected by Western blot for Rpn6, because the CMV promoter on these plasmids contains a cAMP-responsive element, which was induced by the elevation of cAMP by forskolin. (F) Overexpression of constitutively active PKA promotes the phosphorylation of Rpn6-S14 in HEK293 cells. Western blot was performed both on the cell lysates and on 26S proteasomes purified from HEK293 cells or HEK293 cells overexpressing PKA. After 26S proteasomes were purified from PKA-overexpressing cells, they were incubated with PP1. (G) Forskolin, epinephrine, or glucagon increase similarly phosphorylation of Rpn6-S14 in primary mouse hepatocytes. Error bars are SEM for three cells per condition. One-way ANOVA with Bonferroni post hoc against the DMSO control condition. ***P ≤ 0.001. (H) Degradation rates of short-lived proteins increased similarly after treatment with glucagon or forskolin. To follow degradation of short-lived proteins, mouse primary hepatocytes were incubated with [3H]phenylalanine for 10 min and then washed three times with chase medium containing 150 µg/mL cycloheximide and 2 mM nonradioactive phenylalanine. The cells were then resuspended in chase media containing either vehicle control (DMSO), forskolin (10 μM), or glucagon (1 µg/mL), and media samples were collected at the indicated times. The radioactivity released from cell proteins was measured and plotted as a percentage of the total radioactivity incorporated into proteins at time 0. Error bars represent the SEM of four independent samples. (I) PKA activation by forskolin or glucagon does not enhance the degradation of long-lived proteins in hepatocytes. Mouse primary hepatocytes were incubated with [3H]phenylalanine (2 μCi/mL) for 20 h to label cell proteins and then switched to chase medium containing 2 mM nonradioactive phenylalanine for 2 h (15). To dissect the relative rates of lysosomal and proteasomal degradation, cells were pretreated with concanamycin A (100 nM) for 1 h, and then with forskolin, glucagon, or Torin1 for 1 h before collecting medium samples and calculating the rate of proteolysis. The concanamycin A-sensitive portion of total proteolysis was considered lysosomal proteolysis and the concanamycin A-resistant portion of total proteolysis was used as proteasomal proteolysis, as validated previously (15). *P < 0.05.
To further evaluate proteasome activation, 26S proteasomes were gently purified using the UBL domain as the affinity ligand (13), and their activities assayed. The proteasomes purified from the epinephrine-, glucagon-, or forskolin-treated cells all showed greater chymotrypsin-like, caspase-like, and trypsin-like activities than those from control cells (Fig. 1B). Furthermore, the 26S particles from the treated cells also had a greater capacity to degrade their physiological substrates, ubiquitinated proteins, as assayed using 32P-labeled Ub5 dihydrofolate reductase (DHFR) (Fig. 1C). Our prior studies (4) had noted that the activation of proteasomes by cAMP and PKA was accompanied by a modest increase in doubly capped 26S proteasomes. In the hepatocytes, treatment for 1 h with glucagon or epinephrine increased the amount and activity of the singly capped particles (Fig. 1D), which were by far the predominant form in these cells, and the amount of the doubly capped species seemed to increase slightly (Fig. 1D). However, it remains unclear if these increases in either species cause the rapid rise in proteasome activity or are an associated consequence of Rpn6 phosphorylation, which may stabilize the interactions between the 19S and the 20S particles.
To determine if Rpn6 was phosphorylated by PKA under these conditions, we generated a polyclonal antibody specific for phosphorylated-Rpn6-S14 using a phosphopeptide as the immunogen. This antibody detected by Western blot phosphorylation of Serine14 of endogenous Rpn6 and overexpressed Rpn6 in HEK293 cells, and this band was much more prominent after forskolin treatment (Fig. 1E). The antibody did not detect nonphosphorylated Rpn6, nor phosphomimetic (Rpn6-S14D) or phospho-dead (Rpn6-S14A) mutants (4) when overexpressed in HEK293 cells (Fig. 1E), and reacted stronger with 26S proteasomes purified from HEK293 cells overexpressing the catalytic subunit of PKA than with those from control cells (Fig. 1F). The increased signal could be reduced by incubating the proteasomes with protein phosphatase 1 (PP1) (Fig. 1F). Using this phospho-Rpn6-S14–specific antibody (Fig. 1 E and F), we found that treatment of hepatocytes with forskolin, epinephrine, or glucagon increased similarly the phosphorylation of Rpn6-S14 (Fig. 1G and SI Appendix, Fig. S1A).
We previously found that raising cAMP levels with forskolin in HEK293 cells and mouse myotubes promoted the degradation of short-lived cell proteins, but surprisingly not long-lived ones (4). To determine if raising cAMP with glucagon has similar selective effects on the breakdown of short-lived proteins in hepatocytes, we measured the rates of degradation of this class of proteins by pulse-chase methods, as described previously (4). Proteins in the primary hepatocytes were labeled for 10 min with [3H]phenylalanine and then chased with a large excess of nonradioactive phenylalanine. The conversion of prelabeled cell proteins into amino acids (TCA-soluble material in the medium) was then assayed in the presence of glucagon or forskolin. Glucagon and forskolin stimulated similarly the degradation of these short-lived proteins (Fig. 1H), which occurs exclusively by the ubiquitin proteasome pathway. Thus, it is very likely that the increased proteasome activity and the more rapid degradation of short-lived proteins were due to the rise in cAMP, the activation of PKA, and the phosphorylation of Rpn6. These observations also predict that in vivo upon fasting, when hepatic glycogen breakdown is stimulated via cAMP by glucagon, or in exercise by epinephrine, proteasomes are activated (as tested below), and the hepatocytes’ capacity to degrade misfolded and regulatory proteins will also increase.
To test for possible effects of glucagon on the degradation of long-lived proteins, hepatocytes were incubated with [3H]phenylalanine for 20 h and then with chase medium for 2 h to allow turnover of short-lived components, followed by exposure to glucagon or forskolin for 2 h. Neither agent significantly increased the degradation of the long-lived proteins (Fig. 1I). The exact contributions of proteasomes and lysosomes to their breakdown were also measured using selective inhibitors of each process, as previously described (14, 15). The treatment with forskolin or glucagon did not enhance degradation of these proteins by proteasomes, despite the increased proteasome activity (Fig. 1 A–C), and also did not significantly increase lysosomal proteolysis (Fig. 1I). Accordingly, there was no change in levels of lipidated LC3 or p62 (SI Appendix, Fig. S1B). In contrast, inhibition of mammalian target of rapamycin (mTOR) with Torin1, which stimulates autophagy (15), caused a large increase in lysosomal proteolysis in these cells (Fig. 1I).
This failure of glucagon or forskolin to stimulate autophagy and lysosomal degradation of long-lived proteins in hepatocytes was a surprising finding because some of the earliest descriptions of autophagy reported that glucagon enhances this process in liver (16). However, in more recent studies, cAMP has been found to inhibit autophagy in certain cells (17). To clarify the effects of raising cAMP on autophagy in hepatocytes and other cells, we used Torin1 to activate autophagy with or without forskolin to increase cAMP levels. Together, these two treatments should mimic the changes in these signaling pathways seen in vivo in fasting. In the hepatocytes, these combined treatments did not further increase rates of proteolysis beyond what was seen with Torin1 alone (SI Appendix, Fig. S2A), and in mouse C2C12 myotubes (SI Appendix, Fig. S2B), the increase in cAMP caused a small but reproducible inhibition in the autophagic degradation induced by Torin1. Interestingly, in HEK293 cells, raising cAMP caused a large inhibition of the lysosomal degradation induced by Torin1 (SI Appendix, Fig. S2C). Because raising cAMP in none of these cells stimulated autophagy, and in some inhibited this process, these observations emphasize that epinephrine and glucagon via cAMP selectively promote the breakdown of short-lived proteins by the UPS, not the bulk of cell proteins, which are degraded by both autophagy and the UPS, especially in fasting and atrophying muscles (18) (see below).
Epinephrine Enhances Proteasome Activities in Working Hearts by Stimulating Proteasome Phosphorylation.
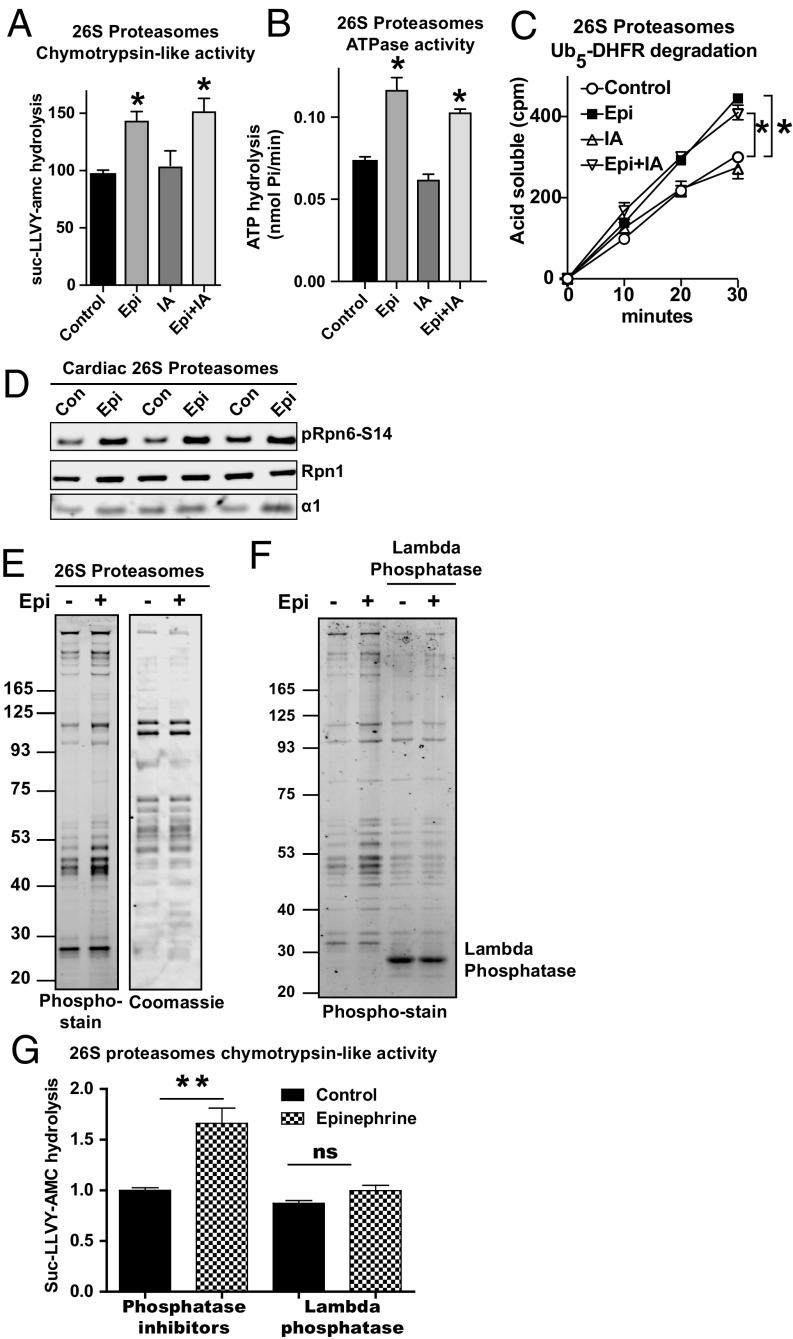
To test if epinephrine also activates proteasomes in other tissues, we examined hearts in which catecholamines via cAMP stimulate cardiac output, as well as glycogen and triglyceride breakdown. Using perfused working rat hearts (19), we examined proteasome function and phosphorylation in three different conditions that enhanced cardiac workload: exposure to epinephrine, increased afterload (i.e., increased aortic pressure), and epinephrine plus afterload (SI Appendix, Fig. S3 A and B). After perfusion for 45 min, the contracting hearts were frozen and then lysed as previously described (19). The 26S proteasomes affinity-purified by the Ubl-method from hearts exposed to epinephrine with or without increased afterload exhibited greater proteasomal chymotrypsin-like peptidase activity than control hearts (Fig. 2A). These effects, which were associated with increased cardiac power output (SI Appendix, Fig. S3A), were due to the exposure to epinephrine and not to the change in contractile work because proteasomal peptide hydrolysis did not change after the hearts pumped against greater pressure in the absence of epinephrine (Fig. 2A), and epinephrine combined with increased afterload resulted in a similar activation as epinephrine alone (Fig. 2A). After epinephrine treatment of the hearts, but not after working against increased pressures, the cardiac 26S proteasomes also exhibited a greater capacity to hydrolyze ATP (Fig. 2B) and the polyubiquitinated protein Ub5-DHFR (Fig. 2C). This enhanced rate of hydrolysis of ubiquitin conjugates, ATP, and small peptides resembles our previous findings obtained by using pharmacological agents to raise cAMP and activate PKA in HEK293 cells and mouse myotubes (4).
Fig. 2.
Epinephrine stimulates 26S proteasome activities in isolated working rat hearts. (A) Epinephrine enhances the peptidase activity of 26S proteasomes purified by the Ubl-method from perfused rat hearts. Working hearts were treated with epinephrine or subjected to increased afterload (IA) or both. Chymotrypsin-like activity of the 26S proteasomes was measured with suc-LLVY-amc. n = 3, *P < 0.05. Error bars represent mean ± SEM. Cardiac work load (power and atrial pressure during perfusion are shown in the SI Appendix, Fig. S3 A and B). (B) Epinephrine enhances ATP hydrolysis by 26S proteasomes purified from rat hearts treated as in A. Basal ATPase activity was measured by following the production of free phosphate using the malachite green. n = 3. *P < 0.05. (C) Epinephrine stimulates degradation by 26S proteasomes of Ub5-DHFR. Rates of degradation were measured by following the conversion of radiolabeled protein to TCA-soluble labeled peptides. n = 3. *P < 0.05. (D) Epinephrine treatment increased phosphorylation of Rpn6-S14 in 26S proteasomes purified from perfused hearts treated as in A. 26S proteasomes were purified from three hearts per condition. (E) 26S proteasomes purified from rat hearts treated with epinephrine exhibited more phosphorylated bands than from control hearts. Proteasomes were run on SDS/PAGE and stained with ProQ Diamond phospho-stain followed by Coomassie blue. The experiment was performed with three hearts per condition, which gave similar results. Representative gels are shown. (F) The 26S proteasomes from rat hearts were incubated with λ-phosphatase or phosphatase inhibitors. ProQ diamond phospho-stain was used to confirm the phosphorylation status of the proteasomes. (G) Dephosphorylation of 26S proteasomes from rat hearts after treatment with epinephrine reversed the increase in proteasome activity.
The lysates from epinephrine-treated hearts, but not those exposed to increased afterload, showed a large increase in PKA-mediated phosphorylation of target proteins by Western blot analysis (SI Appendix, Fig. S3C). Accordingly, only the 26S proteasomes purified from hearts exposed to epinephrine had increased phosphorylation of Rpn6-S14 (Fig. 2D and SI Appendix, Fig. S3D). Further analysis of these 26S proteasomes by SDS/PAGE and a phospho-protein gel stain that can detect phosphorylation of all amino acids showed multiple bands that seemed to also be more phosphorylated than control (Fig. 2E). To test if this increased peptidase activity was due to the greater phosphorylation, these preparations were incubated with λ-phosphatase, which decreased the amounts of phosphorylated proteins to control levels and reversed the epinephrine-mediated increase in proteasomal chymotrypsin-like activity (Fig. 2 F and G). Thus, epinephrine activates 26S proteasomes by stimulating their phosphorylation, including the phosphorylation of Rpn6-S14. The possible significance of the other phosphorylated proteins (which may be proteasome subunits or associated proteins), remains unclear, but the modification of Rpn6-S14 correlated consistently with the enhanced proteasome activity in these various studies.
Vasopressin via PKA Enhances Proteasomal Activity and Selective Breakdown of Short-Lived Proteins in Renal Collecting Duct Cells.
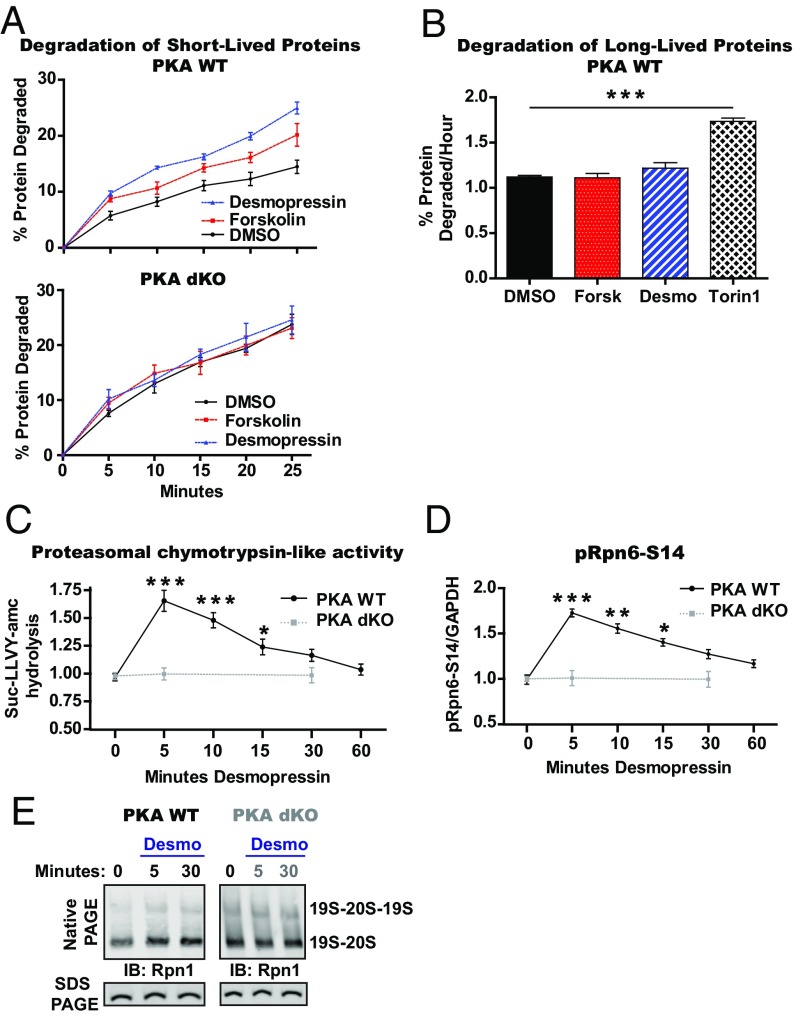
To test if diverse hormones that signal through cAMP and PKA also activate proteasomes and protein degradation, it seemed important to analyze a hormone that has fewer metabolic consequences than epinephrine or glucagon. Vasopressin, also called antidiuretic hormone (ADH), is a peptide hormone that through cAMP and PKA stimulates water reabsorption in the kidneys by enhancing water permeability of the renal collecting duct. We tested if the rise in cAMP caused by the binding of desmopressin, a stable analog of vasopressin, to receptors on immortalized mouse kidney collecting duct cells (mpkCCD) also stimulates proteasomal degradation. Desmopressin and forskolin increased the degradation of short-lived proteins in mpkCCD cells above levels in control cells, as measured by the conversion of [3H]phenylalanine-labeled cell proteins into TCA-soluble amino acids (Fig. 3A). In contrast, desmopressin and forskolin did not alter the rate of degradation of the bulk of cell proteins, which are long-lived components (Fig. 3B).
Fig. 3.
Vasopressin enhances proteasomal activity and intracellular proteolysis of only short-lived proteins in mouse kidney collecting duct cells. (A) Desmopressin, like forskolin, stimulates the degradation of short-lived proteins in a PKA-dependent manner in mpkCCD cells. PKA WT and PKA dKO mpkCCD cells (20) were pulsed with [3H]phenylalanine (2.5 μCi/mL) for 20 min and then washed twice with chase medium containing 150 µg/mL cycloheximide and 2 mM nonradioactive phenylalanine. The cells were then resuspended in chase media containing either DMSO, forskolin (5 μM), or desmopressin (100 nM), and media samples were collected at the indicated times. The TCA-soluble radioactivity in the media was plotted as a percentage of the radioactivity initially incorporated into cell proteins. Error bars are the SEM of four samples. (B) Raising cAMP with desmopressin or forskolin did not enhance the degradation of long-lived proteins in mpkCCD cells, unlike Torin1. To label cell proteins, WT mpkCCD cells were incubated with [3H]phenylalanine (1 μCi/mL) for 20 h and then switched to chase medium containing 2 mM nonradioactive phenylalanine for 2 h. New chase media was added containing either forskolin (5 μM), desmopressin (100 nM), or Torin1 (250 nM). Media samples were collected at 1, 2, and 3 h, and the TCA-soluble radioactivity in the media was plotted as a percentage of the radioactivity incorporated into cell proteins over time. Data shown are the slopes calculated from the linear degradation rates. Error bars are the SEM of three samples. One-way ANOVA with a Bonferroni poshoc analysis against DMSO. ***P < 0.001. (C) Desmopressin increases proteasomal peptidase activity in mpkCCD cell lysates and this effect required PKA. PKA WT and PKA dKO mpkCCD cells were treated for indicated times and proteasomal hydrolysis of suc-LLVY-amc was measured in the cell extracts. Error bars are the SEM of three independent samples. One-way ANOVA with a Bonferroni poshoc analysis against time 0. ***P < 0.001, *P < 0.05. (D) Desmopressin stimulates rapid phosphorylation of Rpn6-S14 in mpkCCD cell lysates. The cell lysates from C were analyzed by SDS PAGE and Western blot for pRpn6-S14 and GAPDH. Error bars are the SEM of three samples. One-way ANOVA with a Bonferroni post hoc analysis against time 0. ***P < 0.001, **P < 0.01, *P < 0.05. (E) Desmopressin treatment caused small, but reproducible increases in the amount of doubly capped and singly capped 26S in mpkCCD WT cells, but not in cells lacking PKA. PKA WT and PKA dKO mpkCCD cells were treated for indicated times, lysed, and lysates were analyzed by Native PAGE and Western blot for Rpn1. The same lysates were also analyzed by SDS PAGE and Western blot for Rpn1 to evaluate levels of proteasome subunits. IB, immunoblot.
To test if PKA is required for this stimulation of proteolysis, we utilized an mpkCCD cell line in which both catalytic subunits of PKA were knocked out via CRISPR-Cas9 gene editing (mpkCCD PKA dKO) (20). In these double-knockout (dKO) cells, desmopressin did not stimulate the phosphorylation of PKA target proteins (SI Appendix, Fig. S4A), and neither desmopressin nor forskolin increased the degradation of short-lived cell proteins (Fig. 3A). Thus, cAMP and PKA are required for the selective increase in the degradation of short-lived proteins by the UPS.
To examine the effects of desmopressin on proteasomal activities, mpkCCD cells were treated with desmopressin for brief periods, and the chymotrypsin-like activity was then measured in cell lysates. The 26S proteasomes’ peptidase activity was increased maximally (by about 60%) after only a 5-min exposure to desmopressin and then returned gradually to control levels between 30 and 60 min (Fig. 3C). This activation of the proteasome occurred without any changes in the levels of proteasome subunits (SI Appendix, Fig. S4B). The levels of pRpn6-S14 in the lysates determined by Western blot analysis also increased very rapidly and then returned to control levels by 60 min (Fig. 3D). This very close correlation of the changes in Rpn6-S14 phosphorylation with those in peptidase activity strongly implies that these two changes are coupled. Accordingly, in the PKA dKO cells, desmopressin did not increase either the proteasomal chymotrypsin-like activity or Rpn6-S14 phosphorylation (Fig. 3 C and D). Interestingly, in the desmopressin-treated WT cells, the levels of other PKA-phosphorylated proteins also increased within 5 min of desmopressin treatment but did not then decrease like pRpn6-S14, and instead remained greater than in untreated cells for at least 60 min (SI Appendix, Fig. S4C). Thus, the termination of desmopressin’s activation of proteasomes is probably due to the preferential dephosphorylation of Rpn6-S14.
Because activation of proteasomes by cAMP and PKA seem to be associated with increased levels of 26S proteasomes (Fig. 1D) (4), we also investigated the levels of doubly and singly capped particles after desmopressin treatment. In WT mpkCCD cells, but not in the PKA dKO cells (Fig. 3E), treatment for 5 min with desmopressin caused a small, but reproducible increase in both singly and doubly capped 26S particles above control, which was still evident at 30 min of treatment (Fig. 3E) when the peptidase activity had returned toward control levels.
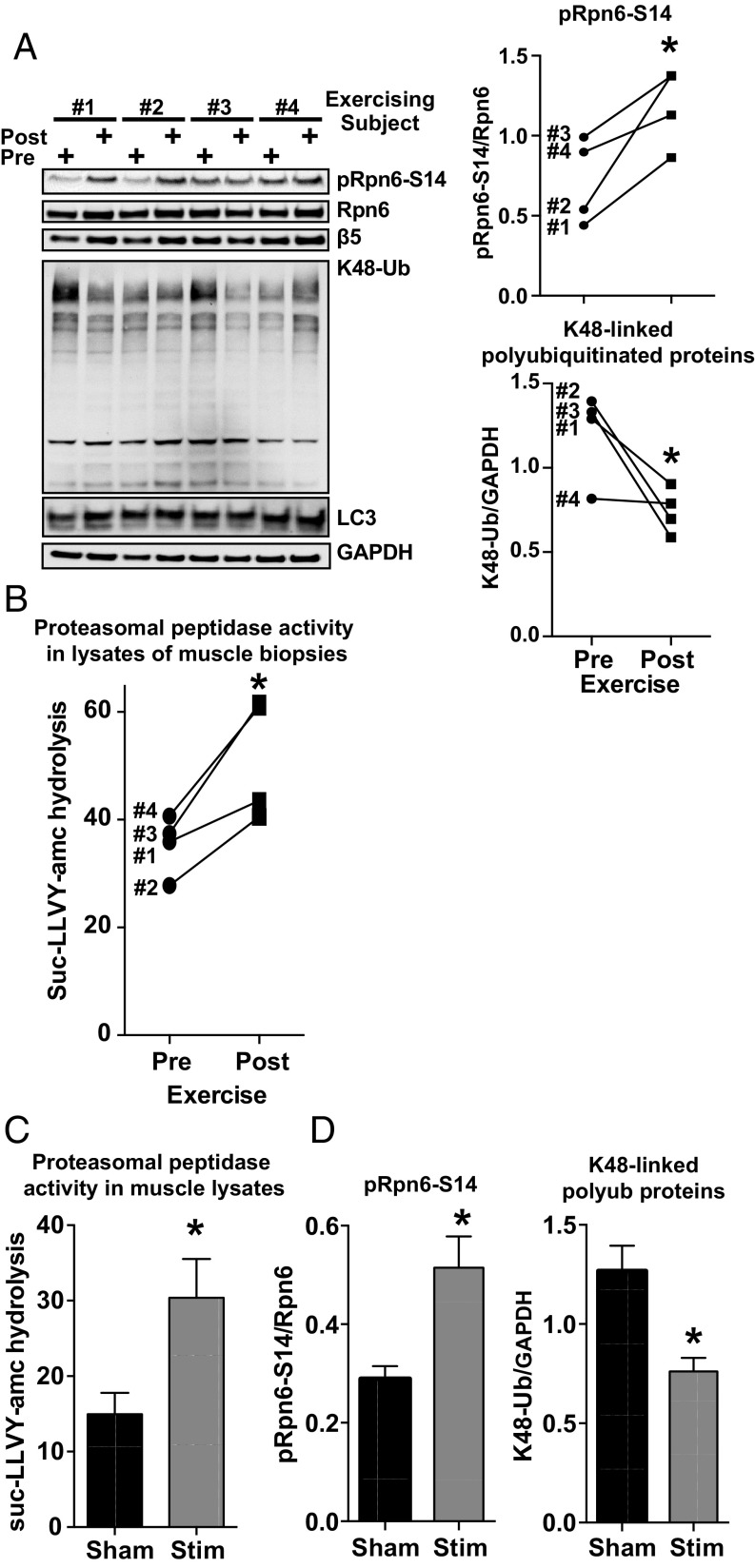
Exercise in Humans and Repetitive Contractions of Rat Muscles Enhances Phosphorylation of Rpn6 and Proteasome Activity in Skeletal Muscles.
Exercise involves multiple hormone-induced metabolic changes, but of particular importance is the secretion by the adrenal medulla and sympathetic neurons of epinephrine and norepinephrine, which triggers a cAMP-mediated increase in the breakdown of glycogen and triglycerides in skeletal muscles. We therefore tested whether Rpn6-S14 is phosphorylated and 26S proteasome activity is activated in human muscle biopsies taken immediately after a single session of high-intensity cycling exercise, as described previously (21). Biopsies were obtained from the vastus lateralis muscle of four human male volunteers before and after the exercise as part of a high-throughput mass spectrometry study of the effects of exercise on muscle protein phosphorylation (21). This phosphoproteomic analysis demonstrated that in all four subjects, exercise increased phosphorylation of several well-characterized PKA substrates, indicating activation of this kinase (21). Phosphorylation of Rpn6-S14, as detected by the TMT phosphoproteomic analysis, also seemed to increase in all four subjects (SI Appendix, Fig. S5A) (21). Using our phospho-specific Rpn6-S14 antibody, we confirmed the increased phosphorylation of Rpn6-S14 in the biopsies taken following the intense exercise (Fig. 4A). These changes were clear in three of the samples and became evident in the fourth after correcting for total amount of Rpn6 in the biopsy lysate (Fig. 4A). As expected, based on the increased levels of phosphorylated Rpn6-S14 in the lysates of all four muscle biopsies, the proteasomes’ peptidase activity was also enhanced after exercise (Fig. 4B) without any changes in the levels of proteasome subunits (Fig. 4A). It is also noteworthy that the muscles’ contents of K48-linked polyubiquitinated proteins (the primary substrate for proteasomal degradation) were decreased by the intense exercise, while the levels of the autophagy marker LC3 were unchanged (Fig. 4A).
Fig. 4.
Intense exercise in humans and repetitive contractions in rat hindlimbs enhances Rpn6-S14 phosphorylation and proteasome activity in skeletal muscles. (A) High-intensity bicycling by human volunteers caused phosphorylation of Rpn6-S14 and reduced the levels of K48-linked ubiquitin conjugates in biopsies of quadricep muscles from four male volunteers. Biopsies are the same ones analyzed previously (21). Both preexercise and postexercise muscle samples were subjected to immunoblot analysis for pRpn6-S14, Rpn6, β5, LC3, and K48-Ub. GAPDH was used as loading control. Line graphs represent the levels of K48-linked polyubiquitinated proteins and pRpn6-S14 determined by densitometry. n = 4. *P < 0.05. Error bars here and below represent mean ± SEM. (B) High-intensity cycling exercise in humans promotes peptidase activity of 26S proteasomes in muscle lysates. Biopsies studied in A were lysed and chymotrypsin-like peptidase activity was measured in muscle extracts using suc-LLVY-amc as the substrate. n = 4, *P < 0.05. (C) High-intensity repetitive contractions of rat anterior tibialis muscles by repetitive stimulation for 5 min of the sciatic nerves in anesthetized rats (Stim) enhances 26S proteasome activity. Chymotrypsin-like peptidase activity (the hydrolysis of suc-LLVY-amc) was measured in muscle extracts. n = 5, *P < 0.05. (D) Repetitive contractions of rat anterior tibialis muscles by stimulation of the sciatic nerves (Stim) as described in C increased phosphorylation of Rpn6-S14 and reduced the levels of K48-linked polyubiquitinated proteins conjugates in muscle lysates. Both sham and stimulated muscle samples were subjected to immunoblot analysis for pRpn6-S14, Rpn6, and K48-Ub. GAPDH was used as the loading control. Bar graphs represent the levels of K48-Ub and pRpn6-S14 determined by densitometry. n = 5, *P < 0.05.
To obtain further evidence that exercise leads to proteasome activation, hindlimb muscles in anesthetized rats were subjected to high-intensity repetitive contractions by stimulation of the sciatic nerve (n = 5; 100 Hz; 1-s on, 3-s off; 5 min). Following the stimulation, there was greater chymotrypsin-like proteasome activity in the muscle lysates (Fig. 4C) and more Rpn6-S14 phosphorylation than in lysates from the sham control muscles (Fig. 4D and SI Appendix, Fig. S5B). Interestingly, there was no change in the autophagy marker LC3 immediately after these intense contractions, as was also observed in human muscles after exercise, but there was a reduction in the muscles’ content of K48-linked polyubiquitinated proteins below control levels (Fig. 4D and SI Appendix, Fig. S5B). Thus, the rise in proteasome activity was accompanied by a decrease in ubiquitin conjugate levels in both rat skeletal muscles after repetitive contractions and in human muscles following intense exercise. Raising cAMP levels and proteasomal activities in myotubes or HEK293 cells with forskolin also leads to a rapid reduction in ubiquitin conjugate levels due to their enhanced degradation by proteasomes (4).
In skeletal muscles, exercise also can activate AMPK and this enzyme was in fact activated (i.e., phosphorylated) in the biopsies from human muscle after a vigorous bout of exercise (21). However, AMPK was not activated in the stimulated rat muscles, as assayed by immunoblot for phosphorylated AMPK substrates (SI Appendix, Fig. S5C). In either case, pharmacological activation of AMPK does not cause Rpn6-S14 phosphorylation in myotubes (21) and cannot account for the changes in proteasome activity observed here with repetitive stimulation.
In Mouse Muscle and Liver During Fasting, cAMP Levels, Rpn6-S14 Phosphorylation, and Proteasome Activities Increase.
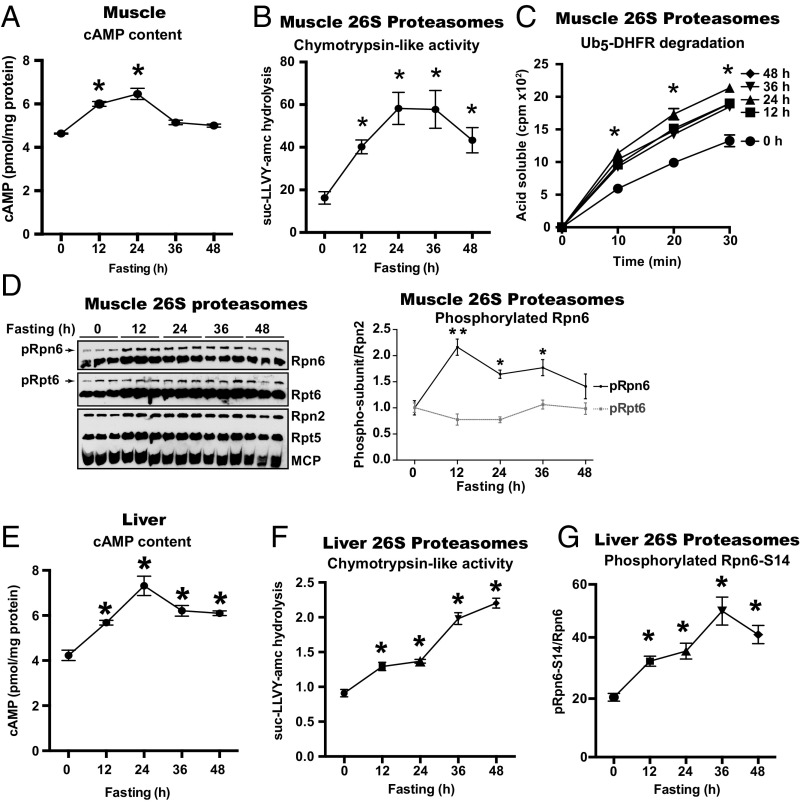
In skeletal muscle during prolonged fasting (e.g., in mice deprived of food for 1–2 d), overall protein degradation rises by activation of both the ubiquitin-proteasome pathway and autophagy, which together cause marked loss of muscle mass (18). At these times, a set of atrophy-related genes (“atrogenes”), including several ubiquitin ligases and most autophagy genes, is induced in muscles by FoxO transcription factors (18). However, with shorter times of food-deprivation when insulin levels decrease, mTOR activity in muscles falls, which stimulates both ubiquitination and autophagy (15), and cAMP levels rise in some tissues (22). To test if food deprivation raises cAMP levels and activates proteasomes in mouse skeletal muscles and liver, male mice were deprived of food for 12, 24, 36, or 48 h. At each time point four mice were killed and their hindlimb muscles separately homogenized. We first assayed cAMP content by ELISA in each lysate of the tibialis anterior (TA) muscles in fed and fasted mice. A clear increase in cAMP levels above that of fed animals was evident at 12 and 24 h after food deprivation, but at 36 and 48 h the cAMP content in the muscles returned toward fed levels (Fig. 5A). At these longer times of food deprivation, proteolysis rises through induction of atrogenes, such as atrophy-specific ubiquitin ligases (18), the proteasome activating protein ZFAND5 (23, 24), and many autophagy genes (14).
Fig. 5.
Food deprivation of mice increases cAMP levels, proteasomal activities, and Rpn6-S14 phosphorylation in skeletal muscle and liver. (A) Depriving mice of food for the indicated times increased at 12 and 24 h the amount of cAMP in skeletal muscles. cAMP content was measured in TA muscle extracts using a cAMP-ELISA kit. Error bars here and below represent mean ± SEM. n = 4, *P < 0.05. (B) Fasting stimulates the peptidase activity of 26S proteasomes purified from hindlimb muscles. Food was removed from the mice at the indicated times, the muscles were homogenized, and 26S proteasomes were affinity-purified from the muscle extracts of the fasted mice and fed controls. The chymotrypsin-like activity was measured with suc-LLVY-amc. n = 4 mice per time point, *P < 0.05. (C) Fasting increases the capacity of muscle 26S proteasomes to degrade Ub5-DHFR. Proteasome preparations were the same as studied in B. Rates of degradation were measured by following the conversion of radiolabeled protein to TCA-soluble 32P-labeled peptides. n = 4 mice per time point, *P < 0.05. (D) Food deprivation enhances phosphorylation of Rpn6 but not the phosphorylation of Rpt6. 26S proteasomes were purified from hindlimb muscles of mice fasted for indicated times as described in A. Samples were subjected to Zn2+-Phos-tag SDS/PAGE and followed by immunoblot analysis for Rpn6 and Rpt6. The same samples were also analyzed by SDS/PAGE and Western blot for Rpn2, Rpt5, and MCP. *P ≤ 0.05, **P ≤ 0.01. (E) After food deprivation of mice levels of cAMP in liver extracts increased. Levels of cAMP were measured by cAMP-ELISA kit. n = 4 mice per time point, *P < 0.05. (F) Peptidase activity of hepatic 26S proteasomes increased after 12–48 h of food deprivation. At each time of food deprivation, livers were removed from the mice studied in A, homogenized, and 26S proteasomes purified via the Ubl-method. Chymotrypsin-like activity was measured as in B. *P < 0.05. (G) Food deprivation of mice stimulates the phosphorylation of Rpn6-S14 in liver. 26S proteasomes purified from livers were subjected to immunoblot analysis for pRpn6-S14 and Rpn6. n = 4 mice per time point, *P < 0.05.
We then affinity-purified 26S proteasomes from the muscle extracts to test whether their activities were enhanced by fasting. The chymotrypsin-like activity of the 26S proteasomes from the muscles of fasted mice was two- to threefold greater than proteasomes from fed mice (Fig. 5B). This activity seemed be maximally increased at 24 h of food deprivation and then appeared to decrease slightly at 48 h, but nevertheless remained higher than in fed controls. Furthermore, the 26S proteasomes from muscles of mice fasted for 12–48 h showed a greater capacity to degrade ubiquitinated proteins, as shown using 32P-labeled Ub5-DHFR as the substrate (Fig. 5C). The amounts of the 20S and 19S proteasome subunits in the muscle lysates, as assayed by Western blot, did not change at any of the tested times of food deprivation (SI Appendix, Fig. S6). Thus, the proteasomes’ activities increase in fasting without any demonstratable increase in proteasome content, and their activity rises before the induction in muscles of atrogenes by FoxO transcription factor, which begins between 24–36 h of food deprivation (14).
This increase in muscle cAMP levels and the proteasome’s degradative capacity after food deprivation coincided with an increase in Rpn6 phosphorylation, as shown by phos-tag SDS/PAGE followed by Western blot analysis of 26S proteasomes purified from the muscles (Fig. 5D). In contrast, in the proteasomes from these muscles (Fig. 5D) no increase was seen in the phosphorylation of Rpt6, which had been reported to be phosphorylated by PKA (25). Furthermore, no change in its phosphorylation was seen in our studies with pharmacological agents that raise cAMP and cause Rpn6 phosphorylation (4).
Among the earliest metabolic responses to food deprivation are the activation of glycogen breakdown and the inhibition of glycogen synthesis in liver, which are signaled primarily by glucagon release and the fall in insulin levels. Because these responses are mediated by cAMP and PKA, we tested if fasting also enhances proteasome activities in liver in vivo, as it does in muscle and as would be predicted by our findings with glucagon treatment of isolated mouse hepatocytes (Fig. 1 A–C). We initially assayed cAMP content by ELISA in lysates of the liver of fed and fasted mice. A clear increase in cAMP levels above that in fed animals was evident by 12 and 24 h after food deprivation (Fig. 5E). At 36 and 48 h of food deprivation, the liver cAMP content decreased somewhat, but remained higher than in fed mice (Fig. 5E). At each time point, the 26S proteasomes were affinity-purified from the liver extracts, and between 12 and 48 h of food deprivation, their chymotrypsin-like activity increased almost in a linear fashion above the activity measured in fed mice (Fig. 5F). This increase in the proteasomes’ activity and the rise in hepatic cAMP levels after food deprivation coincided with an increase in Rpn6-S14 phosphorylation, as shown by Western blot analysis of the liver lysate and the purified 26S proteasomes (Fig. 5G and SI Appendix, Fig. S7). Thus, cAMP-mediated Rpn6-S14 phosphorylation coincides with the rapid rise in proteasomal activities upon food deprivation in liver and muscle. In the liver, however, there was a further apparent rise in proteasomal activities after 1 d of fasting when cAMP levels seemed to fall slightly, which suggests additional adaptations to increase proteasome activity in liver during prolonged fasting, as occurs in muscle (23, 24).
It is noteworthy that the content of 20S and 19S proteasome subunits, as measured by Western blot, did not change in the liver or muscle lysates at any time after food deprivation, despite the large increases in 26S activity. Thus, proteasome activation is occurring through postsynthetic modifications of existent particles and not through production of new proteasomes, as had been reported to occur in yeast and cultured mammalian cells upon nutrient deprivation (26).
Discussion
Regulation of Proteasomal Function by Phosphorylation.
This ability of PKA and other kinases (e.g., DYRK2 and PKG) to enhance rates of proteolysis through increases in proteasome activity represents an under appreciated mode of regulation of protein breakdown (2, 3). While control of ubiquitination rates influences the levels of individual proteins or small groups of related proteins, control of proteasome function allows more global, coordinated regulation of the degradation of large classes of proteins. These two mechanisms to regulate degradation by the UPS do not necessarily function independently. In fact, in addition to enhancing proteasomal activity, cAMP can stimulate the ubiquitination of some cell proteins (4), suggesting these two mechanisms to increase proteolysis are activated simultaneously by PKA. The selective ubiquitination of short-lived proteins may account for their preferential destruction when cAMP levels rise.
This control of proteolysis by ubiquitination and proteasomal activities seems analogous to the two levels for the control of protein production. Transcriptional regulation of gene expression allows the highly selective control of the levels of specific proteins or groups of related proteins (like ubiquitination), while the global regulation of ribosomal translation influences the rates of the production and accumulation of large classes of cell proteins (like regulating proteasomal function).
These studies clearly demonstrate not only that proteasome activity can rise and fall in various tissues (from renal collecting ducts to skeletal muscle) upon exposure to diverse hormones that raise cAMP, but also that these responses are surprisingly rapid. After ADH addition to renal epithelial cells, proteasome activity and Rpn6-S14 phosphorylation rose maximally within 5 min and both returned to control levels by 60 min (Fig. 3 C and D). Thus, the cell’s capacity for degradation by the UPS can change through postsynthetic modifications of proteasomes in a highly dynamic fashion that has not been widely appreciated.
These findings also confirmed our earlier conclusion that Rpn6-S14 is a bona fide PKA target (4) and show that it is phosphorylated in vivo under very different physiological conditions, including fasting, intense exercise, and in response to many (perhaps most) hormones that activate adenylate cyclases. Under the various conditions studied here, in our prior study (4), and in prior mass spectrometry studies of exercising human muscles (21), similar changes were not seen in the phosphorylation of the ATPase subunit Rpt6 (Fig. 5D), which had been reported to be modified by PKA (25), CaMKII (8), and PKG (9, 10). Although other proteasome-associated proteins or 26S subunits were also phosphorylated in these 26S preparations, the levels of pRpn6-S14 increased and decreased together with proteasome activities upon desmopressin treatment. Moreover, our mutagenesis studies indicated that a phosphomimetic mutation of Rpn6-S14 causes 26S activation (4).
Phosphorylation of Rpn6 somehow enhances multiple proteasomal activities, including peptide hydrolysis by the 20S core particle and ATP hydrolysis by the 19S regulatory particle, leading to a greater capacity to degrade polyubiquitinated proteins. In fact, the rate of breakdown of ubiquitinated conjugates by proteasomes is proportional to their rates of ATP hydrolysis (27), and thus the increased ATPase activity following PKA treatment should by itself stimulate the hydrolysis of ubiquitinated proteins. The simultaneous increases in peptide hydrolysis by the 20S’s three types of active sites probably reflects enhanced peptide entry into the core particle, as occurs during proteasome activation induced by binding of ubiquitinated proteins or ATPγS, which causes an enlargement of the substrate entry channel through the ATPase ring and opening of the gate in the 20S’s outer ring (1, 28). Presumably, Rpn6 phosphorylation facilitates similar structural changes in these particles. The phosphorylation of Rpn6-S14 also seems to cause rapid, but small increases in the content of assembled 26S particles (Figs. 1D and 3E) (4), which suggests a PKA-mediated increase in the formation or stability of singly capped and doubly capped complexes. However, it remains unclear if these small increases in 26S content account for the associated large changes in proteasome activity (26).
Proteasome Activity in Fasting.
These findings imply that the enhanced proteasomal activity and the ability to degrade short-lived cell proteins accompany the increased glycogenolysis and triglyceride breakdown that are induced in liver, muscle, and heart by epinephrine during the fight-or-flight response and the hepatic gluconeogenesis stimulated by glucagon in fasting. In the tissues of fasted mice, these proteasomal activities increased without any changes in 26S content. Even in the mice fasted for 2 d, where degradation rates are exceptionally high, there was also no significant change in proteasome subunit levels in muscles or liver. Surprisingly, there has been appreciable controversy about how starvation affects proteasome content in cultured mammalian cells. Levels of proteasomes have been reported to rise very rapidly upon nutrient deprivation (29), to fall slowly due to accelerated autophagy (30), or to remain unchanged for many hours (15). Whatever the basis for these divergent in vitro results, clearly the physiological mechanism to increase the capacity for protein degradation in liver and muscles in fasting mice is through postsynthetic modification of proteasomes, not through production of new proteasomes. Upon nutrient deprivation or mTOR inhibition of cultured cells, total protein degradation by the UPS rises within minutes through a general increase in ubiquitination, without any change in proteasome activity (15), in sharp contrast to the rapid activation of proteasomes by cAMP, which also occurs within minutes after glucagon, forskolin, or vasopressin. However, in tissues of fasting organisms, where insulin levels fall, cAMP rises, and mTOR activity decreases, both protein ubiquitination (15) and 26S activity probably increase simultaneously and synergize to enhance proteolysis.
The increase in proteasome activity in mouse muscles and liver were clearly evident by 12 h after food was removed from fed animals and thus represents a rather rapid metabolic response to food deprivation. This timing suggests that a similar enhancement of proteolysis should also occur in humans in these tissues after an overnight fast (i.e., between dinner and breakfast). This rapid response long precedes the FoxO-mediated induction of ubiquitin ligases and autophagy genes that leads to muscle wasting, especially the breakdown of myofibrils, which is evident in rat and mouse muscles at 1–2 d after food deprivation (18). The FoxO-mediated response stimulates the breakdown of long-lived proteins, the great bulk of cell proteins, to provide the starving organism with amino acids for gluconeogenesis and energy production and thus serves distinct physiological functions from the PKA-mediated enhancement of the degradation of short-lived proteins. In fact, raising cAMP levels was shown to inhibit the FoxO-mediated induction of ubiquitin ligases and autophagy genes, upon denervation or fasting for 2 d (31, 32, 33). PKA activation thus suppresses the degradation of the bulk of muscle proteins and the resulting atrophy. By this mechanism β-adrenergic agents can reduce the degradation of long-lived components and increase muscle mass, especially in atrophy, while still enhancing the degradation of the small fraction of cell proteins that are damaged, misfolded, or serve regulatory functions (4). Accordingly, in the present studies, raising cAMP did not enhance autophagy and even inhibited its activation in myotubes and HEK293 cells (SI Appendix, Fig. S2), when it promotes breakdown of short-lived proteins (4).
Physiological Significance of the Increased Degradation of Short-Lived Proteins.
Our previous studies showed that the PKA-mediated activation of proteasomes increases the cell’s capacity to eliminate various misfolded proteins that cause neurodegenerative diseases (including mutant tau, SOD1, FUS), as well as short-lived regulatory proteins (4, 5). The present studies strongly suggest that fasting, exercise, or other conditions that raise cAMP levels might also be beneficial in promoting the clearance of potentially toxic proteins and stimulating protein degradation in diseases in which proteasome function is impaired (5, 34, 35). A fundamental question raised by these findings is why the cells’ degradative capacity, especially its ability to destroy misfolded or damaged proteins, is not normally maintained at maximal levels to provide cell protection, and why in exercise and fasting is the capacity to destroy such proteins activated by cAMP and PKA. Presumably, maintaining proteasomes in an activated state continually has negative consequences and leads to excessive degradation of some critical regulatory proteins.
The rapid enhancement of the cell’s capacity to degrade short-lived proteins in exercise is of particular physiological interest. Such components represent only a minor fraction of cell constituents, and therefore even their complete hydrolysis and metabolism of the constituent amino acids cannot provide a significant source of energy to a fasting or exercising organism. Perhaps after exercise, the enhanced capacity to degrade misfolded proteins enables the muscles to eliminate proteins damaged mechanically by the repeated contractions or by free radicals generated by mitochondrial metabolism. Another attractive possibility would be that the PKA-accelerated degradation of short-lived regulatory proteins with exercise, early in fasting, or in various hormonal responses facilitates adaptive changes in the cell’s protein composition. As these cells adapt to new physiological conditions, it would seem advantageous to degrade certain preexistent regulatory proteins or critical enzymes that are not advantageous under the new conditions. In such cells, cAMP also stimulates via a cAMP-responsive element the expression of new proteins that are more appropriate for these new conditions. More rapid elimination of some preexistent regulatory proteins could synergize with this enhancement of new gene expression to promote the cellular adaptation to the new physiological states.
Materials and Methods
Reagents and Plasmids.
Forskolin, glucagon, and epinephrine were purchased from Sigma, desmopressin from Cayman Chemical, glutathione Sepharose 4B from GE Healthcare Life sciences, the direct cAMP ELISA kit from Enzo Life sciences, PP1 and λ-phosphatase from New England Biolabs, and L-[3,4,5-3H]-phenylalanine from American Radiolabeled Chemicals. Ub5-DHFR and Bortezomib were kindly provided by Millennium pharmaceuticals. Information pertaining to the plasmids and their origins is presented in ref. 4.
Human Subjects and Exercise Protocol.
The human male volunteers, the single bout of high-intensity cycle exercise, and biopsies were described previously (21). The assurances of human consent were published previously (see ref. 21).
Repetitive Contractions of Rat Muscle by Stimulation of the Sciatic Nerve.
All experiments with rats were approved by the Animal Ethics Committee of the University of Melbourne and were conducted in accordance with the Australian code of practice for the care and use of animals for scientific purposes, as stipulated by the National Health and Medical Research Council (Australia). Male Wistar rats were group-housed, kept on a 12:12-h light-dark cycle and had free access to standard rodent chow diet and water. Following a semifasting period overnight, 10 Wistar rats were anesthetized by intraperitoneal administration of pentobarbital and either sham surgery (n = 5) or high-intensity stimulation of the sciatic nerve in situ (n = 5; 100 Hz; 1-s on, 3-s off; 5 min) using a custom-built force transducer to monitor contractile work of the tibialis anterior muscles for a total of 5 min.
Isolated Rat Heart Perfusions.
All heart perfusions were performed by Heinrich Taegtmeyer and Giovanni Davogustto (McGovern Medical School of the University of Texas, Houston) in accordance with the NIH’s Guide for the Care and Use of Laboratory Animals (36) with an animal protocol approved by the Institutional Animal Care and Use Committee at the McGovern Medical School. Male Sprague–Dawley rats (287–300 g) were obtained from Charles River Laboratories and housed in the Center for Laboratory Animal Medicine and Care of the McGovern Medical School of The University of Texas at Houston under controlled conditions (23 ± 1 °C; 12-h light/12-h dark cycle). Hearts were perfused ex vivo as previously described (19) at 37 °C with nonrecirculating Krebs–Henseleit buffer equilibrated with 95% O2, 5% CO2 and containing glucose (5 mM) and sodium lactate (0.5 mM). Mean aortic pressure was continuously monitored using a 3 French pressure transducer catheter (Millar Instruments) connected to a PowerLab 8/30 recording system (AD Instruments). After a 5-min stabilization period, hearts were perfused at normal workload (preload of 15 cm of H2O and afterload of 100 cm H2O) for 15 min. At this point, the conditions were modified according to the experimental group assignment regarding addition of epinephrine bitartrate (1 μM) to the perfusion buffer and increased afterload (afterload set to 140 cm H2O) and hearts were perfused for another 45 min. At 60 min, beating hearts were freeze-clamped with aluminum tongues and cooled in liquid N2. A portion of each frozen heart tissue was weighed and dried to constant weight. The remainder was stored at −80 °C for further analysis.
Cell Culture and Treatments.
Mouse primary hepatocytes were kindly provided by Pere Puigserver, Harvard Medical School, Boston, MA, and isolated and maintained as described previously (37). For all our studies, mouse primary hepatocytes were treated with vehicle control (DMSO), forskolin (10 μM), epinephrine (1 μg/mL), or glucagon (1 µg/mL) for 1 h unless otherwise stated. HEK293A cells were grown in DMEM containing 10% FBS and 100 U/mL penicillin and streptomycin. HEK293A cells cell were treated with control, forskolin (10 μM) for the times indicated in Results. PKA WT and PKA dKO mpkCCD cells were kindly provided by Mark Knepper, NIH, Bethesda, MD, cultured as previously described (20), and treated with desmopressin (100 nM) for the times indicated.
Food Deprivation of Mice.
All fasting studies on mice were performed according to ethical guidelines of the NIH Guide for Care and Use of Laboratory Animals (36) on adult male CD-1 mice initially weighing 25–26 g. Beginning in the morning, food stores were removed from cages for up to 48 h while fed littermate controls were given ad libitum access to food. All mice had ad libitum access to water.
cAMP ELISA.
cAMP ELISA was performed on mouse TA muscle and liver extracts according to the instructions of the manufacturer, Enzo Life Sciences.
Degradation of Long- or Short-Lived Cell Proteins.
The degradation rate of long- or short-lived cell proteins was measured after labeling with [3H]phenylalanine and the conversion of radiolabeled cell proteins to TCA-soluble radiolabeled amino acids in the media was assayed as described previously (15, 38).
Cell Lysis and Western Blotting.
Before lysing, cells were washed with ice cold 1× PBS and collected by scraping. Cells were pelleted by centrifuging at 1,000 × g at 4 °C for 5 min. The resultant cell pellet was resuspended in lysis buffer containing 50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% Nonidet P-40, Complete protease inhibitor mixture (Roche), 2 mM NaF, 1 mM Na3VO4, 1 mM PMSF, and 1× Phosphatase inhibitor mixture (Thermo Scientific). The lysate was cleared by centrifugation at 20,800 × g for 15 min at 4 °C. The total protein content was measured using Coomassie Plus staining (Thermo Scientific). Immunoblotting was performed using various antibodies whose sources were described previously (4), and total protein was determined after transfer with the REVERT stain (LiCor).
Affinity Purification of 26S Proteasomes.
After in vivo or in vitro treatments, 26S proteasomes were purified from mouse hindlimb muscles, livers, or hepatocytes, and rat hearts using the ubiquitin-like domain (Ubl) method described previously (13). Tissues and cells were lysed in the presence of phosphatase inhibitors (Calyculin A, 25 nM; NaF, 1 mM; β-glycerophosphate, 1 mM).
Measuring Activities of 26S Proteasome.
Peptidase activity of 26S proteasomes in the cell extracts was assayed as described previously (4). Degradation of ubiquitinated proteins by purified 26S was assayed using, ubiquitinated dihydrofolate reductase, Ub5-DHFR, kindly provided by Takeda Oncology Company (formerly Millennium Pharmaceuticals), Cambridge, MA, which we radiolabeled using PKA and [γ-32P]ATP, as described previously (4). Degradation of Ub5-DHFR and ATPase activity were assayed were performed as described previously (27). The three peptidase activities were measured as described previously (12).
Generation of Phospho-Specific Rpn6 Antibody.
Rabbit anti-pRpn6-S14 polyclonal antibody was raised using as immunogen the phospho-peptide: EFQRAQ(pS)LLSTDR, by AMSBIO facility at Cambridge, MA.
Detecting Assembled 26S Proteasomes by Native PAGE.
Cells were lysed by sonication in APB (25 mM Hepes-KOH pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 1 mM ATP) and run on 3–8% gradient NuPAGE Tris Acetate Gels (Life Technologies) in Tris-Glycine Native buffer (Thermo) supplemented with 5 mM MgCl2 and 1 mM ATP for 4 h at 135 V at 4 °C. To measure peptidase activity, the gels were incubated in activity buffer containing 20 μM suc-LLVY-amc for 30 min at 37 °C. For Western analysis, gels were soaked for 15 min at room temperature in a buffer containing 25 mM Tris,190 mM glycine and 1% SDS and then transferred to PVDF membranes in transfer buffer (25 mM Tris and 190 mM glycine) containing 1% MeOH overnight at 15 V at 4 °C.
Dephosphorylation of 26S Proteasomes with PP1.
Approximately 50 nM of Ubl-purified 26S proteasomes were incubated with PP1 for 60 min at 30 °C. The reaction was stopped by the addition of sample buffer and boiling, and then Western blot analysis was performed.
Dephosphorylation of 26S Proteasomes with λ-Phosphatase.
The 26S proteasomes were purified by the Ubl-method and incubated with either phosphatase inhibitors (Calyculin A, 25 nM; NaF, 1 mM; β-glycerophosphate, 1 mM) or λ-phosphatase for 60 min at 30 °C, as previously described (39). Peptidase activity was then assayed as described previously (12) with the addition of phosphatase inhibitors (Calyculin A, 25 nM; NaF, 1 mM; β-glycerophosphate, 1 mM) to the activity buffer.
Detecting of Phosphorylated Residues of 26S Proteasomes.
The 26S proteasomes were purified by the Ubl-method in the presence of phosphatase inhibitors (Calyculin A, 25 nM; NaF, 10 mM; 25 mM β-glycerophosphate), and then analyzed by SDS/PAGE and ProQ Diamond phospho-stain (Thermo) according to the manufacturer’s instructions. Gels were imaged on a Typhoon 5 (GE) and then subsequently stained with Coomassie blue to detect total protein.
Zn2+-Phos-tag SDS/PAGE.
To identify phosphorylated Rpn6 or Rpt6 by Western blotting, we performed on 26S proteasomes purified by the Ubl-method Zn2+-Phos-tag SDS gels (39) (Wako) as per the manufacturer’s instructions.
Statistical Analysis.
Was performed using Student’s t tests, one-way post hoc Tukey honestly significant diffefrence and one-way ANOVA post hoc Bonferroni’s comparison. All values are expressed as means ± SEM. Asterisks represent P values, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Supplementary Material
Acknowledgments
We thank Megan LaChance for her valuable assistance in preparing this manuscript; Prof. Pere Puigserver (Harvard Medical School) for providing mouse primary hepatocytes; Prof. Heinrich Taegtmeyer and Dr. Giovanni Davogustto (McGovern Medical School, University of Texas) for the working rat hearts and characterizing their physiological properties; Prof. David James and Dr. Benjamin Parker (University of Sydney), for providing the stimulated rat muscles and human muscle biopsies; Prof. Erik Richter, Dr. Bente Kiens, and Dr. Jorgen Wojtaszewski (University of Copenhagen), for generating the human muscle biopsies; and Dr. Mark Knepper (NIH National Heart, Lung, and Blood Institute) for the PKA WT and PKA double knockout mpkCCD cell lines. Our research has been supported by NIH National Institute of General Medical Sciences Grant R01 GM051923-20, Cure Alzheimer’s Fund, Muscular Dystrophy Association Grant MDA-419143, a Genentech and Project ALS grant (to A.L.G.), and NIH National Institute of General Medical Sciences Grant F32 GM128322 (to J.J.S.V.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809254116/-/DCSupplemental.
References
- 1.Collins GA, Goldberg AL. The logic of the 26S proteasome. Cell. 2017;169:792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.VerPlank JJS, Goldberg AL. Regulating protein breakdown through proteasome phosphorylation. Biochem J. 2017;474:3355–3371. doi: 10.1042/BCJ20160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo X, Huang X, Chen MJ. Reversible phosphorylation of the 26S proteasome. Protein Cell. 2017;8:255–272. doi: 10.1007/s13238-017-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lokireddy S, Kukushkin NV, Goldberg AL. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc Natl Acad Sci USA. 2015;112:E7176–E7185. doi: 10.1073/pnas.1522332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myeku N, et al. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat Med. 2016;22:46–53. doi: 10.1038/nm.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo X, et al. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat Cell Biol. 2016;18:202–212. doi: 10.1038/ncb3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerjee S, et al. Ancient drug curcumin impedes 26S proteasome activity by direct inhibition of dual-specificity tyrosine-regulated kinase 2. Proc Natl Acad Sci USA. 2018;115:8155–8160. doi: 10.1073/pnas.1806797115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djakovic SN, Schwarz LA, Barylko B, DeMartino GN, Patrick GN. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. J Biol Chem. 2009;284:26655–26665. doi: 10.1074/jbc.M109.021956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ranek MJ, Terpstra EJ, Li J, Kass DA, Wang X. Protein kinase G positively regulates proteasome-mediated degradation of misfolded proteins. Circulation. 2013;128:365–376. doi: 10.1161/CIRCULATIONAHA.113.001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranek MJ, Kost CK, Jr, Hu C, Martin DS, Wang X. Muscarinic 2 receptors modulate cardiac proteasome function in a protein kinase G-dependent manner. J Mol Cell Cardiol. 2014;69:43–51. doi: 10.1016/j.yjmcc.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krebs EG. Historical perspectives on protein phosphorylation and a classification system for protein kinases. Philos Trans R Soc Lond B Biol Sci. 1983;302:3–11. doi: 10.1098/rstb.1983.0033. [DOI] [PubMed] [Google Scholar]
- 12.Kim HT, Collins GA, Goldberg AL. Measurement of the multiple activities of 26S proteasomes. Methods Mol Biol. 2018;1844:289–308. doi: 10.1007/978-1-4939-8706-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Besche HC, Goldberg AL. Affinity purification of mammalian 26S proteasomes using an ubiquitin-like domain. Methods Mol Biol. 2012;832:423–432. doi: 10.1007/978-1-61779-474-2_29. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci USA. 2015;112:15790–15797. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deter RL, De Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol. 1967;33:437–449. doi: 10.1083/jcb.33.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams A, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: Molecular mechanisms and promising therapies. Nat Rev Drug Discov. 2015;14:58–74. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin GW, Taylor CS, Taegtmeyer H. Regulation of energy metabolism of the heart during acute increase in heart work. J Biol Chem. 1998;273:29530–29539. doi: 10.1074/jbc.273.45.29530. [DOI] [PubMed] [Google Scholar]
- 20.Isobe K, et al. Systems-level identification of PKA-dependent signaling in epithelial cells. Proc Natl Acad Sci USA. 2017;114:E8875–E8884. doi: 10.1073/pnas.1709123114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman NJ, et al. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab. 2015;22:922–935. doi: 10.1016/j.cmet.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): Transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 23.Hishiya A, et al. A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J. 2006;25:554–564. doi: 10.1038/sj.emboj.7600945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee D, Takayama S, Goldberg AL. ZFAND5/ZNF216 is an activator of the 26S proteasome that stimulates overall protein degradation. Proc Natl Acad Sci USA. 2018;115:E9550–E9559. doi: 10.1073/pnas.1809934115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, et al. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J Biol Chem. 2007;282:22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- 26.Rousseau A, Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol. 2018;19:697–712. doi: 10.1038/s41580-018-0040-z. [DOI] [PubMed] [Google Scholar]
- 27.Peth A, Nathan JA, Goldberg AL. The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J Biol Chem. 2013;288:29215–29222. doi: 10.1074/jbc.M113.482570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bard JAM, et al. Structure and function of the 26S proteasome. Annu Rev Biochem. 2018;87:697–724. doi: 10.1146/annurev-biochem-062917-011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rousseau A, Bertolotti A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature. 2016;536:184–189. doi: 10.1038/nature18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen-Kaplan V, Ciechanover A, Livneh I. Stress-induced polyubiquitination of proteasomal ubiquitin receptors targets the proteolytic complex for autophagic degradation. Autophagy. 2017;13:759–760. doi: 10.1080/15548627.2016.1278327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lira EC, et al. Phosphodiesterase-4 inhibition reduces proteolysis and atrogenes expression in rat skeletal muscles. Muscle Nerve. 2011;44:371–381. doi: 10.1002/mus.22066. [DOI] [PubMed] [Google Scholar]
- 32.Gonçalves DA, et al. Clenbuterol suppresses proteasomal and lysosomal proteolysis and atrophy-related genes in denervated rat soleus muscles independently of Akt. Am J Physiol Endocrinol Metab. 2012;302:E123–E133. doi: 10.1152/ajpendo.00188.2011. [DOI] [PubMed] [Google Scholar]
- 33.Graça FA, et al. Epinephrine depletion exacerbates the fasting-induced protein breakdown in fast-twitch skeletal muscles. Am J Physiol Endocrinol Metab. 2013;305:E1483–E1494. doi: 10.1152/ajpendo.00267.2013. [DOI] [PubMed] [Google Scholar]
- 34.VerPlank JJS, Lokireddy S, Feltri ML, Goldberg AL, Wrabetz L. Impairment of protein degradation and proteasome function in hereditary neuropathies. Glia. 2018;66:379–395. doi: 10.1002/glia.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kristiansen M, et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell. 2007;26:175–188. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 36.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 37.Rines AK, Sharabi K, Tavares CD, Puigserver P. Targeting hepatic glucose metabolism in the treatment of type 2 diabetes. Nat Rev Drug Discov. 2016;15:786–804. doi: 10.1038/nrd.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sha Z, Zhao J, Goldberg AL. Measuring the overall rate of protein breakdown in cells and the contributions of the ubiquitin-proteasome and autophagy-lysosomal pathways. Methods Mol Biol. 2018;1844:261–276. doi: 10.1007/978-1-4939-8706-1_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.VerPlank JJS, Goldberg AL. Exploring the regulation of proteasome function by subunit phosphorylation. Methods Mol Biol. 2018;1844:309–319. doi: 10.1007/978-1-4939-8706-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.