Significance
Activity-dependent translation, the reversible activation of translation in response to synaptic activity, is important for synaptic plasticity and long-term memory formation. How synaptic activity triggers translation of silenced mRNAs within neuronal granules, which are membraneless RNA–protein assemblies, is not clear. We show that the C-terminal region of the Fragile X Mental Retardation Protein (FMRP), an abundant neuronal granule protein, phase separates in vitro with RNA and that its phase separation is fine-tuned by posttranslational modifications controlled in neurons by synaptic activity. We observe that phase separation is correlated with translation inhibition in vitro, providing a conceptual framework to unify FMRP’s diverse roles. Experimental and bioinformatic data suggest that phase separation may be a general mechanism linked to activity-dependent translation.
Keywords: phase separation, activity-dependent translation, translational regulation, posttranslational modifications, RNA granules
Abstract
Activity-dependent translation requires the transport of mRNAs within membraneless protein assemblies known as neuronal granules from the cell body toward synaptic regions. Translation of mRNA is inhibited in these granules during transport but quickly activated in response to neuronal stimuli at the synapse. This raises an important question: how does synaptic activity trigger translation of once-silenced mRNAs? Here, we demonstrate a strong connection between phase separation, the process underlying the formation of many different types of cellular granules, and in vitro inhibition of translation. By using the Fragile X Mental Retardation Protein (FMRP), an abundant neuronal granule component and translational repressor, we show that FMRP phase separates in vitro with RNA into liquid droplets mediated by its C-terminal low-complexity disordered region (i.e., FMRPLCR). FMRPLCR posttranslational modifications by phosphorylation and methylation have opposing effects on in vitro translational regulation, which corroborates well with their critical concentrations for phase separation. Our results, combined with bioinformatics evidence, are supportive of phase separation as a general mechanism controlling activity-dependent translation.
Local translation at the synaptic region of neurons occurs in response to neuronal activity and is referred to as activity-dependent translation (1). This process regulates synaptic strength and facilitates synaptic plasticity and long-term memory formation (2). Dysfunction in activity-dependent translation is a common pathological mechanism for neurodegenerative (3) and neurodevelopmental disorders (4). Therefore, understanding this process will provide important insights into a diverse set of diseases.
Neurons control activity-dependent translation by sorting and packaging mRNAs into non–membrane-bound protein assemblies known as neuronal granules (5, 6). These granules transport mRNAs from the neuronal cell body toward the synaptic terminals; mRNA translation is inhibited during transport and then activated in response to stimuli at the synapse (1, 2). It is currently not well understood how mRNAs that were once translationally silent within neuronal granules become translationally active in an activity-dependent manner. Experiments performed by the Singer laboratory showed that β-actin mRNA is packaged into neuronal granules, which undergo cycles of assembly and disassembly in response to neuronal stimuli (7, 8); assembly of granules “masks” mRNAs from translation, while disassembly of granules releases mRNAs for localized translation (7, 8). Thus, activity-dependent translation is hypothesized to require reversible granule formation (7, 8). Although different signaling pathways have been shown to be involved in activity-dependent translation (9, 10), mechanistic details explaining how neuronal granules can reversibly assemble in a stimulus-dependent manner remain unclear.
Intracellular RNA granules, including neuronal granules, are formed by phase separation (11–13). This process concentrates RNA-binding proteins and RNAs into condensed membraneless compartments, which have been described as intracellular puncta, foci, granules, membraneless organelles, or ribonucleoprotein (RNP) particles. Critical to phase separation are synergistic multivalent interactions, which are facilitated by the modular structure of RNA-binding proteins containing multiple folded RNA-binding domains along with intrinsically disordered regions (IDRs) with low-complexity sequences (14, 15). Between RNA-binding proteins and RNAs, multivalent interactions can arise in multiple ways (14–18): folded RNA-binding domains, such as RRM (RNA Recognition Motif) or KH (K Homology) domains, binding to RNA; RNA interacting with RNA; IDRs containing short linear motifs, such as arginine-glycine–rich (RGG) motifs, interacting with RNA; and IDRs interacting with other IDRs. By using these principles, different RNA granules have been modeled in vitro by phase separation of specific RNA-binding proteins (19–22). For example, studies examining phase separation of stress granule-associated RNA-binding proteins, including FUS (23–28), hnRNPA1 (29), TDP-43 (30–32), and TIA-1 (33), have provided valuable information regarding stress granule formation, dynamics, pathological disease states, and therapeutic reversal of pathologic processes. However, to our knowledge, this approach has not been applied to study the functional role of neuronal granules, including granule assembly and disassembly, the regulation of activity-dependent translation, and neuronal granule dysfunction related to neurodevelopmental disorders.
The Fragile X Mental Retardation Protein (FMRP) is an abundant RNA-binding protein found within neuronal granules (34–36), and is predicted from its sequence composition to phase separate (37). Previously, smaller FMRP protein fragments have been demonstrated to phase separate in vitro (37, 38). FMRP is multifunctional and is involved in numerous processes, including pre-mRNA processing (39), neuronal granule transport (40), translational regulation (41), and ion channel binding (42). However, its canonical and best-studied role is to repress translation in a phosphorylation-dependent manner via activity from neuronal G protein-coupled receptors, such as the metabolic glutamate receptor (mGluR) (43). One possible mechanism of mGluR stimulation involves activation of protein phosphatase 2A (PP2A) dephosphorylating FMRP and triggering translation (44). Following mGluR stimulation (>5 min), PP2A activity is inhibited and ribosomal protein S6 kinase 1 (S6K1) is activated, enabling phosphorylation of FMRP and translational repression via ribosomal stalling (45). Furthermore, casein kinase II (CKII) phosphorylates FMRP and promotes dynamic hierarchal multisite phosphorylation, suggesting that multiple pathways control FMRP phosphorylation (46–48). Besides phosphorylation, methylation of RGG motifs within FMRP has been suggested to prevent ribosomal stalling by decreasing FMRP association with polyribosomes (49, 50) and decreasing its binding affinity toward G-quadruplex–containing RNAs, a subset of FMRP targets (51–53). Loss of FMRP expression results in translational dysregulation and defective synaptic maturation leading to fragile X syndrome, one of the most common inherited forms of intellectual disability and autism spectrum disorders (ASDs) (54). Thus, FMRP is an integral neuronal granule component and plays a critical role in activity-dependent translation via posttranslational modifications.
Despite our understanding of the translational control by FMRP, FMRP-mediated translational regulation within the context of phase-separated neuronal granules remains enigmatic. Here, we show that FMRP forms membraneless foci in cells and liquid droplets with RNA in vitro. We identify that the C-terminal 188-residue low-complexity region (i.e., FMRPLCR) is necessary and sufficient for this behavior in vitro. We further demonstrate that translational repressors functioning with FMRP, including neuronal eukaryotic initiation factor 4E binding protein (4E-BP) 2 (55, 56) and microRNA (miRNA) 125b (57, 58), are sequestered into FMRPLCR-RNA droplets, which may complement their inhibitory activity. Most strikingly, we observe highly cooperative in vitro translation inhibition with increasing FMRPLCR concentration, which correlates well with its critical concentration for phase separation. This effect is tuned by posttranslational modifications, with FMRPLCR phosphorylation enhancing phase separation and translation inhibition while methylation opposes them. These results, combined with bioinformatics prediction of phase separation (37), suggest that phase separation may be a general mechanism involved in activity-dependent translation and therefore synaptic plasticity.
Results
FMRP Forms Membraneless Foci in Cells and Liquid Droplets with RNA in Vitro Mediated by Its Low-Complexity Region.
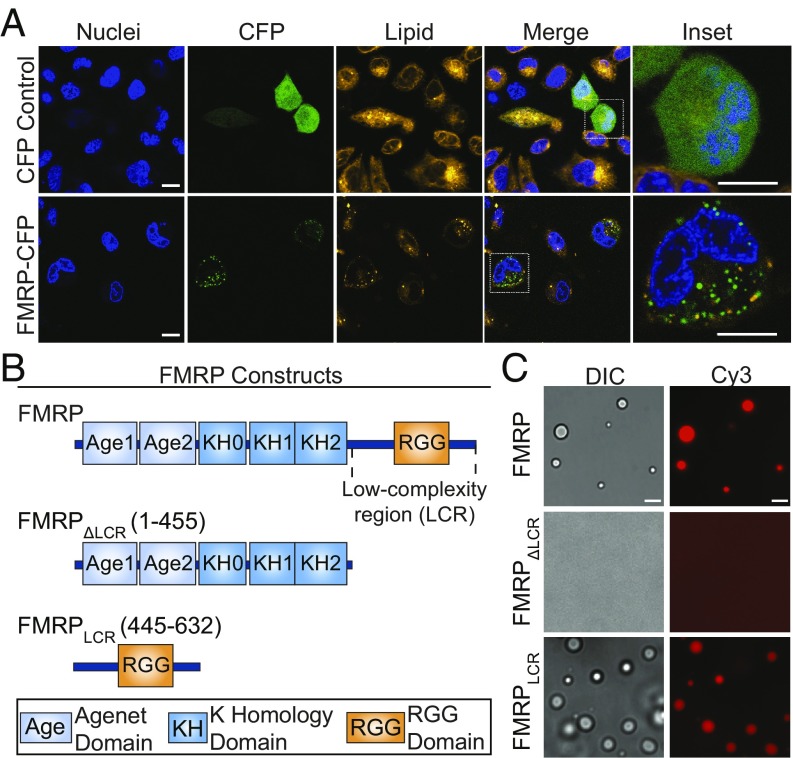
FMRP is a primary component of dynamic neuronal granules trafficking from the soma to dendrites (34–36) and is predicted to phase separate (37), suggesting that FMRP phase separation could play a role in neuronal granule formation and dynamics. Thus, we first tested whether FMRP can form membraneless assemblies, akin to neuronal granules, in cells, by transfecting CHO cells with a CFP fused to FMRP (FMRP-CFP) or CFP alone. FMRP-CFP induces the formation of distinct micrometer-sized foci in the cytoplasm, whereas transfection of CFP alone results in diffuse fluorescence (Fig. 1A). To test if FMRP-CFP foci are compartmentalized in a lipid-bound membrane, we used DiI as a lipid membrane marker (59). Within 30 min of treatment with DiI, live cells endocytose the dye, enabling diffusion throughout intracellular compartments via normal membrane dynamics. Lack of significant colocalization between FMRP-CFP with DiI is consistent with the observed foci being membraneless bodies that form independently of, and are not encapsulated by, lipid bilayers (Fig. 1A and SI Appendix, Fig. S1A). Although these experiments were performed in nonneuronal cells and by protein overexpression, they still highlight the ability of FMRP to form cellular non–membrane-bound cytoplasmic foci, similar to neuronal granules observed in neuronal cells.
Fig. 1.
FMRP forms membraneless foci in cells and liquid droplets in vitro mediated by its low-complexity region. (A) Spinning-disk confocal photomicrographs of CHO cells expressing CFP or FMRP-CFP. Images are stained with the lipophilic membrane dye DiI representing lipid membranes and Hoechst stain representing the nucleus. FMRP-CFP fluorescence is punctate and does not significantly localize with DiI. (Scale bar, 10 μm.) (B) Schematic of the three constructs studied: (i) the full-length FMRP, (ii) the N-terminal region (FMRPΔLCR) comprising two folded Agenet domains and three folded KH RNA-binding domains, and (iii) the intrinsically disordered low-complexity region (FMRPLCR) containing the Arg-Gly–rich RGG domain. (C) DIC and fluorescent images of FMRP, FMRPΔLCR, and FMRPLCR protein (50 μM) mixed with Cy3-labeled sc1 RNA (5 μM) in 25 mM Na2PO4, pH 7.4, 30 mM NaCl, 2 mM DTT. Images show that only FMRP and FMRPLCR form droplets with sc1 RNA. (Scale bar, 10 μm.)
To determine if purified recombinant human FMRP can phase separate in vitro and identify important protein regions responsible for this behavior, we prepared three different constructs: full-length FMRP, FMRP lacking the low-complexity region (FMRPΔLCR), and the 188-residue C-terminal low-complexity region of FMRP containing the RGG motifs (FMRPLCR; Fig. 1B). To simplify our understanding of the molecular interactions driving phase separation, no crowding reagents were added to any experiment unless stated. Differential interference contrast (DIC) and fluorescence microscopy were used to examine the formation of phase-separated protein-rich condensates, here simply referred to as droplets. At low in vitro protein concentrations (50 μM), droplet formation was not observed for any construct. As FMRP is an RNA-binding protein, we tested phase separation in the presence of an RNA binding partner, sc1 RNA. Sc1 is an extensively characterized in vitro selected 36-mer RNA molecule that forms a three-layered G-quadruplex linked to a stem duplex and binds to the RGG region of FMRP (52, 60). Upon addition of Cy3-labeled sc1 RNA, droplets were formed with FMRP and FMRPLCR, but not with FMRPΔLCR (Fig. 1C). Our results show that FMRPLCR alone is necessary and sufficient to drive phase separation with sc1 RNA in vitro. Given that FMRPLCR is known to specifically bind to the sc1 sequence, we wondered if FMRPLCR phase separation requires a specific RNA sequence or structure. By mixing different RNA sequences with FMRPLCR, we showed that FMRPLCR-RNA phase separation is not sequence-specific and does not require sequences capable of G-quadruplex formation (SI Appendix, Fig. S2). Hence, FMRPLCR-RNA phase separation is likely driven by a combination of protein:RNA electrostatic and pi–pi interactions (37), the latter involving pi electron orbitals of double bonds such as those found in RNA bases, protein backbone, and protein side-chain groups including aromatic, amide, guanidinium, and carboxyl groups. Given this behavior, we used FMRPLCR as a model system to recapitulate properties of FMRP-containing neuronal granules as a result of its importance in driving phase separation, its well-characterized role in RGG-mediated RNA binding (52, 53, 60), its numerous reported regulatory posttranslational modifications, and its stability at higher protein concentrations compared with the aggregation-prone full-length protein (SI Appendix, Fig. S1 B and C).
FMRPLCR and sc1 RNA Form Dynamic Liquid Droplets.
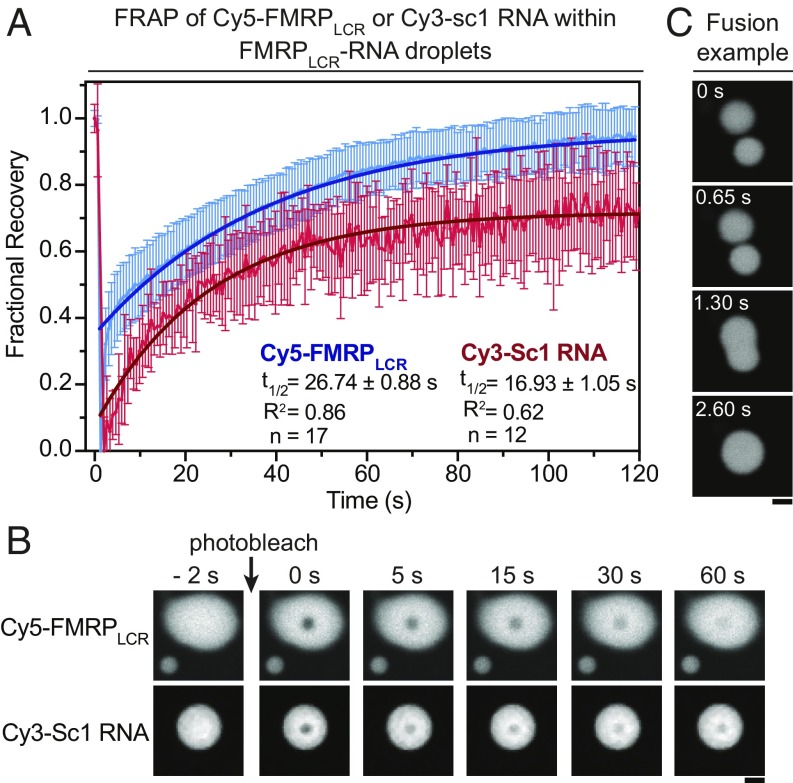
Phase separation produces RNA granules with diverse properties, including dynamic liquid droplets, hydrogels, and irreversible fibers and aggregates (extensively reviewed in refs. 11–13). As some RNA granules, including neuronal granules, have been described as liquid-like (61, 62), we next characterized the liquid properties of fluorescently labeled FMRPLCR-RNA droplets. By using fluorescence recovery after photobleaching (FRAP), we observed recovery of fluorescence of Cy5-tagged FMRPLCR (t1/2 = 26.74 ± 0.88 s) and Cy3-tagged sc1 RNA (t1/2 = 16.93 ± 1.05 s; Fig. 2 A and B), which demonstrates dynamic intradroplet molecular mobility. We also saw droplet fusion upon contact, which is consistent with the fusion of RNA granules observed throughout neuronal processes (36, 62) (Fig. 2C). These results demonstrate that FMRPLCR-RNA droplets are dynamic and liquid-like.
Fig. 2.
Phase separation of FMRPLCR with RNA leads to dynamic liquid droplets. (A) Fluorescence recovery after photobleaching of Cy5-FMRPLCR (150 μM) or Cy3-sc1 RNA (5 μM) within FMRPLCR-sc1 RNA droplets in buffer containing 25 mM Na2PO4, pH 7.4, 30 mM NaCl, and 2 mM DTT at room temperature. Fractional recovery of FMRPLCR (n = 17) and sc1 RNA (n = 12) is represented as the average ± SD and was fitted to a single exponential function. (B) Representative images of photobleaching. (Scale bar, 5 μm.) (C) FMRPLCR-RNA droplets fuse over time, demonstrating liquid-like properties. (Scale bar, 5 μm.)
To further characterize FMRPLCR phase separation, we used solution NMR spectroscopy. By concentrating FMRPLCR at physiological pH (7.4) and low salt conditions, the spontaneous fusion of FMRPLCR droplets creates a bulk two-phase partitioned liquid system: on the top, a protein-depleted phase, and on the bottom, a protein-condensed phase, representing a macroscopic FMRPLCR “droplet” (SI Appendix, Fig. S3A). Protein concentrations in each phase were measured by pipetting and diluting the solution and using absorbance at 280 nm. By producing a bulk two-phase system, we were able to record NMR spectra on different protein phases. Overlay of 1H-15N heteronuclear single-quantum coherence spectra of the protein-depleted (i.e., monomeric) and condensed phases shows almost identical chemical shifts with narrow amide proton dispersion (∼8.0–8.7 ppm), reflecting similar global disordered character (SI Appendix, Fig. S3B). The chemical shifts are the averaged result of the conformational exchange between distinct structures. The resonances of the condensed phase are significantly broadened compared with the dilute phase, which reflects the higher viscosity of a protein-condensed phase and agrees with NMR characterization of other phase-separating proteins (24, 63). Although this approach can be insensitive to the fractional population of large oligomers such as amyloid structures, in principle, it can detect the presence of partially or significantly stabilized helical or β-sheet structures that have been reported in other phase-separating proteins (25, 32). In contrast, our solution NMR data show FMRPLCR to be primarily structurally disordered in the condensed phase.
Translational Repressors and miRNAs Partition into, and Concentrate Within, FMRPLCR-RNA Droplets.
A primary mechanism of translational repression by FMRP involves ribosomal stalling through FMRP interacting with mRNA (64) or directly binding to the ribosome (65). However, FMRP can also interact with numerous other proteins and RNAs to inhibit translation through nonexclusive and complementary mechanisms (43). These include inhibiting cap-dependent translation initiation through interacting with cytoplasmic FMRP-interacting protein (CYFIP1), a eukaryotic initiation factor 4E-BP (56), or by associating with miRNAs that have near-perfect complementary base pairing with target mRNAs (57). To understand how FMRP may interact with these diverse components in the context of a neuronal granule, we tested if FMRPLCR-RNA phase separation can concentrate two representative components in these pathways: (i) 4E-BP2, a neuronal cap-dependent translation initiation inhibitor (55, 56); and (ii) miRNA-125b, an miRNA that requires FMRP to regulate neuronal spine morphogenesis (57). We labeled 4E-BP2 and miRNA-125b with a Cy3 dye and mixed a small concentration (500 nM) of each component with FMRPLCR-RNA droplets. By using a spin-down assay, we detected Cy3 fluorescence in the protein-depleted phase (after phase separation) and compared it to Cy3 fluorescence in the total mixture (before phase separation; Fig. 3A). A high Cy3 percentage remaining in the protein-depleted phase is interpreted as exclusion from the droplet, whereas a low percentage is interpreted as favorable partitioning into the droplet. As expected based on the general RNA-interacting properties of FMRPLCR, Cy3-labeled sc1 RNA and miRNA-125b are nearly completely sequestered into droplets (>90%), whereas 4E-BP2 strongly partitions into the droplet (∼60%; Fig. 3B). In contrast, the free Cy3 dye and Cy3-BSA exhibit negligible partitioning (Fig. 3B). To confirm our results, we used fluorescence microscopy to visualize the partitioning of labeled Cy3 species into FMRPLCR-RNA droplets (Fig. 3C). Images were taken at the focal plane of the coverslip where droplets settled; only droplets of similar sizes (radius of 1–5 μm) were used in the analysis. In agreement with the experiments described here previously, partition coefficients, calculated from the mean fluorescence intensities in the droplet compared with the bulk solution, show that sc1 RNA (21.4 ± 3.7) and miR-125b (19.2 ± 3.1) are almost exclusively partitioned into FMRPLCR-RNA droplets, whereas 4E-BP2 (6.2 ± 1.1) partitions reasonably well (Fig. 3D). The Cy3 dye alone (1.92 ± 0.58) and Cy3-BSA control (1.41 ± 0.31) show negligible partitioning (Fig. 3D). Differences in partitioning between the RNAs and 4E-BP2 are likely a result of the differences in interactions of available chemical groups of the partitioning molecule with those in the FMRPLCR-sc1 RNA mixture. For instance, 4E-BP2 could interact with either or both FMRPLCR and sc1 RNA, which together may function to capture it into the FMRPLCR-RNA droplet. These physical interactions appear to drive components known to repress translation, including 4E-BP2 and miRNA-125b, into FMRPLCR-RNA droplets to variable degrees, concentrating and localizing them, which may potentially function to enhance their inhibitory activity.
Fig. 3.
Translational repressors 4E-BP2 and miR-125b partition into and concentrate within FMRPLCR-RNA droplets. (A) Schematic illustration depicting spin-down assay. (B) Partitioning of 500 nM Cy3-labeled dye, BSA, sc1-RNA, miR-125b, and 4E-BP2 into condensed FMRPLCR-RNA droplets quantified from the percentage of Cy3 fluorescence remaining in the dilute phase. No crowding reagents were used. Plot represents mean percentage of Cy3 fluorescence, and error bars represent SD from three independent experiments. (C) Example of a droplet observed from DIC and fluorescence microscopy of mixtures from B confirms the results of the spin-down assay. Differing background intensities are inherent to the different partitioning of dye-labeled molecule in each mixture. (Scale bar, 5 μm.) (D) Partition coefficients are quantified by the fluorescence intensity within droplets compared with the background solution intensity from microscopy images. Plot represents mean values, and error bars represent the SD.
Ionic Strength Modulates FMRPLCR-RNA Phase-Separation Propensity.
As FMRPLCR-RNA phase separation can capture and concentrate different proteins and RNAs involved in FMRP-mediated translational repression, we wondered how phase separation could be reversed to alleviate translational inhibition by releasing mRNAs for active translation. First, we confirmed that FMRPLCR phase separates without any additives and that addition of RNA increases its phase-separation propensity at protein concentrations closer to physiological levels (SI Appendix, Fig. S4A). We then tried perturbing electrostatic interactions present within FMRPLCR-RNA droplets. By using DIC microscopy to map a phase diagram in varying protein and salt concentrations, we found that increasing salt concentrations disfavor FMRPLCR droplet formation in the presence of sc1 RNA (SI Appendix, Fig. S4B). To test whether this result is caused by a general ionic strength effect or a specific salt ion effect, we incubated FMRPLCR-RNA droplets in salt solutions with differing valences (KCl, MgCl2, CaCl2) and found that identical ionic strengths of different salt solutions disrupt droplet formation to a similar degree (SI Appendix, Fig. S4C). However, the divalent ions Mg2+ and Ca2+ qualitatively appear to have a slightly stronger ability to disrupt droplet formation. Thus, ionic strength can regulate FMRPLCR-RNA droplet formation in vitro, likely as a result of modulating electrostatic interactions, including between protein and RNA.
FMRPLCR Phosphorylation Increases Its Phase-Separation Propensity Possibly Through Increasing Negative Charge Densities of Glutamic/Aspartic Acid-Rich Clusters.
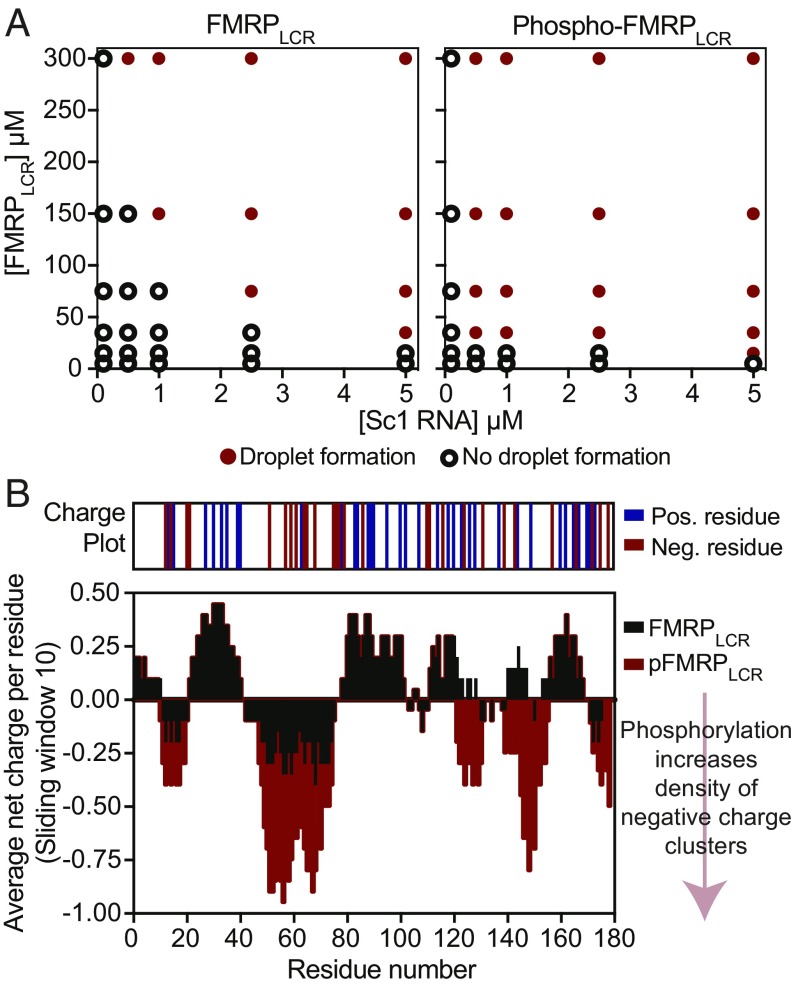
Besides the effect of ionic strength, posttranslational modifications can alter the biophysical properties of IDRs, which in turn can regulate their phase-separation behavior (66). As activity-dependent phosphorylation is known to regulate neuronal granule trafficking (34) and FMRP-controlled translation (67), we next investigated its role in FMRP phase separation. We first performed in vitro phosphorylation of recombinant FMRPLCR (phospho-FMRPLCR) by using CKII, a kinase that phosphorylates FMRP in vivo (46) and mediates dynamic multisite phosphorylation by other kinase/phosphatase systems, including S6K1/PP2A (47). Prolonged in vitro phosphorylation by CKII results in an increase of ∼640, 720, or 800 Da, which is equivalent to the addition of 8, 9, or 10 phosphate groups, respectively (SI Appendix, Fig. S5A). After the phosphorylation reaction, we removed the kinase via gel filtration and dialyzed the protein into our assay buffers. At room temperature, and in the absence of RNA and crowding reagents, we observed that phospho-FMRPLCR (containing 8–10 phosphate groups added) readily phase separates at ∼250 μM, whereas FMRPLCR does not. Furthermore, by using microscopy to map and compare the phase diagrams of phospho-FMRPLCR and FMRPLCR (pH 7.4 and 150 mM KCl), we found that moderately phosphorylated FMRPLCR (ranging from three to five groups, reflective of the phosphorylation state in vivo; ref. 47) increases its propensity to form liquid-like droplets with sc1 RNA at lower protein concentrations (Fig. 4A). Therefore, phosphorylation promotes phase separation in vitro, suggestive of a role in promoting FMRP-containing neuronal granule formation in cells.
Fig. 4.
Phosphorylation of FMRPLCR enhances FMRPLCR phase separation with sc1 RNA. (A) Phase diagrams as a function of different sc1 RNA and protein concentrations are shown for nonphosphorylated and phosphorylated FMRPLCR (range of three to five phosphate groups added) in 25 μM Na2PO4, pH 7.4, 100 mM KCl, and 2 mM DTT at room temperature. A red circle represents droplet formation, and an open circle represents no droplet formation detected. Phosphorylation enhances phase separation. (B) Average net charge plot (10-residue sliding window) of FMRPLCR in black, overlaid with the net charge plot of phospho-FMRPLCR in red, assuming all 12 serine residues that fit the CKII phosphorylation motif are phosphorylated. Phosphorylation increases the local negative charge densities in FMRPLCR sequence.
To gain insights into how phosphorylation enhances FMRPLCR phase separation, we analyzed the charge pattern in FMRPLCR in view of its role in regulating phase separation (21, 68, 69). The average net charge per residue plot of FMRPLCR reveals an alternating pattern of negatively charged glutamic/aspartic acid (E/D) clusters separated by positively charged arginine/lysine clusters (Fig. 4B). We identified 12 potential serine phosphorylation sites in FMRPLCR that fit the general CKII phosphorylation motif, which contains acidic E/D residues C-terminal to the serine phosphoacceptor site (SX1–5E/D) (70, 71) (SI Appendix, Fig. S5B). Assuming phosphorylation can occur at all 12 sites, we compared calculated average charge plots for FMRPLCR and phospho-FMRPLCR and found that phosphorylation increases the negative charge density around E/D clusters (Fig. 4B). This suggests that multisite phosphorylation may promote phase separation by exaggerating the negative charge densities and increasing their potential for forming multivalent electrostatic interactions between other charge clusters.
FMRPLCR Methylation Decreases Phase-Separation Propensity Without Notably Altering sc1 RNA-Binding Affinity.
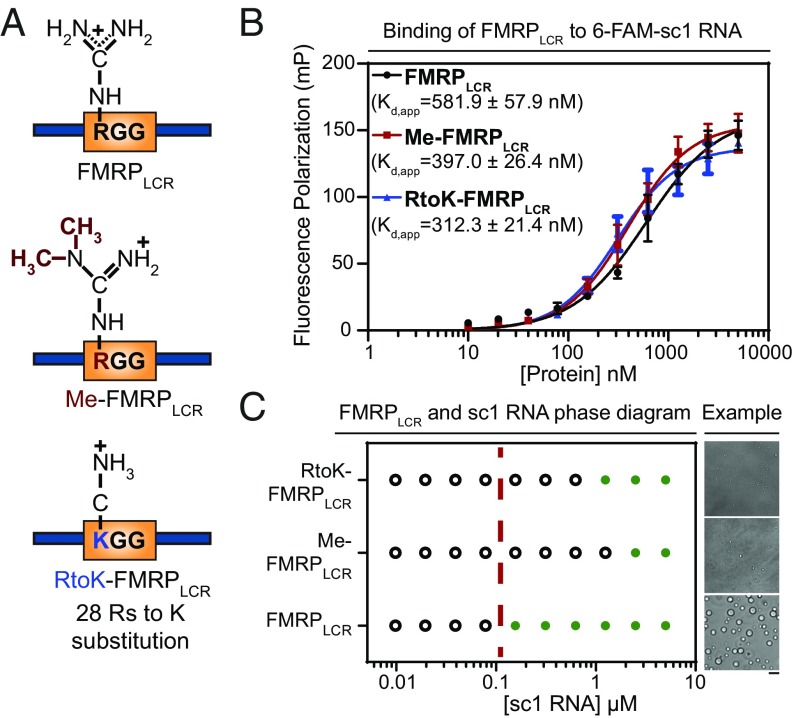
Arginine methylation is another posttranslational modification that can modulate phase-separation propensities (66). Previous work has shown that the RGG motif of FMRP is methylated in vivo, which has been reported to weaken its binding to G-quadruplex mRNAs and oppose translational repression as a result of ribosome stalling (49, 50). Thus, we hypothesized that methylation may regulate phase separation by modulating RGG-mediated RNA-binding interactions. To generate methylated FMRPLCR (Me-FMRPLCR), we used protein arginine N-methyltransferase 1 (PRMT1), an enzyme known to methylate RG motifs of FMRP in vivo (51). PRMT1 catalyzes the addition of two methyl groups to the same arginine guanidine nitrogen atom, forming asymmetric N,N-dimethyl-l-arginine (Fig. 5A and SI Appendix, Fig. S6A). Coexpression of PRMT1 and FMRPLCR yields a protein with a ∼196- or 224-Da increase in FMRPLCR molecular weight, consistent with asymmetrical dimethylation of seven or eight RG sites, respectively, or possibly incomplete dimethylation (i.e., monomethylation) on a large number of sites (SI Appendix, Fig. S6 B and C). PRMT1 was removed from the samples via gel filtration before any assays were performed. To evaluate differences in binding affinity between methylated and nonmethylated FMRPLCR, we used fluorescence polarization (FP) with 6-FAM–labeled sc1 RNA. Surprisingly, FMRPLCR and Me-FMRPLCR bind to sc1 RNA with similarly high nanomolar affinity (Fig. 5B). This suggests that binding between FMRPLCR and sc1 RNA may be more dynamic than the stable interaction depicted by the crystal structure of a small RGG peptide in complex with sc1 (Protein Data Bank ID codes 5DE5, 5DE8, and 5DEA) (72). Thus, IDR-mediated interaction with RNA may be more dependent on broad electrostatic properties than specific binding geometries. Confirming this, we showed that a charge-conserved mutant that abolishes RGGs by substituting all 28 arginine residues in the sequence with lysines, RtoK-FMRPLCR, has a comparable binding affinity (Fig. 5 A and B).
Fig. 5.
Arginine perturbations disrupt FMRPLCR-sc1 RNA phase separation but do not notably affect FMRPLCR binding to sc-1 RNA. (A) Schematic illustration of the three constructs used. (B) Similar binding affinities for interactions between FMRPLCR, Me-FMRPLCR, and RtoK-FMRPLCR with 6-FAM–labeled sc1 RNA (25 nM) are shown using FP assayed in 25 mM Na2PO4, pH 7.4, 100 mM KCl, 2 mM DTT, 0.02 mg/mL E. coli tRNA, and 0.01% Nonidet P-40. Apparent dissociation constants (kd,app ± SD) determined through Hill binding equation (SI Appendix, Eq. S2) from three experimental replicates. (C) Phase diagram of FMRPLCR, Me-FMRPLCR, and RtoK-FMRPLCR at a constant protein concentration of 100 μM with increasing sc1 RNA concentrations in buffer containing 25 mM Na2PO4, pH 7.4, 30 mM KCl, and 2 mM DTT. Green circles represent an observation of droplet formation, and open circles represent no droplet formation detected. The dotted red line is a visual aid for a qualitative phase boundary of FMRPLCR. (Right) Representative DIC microscopy images of droplets for the addition of 2.5 μM of RNA in the conditions described. (Scale bar, 5 μm.) Phase-separation propensity is dramatically decreased for methylated and RtoK mutant proteins, with smaller droplet sizes observed.
As FMRLCR phase separates at a ∼20-fold lower concentration in the presence of sc1 RNA (SI Appendix, Fig. S4A), and all three constructs, FMRPLCR, Me-FMRPLCR, and RtoK-FMRPLCR, bind sc1 RNA with similar affinity, we surmised that their phase-separation propensities in the presence of RNA would be similar. However, contrary to our expectations, Me-FMRPLCR and RtoK-FMRPLCR have a dramatic decrease in phase-separation propensity compared with FMRPLCR (Fig. 5C). This suggests that arginine perturbations, either by methylation or reduction in pi character by substitution of arginine for lysine residues, perturb important interactions that facilitate the general phase-separation behavior of FMRPLCR, as previously found for Ddx4 and the low-complexity regions of FMRPLCR (21, 37). Thus, methylation represents a posttranslational modification that decreases FMRP phase-separation propensity in vitro, and, by extension, may be important for facilitating neuronal granule disassembly in cells.
In Vitro Translation Inhibition Coincides with FMRPLCR Phase Separation and Is Regulated by Posttranslational Modifications.
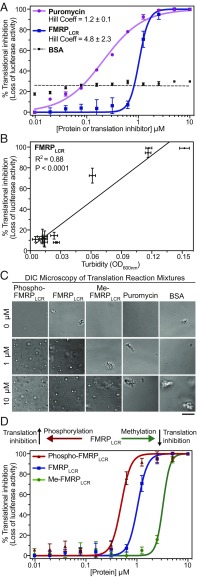
To test the proposed model whereby neuronal granule formation represses translation by “masking” mRNAs and granule disassembly allows translation to occur by liberating mRNAs from translational repressors within a neuronal granule (8), we adapted an in vitro rabbit reticulocyte lysate translation assay measuring firefly luciferase activity (73) (full details are provided in SI Appendix, SI Materials and Methods). In this assay, translational efficiency is determined by the amount of luciferase mRNA translation measured by luminescence (i.e., luciferase activity). Addition of increasing amounts of FMRPLCR to a rabbit reticulocyte lysate mixture with luciferase mRNA inhibits translation in a cooperative manner (Hill coefficient = 4.8 ± 2.3), whereas the protein synthesis inhibitor puromycin inhibits translation in a noncooperative dose-dependent manner (Hill coefficient = 1.2 ± 0.1); as a control, the addition of BSA only weakly reduced overall translation in a nonspecific manner (Fig. 6A). Thus, puromycin and FMRPLCR inhibit translation via different mechanisms. Moreover, we noticed a significant positive correlation (R2 = 0.88, P < 0.0001) between the turbidity of the FMRPLCR translation assay mixture from Fig. 6A and translation inhibition (Fig. 6B). The increasing turbidity of the mixture corresponds to liquid droplet formation, as detected by DIC microscopy of the assay mixture (Fig. 6C). Note that the concentrated lysate mixture alone contains some inherent aggregates, for which we have accounted in our turbidity measurements (Fig. 6C). The cooperative switch-like appearance of the FMRPLCR curve, instead of the noncooperative trend observed with puromycin, indicates that the “all-or-nothing” property of phase separation, i.e., droplet formation or no droplet formation, represents a mode of translation inhibition different from puromycin-induced translation inhibition, which involves premature chain termination during translation on the ribosome (74).
Fig. 6.
In vitro translation inhibition by FMRPLCR is mechanistically distinct from inhibition by puromycin, and posttranslational modifications in FMRPLCR correlate with phase-separation propensities. (A) Translation inhibition of luciferase mRNA is determined by percent decrease in luminescence in the presence of FMRPLCR, puromycin, or BSA. The rabbit reticulocyte lysate, pH 7.4, is nuclease-treated. (B) Correlation between translation inhibition and the turbidity of the FMRPLCR translation assay mixture from A is suggestive of droplet formation. (C) Representative DIC microscopy images of in vitro translation assay mixtures at 0, 1, and 10 μM protein or puromycin concentrations correlate the presence of droplet formation with inhibitory translation response in A and D. Some aggregates inherent to the concentrated lysate mixture are observed. (Scale bar, 5 μm.) (D) Phospho-FMRPLCR (containing three to five phosphates), FMRPLCR, and Me-FMRPLCR were titrated into the luciferase translation reaction. A switch-like inhibitory response is observed and is shifted left or right with phosphorylation and methylation, respectively. Error bars show the SD from two independent experimental replicates.
To examine the role of posttranslational modifications in regulating translation via phase separation, we tested phospho-FMRPLCR and Me-FMRPLCR in our assay and observed opposite effects. Phospho-FMRPLCR (containing three to five phosphate groups) shifted the curve to the left, representing a lower concentration threshold necessary for translation inhibition, whereas Me-FMRPLCR shifted the curve to the right, representing a higher concentration threshold (Fig. 6D). Importantly, DIC microscopy of the assay mixtures from variously modified FMRPLCR correlates droplet formation to the apparent switch-like nature of translation inhibition (Fig. 6C). In our assay, phase separation of FMRPLCR is significantly enhanced in the presence of rabbit reticulocyte lysate compared with our earlier experiments, which were all performed in the absence of crowding reagents. The relatively large concentration required for isolated FMRPLCR phase separation is far from cellular levels, with addition of RNA decreasing the FMRPLCR concentration required. The decreased FMRPLCR concentrations seen to phase separate in our translation assays, however, particularly with highly phosphorylated protein (∼1 μM), are close to what could be expected in cells. Thus, FMRPLCR phase separation in vitro, with posttranslational modifications potentially acting as a switch at near physiological concentrations, represents a distinct mechanism for translation inhibition.
Translation Inhibition by Other Phase-Separating Proteins.
Neuronal granules are not the only intracellular RNA granules linked to inhibition of translation (75). Moreover, a correlation between negative regulation of translation and predicated phase-separation propensities has been previously reported (37). Thus, to further explore the relationship between translational inhibition and phase separation of RNA granules, we undertook a bioinformatic assessment of proteins involved in different RNA granule types, including stress granules and processing bodies (full details are provided in SI Appendix, SI Materials and Methods). We based our analysis on a set of 144 proteins identified as the core cytoplasmic RNA granule proteome by Bio-ID interactions (76). Within this select proteome, we find that some proteins, including FMRP, interact broadly with the entire core set rather than exclusively belonging to a single granule type. Furthermore, we find that these highly interacting proteins are more likely to be involved in negative regulation of translation [by Gene Ontology (GO) annotation, 0017148] (77, 78) and are more likely to be predicted to phase separate (37) (SI Appendix, Fig. S7 A and B). Within the 144 core protein set, 14 proteins are predicted to phase separate and are annotated to negatively regulate translation. Of these 14 proteins, 5 are associated, either directly or by homology, to synaptic plasticity by functioning as translational repressors, including FMRP, CPEB4, GIGYF2, Caprin1, and FXR2 (SI Appendix, Table S1). Therefore, our bioinformatics analysis supports phase separation as a more general mechanism used by the cell to modulate translation with functional consequences impacted by synaptic plasticity.
Discussion
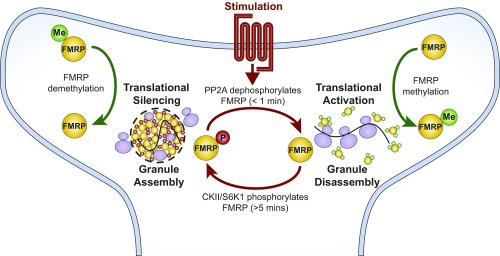
A detailed mechanistic understanding of activity-dependent translation is important for understanding fundamental synapse biology and neurological disorders rooted in translational dysregulation. In our present work, we show that the low-complexity region of FMRP is sufficient to interact with RNA to form dynamic liquid droplets in vitro and that this process coincides with in vitro translation inhibition. We also find that FMRPLCR phase separation with RNA could potentially inhibit translation by multiple nonexclusive mechanisms, including concentrating and forming inhibitory complexes with translational repressors 4E-BP2 and miRNA-125b. In terms of regulating phase separation, FMRPLCR phosphorylation increases its phase-separation propensity, favoring granule assembly, whereas FMRPLCR methylation decreases its phase-separation propensity, favoring granule disassembly. Compellingly, in vitro translation inhibition correlates with the changes of phase-separation propensities by posttranslational modifications, supporting the model that posttranslational modifications arising from synaptic signaling may control neuronal granule formation, which regulates activity-dependent translation (Fig. 7). Moreover, the mechanism of simply capturing and releasing target mRNAs during reversible granule formation allows the advantage of repeated local translation of the same mRNA transcript. Although Fig. 7 is evocative of a dendritic spine, neuronal granules containing FMRP are present in dendritic spines and shafts, consistent with the significant literature on postsynaptic mRNA translational regulation by FMRP; they are also present and act to regulate translation in axons, including during development (79). In summary, our findings suggest that protein and RNA phase transitions provide a potential mechanism for how mRNAs move from a repressed state (within granules) into an active state (translating polyribosomes) (5) by cycles of mRNA “masking” and “unmasking” (8) (Fig. 7). Importantly, the results of this study raise several key points regarding synaptic regulation of phase separation, mechanistic insights into FMRP function, and physiological and disease implications.
Fig. 7.
Model for reversible neuronal granule assembly during activity-dependent translation. During neuronal stimulation, such as mGluR1/5 activation, PP2A dephosphorylates FMRP, which promotes neuronal granule disassembly and activates translation. FMRP methylation complements granule disassembly and translational activation by reducing its propensity to form higher-order assemblies with mRNA and polyribosomes. Poststimulation (>5 min), PP2A activity is inhibited, and S6K1 phosphorylates FMRP, which promotes neuronal granule assembly and facilitates translational silencing. FMRP demethylation complements granule formation by promoting higher-order interactions with other proteins, mRNA, and polyribosomes.
Activity-Dependent Regulation of FMRP Phase Separation via Posttranslational Modifications and Ionic Strength.
Regulating FMRP phosphorylation is important for facilitating its ability to repress translation (41). It is well established that signaling through mGluR and other related neuronal GPCRs can regulate the phosphorylation status and in turn translational activity of FMRP via the PP2A/S6K1 system (43). In addition, CKII is another kinase identified to phosphorylate FMRP and promote dynamic hierarchal multisite phosphorylation by other kinases (47), demonstrating that there are multiple activity-dependent phosphorylation pathways regulating FMRP. To date, there have been eight phosphorylation sites identified in the C-terminal region of FMRP (80), but only phosphorylation of Ser500 has been extensively characterized and linked to translational regulation (67). Our results emphasize that an important consequence of hierarchal phosphorylation could be to regulate phase separation, which links reversible granule formation to activity-dependent translation. More broadly, phosphorylation regulates phase separation of other neuronal proteins including FUS (25, 81), Tau (82, 83), and CPEB4 (84), highlighting the importance of phosphorylation as a regulatory switch for controlling phase separation.
Our data show that arginine methylation decreases FMRP phase-separation propensity, consistent with previous studies investigating the importance of RGGs in regulating phase separation (38). Specifically for FMRP, methylation inhibits FMRP self-interactions, reported as dimerization (50), but may, in fact, represent higher-order associations and phase separation. Methylation of other RNA-binding proteins has been demonstrated to perturb phase separation, including Ddx4 (21), FUS (28), and HnRNP A2 (85), supporting the crucial role of methylation in modulating phase separation. In contrast to phosphorylation, the dependence of FMRP methylation on synaptic activity and associated signaling pathways is somewhat less clear. However, multiple lines of evidence support the role of arginine methylation in controlling activity-dependent translation. Protein arginine methyltransferase (PRMT) 8 has been shown to localize to the synapse, and loss of PRMT8 alters synaptic protein levels, causing behavioral defects in mice (86). Inhibition of coactivator-associated arginine methyltransferase 1 (CARM1 or PRMT4), another postsynaptic protein, results in a substantial increase of synaptic proteins, suggesting a regulatory role for translational regulation (87). Importantly, inhibiting methylation of FUS, which enhances its phase separation, reduces protein synthesis in axon terminals (28). In agreement, our results indicate that FMRP methylation decreases its overall phase-separation propensity with RNA and that this effect is correlated with the relative decrease of in vitro translation repression. In other words, our work suggests that FMRP methylation opposes the formation of higher-order protein assemblies, i.e., phase separation, that would otherwise repress translation. Based on published observations coupled with our findings, we speculate that FMRP arginine methylation contributes to the disassembly of neuronal granules (Fig. 7).
Although posttranslational modifications are a downstream effect of neuronal stimulation that can modulate phase-separation propensities, neuronal stimulation is also coupled to transient ion fluxes localized to tiny dendritic compartments that may also affect phase separation. For example, transient bursts of calcium concentrations in combination with synergistic posttranslational modifications may play a role in reversible granule formation. This is supported by our data on the sensitivity of FMRPLCR phase separation to ionic strength. We speculate that, during neuronal stimulation, the transient influx and transient release of calcium ions stored within the endoplasmic reticulum (88, 89) may disrupt protein–RNA and protein–protein interactions within neuronal granules to favor granule disassembly. Moreover, FMRP interacts directly with ion channels and regulates calcium signaling via mechanisms that are not well-understood (42, 90). Thus, a potential novel role for the low-complexity region of FMRP is to act as a direct biophysical calcium sensor within a neuronal granule.
Phase Separation Provides Unifying Insights into FMRP’s mRNA Binding Affinity and Role as a Translational Regulator.
FMRPLCR phase separation represents a simplified system lacking the folded regions that may further contribute to phase separation. Effects of synergistic interactions between the folded and low-complexity regions and interactions between the RNA-binding domains and RNA on FMRP phase-separation behavior remain untested here; previous data suggest that the disordered region is required to work with the folded KH domains to regulate protein synthesis and synaptic activity (91). Nonetheless, our reductionist approach provides important insights into the diverse functions of FMRP and may clarify a number of perplexing questions present in the FMRP literature. Previous work has shown that FMRP has more than 800 mRNA targets, estimated to be ∼4% of total brain mRNAs (64, 92). This vast number of mRNA targets cannot be accounted for by specific binding to RNA-binding domains and/or RGG motifs. Hence, the lack of FMRP-mRNA specificity has been a longstanding enigma. The observation that condensed FMRPLCR concentrates mRNAs in a dynamic manner that does not depend on rigid RGG-driven structural interactions provides a potential explanation. However, because there are thousands of mRNAs found localized to dendritic and axon regions (64, 92), this raises the questions of how neuronal granules recognize specific mRNA targets. We speculate that targeting of a specific subset of mRNAs occurs by cophase separation of FMRP with other RNA-binding proteins in neuronal granules. This is consistent with proteomic data showing heterogeneous protein populations in distinct neuronal granule populations (93). Moreover, additional FMRP specificity for mRNAs may also emerge from a combination of interactions with disordered regions and the folded KH domains not present in our experimental system.
Following decades of research, the mechanism responsible for FMRP’s translation inhibition activity remains controversial (43). FMRP has been suggested to have multiple repression activities, including inhibition of translational initiation (56), stalling of translocating ribosomes (64, 65), and interacting with miRNA (57) and the RNA-induced silencing complex (57). Our results suggest that reversible FMRP-RNA phase separation may be a molecular mechanism that unifies these diverse regulatory models. FMRP phase separation can partition 4E-BP2 to inhibit translation initiation, concentrate FMRP and mRNA to block ribosome translocation, and facilitate interactions with miRNAs even though FMRP lacks a canonical miRNA-binding domain. Thus, FMRP phase separation can prime translational repression by concentrating target mRNAs into a localized environment enriched with FMRP and other translational repressors. However, determining which translational mechanism may be dominant under different circumstance requires further investigation, and will likely depend on multiple molecular partners and signaling pathways.
Interestingly, Drosophila FMRP (dFMRP) has been shown to enhance the translation of long mRNAs in Drosophila melanogaster oocytes (94), in contrast to FMRP’s established role as a translational inhibitor. FMRP/dFMRP behavior may vary between tissues and oocytes and depend on cell type, developmental stage, and available protein interactions. In addition, the low-complexity disordered regions of FMRP and dFMRP differ in net charge, enrichment of RGG motifs, and predicted CKII phosphorylation sites. Thus, we speculate that differences in regulation of phase separation could lead to distinct dFMRP and FMRP activities.
General Physiological and Disease Implications.
Although we use FMRP as a model representing neuronal granules to investigate the relationship between phase separation and translation inhibition, our findings may be applicable to many other proteins and different cellular RNA granules involved in translational repression. A wide range of proteins has been shown to repress translation, with more than 100 human genes annotated to have related roles in the GO database (annotation 0017148) (77, 78). Moreover, this category has been previously found to be enriched in proteins predicted to phase separate (37). Together, our analysis supports our conclusion that the RNA-masking properties of phase separation may be a distinct and ubiquitous mechanism of translational inhibition.
From our bioinformatics assessment, we found that the regulation of synaptic plasticity involved a much smaller set of proteins, such that the statistical significance of observed enrichment cannot be gauged, but we can identify other proteins involved in synaptic plasticity that are known or predicted to phase separate (SI Appendix, Table S1). Specific mechanisms may differ between proteins, but the basic property of phase separation appears to be common in pathways involved in activity-dependent translation.
As a result of the widespread importance of translational regulation, dysregulation of translation, specifically at the neuronal synapse, is a feature of many neurodevelopmental disorders, including fragile X syndrome and ASDs (9, 95). Cell-based and mouse model experiments support the concept that expressing more or less FMRP is linked to neurological disorders; in other words, too much or too little FMRP is deleterious (96). Intriguingly, there is a strong overlap between FMRP-regulated gene expression and genes encoding proteins that control synaptic structure and that have rare mutations implicated in ASDs. For example, SynGAP and PSD-95 are proteins that phase separate to maintain synapse architecture (97), contain mutations linked to ASDs (98, 99), and have their mRNA translation regulated by FMRP (41). Questions emerge now regarding how the condensed and crowded postsynaptic environment affects phase-separation propensities of FMRP and other phase-separating proteins in normal and disease states. Future work building on understanding the regulation of phase separation at the neuronal synapse will provide important details regarding translational regulation, RNA localization, and neurodevelopmental disorders.
Materials and Methods
A brief summary of the methods is provided here. Full details are provided in SI Appendix, SI Materials and Methods.
Protein Expression and Purification.
All proteins, including FMRP, FMRPΔLCR, FMRPLCR, and RtoK-FMRPLCR were expressed and purified in Escherichia coli BL21 (DE3) Codon Plus Cells (Agilent). FMRPLCR (37) and RtoK-FMRPLCR (37) were purified as previously described.
Live-Cell Experiments and Imagining.
CHO cells were grown at 37 °C (5% CO2) in F-12K Nutrient Mixture (Gibco) supplemented with 1% penicillin/streptomycin (10,000 U/mL; Thermo Fisher Scientific) and 10% FBS (HyClone). Cells were transfected with 2.4 µg of CFP or FMRP-CFP plasmid DNA by using Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer’s protocol. Images were taken on a Quorum spinning-disk confocal microscope (Olympus IX81) with a 60×/1.35 (oil immersion) lens and a Hamamatsu C9100-13 EM-CCD camera.
RNA Oligonucleotide Sample Preparation.
Sc1 RNA, 5′-labeled Cy3-sc1 RNA, 5′-labeled Cy3-miRNA-125b, and 5′-labeled 6-FAM-sc1 RNA were purchased from Sigma as lyophilized samples. Stocks of 100 μM were made by resuspending in Tris⋅EDTA (TE buffer, pH 7.4). Working stocks of 10 μM were made by further dilutions into TE buffer and stored at −20 °C until use. Stock solutions of total yeast RNA or homopolymers (polyA, polyU, polyC, polyG; Sigma) were prepared by resuspending lyophilized samples in TE buffer into 1-mg/mL stocks and diluted into appropriate concentrations.
FRAP Method and Analysis.
FRAP experiments were performed on a Leica DMI6000 SP8 confocal microscope with a 63×/1.4 oil objective by using a Hamamatsu C9100-13 EM-CCD camera. The recovery intensities were corrected for overall background photobleaching during image acquisition, normalized to the same “prebleach” spot intensity, and fitted to a single exponential function.
Preparation of Condensed Phase-Separated FMRPLCR for NMR Spectroscopy.
Condensed phase-separated FMRPLCR NMR samples were prepared as previously described for Ddx4LCR (63).
In Vitro Partitioning Assay.
Partitioning was determined by Cy3 concentrations in the protein-depleted or condensed phases by measuring absorbance at 552 nm with an extinction coefficient of 0.15 µM−1⋅cm−1.
DIC and Fluorescence Microscopy of Phase-Separated Samples.
DIC and fluorescence images were obtained on an Axio Observer 7 bright-field and fluorescence inverted microscope (Carl Zeiss) with a 40× (air) or 63× objective (air) by using an Axiocam 702 mono CMOS camera.
Phosphorylation and Methylation of FMRPLCR.
CKII phosphorylation of FMRPLCR (phospho-FMRPLCR) was performed on purified FMRPLCR and verified by using intact MS. Methylation of FMRPLCR (Me-FMRPLCR) by PRMT1 was performed by using a previously described method (21).
Binding Assay Using FP.
The SpectraMax i3x Multi-Mode Plate Reader was used to determine apparent dissociation constants between 5′-labeled 6-FAM sc1 RNA and different FMRPLCR constructs.
In Vitro Translation and Luciferase Detection Assay.
In vitro translation of luciferase mRNA (Promega) was performed with a nuclease treated Rabbit Reticulocyte Lysate System (Promega) with slight modifications (SI Appendix, SI Materials and Methods) and monitored with a standard luciferase assay (Promega) using a SpectraMax i3x Multi-Mode Plate Reader (Molecular Devices).
Supplementary Material
Acknowledgments
We thank Jacob Brady, Tae Hun Kim, P. Andrew Chong, Peter St. George-Hyslop, Avi Chakrabartty, and Michael Salter for useful discussions and comments on the manuscript; Iva Pritisanac for helpful discussion and analysis of FMRP/dFMRP properties; and Ranjith Muhandiram in the laboratory of Lewis Kay for help in setting up NMR experiments. This work was supported by the Canadian Institutes of Health Research (CIHR; to J.D.F.-K. and L.-Y.W.), a SickKids Center for Brain and Mental Health Chase-an-Idea Grant (to L.-Y.W.), and a CIHR Vanier Scholarship (to B.T.). The authors acknowledge the use of The Hospital for Sick Children Structural & Biophysical Core Facility.
Footnotes
Conflict of interest statement: J.D.F-K. and R.W.K. were coauthors on a 2016 review article. J.D.F-K., R.W.K., and G.S. were coeditors of a 2018 special issue of the Journal of Molecular Biology, and they coauthored an introduction to the issue.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814385116/-/DCSupplemental.
References
- 1.Jung H, Gkogkas CG, Sonenberg N, Holt CE. Remote control of gene function by local translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu-Yesucevitz L, et al. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krichevsky AM, Kosik KS. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 6.Kiebler MA, Bassell GJ. Neuronal RNA granules: Movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 7.Park HY, et al. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 2014;343:422–424. doi: 10.1126/science.1239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buxbaum AR, Wu B, Singer RH. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343:419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bramham CR, Wells DG. Dendritic mRNA: Transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 11.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 12.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: Organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boeynaems S, et al. Protein phase separation: A new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calabretta S, Richard S. Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem Sci. 2015;40:662–672. doi: 10.1016/j.tibs.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Lin Y, Protter DSW, Rosen MK, Parker R. formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banani SF, et al. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha S, et al. Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell. 2016;166:1572–1584.e16. doi: 10.1016/j.cell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Treeck B, Parker R. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell. 2018;174:791–802. doi: 10.1016/j.cell.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feric M, et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith J, et al. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. eLife. 2016;5:e21337. doi: 10.7554/eLife.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nott TJ, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitrea DM, et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife. 2016;5:e13571. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel A, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 24.Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray DT, et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627.e16. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakami T, et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo L, et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell. 2018;173:677–692.e20. doi: 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qamar S, et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 2018;173:720–734.e15. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang A, et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 2018;37:e97452. doi: 10.15252/embj.201797452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H-R, Chiang W-C, Chou P-C, Wang W-J, Huang J-R. TAR DNA-binding protein 43 (TDP-43) liquid-liquid phase separation is mediated by just a few aromatic residues. J Biol Chem. 2018;293:6090–6098. doi: 10.1074/jbc.AC117.001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conicella AE, Zerze GH, Mittal J, Fawzi NL. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackenzie IR, et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron. 2017;95:808–816.e9. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Diego Otero Y, et al. Transport of fragile X mental retardation protein via granules in neurites of PC12 cells. Mol Cell Biol. 2002;22:8332–8341. doi: 10.1128/MCB.22.23.8332-8341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Fatimy R, et al. Tracking the fragile X mental retardation protein in a highly ordered neuronal riboNucleoParticles population: A link between stalled polyribosomes and RNA granules. PLoS Genet. 2016;12:e1006192. doi: 10.1371/journal.pgen.1006192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vernon RM, et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife. 2018;7:e31486. doi: 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boeynaems S, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65:1044–1055.e5. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou L-T, et al. A novel role of fragile X mental retardation protein in pre-mRNA alternative splicing through RNA-binding protein 14. Neuroscience. 2017;349:64–75. doi: 10.1016/j.neuroscience.2017.02.044. [DOI] [PubMed] [Google Scholar]
- 40.Kao D-I, Aldridge GM, Weiler IJ, Greenough WT. Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc Natl Acad Sci USA. 2010;107:15601–15606. doi: 10.1073/pnas.1010564107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darnell JC, Klann E. The translation of translational control by FMRP: Therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferron L. Fragile X mental retardation protein controls ion channel expression and activity. J Physiol. 2016;594:5861–5867. doi: 10.1113/JP270675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu Rev Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 44.Narayanan U, et al. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narayanan U, et al. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siomi MC, Higashijima K, Ishizuka A, Siomi H. Casein kinase II phosphorylates the fragile X mental retardation protein and modulates its biological properties. Mol Cell Biol. 2002;22:8438–8447. doi: 10.1128/MCB.22.24.8438-8447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartley CM, et al. Mammalian FMRP S499 is phosphorylated by CK2 and promotes secondary phosphorylation of FMRP. eNeuro. 2016;3:ENEURO.0092-16.2016. doi: 10.1523/ENEURO.0092-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ceman S, et al. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 49.Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum Mol Genet. 2010;19:1314–1323. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dolzhanskaya N, Merz G, Aletta JM, Denman RB. Methylation regulates the intracellular protein-protein and protein-RNA interactions of FMRP. J Cell Sci. 2006;119:1933–1946. doi: 10.1242/jcs.02882. [DOI] [PubMed] [Google Scholar]
- 51.Stetler A, et al. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum Mol Genet. 2006;15:87–96. doi: 10.1093/hmg/ddi429. [DOI] [PubMed] [Google Scholar]
- 52.Ramos A, Hollingworth D, Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9:1198–1207. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 54.De Rubeis S, Bagni C. Regulation of molecular pathways in the fragile X syndrome: Insights into autism spectrum disorders. J Neurodev Disord. 2011;3:257–269. doi: 10.1007/s11689-011-9087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 56.Napoli I, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 57.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muddashetty RS, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diao J, et al. A single vesicle-vesicle fusion assay for in vitro studies of SNAREs and accessory proteins. Nat Protoc. 2012;7:921–934. doi: 10.1038/nprot.2012.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phan AT, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 62.Gopal PP, Nirschl JJ, Klinman E, Holzbaur ELF. Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc Natl Acad Sci USA. 2017;114:E2466–E2475. doi: 10.1073/pnas.1614462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brady JP, et al. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc Natl Acad Sci USA. 2017;114:E8194–E8203. doi: 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell. 2014;54:407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niere F, Wilkerson JR, Huber KM. Evidence for a fragile X mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. J Neurosci. 2012;32:5924–5936. doi: 10.1523/JNEUROSCI.4650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pak CW, et al. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin Y-H, Forman-Kay JD, Chan HS. Theories for sequence-dependent phase behaviors of biomolecular condensates. Biochemistry. 2018;57:2499–2508. doi: 10.1021/acs.biochem.8b00058. [DOI] [PubMed] [Google Scholar]
- 70.Bian Y, et al. Global screening of CK2 kinase substrates by an integrated phosphoproteomics workflow. Sci Rep. 2013;3:3460. doi: 10.1038/srep03460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 72.Vasilyev N, et al. Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proc Natl Acad Sci USA. 2015;112:E5391–E5400. doi: 10.1073/pnas.1515737112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z, et al. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Azzam ME, Algranati ID. Mechanism of puromycin action: Fate of ribosomes after release of nascent protein chains from polysomes. Proc Natl Acad Sci USA. 1973;70:3866–3869. doi: 10.1073/pnas.70.12.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Youn J-Y, et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell. 2018;69:517–532.e11. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 77.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.The Gene Ontology Consortium Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Banerjee A, Ifrim MF, Valdez AN, Raj N, Bassell GJ. Aberrant RNA translation in fragile X syndrome: From FMRP mechanisms to emerging therapeutic strategies. Brain Res. 2018;1693:24–36. doi: 10.1016/j.brainres.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monahan Z, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–2967. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wegmann S, et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 2018;37:e98049. doi: 10.15252/embj.201798049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ambadipudi S, Biernat J, Riedel D, Mandelkow E, Zweckstetter M. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat Commun. 2017;8:275. doi: 10.1038/s41467-017-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guillén-Boixet J, Buzon V, Salvatella X, Méndez R. CPEB4 is regulated during cell cycle by ERK2/Cdk1-mediated phosphorylation and its assembly into liquid-like droplets. eLife. 2016;5:e19298. doi: 10.7554/eLife.19298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ryan VH, et al. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol Cell. 2018;69:465–479.e7. doi: 10.1016/j.molcel.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Penney J, et al. Loss of protein arginine methyltransferase 8 alters synapse composition and function, resulting in behavioral defects. J Neurosci. 2017;37:8655–8666. doi: 10.1523/JNEUROSCI.0591-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lim CS, Alkon DL. Inhibition of coactivator-associated arginine methyltransferase 1 modulates dendritic arborization and spine maturation of cultured hippocampal neurons. J Biol Chem. 2017;292:6402–6413. doi: 10.1074/jbc.M117.775619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brini M, Calì T, Ottolini D, Carafoli E. Neuronal calcium signaling: Function and dysfunction. Cell Mol Life Sci. 2014;71:2787–2814. doi: 10.1007/s00018-013-1550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bading H. Nuclear calcium signalling in the regulation of brain function. Nat Rev Neurosci. 2013;14:593–608. doi: 10.1038/nrn3531. [DOI] [PubMed] [Google Scholar]
- 90.Yang Y-M, et al. Identification of a molecular locus for normalizing dysregulated GABA release from interneurons in the fragile X brain. Mol Psychiatry. 2018 doi: 10.1038/s41380-018-0240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pfeiffer BE, Huber KM. Fragile X mental retardation protein induces synapse loss through acute postsynaptic translational regulation. J Neurosci. 2007;27:3120–3130. doi: 10.1523/JNEUROSCI.0054-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ascano M, Jr, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fritzsche R, et al. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 2013;5:1749–1762. doi: 10.1016/j.celrep.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 94.Greenblatt EJ, Spradling AC. Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science. 2018;361:709–712. doi: 10.1126/science.aas9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kelleher RJ, 3rd, Bear MF. The autistic neuron: Troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 96.Arsenault J, et al. FMRP expression levels in mouse central nervous system neurons determine behavioral phenotype. Hum Gene Ther. 2016;27:982–996. doi: 10.1089/hum.2016.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zeng M, et al. Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell. 2016;166:1163–1175.e12. doi: 10.1016/j.cell.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berryer MH, et al. Mutations in SYNGAP1 cause intellectual disability, autism, and a specific form of epilepsy by inducing haploinsufficiency. Hum Mutat. 2013;34:385–394. doi: 10.1002/humu.22248. [DOI] [PubMed] [Google Scholar]
- 99.Feyder M, et al. Association of mouse Dlg4 (PSD-95) gene deletion and human DLG4 gene variation with phenotypes relevant to autism spectrum disorders and Williams’ syndrome. Am J Psychiatry. 2010;167:1508–1517. doi: 10.1176/appi.ajp.2010.10040484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.