Significance
The development of the peripheral T cell pool is typically described based on the maturation and survival of cells in the periphery, assuming all cells are created equal and have similar lifespans. We have developed a mouse model in which we can “timestamp” CD8+ T cells produced at different ages and track their survival. We find that cells produced early in life persist and contribute to the adult pool. However, the dynamics of cell survival are dependent on the age at which they are produced, leading to a shifting population of cells with age. We propose that heterogeneity in the developmental origins of cells contributes to the phenotypic heterogeneity of individual cells observed in adult animals.
Keywords: CD8+ T cells, ontogeny, T cell homeostasis, mathematical modeling, immunology
Abstract
Accumulating evidence indicates that the immune system does not develop in a linear fashion, but rather as distinct developmental layers formed from sequential waves of hematopoietic stem cells, each giving rise to unique populations of immune cells at different stages of development. Although recent studies have indicated that conventional CD8+ T cells produced in early life persist into adulthood and exhibit distinct roles during infection, the developmental architecture of the peripheral T cell compartment remains undefined. In this study, we used a mouse model to permanently label CD8+ T cells produced during distinct windows of development and traced their history to generate fate maps of CD8+ T cells produced during different stages of life. We then used mathematical modeling to understand the age structure of the CD8+ T cell compartment across the lifespan. Interestingly, we found that survival rate of CD8+ T cells depends on both the age and developmental origin of the cells. Recently produced cells show an initial rapid decay rate, which slows with age of the animal at which the cells were produced. For cells produced at any age, the rate of decay also slows with the age of the cell. We derive a function to describe this and predict the “age distribution” of the CD8+ T cell pool for animals of any given age. These data provide a quantitative framework for understanding the ontogeny of the CD8+ T cell compartment and help to contextualize age-related changes in the CD8+ T cell response to infection.
Immune protection against a wide array of intracellular pathogens requires the generation and maintenance of large numbers of CD8+ T cells that arise from hematopoietic stem cells (HSCs). During immune ontogeny, the thymus is colonized by successive waves of HSCs. The first major wave of HSCs is derived from the fetal liver, and colonization of the thymus occurs around midgestation (approximately embryonic day 13, in mice) (1). These fetal HSCs give rise to fetal and neonatal CD8+ T cells, which have an inherent propensity to rapidly proliferate and quickly become terminally differentiated after antigenic stimulation (2). The second wave of HSCs originates from the bone marrow. These cells seed the thymus just before birth (approximately embryonic day 20) and produce adult CD8+ T cells (1, 3). The adult lineage of CD8+ T cells respond less vigorously in the early phases of infection compared with fetal-derived CD8+ T cells but have an enhanced ability to transition into the long-lived memory pool (2). We recently discovered that fetal-derived CD8+ T cells persist into adulthood as a distinct developmental layer and maintain their unique functional properties (4). Importantly, these fetal-derived CD8+ T cells are the first to respond to infection and become effector cells, whereas the cells produced later in life respond more slowly but are more efficient at forming memory cells. This leads to the important and unanswered question of how many fetal- and adult-derived cells are present in healthy animals at various stages of life.
Understanding how the entire CD8+ T cell compartment is constructed and maintained requires an accurate estimate of the lifespan of T cells produced at different stages of life. Experimental studies have used a variety of techniques, including studies of the proportion of replicating or dying cells (using staining for markers such as Ki67, annexin V, or TUNEL), or labeling of dividing cells using BrdU or deuterated water or glucose (5–7). Modeling of this data has then been used to predict average cell turnover and survival rates. For example, the average lifespan of naïve CD8+ T cells in mice is predicted to be ∼11 wk (8), whereas naïve CD8+ T cells in humans have been estimated to persist for ∼5 to 10 y (9). However, a limitation of this approach is that the survival rate is inferred rather than measured directly. Alternative approaches have used adoptive transfer or studies of cell numbers under different manipulations (such as thymectomy or busulfan treatment) to infer cell survival (8, 10–12). These studies have often assumed a homogenous population of cells, although recent studies have hinted at adaptation or selection of cells over time (12).
We have recently developed an experimental method to permanently label, or “timestamp,“ CD8+ T cells during thymic development. This utilizes a CD4 promoter-driven, tamoxifen-inducible cre (CD4cre-ERT2) (4, 13) to switch on expression of red fluorescent protein (RFP) in CD8+ T cells. Since CD8+ T cells only express CD4 briefly during the double-positive stage of T cell development in the thymus, this approach can be used to selectively label CD8+ T cells undergoing thymic development during a brief window of tamoxifen administration. Moreover, once switched on this RFP expression is permanent, and thus the survival of a cohort of cells produced during a brief window can be tracked for the life of the animal. The studies of timestamped cells have found that the diversity in cell phenotype and behavior during infection is due to the recruitment of cells with different developmental origins into the response (2, 4, 14). For example, cells produced in the neonatal period show faster responsiveness to infection but poor memory formation compared with cells produced later in life. These cells also persist into adulthood, maintaining their neonatal phenotype due to a unique epigenetic landscape and transcription profile (4). These phenotypic differences among cells produced at different ages suggest that cell survival may also be dependent on the age at which cells were produced. Understanding the phenotype of cells produced at different ages and their subsequent survival in the periphery may provide insights into immune responsiveness in later life.
In this study, the timestamp mice were used to directly analyze the survival of cells produced at different ages over time, with minimal manipulation of the animal. We applied a mathematical modeling approach to the data to obtain a comprehensive picture of how the developmental layers in the CD8+ T cell compartment change over the life of the animal. Our study revealed that subsets of cells produced early in life persist across the lifespan. Consistent with previous reports (10, 12), we found that cells showed increased survival the longer they persisted in the periphery. However, our data also revealed that the developmental origin of the cells (the age at which they were produced from the thymus) played an important role in the survival of CD8+ T cells. Our modeling demonstrates that the survival of CD8+ T cells in the periphery can be predicted from a combination of age of cell production plus time since cell production, producing a shifting landscape of cell age and origin over time.
Results
Labeling and Tracking of Timestamped Cells over Time.
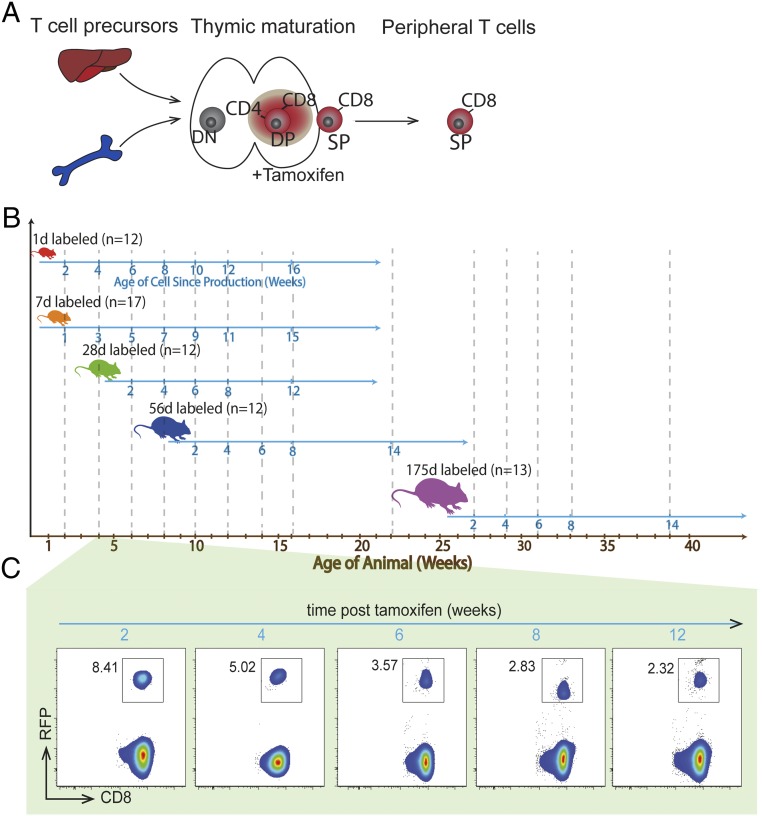
To study the survival of CD8+ T cells produced at different ages, we labeled cells by administration of tamoxifen at ages 1, 7, 28, 56, and 175 d (we refer to these as “1-d-labeled,” “7-d–labeled,” etc.; Fig. 1B). This resulted in expression of RFP in the thymus and the emergence of RFP-labeled cells in the periphery (Fig. 1C). Because CD8+ T cells are labeled in the double-positive stage in the thymus, we waited 2 wk after labeling for cells to emerge from the thymus before commencing our tracking of labeled cells in peripheral blood. The initial proportion of timestamped CD8+ T cells observed at 2 wk after tamoxifen administration varied with the age of the animal, reflecting both the duration of labeling (which is likely not constant with age, due to variation in animal size, dosage, and metabolism of tamoxifen) and also variation in thymic production with age. To understand cell survival, we analyzed and modeled the persistence of stamped CD8+ T cells in the periphery for 12 to 16 wk after labeling at different ages.
Fig. 1.
Tracking the production and survival of CD8+ T cells across the lifespan. (A) The timestamp approach uses tamoxifen to label CD8+ T cells by inducing reporter protein expression in CD4+ CD8+ (double-positive DP) T cells developing in the thymus. Single-positive (SP) CD8+ T cells then retain reporter protein expression for life. (B) Schematic of the experimental procedure. Mice were given tamoxifen at various ages (1, 7, 28, 56, and 175 d). Mice were then bled at various times as indicated by dashed vertical lines. (C) The expression of RFP over time in a representative sample of a 28-d stamped mouse.
For animals labeled with tamoxifen on day 1 of life (1-d-labeled) we tracked the proportion of labeled cells in peripheral blood from 2 wk of life onward. The persistence of cells presents the balance of cell replication and death, since the label is not diluted by cell division. Cells labeled at 1 d of life declined rapidly soon after labeling, and then their decay slows over time, suggesting that they are approaching a plateau beyond 100 d (red line in SI Appendix, Fig. S1C), consistent with the recent observation that cells from young animals seemed “resistant” to replacement after bone marrow transplantation (11, 15). However, this steep decrease in the proportion of cells produced early in life may be at least in part because of the degree of expansion in the total lymphocyte population during this early period of life (SI Appendix, Fig. S1A). Thus, even if the stamped cells were at a constant number, the fraction of total CD8+ T cells would still decrease with time. To correct for this dilutional effect, we account for changes in total CD8+ T cells with age. Similarly, the “plateau” or reduced decay of cells at older ages may be in part because of cells reaching the background level of RFP expression (due to “leakiness” of the timestamp label), and thus we also correct for the background level of RFP expression at different ages.
Correcting for Increasing CD8+ T Cell Number.
The change in total CD8+ T cell number with age was estimated from analysis of pooled spleen and lymph node cells of mice at ages 14, 28, 56, and 245 d. Using a logistic function (SI Appendix, Eq. S2) we obtained a function for the total CD8+ T cell number with age of the host (SI Appendix, Fig. S1A). As these mice were not treated with tamoxifen, we also checked the background levels of RFP in the CD8+ T cell population in the absence of treatment (∼0.1% of CD8+ T cells). A smooth logistic function was used to fit the raw background of RFP expression (SI Appendix, Fig. S1B). Using these functions for total CD8+ T cell number and background staining we can then correct for these factors (SI Appendix, Eqs. S3 and S4), to analyze labeled cell numbers over time.
Dynamics of Timestamped CD8+ T Cells with Age.
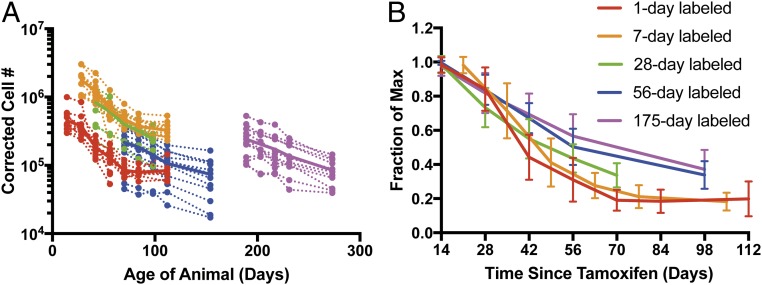
The total number of stamped cells over time was estimated after correcting for the total number of CD8+ T cells and the small amount of background RFP staining (Fig. 2A). This revealed an age-dependent variation in the initial number of stamped cells observed 2 wk after labeling, as well as the subsequent kinetics of survival of these cells. Fig. 2A shows the corrected number of cells produced at different ages. Cells labeled on the first day of life (red line) are lost rapidly soon after production, but this rate of loss slows over time to leave a fairly stable population by 100 d. Cells produced later in life appear to have a slightly slower initial rate of decline, but this rate also slows over time. To directly investigate whether cellular age affects cell survival, we plot the fraction of labeled cells remaining (as proportion of the peak number) against time since cellular production (Fig. 2B). From this it can be seen that cells produced later in life tend to have a slower initial decline in number. However, regardless of age of production, the rate of loss of cells seems to slow with time since production.
Fig. 2.
Dynamics of timestamped CD8+ T cells with age. (A) Corrected cell numbers of stamped CD8+ T cells based on the percentage of labeled cells in blood. Dashed lines depict trajectories of individual animals (n = 12–17 per group), while the solid lines represent the arithmetic mean trajectory for each age group. (B) Survival of CD8+ T cells based on time since production. The fraction of the maximum labeled CD8+ T cells that persist at different times after tamoxifen administration (error bars represent SD from each group). The rate of decay of labeled CD8+ T cells is not constant with age but rather it is decreasing with the age at stamping.
Modeling the Survival of CD8+ T Cells Produced at Different Ages.
To understand the dynamics of survival of cells produced at different ages, we fitted the data with a variety of mathematical models of increasing complexity (SI Appendix, Dynamics of Labeled Cells). The simplest of these models describes cell decline that is at a constant exponential rate regardless of the age of the animal, and time since production of the cell (model 1). The models then increase with complexity so that different hypotheses relating cell loss rates to the age of the animal, and/or the time since production of the cell can be incrementally tested and compared (SI Appendix, Table S1) using the Akaike information criterion (AIC).
Comparison of these models suggests that cell survival changes with both the age of the animal at which the cells were produced and the time since production of the cells. The model with the best fit is one where cells that were timestamped at different ages have different initial rates of decay, and this rate of decay slows with time since production (the slowing of decay rate being the same regardless of age of the animal) (SI Appendix, Table S1, model 5). Furthermore, a model with an equivalently good fit describes a process whereby the initial rate of decay reduces exponentially as a function of age of the animal, rather than fitting individual initial decay rates as in model 5. Although this model has very marginally poorer fit (AIC difference of 0.114 to model 5; SI Appendix, Table S1), it has two practical benefits in that it is a simpler model with fewer parameters and also describes the way in which initial decay rates decrease as a continuous function of animal age—allowing predictions for ages not sampled in this study. Models 7 and 8 also had similar (but worse) fits to models 5 and 9 and describe initial cellular death rates to be dependent on the age of the animal, and those decay rates reduce with time (as a function of age of the animal rather than time since production of the cell).
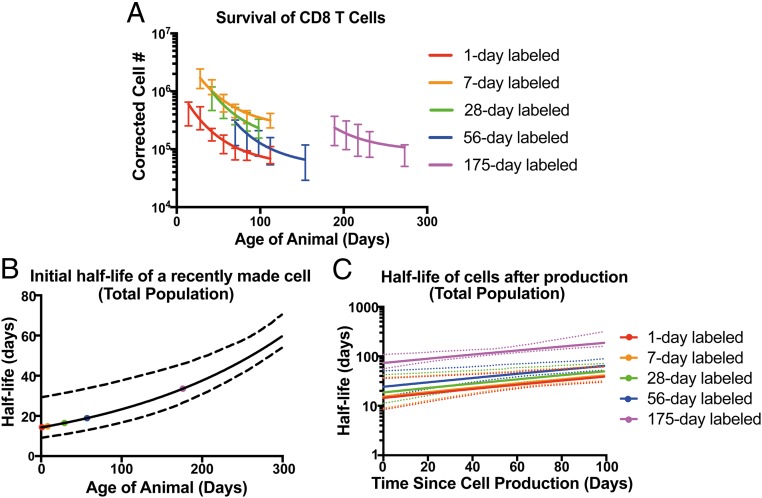
The model with best interpretation and equivalently best fit (model 9) corresponds to CD8+ T cells produced at birth having an initial half-life of ∼14.3 d (95% CI = 10.2 to 22 d). The initial rate of decay for recently produced cells decreases as a function of the age of the host at the time of cell production, halving every ∼150 d (Fig. 3B; 95% CI = 72 to 220 d). For cells produced at any given age, this initial (age-dependent) decay rate of cells decreases with time since production of the cell, with the decay rate halving every ∼50 d (Fig. 3C; 95% CI = 40 to 95 d). This model fitted to the data can be seen in Fig. 3A.
Fig. 3.
Modeling the production and survival of CD8+ T cells. (A) The parsimonious best fit (model 9) to the corrected number of CD8+ T cells (total population) over time since production for five groups of mice that were stamped at different ages (1, 7, 28, 56, and 175 d). Error bars represent SD from each group. (B) Estimated initial half-life of cells made at various ages. The dashed lines indicate 95% CI, and the solid line represents the estimated initial half-life of a recently made cell. (C) Estimated half-life over the lifespan of cells made at various ages (solid lines, estimated half-life of cells after production; dashed lines, 95% CI).
Persistence of Naïve Cells also Varies with Age.
We have previously investigated the phenotype of CD8+ T cells produced at different ages, which showed that neonatal-produced cells have a high proportion of memory phenotype cells (∼60%) by 2 wk after labeling. However, in older animals the newly produced cells have a much lower fraction of memory phenotype cells (9% in adult mice) (16). A simple explanation for changing survival trajectories of cells produced at different ages might be this changing initial proportion of naïve and memory phenotype cells with age. That is, naïve and memory phenotype cells may show the same survival regardless of what age they were produced, but their changing proportion drives more rapid loss in younger animals.
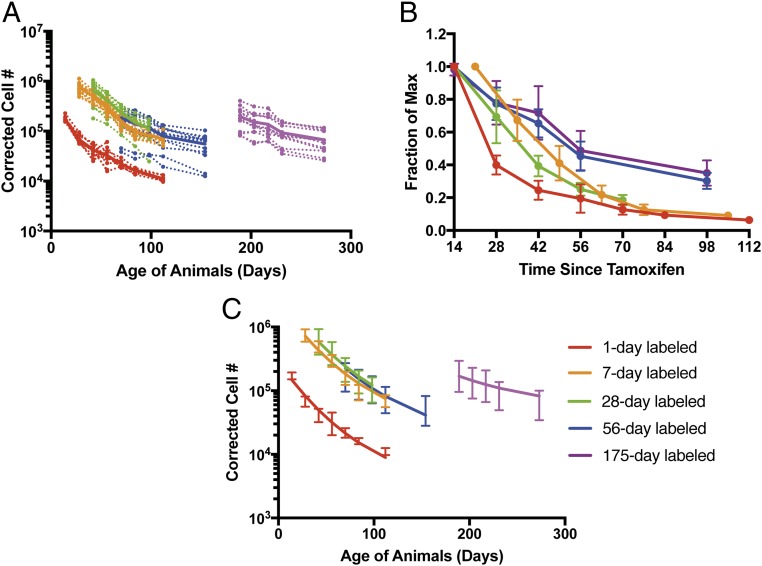
To investigate this, we extended our analysis to track the dynamics of persistence of naïve (CD44low, CD122low) CD8+ T cells over time in individual animals from our cohorts (Fig. 4A). Importantly, naïve cell numbers reflect the balance of cell proliferation, death, and differentiation to memory phenotype. We applied the same correction for cell number and background (Fig. 4 A and B) and used the same model comparison used for total cell number to also analyze naïve cell behavior (SI Appendix, Table S1). These results suggest that the naïve CD8+ T cell population behaves similarly to the total CD8+ T cell population. That is, naïve cells produced early in life show more rapid decay than cells produced later in life (initial half-life of 10 d, 95% CI = 8.8 to 11 d, and halving every 98 d, 95% CI = 70 to 168 d) and for cells produced at any age, the rate of decay slows with time since production (decay rate halving every 102 d, 95% CI = 91 to 116 d; Fig. 4C and SI Appendix, Table S1). We also considered the variation in both total cell number and background values and repeated the fitting 1,000 times (SI Appendix, Figs. S3 and S4).
Fig. 4.
Naïve cells (CD44lowCD122low) show increased persistence with age. (A) Corrected cell number of naïve (CD44low, CD122low) cells stamped at different ages in peripheral blood. The dashed lines represent the trajectories of individual animals, and the solid lines represent the mean trajectory for each age group. (B) Survival of naïve CD8+ T cells based on time since production, expressed as the fraction of the maximum labeled CD8+ T cells that persist at different times after tamoxifen administration (error bars represent SD from each group). (C) The best fit (model 9) to the naïve CD8+ T cell number over time for five groups of mice labeled at different ages (error bars represent SD from each group).
Overall, these results support the contention that the faster decay of total cells produced at younger ages is not an artifact of the varying proportions of naïve and memory cells with age. Further, the analysis of naïve cells allows a more direct comparison with the previous literature on cell turnover, which has often focused on naïve phenotype rather than total cells (11).
Cell-Intrinsic vs. Environmental Factors in Cell Survival.
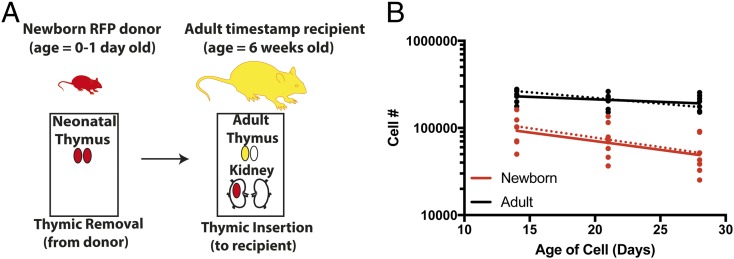
The modeling above demonstrates that cells produced at different ages have different survival. However, it is not clear if these differences arise from the cells themselves (cells produced at different ages have a different intrinsic “program” of survival) or because cells produced at different ages enter different immunological environments, which alters their subsequent trajectory. To explore this, we used data from a thymic transplant model, in which a thymus from a newborn RFP-timestamp mouse is transplanted into a YFP-timestamp adult animal, followed by tamoxifen administration, as described in ref. 4. In this circumstance, labeled neonatal-derived cells and adult-derived cells emerge from the thymus into the same peripheral environment, and so the only difference is that RFP-labeled cells are derived from a neonatal thymus. Recipient mice were bled and analyzed at days 14, 21, and 28 posttransplant to compare cell survival (Fig. 5A).
Fig. 5.
Neonatally derived CD8 T cells are intrinsically different from adult cells. (A) Schematic of thymic transfer experiment. A single thymic lobe from a neonatal RFP (red) timestamp mouse was placed under the kidney capsule of an adult YFP (yellow) timestamp mouse and the animal treated with tamoxifen (n = 7). Thus, RFP-labeled neonatally derived cells and YFP-labeled adult cells emerged simultaneously into the adult host environment. (B) Cells made from the neonatal thymus show a higher decay rate. The solid line represents the best-fit model (linear mixed effect model) of decay of neonatal derived vs. adult-derived stamped cells. The dashed line shows the expected survival from the basic model (model 9; SI Appendix, Table S1), using our previously estimated parameters of initial decay rate of CD8+ T cells (0.048 per day for neonatal-produced cells vs. 0.039 per day for cells produced in a 6-wk-old adult host).
Using this approach, we observed that cells made from the neonatal thymus decay faster than cells made from adult thymus [comparing the slope from a mixed-effect log-linear regression, P = 0.031 (half-life of 15 d vs. 53 d for neonatal vs. adult cells, respectively)], indicating that the developmental origins of the cells, rather than peripheral environment, drives initial decay rates. Importantly, when we used our best model (model 9) with the parameters estimated previously to predict the decay of cells in this adoptive transfer setup we observed a good fit to the experimental data (dashed lines in Fig. 5B). This independently confirms the validity of our best model and also demonstrates that adult and neonatal cells behave differently, even when in the same host environment, suggesting the observed differences in survival are largely cell-intrinsic.
Predicting the Composition of CD8+ T Cells in a Host.
The analysis above demonstrates that cells produced early in life persist for long periods, making a small but stable contribution to the adult T cell population. Thus, at any age the CD8+ T cell pool consists of a spectrum of cells produced at different ages, which have different lifespans. This spectrum cannot be viewed experimentally, as we cannot administer tamoxifen to mice throughout entire stages of development. However, from the analysis above, we have estimates of the survival rate of cells made at various ages, which we can use to mathematically predict the composition of the CD8+ T cell population in a host at any given age. This prediction also requires rates of thymic production at different ages. Although this has been estimated previously, based on average cell turnover numbers, or persistence of RAG-GFP–expressing cells (8, 17), these approaches could be problematic if recent thymic emigrants have different rates of cell survival with the age of the animal. However, as we measure thymic output via total cell number, we can directly estimate the thymic production necessary to maintain the observed CD8+ T cell population over time (SI Appendix, Fig. S1D).
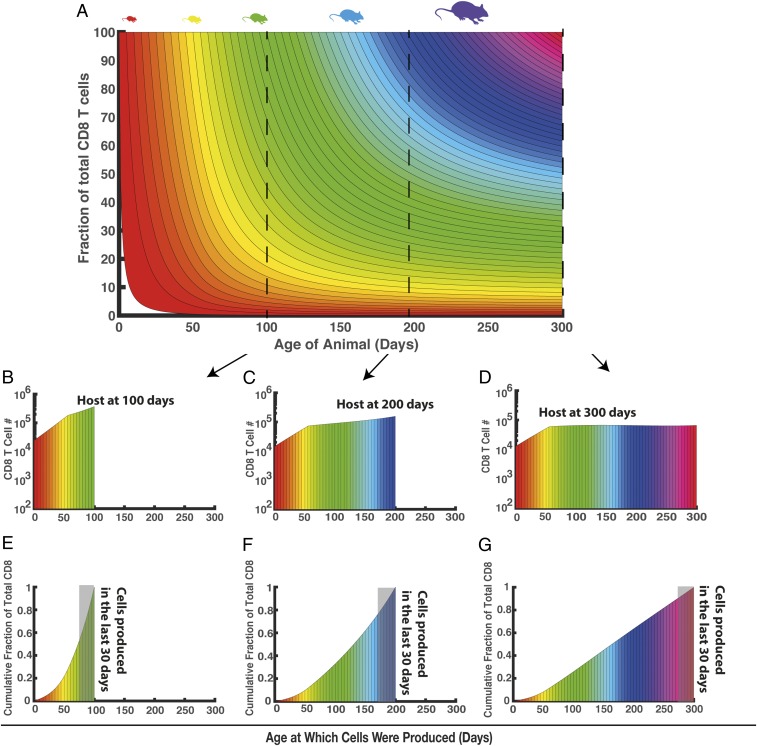
This modeling can then be used to understand the “layering” of cells produced at different ages, and how this contributes to the total CD8+ T cell pool, and changes with the age of the animal (Fig. 6A). In Fig. 6A, cells produced at different ages are colored differently. At any given age of the animal (x axis), the proportion of the total CD8+ T cell pool made up by cells produced at a given previous age (color-coded) is indicated on the y axis. Thus, for example, a cross-section taken at day 100, 200, or 300 reveals the number of cells present that were produced at different ages (Fig. 6 B–D). In a 100-d-old animal, ∼3 × 104 cells remain that were produced in the first week of life and ∼105 cells are present that were produced in the sixth week of life. In a 200-d-old animal, the number of cells produced at these times has declined (Fig. 6C). However, they remain significant contributors to the total T cell population, as decreased thymic production reduces the contribution of more recently produced cells. The cumulative proportion of the total CD8+ T cells made up of cells produced at different ages is shown in Fig. 6 E–G. This shows, for example, that in an animal aged 100 d, 55.8% of its CD8+ T cells were made within the last 30 d, and the median age of cells (i.e., median time since cell production) is 25 d (Fig. 6E). By contrast, for animals aged 300 d, only 11.7% of cells were made within the last month, the median age of cells is 135 d, and 36% of the total CD8+ T cells are >200 d old (Fig. 6G).
Fig. 6.
Predicting the composition of the CD8+ T cell population at any given age. (A) Composition of cells (as fraction of total cells) made at various ages [indicated by different colors for each week of age in which cells were produced. based on the parsimonious best fit (model 9)]. (B–D) The composition of CD8+ T cells in a 100-, 200-, and 300-d-old host. (E–G) Cumulative distribution of cells produced at different ages. The gray shaded area indicates cells made in the last 30 d.
Discussion
The T cell compartment is characterized by high levels of new cell production early in life, followed by cell persistence in the face of waning production with age. Thus, the dynamics of immune cell survival is essential for understanding the maturation and maintenance of immunity over the lifespan. Previous work analyzing immune cell survival has often relied on proliferation labels to measure cell division and death rates to predict cell survival, analysis of cell replacement after bone marrow transplantation, or adoptive transfer of cells into new hosts (5, 8, 10, 11). These approaches have often assumed that the population of cells is homogeneous, and thus if we know the average division and death times we can estimate how long individual cells survive. However, studies have also shown that cell survival changes with age (10, 11), and modeling has been used to compare mechanisms of cell selection or adaptation to explain these changes over time (12). However, these models assume that recent thymic emigrants behave the same with age, and that changes in phenotype occur in the periphery.
Our timestamp approach allows us to directly track the survival of cohorts of cells produced at different ages. That is, whereas labeling studies typically infer clonal survival from the balance of birth and death rates, our approach follows a cohort of individual cells, labeled at a given time. Importantly, since the label is not diluted by cell division, we measure the net effect of cell division and death (or differentiation, in the case of naïve cells). This labeling approach demonstrates that far from a linear pathway of development and death (11), the CD8+ T cell pool exhibits an age-dependent heterogeneity in survival of cells, leading to a shifting palette of cells of different ages vying to contribute to the mature T cell pool. Cells made very early in life exhibit a rapid early rate of decline, but this rapid loss of cells slows down with the time since production of the cell such that some of these cells produced early in life persist into the adult T cell pool. In contrast, cells produced later in life show a slower initial rate of decay immediately after thymic production, but this decay also decreases with the time since production. At any given age, recently produced cells show a higher rate of decay than cells produced earlier in life, consistent with the recent observation of a pool of long-lived cells formed in the host before 6 to 8 wk of age (11) and previous work suggesting increased CD4+ T cell longevity with age (10). The net result of these findings is that with time the composition of cellular ages of the CD8+ T cell population becomes more diverse while maintaining cellular representation from all ages at some level. Coupled with the phenotypic heterogeneity of cells produced at different times (4, 14), this paints a picture of a dynamic and changing T cell population that is difficult to capture with cross-sectional labeling approaches.
One caveat of this work is that we followed the neonatal cell population up to 16 wk of age and used modeling to extrapolate from this time to later ages. Previous work using a TCR-δ driven marker to study CD4 T cells has shown that CD4 T cells labeled in young animals are detectable at 1.5 y of age (18). There have also been adoptive transfer studies, whereby 10% of the donor cells are detected in the recipient 1 y later (19). However, these studies involve a different cell type (CD4+ T cells), and their survival was not analyzed in detail. It would be interesting to study the persistence, phenotype, and role of neonatal T cells in advanced age, and future work should focus on this.
Any approach to modeling T cell persistence requires choices on the modeling framework to be used to capture the experimentally observed behavior. In our case, we compared a series of exponential models of cell survival. However, alternative frameworks such as a gamma or log-normal distribution of cell lifetimes are likely also possible and could also capture the behavior of cell loss slowing with age. It may also be possible to compare mechanisms of cell persistence such as cellular adaptation or selection (10, 12). To facilitate alternative explorations of the data, we have included the dataset in Datasets S1–S3.
Heterogeneity in individual CD8+ T cell behavior following stimulation has been previously demonstrated in vivo, with individual cells leading to widely different numbers and phenotypes of their progeny (20, 21). The prevailing notion in the field is that it is impossible to predict how individual cells will respond, because it assumes that all naïve CD8+ T cells have the same potential to become either a short-lived effector or long-term memory cells and that cell fate decisions arise from stochastic events that occur during priming (20, 21). However, our recent data demonstrating that individual CD8+ T cells are, in fact, programmed differently raises the possibility that we may be able to predict cell behavior based on their developmental origins. Moreover, the immune system as a whole may exhibit different infection outcomes based on the ratio of fetal- and adult-derived cells that are present in the starting population. The results in this report suggest that many age-related differences in the immune responses to infection may relate to changes in the developmental architecture of the peripheral T cell compartment.
Our findings provide insight into individual variations observed in immune system responses by demonstrating how the CD8+ T cell composition is governed by the cumulative life history of an individual, rather than only immediate history as might be the case in a more linear development system (4). Thus, replacement of cells after depletion by bone marrow transplantation, chemotherapy, or HIV infection in later life likely cannot recapitulate the full diversity of phenotypes that would have arisen during ontogeny in the unaffected host. Similarly, the cells that persist after thymectomy may follow “rules” different from those of cells that would otherwise have been made later in life. A more comprehensive understanding of the influence of genetic and environmental factors on the layered development of T cells would provide much greater predictive power of how individuals respond to infection and is a promising avenue of future research.
Methods
Mice.
Ai9 and floxed-STOP eYFP mice were purchased from The Jackson Laboratory. The CD4cre-ERT2 mice, commercially available from Jackson, were provided by Fotini Gounari, The University of Chicago, Chicago (13). Male and female mice were used in experiments and were maintained under specific pathogen-free conditions at Cornell University College of Veterinary Medicine, accredited by the American Association of Accreditation of Laboratory Animal Care. The experiments in this study were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (22). The protocols were approved by the Institutional Animal Care and Use Committee at Cornell University.
Timestamp Mouse Model.
We crossed Ai9 RFP or floxed-STOP yellow fluorescence protein (eYFP) reporter mice to CD4cre-ERT2 mice in large timed-mating cohorts. At birth, litters were divided into groups for marking at different ages. We administered tamoxifen by oral gavage to induce RFP expression. To mark the cells of newborns, 2.5 mg tamoxifen was administered to dams by oral gavage on days 0 and 1 (2.5 mg per mouse two to three times in a 24-h period) and pups received tamoxifen through lactation. To mark 7-d-old mice, animals were given 0.25 mg (single dose). To mark the 28-d group, 1 to 2 mg tamoxifen (one to two doses in a 24-h period) was administered. For the 56-d and 175-d groups, we gave daily injections of 5 mg tamoxifen to mark cells (three doses in a 72-h period). Administration of tamoxifen results in the excision of a stop codon upstream of the reporter fluorescent protein in cells expressing CD4, including CD4+ CD8+ (DP) thymocytes. Cells expressing CD4 at the time of tamoxifen exposure are permanently “marked” by the fluorescent protein (Fig. 1A). A separate cohort of mice was maintained without tamoxifen treatment, to ascertain the background (noninduced) level of RFP expression with age, as well as to estimate total CD8+ T cell numbers by analysis of cell numbers in the spleen and pooled lymph nodes (cervical, mesenteric, and inguinal).
Collection of Blood Samples and Flow Cytometry.
Serial blood samples were collected from timestamp cohorts by retroorbital bleed. Two rounds of hypotonic lysis were performed to lyse red blood cells and cells were labeled with fluorescent antibodies CD8-e450 (clone: 53-6.7) and CD4-A700 (clone: GK-1.5) (Thermo Fisher) using the IC fixation buffer set from Thermo Fisher according to the manufacturer’s instructions. Samples were run on a LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo software (TreeStar).
Thymic Transplantation Procedure.
Thymic transplants were performed using the protocol described in ref. 23. Briefly, thymi were isolated from 0- to 1-d-old RFP timestamp reporter mice. Thymi were then separated into individual lobes and one lobe was inserted under the kidney capsule of recipient floxed-STOP eYFP timestamp reporter mice that were 6 wk old. To simultaneously mark CD8+ T cells being produced in the newborn and adult thymi, 5 mg of tamoxifen was given by oral gavage on days 0 to 2 (three total). At days 14, 21, and 28 postsurgery, recipients were bled and the phenotype of marked cells was assessed by flow cytometry as described above (Fig. 5A).
Estimating Cell Number over Time.
Our experimental procedure involves sampling blood and measuring the proportion of labeled cells over time. To estimate the total labeled cells remaining in the animal, we need to correct for both increases in total cell number with age (leading to dilution of labeled cells; SI Appendix, Fig. S1A) as well as the background level of RFP expression (which also increases with age; SI Appendix, Fig. S1B).
Modeling the Survival of Labeled CD8+ T Cells.
The survival of labeled cells (bulk and naïve phenotype) was modeled using a series of mathematical models (SI Appendix, Dynamics of Labeled Cells). The models were fitted with the nonlinear mixed effect model R (version 3.3.3) function nlme in library nlme (version 3.1-122), Nonlinear mixed models are a longitudinal data analysis method that models the trajectory of individuals animals over time while allowing for variation across animals in the initial corrected cell number to model the variation in individual animals. The fit was weighted using varPower and we assumed a diagonal variance-covariance matrix for the random effect using the pdDiag option in R. To find the 95% CI, we use the R function intervals in library nlme.
Furthermore, to check if our parameters and ranking of models is robust to variation in total cell number and background staining between animals, we repeated our analysis with 1,000 bootstraps of the control mice, picking a single mouse value at each time point, randomly leaving one time point out, and repeating our model fitting and comparison for each newly sampled dataset (SI Appendix, Figs. S2–S4)
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Awards R01AI105265 (to B.D.R. and M.P.D.) and R01AI110613 (to B.D.R.) from the National Institute of Allergy and Infectious Disease. M.P.D. is a National Health and Medical Research Council (Australia) Senior Research Fellow (1080001). V.V. is a National Health and Medical Research Council Career Development Fellow (1067590).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811634116/-/DCSupplemental.
References
- 1.Jotereau F, Heuze F, Salomon-Vie V, Gascan H. Cell kinetics in the fetal mouse thymus: Precursor cell input, proliferation, and emigration. J Immunol. 1987;138:1026–1030. [PubMed] [Google Scholar]
- 2.Wang J, et al. Fetal and adult progenitors give rise to unique populations of CD8+ T cells. Blood. 2016;128:3073–3082. doi: 10.1182/blood-2016-06-725366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foss DL, Donskoy E, Goldschneider I. The importation of hematogenous precursors by the thymus is a gated phenomenon in normal adult mice. J Exp Med. 2001;193:365–374. doi: 10.1084/jem.193.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith NL, et al. Developmental origin governs CD8+ T cell fate decisions during infection. Cell. 2018;174:117–130.e14. doi: 10.1016/j.cell.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 5.Ribeiro RM, Mohri H, Ho DD, Perelson AS. In vivo dynamics of T cell activation, proliferation, and death in HIV-1 infection: Why are CD4+ but not CD8+ T cells depleted? Proc Natl Acad Sci USA. 2002;99:15572–15577. doi: 10.1073/pnas.242358099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Boer RJ, Mohri H, Ho DD, Perelson AS. Turnover rates of B cells, T cells, and NK cells in simian immunodeficiency virus-infected and uninfected rhesus macaques. J Immunol. 2003;170:2479–2487. doi: 10.4049/jimmunol.170.5.2479. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed R, et al. Reconciling estimates of cell proliferation from stable isotope labeling experiments. PLOS Comput Biol. 2015;11:e1004355. doi: 10.1371/journal.pcbi.1004355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Braber I, et al. Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity. 2012;36:288–297. doi: 10.1016/j.immuni.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Vrisekoop N, et al. Sparse production but preferential incorporation of recently produced naive T cells in the human peripheral pool. Proc Natl Acad Sci USA. 2008;105:6115–6120. doi: 10.1073/pnas.0709713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsukamoto H, et al. Age-associated increase in lifespan of naive CD4 T cells contributes to T-cell homeostasis but facilitates development of functional defects. Proc Natl Acad Sci USA. 2009;106:18333–18338. doi: 10.1073/pnas.0910139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan T, Gossel G, Yates AJ, Seddon B. Temporal fate mapping reveals age-linked heterogeneity in naive T lymphocytes in mice. Proc Natl Acad Sci USA. 2015;112:E6917–E6926. doi: 10.1073/pnas.1517246112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rane S, Hogan T, Seddon B, Yates AJ. Age is not just a number: Naive T cells increase their ability to persist in the circulation over time. PLoS Biol. 2018;16:e2003949. doi: 10.1371/journal.pbio.2003949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aghajani K, Keerthivasan S, Yu Y, Gounari F. Generation of CD4CreER(T2) transgenic mice to study development of peripheral CD4-T-cells. Genesis. 2012;50:908–913. doi: 10.1002/dvg.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith NL, et al. Rapid proliferation and differentiation impairs the development of memory CD8+ T cells in early life. J Immunol. 2014;193:177–184. doi: 10.4049/jimmunol.1400553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gossel G, Hogan T, Cownden D, Seddon B, Yates AJ. Memory CD4 T cell subsets are kinetically heterogeneous and replenished from naive T cells at high levels. eLife. 2017;6:e23013. doi: 10.7554/eLife.23013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith NL, et al. Developmental origin governs CD8(+) T cell fate decisions during infection. Cell. 2018;174:117–130.e14. doi: 10.1016/j.cell.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 17.Hale JS, Boursalian TE, Turk GL, Fink PJ. Thymic output in aged mice. Proc Natl Acad Sci USA. 2006;103:8447–8452. doi: 10.1073/pnas.0601040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, et al. Glimpse of natural selection of long-lived T-cell clones in healthy life. Proc Natl Acad Sci USA. 2016;113:9858–9863. doi: 10.1073/pnas.1601634113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 20.Stemberger C, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27:985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Plumlee CR, Sheridan BS, Cicek BB, Lefrançois L. Environmental cues dictate the fate of individual CD8+ T cells responding to infection. Immunity. 2013;39:347–356. doi: 10.1016/j.immuni.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 23.Morillon YM, 2nd, Manzoor F, Wang B, Tisch R. Isolation and transplantation of different aged murine thymic grafts. J Vis Exp. 2015:e52709. doi: 10.3791/52709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.