Significance
Immunotherapies targeting the programmed death-1 (PD-1) and its ligand PD-L1 have recently been combined with standard chemotherapy to potentiate the treatment of solid tumors, including triple negative breast cancer (TNBC). Reactive oxygen species (ROS) have been directly linked to the cytotoxic effects of chemotherapy. Here we report that ROS induced either by chemotherapy (paclitaxel) or antioxidant depletion induce PD-L1 expression in macrophages. PD-L1 positive macrophages have immune-suppressive and angiogenic properties that interfere with the efficacy of paclitaxel in vivo. Indeed, PD-L1 blockade reverts this effect and synergizes with paclitaxel to reduce tumor growth. Our work reveals a pathway that further supports the importance of combining taxane and PD-L1 inhibitors as promising anticancer strategy in TNBC.
Keywords: reactive oxygen species, macrophages, programmed death ligand-1, chemotherapy, immune suppression
Abstract
The combination of immune checkpoint blockade with chemotherapy is currently under investigation as a promising strategy for the treatment of triple negative breast cancer (TNBC). Tumor-associated macrophages (TAMs) are the most prominent component of the breast cancer microenvironment because they influence tumor progression and the response to therapies. Here we show that macrophages acquire an immunosuppressive phenotype and increase the expression of programmed death ligand-1 (PD-L1) when treated with reactive oxygen species (ROS) inducers such as the glutathione synthesis inhibitor, buthionine sulphoximine (BSO), and paclitaxel. Mechanistically, these agents cause accumulation of ROS that in turn activate NF-κB signaling to promote PD-L1 transcription and the release of immunosuppressive chemokines. Systemic in vivo administration of paclitaxel promotes PD-L1 accumulation on the surface of TAMS in a mouse model of TNBC, consistent with in vitro results. Combinatorial treatment with paclitaxel and an anti-mouse PD-L1 blocking antibody significantly improved the therapeutic efficacy of paclitaxel by reducing tumor burden and increasing the number of tumor-associated cytotoxic T cells. Our results provide a strong rationale for the use of anti–PD-L1 blockade in the treatment of TNBC patients. Furthermore, interrogation of chemotherapy-induced PD-L1 expression in TAMs is warranted to define appropriate patient selection in the use of PD-L1 blockade.
Triple negative breast cancer (TNBC) is an aggressive heterogeneous disease, which includes up to 20% of breast cancers (BCs). Clinical treatment of this disease is particularly challenging and is currently limited to standard chemotherapy (1). Although TNBCs are particularly sensitive to neoadjuvant chemotherapy with pathological complete response (pCR) rates of about 40%, these cancers maintain a high rate of relapse (2).
TNBC was initially classified among the nonimmunogenic “cold” tumors but recent studies have proved that the expression of immune-related genes and the presence of immune infiltrates in primary lesions are associated with a better clinical outcome (3, 4). TNBC is also characterized by genomic instability and high rates of genetic mutations, which implicate production of more neoantigens and increased immunogenicity (5). These findings have encouraged the development of new combinatory strategies between chemotherapy and immune checkpoint inhibitors targeting the programmed death-1 (PD-1) and its ligand (PD-L1) (6). In these settings, the use of PD-1/PD-L1 inhibitors could elicit or potentiate the antitumor response induced by chemotherapy (6). Indeed, ongoing clinical trials have shown that immune checkpoint blockade in combination with neoadjuvant chemotherapy correlates with increased pathological complete response rates in TNBC patients (7). Although these clinical studies are reporting encouraging data on the efficacy of chemoimmunotherapy, basic understanding of the interplay between chemotherapy and immunotherapy is limited.
Among the most urgent needs, it is the characterization of biomarkers for a better stratification of TNBC patients in the response to these combinatorial strategies (8, 9). In some studies, PD-L1 expression has been evaluated by immunohistochemistry (IHC) in both cancer and stromal cells with the indication that PD-L1 expression among tumor-infiltrating immune cells may be a better robust predictor (7). Indeed, a recent work has found that analysis of PD-L1 levels on both cell types is necessary for predicting best response to atezolizumab (PD-L1 inhibitor) in nonsmall cell lung cancer (10). Being a cell surface protein, it is conceivable that the expression of PD-L1 is regulated by external stresses in the tumor microenvironment (TME) and may represent a key node at the interface of extracellular and intracellular cancer signaling pathways.
A well-characterized mediator of chemotherapy-induced cytotoxicity is the accumulation of reactive oxygen species (ROS) in cancer and stromal cells (11, 12). In TNBC, tumor-associated macrophages (TAMs) support tumor progression and are a potent regulator of therapeutic response in BC because they can suppress the immune-based mechanism of cytotoxic chemotherapy (13, 14). Based on these considerations, we speculated that chemotherapy-induced ROS could affect the expression of PD-L1 in macrophages and the immune properties of the TME.
Here we report that ROS induced by the glutathione synthesis inhibitor, buthionine sulphoximine (BSO) positively regulates mRNA and protein surface expression of PD-L1 in human and mouse macrophages in vitro. These macrophages also produce immunosuppressive cytokines, including IL-4, IL-10, and IL-17 and the angiogenic factor, vascular endothelial growth factor-A (VEGF-A). Interestingly, the chemotherapeutic drug and ROS inducer, paclitaxel, reproduced all BSO-mediated effects in macrophages. Furthermore, in mouse BRCA1/p53-deleted mammary tumors, a model which resembles spontaneous TNBC, in vivo administration of paclitaxel induced PD-L1 expression in TAM as soon as 24 h after treatment, leading to an immunosuppressive TME. Consequently, the in vivo combination of paclitaxel and an anti–PD-L1 blocking antibody reduced mammary tumor burden and reverted the immune properties of TME. Our data show that ROS are regulators of PD-L1 expression, immune suppressive, and angiogenic features of macrophages. This study emphasizes the importance of evaluating PD-L1 expression in TAMs as a predictive biomarker of chemoimmunotherapy response in TNBC patients.
Results
ROS Regulate PD-L1 Expression and Secretion of Immunosuppressive Cytokines in Macrophages in Vitro.
Recent work has shown that in tumor-bearing mice, TAMs expressed much higher surface levels of PD-L1 than circulating monocytes, implying up-regulation of PD-L1 by the tumor microenvironment (15). ROS generation and accumulation in the TME have important implications in the initiation and progression of cancer (12). To elucidate if ROS could regulate the expression of PD-L1 in macrophages, we treated bone marrow-derived macrophages (BMDMs) with BSO, which increases ROS by depleting reduced glutathione (GSH) (16). BSO positively induced Pdl1 mRNA levels in a ROS-dependent manner since its effect was reverted by cotreatment with the antioxidant and ROS quencer N-acetylcysteine (NAC) (Fig. 1A). This change coincided with modulation of intracellular ROS levels as shown by quantification of CM-H2DCFDA (DCF-DA) staining by flow cytometry (SI Appendix, Fig. S1A). BSO-mediated effect on Pdl1 expression was greater when BMDMs were first treated with IL-4 and M-CSF that polarize them toward alternatively activated macrophages with features similar to TAM (17) (SI Appendix, Fig. S1B). The polarization of these BMDMs was confirmed by the elevated expression of arginase-1 (Arg1) (17) (SI Appendix, Fig. S1C).
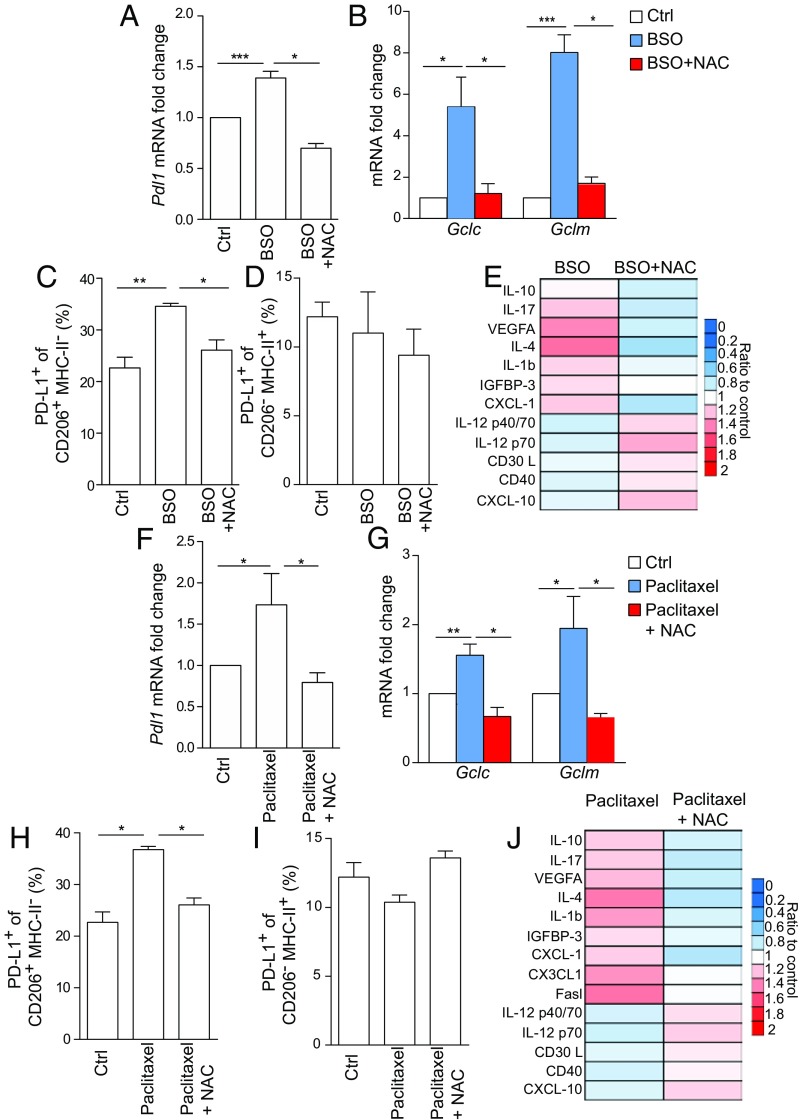
Fig. 1.
ROS regulate PD-L1 expression and secretion of immunosuppressive cytokines in BMDMs. (A) Pdl1 mRNA levels in BMDMs obtained from FVB mice (n = 4 per group). Cells were left untreated (Ctrl) or treated for 24 h with BSO (200 μM) ± NAC (1 mM). (B) Gclc and Gclm mRNA levels in BMDMs treated as in A. n = 4 per group. (C and D) Percentage of PD-L1 positive BMDMs gated on live CD45+CD11b+F4/80+ within CD206+MHC-II− (C) or CD206−MHC-II+ (D) populations. Cells were isolated from n = 3 FVB mice and analyzed after being treated as in A. (E) Levels of indicated chemokines in the supernatant of BMDMs, as determined by cytokine antibody array after treatment with BSO (200 μM) ± NAC (1 mM) for 24 h. Values are the mean of biological duplicates and are represented as ratio to untreated control. (F) Pdl1 mRNA levels in BMDMs that were exposed to DMSO (Ctrl) or paclitaxel (100 nM) ± NAC (1 mM) for 24 h. n = 4 per group. (G) Gclc and Gclm mRNA levels in BMDMs treated as in F. (H and I) Percentage of PD-L1 positive BMDM gated on live CD45+CD11b+F4/80+ within CD206+MHC-II− (H) or CD206−MHC-II+ (I) populations. Cells were isolated from n = 3 mice treated as in F. (J) Levels of indicated chemokines in the supernatant of BMDMs as determined by cytokine antibody array and treated as in F. Values are the mean of biological duplicates and are represented as ratio to DMSO-treated control cells. Data in A–D and F–I are presented as mean ± SEM of biological replicates. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
BSO also triggered the expression of the NRF2 antioxidant targets, Gclc, Gclm, Nqo1, and Hmox-1 as a response to the intracellular redox imbalance (Fig. 1B and SI Appendix, Fig. S1D). Data in mouse BMDMs were validated in human monocyte-derived macrophages treated with BSO with or without NAC. Human macrophages increased PD-L1 mRNA levels as well as the NRF2 target, NQO1, as a response to different ROS conditions (SI Appendix, Fig. S1 E and F).
Next, we investigated which population among BMDMs was mainly affected by BSO in terms of PD-L1 protein surface expression. In BSO-treated mouse BMDMs, defined as CD11b and F4/80 double-positive cells (CD11b+F4/80+), PD-L1 expression was not affected by the presence of BSO (SI Appendix, Fig. S1G), but it completely coincided with a population characterized by high positivity of mannose receptor C type 1 (MRC1/CD206) and absence of expression of major histocompatibility complex class II (MHC-II) molecules (Fig. 1C and SI Appendix, Fig. S1 H and I). Of note, the CD206+MHC-II− macrophages are usually defined as alternatively activated macrophages (14, 18). By contrast, PD-L1 expression was almost unchanged in a population highly expressing MHC-II (CD206−MHC-II+) (Fig. 1D and SI Appendix, Fig. S1 H and I). In human macrophages, PD-L1 surface staining increased in CD11b+ cells upon BSO and decreased when NAC was added to the culture (SI Appendix, Fig. S1J).
PD-L1 expression has been previously associated with the immune-suppressive features of macrophages (19). Therefore, we analyzed which cytokines were present in the media of BSO- and BSO ± NAC-treated mouse BMDMs by applying a mouse cytokine antibody array. We found that, compared with untreated cells, BSO stimulated production of interleukin-10 (IL-10), interleukin-17 (IL-17), interleukin-4 (IL-4), interleukin-1 beta (IL-1b), insulin-like growth factor-binding protein 3 (IGFBP-3), and chemokine (C-X-C motif) ligand 1 (CXCL1) (Fig. 1F). IL-10, IL-4, IGFBP-3, and CXCL1 are usually associated with an immune-suppressive phenotype of macrophages (20–23). IL-1b is a well-known proinflammatory cytokine that has been associated with breast cancer progression and ability to metastasize, especially to extravasate when induced by neutrophils with metalloproteases and other proinflammatory cytokines (24, 25). IL-17 is another inflammatory cytokine but it can enhance immunosuppression in several systems, including macrophages (26). On the other hand, BSO-treated BMDMs produced low levels of interleukin-12 isoform (IL-12) p40/p70 heterodimer and p40 monomer, as well as CD30L (TNFRSP8), CD40, and C-X-C motif chemokine 10 (CXCL-10) (Fig. 1E). Overall, these data suggest that ROS drive a phenotypic change in macrophages characterized by reduced antigen presenting function and costimulatory ability (27–29). In the same cells, ROS also up-regulated the production of VEGF-A, indicative of angiogenic macrophages (30). Notably, NAC completely reverted the production of cytokines and VEGF-A induced by BSO, indicating a key role of ROS in these changes (Fig. 1E).
ROS induction is a key component of the cytotoxic properties of chemotherapy (11). We compared three chemotherapeutic drugs for their ability to increase ROS in BMDMs: the antimitotic agent paclitaxel, the polyADP-ribose polymerase (PARP) inhibitor, olaparib, and the platinum-based drug cisplatin (1). Compared with cisplatin and olaparib, paclitaxel induced the highest ROS levels in BMDMs (SI Appendix, Fig. S2A). In contrast to cisplatin and olaparib, paclitaxel did not cause any DNA damage as measured by intracellular accumulation of phosphorylated H2AX (γH2AX) (SI Appendix, Fig. S2B) (31). Paclitaxel-induced ROS levels were not cytotoxic, since BMDMs had similar cellular viability in both untreated and treated conditions as measured by the sulforhodamine B (SRB) assay (SI Appendix, Fig. S2C). Given its ability to elevate ROS, paclitaxel triggered the expression of Pdl1 compared with control cells, which was reverted when ROS were scavenged by NAC (Fig. 1F and SI Appendix, Fig. S2D). Similarly to BSO, the paclitaxel-mediated effect on Pdl1 expression was augmented by polarization of BMDMs toward alternatively activated macrophages (SI Appendix, Fig. S2E). NRF2-regulated antioxidant genes, Gclc, Gclm, and Hmox1, were also elevated in paclitaxel-treated BMDMs (Fig. 1G and SI Appendix, Fig. S2F). Flow cytometry analysis also showed increased levels of PD-L1 specifically on the surface of CD206+MHC-II− upon treatment with paclitaxel that was reduced by adding NAC (Fig. 1H and SI Appendix, Fig. S2G). On the contrary, CD206−MHC-II+ BMDMs did not show any change (Fig. 1I and SI Appendix, Fig. S2G). Analysis of human macrophages recapitulated the elevation of both PD-L1 mRNA and surface protein after exposure to paclitaxel that was reverted by NAC (SI Appendix, Fig. S2 H and I). The expression of the NRF2 antioxidant enzyme NQO1 was similarly regulated (SI Appendix, Fig. S2J). Then, we analyzed the cytokine and growth factor production of BMDMs treated with paclitaxel with or without NAC. Interestingly, media from paclitaxel-stimulated BMDMs contained the same profile of cytokines observed after BSO treatment (Fig. 1J). In addition, we detected Fas ligand (Fasl) and C-X3-C motif chemokine ligand (CX3CL1) (Fig. 1J). Fasl is a common mediator of apoptosis in T cells expressing the receptor Fas, whereas CX3CL1 functions as an adhesion molecule (32, 33). The production of these cytokines was significantly reverted by cotreatment with NAC (Fig. 1J).
ROS-Induced PD-L1 Expression Is Mediated by the Transcription Factor NF-κB.
We noted that most of the cytokines induced by BSO and paclitaxel treatments have been previously characterized as transcriptional targets of the transcription factor NF-κB (https://www.bu.edu/nf-kb/gene-resources/target-genes/). Furthermore, it is known that activation of NF-κB can promote cell survival and prevent oxidative damage in response to ROS (34). Thus, we hypothesized that ROS might regulate PD-L1 expression through NF-κB activation.
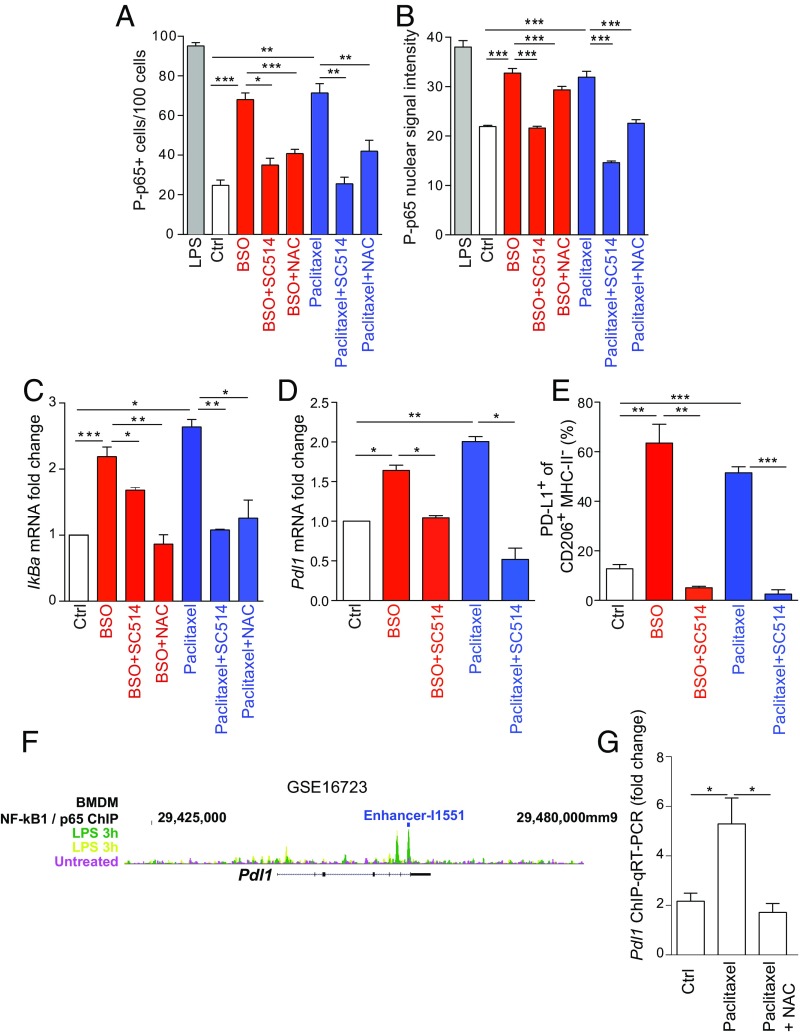
NF-κB molecular forms are usually dimers and the dimer formation is necessary for DNA binding. The most abundant form of NF-κB dimer is p50/p65 heterodimer that mediates the canonical activation of the pathway (35). Upon phosphorylation on the two key residues, S276 and S536, p65 undergoes a conformational change that triggers its transcriptional activity (35). In BSO- and paclitaxel-treated BMDMs, we found an increased frequency of cells positive for the phosphorylation of p65 at S536 residue by immunofluorescence staining (Fig. 2A and SI Appendix, Fig. S3A). The number of these cells was reduced by cotreatment with either the ROS quencher NAC or the nuclear factor kappa-B kinase-2 (IKK-2) inhibitor SC514 (Fig. 2A and SI Appendix, Fig. S3A) (36). Treatment with lipopolysaccharides (LPS) was used as positive control of p65 phosphorylation in BMDMs (Fig. 2A) (37). These results were validated by an overall increase of P-p65 nuclear intensity in the same conditions (Fig. 2B and SI Appendix, Fig. S3B). We further verified the activation of the NF-κB pathway by analyzing the expression of the NF-κB target gene, IkBa. IkBa mRNA was up-regulated by BSO and paclitaxel treatments and the effect was reverted by NAC and SC514 cotreatments (Fig. 2C). Strikingly, SC514 also reduced Pdl1 mRNA in BSO- or paclitaxel-treated BMDMs (Fig. 2D). Then we sought to validate that the SC514-mediated effect on PD-L1 was indeed mainly NF-κB dependent. To do so, we analyzed the expression of IkBa, vascular endothelial factor-A (Vegfa) and Pdl1 in BMDMs treated with BSO and paclitaxel combined with an inhibitor of aryl-hydrocarbon receptor (AhRi). AhR is a transcription factor involved in ROS detoxification and growth factor signaling and can cross-talk with the NF-κB pathway (38). AhR inhibition impaired BSO- and paclitaxel-regulated Vegfa as previously described (39, 40) but did not affect IkBa or Pdl1 increased levels (SI Appendix, Fig. S3C). SC514 also affected PD-L1 cell surface expression in CD206+MHC-II− BMDMs (Fig. 2E and SI Appendix, Fig. S3D). Our data showed a mode of regulation of PD-L1 by NF-κB via ROS. We found that ROS induced p65 phosphorylation at a level similar to LPS (Fig. 2 A and B). PD-L1 has been previously identified to be regulated in BMDMs by LPS-induced NF-κB activation (41). By analyzing the same gene dataset, we confirmed that Pdl1 expression increased in LPS-treated BMDMs and positively correlated with Nfkb1/p65 and Rela/p50 mRNA levels (SI Appendix, Fig. S3E). Moreover, the link between NF-κB and mouse PD-L1 gene transcription was reinforced by the identification of a Nfkb1/p65 binding enhancer (I1551) through bioinformatics analysis of the inflammatory gene expression program in macrophages (42) (Fig. 2F). By chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) we found a significant enrichment of p65 at the I1551 site in the Pdl1 promoter at 1 h after paclitaxel treatment that was reverted by NAC (Fig. 2G). In the same conditions, p65 failed to bind the promoter of the NF-κB target, interleukin-6 (Il6), suggesting a specificity in gene transcription activation by NF-κB upon high ROS (SI Appendix, Fig. S3F).
Fig. 2.
ROS-induced PD-L1 expression is mediated by the transcription factor NF-κB. (A) Relative number of cells containing p65 S536 phosphorylation (P-p65) as assessed by immunofluorescence staining of BMDMs that were treated with DMSO (Ctrl) and BSO (200 μM) or paclitaxel (100 nM) ± SC514 (50 μM) or NAC (1 mM). Cells stimulated with LPS for 30 min were used as positive control. n = 4 slides per group. A total number of 100 cells were counted in each slide. The bar graph represents the mean of all values ± SEM. (B) Nuclear signal intensity of P-p65 in cells treated and presented as in A. See Materials and Methods for additional details. (C) IkBa mRNA levels in BMDMs treated as in A. (D) Pdl1 mRNA levels in BMDMs left untreated or treated with DMSO (Ctrl) and BSO (200 μM) or paclitaxel (100 nM) ± SC514 (50 μM). (E) Percentage of PD-L1 positive BMDMs gated on live CD45+CD11b+F4/80+ within CD206+MHC-II− cells after being treated as in D. (F) Scheme showing NF-kB1/p65 binding sites on mouse Pdl1 promoter region as found through bioinformatic analysis of GSE16723 and Ghisletti et al. (42) datasets. Yellow and green indicates two biological replicates of LPS-treated BMDMs. The location of NF-kB1/p65 binding enhancer from Ghisletti et al. (42) is indicated in blue. (G) ChIP-qPCR showing a peak of enrichment of p65 at I1551 enhancer in Pdl1 promoter region in BMDMs treated with BSO for 1 h. NAC reverted the BSO-mediated effect. n = 3. Data in C–E and G are presented as mean ± SEM of biological replicates. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Paclitaxel Promotes PD-L1 Expression in Tumor-Associated Macrophages in Vivo.
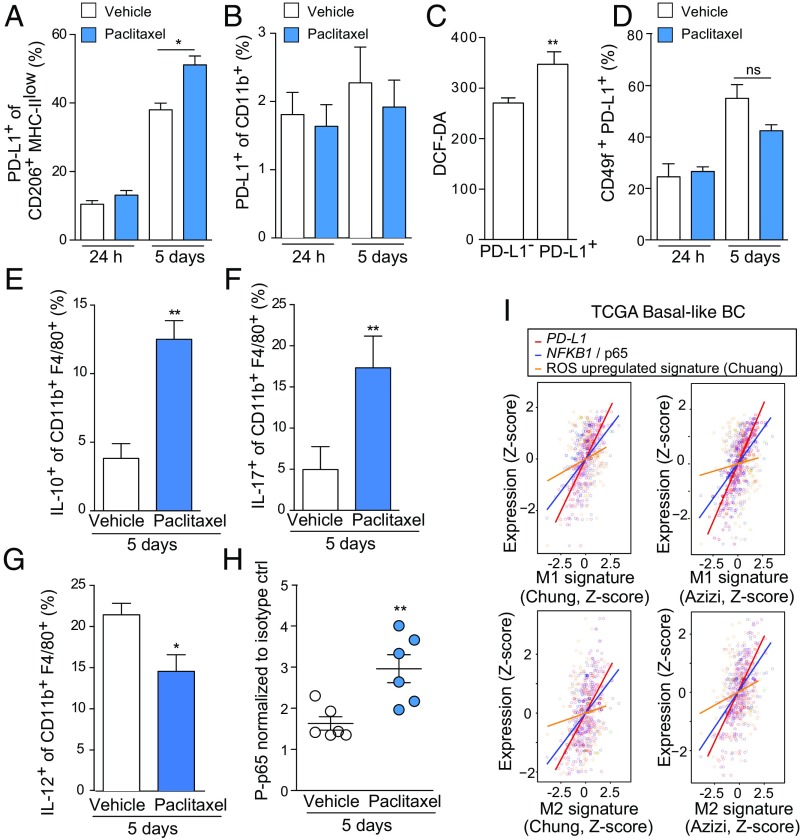
Through bioinformatics analysis of The Cancer Genome Atlas (TCGA) human database of both basal BC and BC with homologous recombination DNA repair defects (HR-defective BC, see Materials and Methods for additional details), we found that cancer-associated PD-L1 positively correlated with an elevated infiltration of monocytic lineage cells (monocytes and macrophages) in the TME (SI Appendix, Fig. S4A) (43). To test if paclitaxel could induce PD-L1 expression in TAMs, we took advantage of a mouse mammary tumor cell line carrying BRCA1/Trp53 deletion and resembling human TNBC (K14cre BRCA1f/f p53f/f, hereafter referred to as KBP) (44). These cells form palpable tumors once transplanted in the mammary fat pad of immune-proficient female mice, allowing analysis of both tumor and immune cell populations in the TME, including TAMs. We administrated vehicle and paclitaxel i.v. at 20 mg/kg to mice bearing tumors at a palpable and measurable size (70 mm3). Tumors were harvested and dissociated for flow cytometry analysis both at 24 h and 5 d posttreatment. In CD206+MHC-IIlow TAMs, PD-L1 surface expression did not change at 24 h posttreatment but showed a significant increase at 5 d after paclitaxel injection, even if we noticed a slight increase of PD-L1 in tumors from vehicle-treated mice, likely due to the tumor mass progression from 70 mm3 to about 200–300 mm3 (Fig. 3A and SI Appendix, Fig. S4B). Indeed, in vitro cocultured BMDMs with KBP cells displayed an increase in Pdl1 and Arg1 expression after being in contact with tumor cells (SI Appendix, Fig. S4 C and D). These results postulate that TAMs are instructed in situ by tumor cells to express Pdl1 during tumor progression. We found that circulating monocytes in tumor-bearing mice either untreated or paclitaxel treated expressed very low to undetectable levels of PD-L1 (Fig. 3B and SI Appendix, Fig. S4E).
Fig. 3.
Paclitaxel promotes PD-L1 expression in tumor-associated macrophages in vivo. (A) Percentage of PD-L1 positive cells gated on live CD49f−CD45+CD11b+F4/80+CD206+ cells isolated from KBP tumors (n = 5 per group) after 24 h and 5 d of treatment with paclitaxel (20 mg/kg) or vehicle (saline). (B) Percentage of PD-L1 positive cells gated on live CD49f−CD45+CD11b+F4/80− cells from peripheral blood of KBP-bearing mice treated as in A. n = 5 per group. (C) ROS intracellular measurement by DCF-DA staining of live CD45+CD11b+F4/80+ cells, gated according to PD-L1 negative (PD-L1−) or positive surface expression (PD-L1+). Cells were isolated from KBP tumors after 5 d of treatment with paclitaxel (20 mg/kg). n = 6 per group. (D) Percentage of PD-L1+ cells gated on live CD49f+CD45− cells isolated from KBP tumors (n = 5 per group) after 24 h and 5 d of treatment with paclitaxel or its vehicle. (E–G) Analysis of intracellular production of IL-10 (E), IL-17 (F), and IL-12 (G) in CD11b+F4/80+ isolated from vehicle- and paclitaxel-treated tumors at 5 d posttreatment. n = 5. (H) Flow cytometric analysis of S536 phosphorylation in p65 in TAMs isolated from tumor-bearing mice (CD49flow/−CD45+CD11b+F4/80+CD206+PD-L1+) at 5 d after treatment with paclitaxel or vehicle (n = 6 per group). Values are normalized on P-p65 levels in isotype control in both groups. (I) Positive correlation between M1 or M2 gene expression signatures [as determined by Chung et al. (48) and Azizi et al. (49)] and the expression levels of CD274, p65/NFKB1, and the “Chuang oxidative stress response” gene signature in the TCGA human basal-like BC cohort. See Material and Methods for details. Data in A–H are presented as mean ± SEM of biological replicates. *P ≤ 0.05, **P ≤ 0.01. ns, not significant.
Then, we asked the question if PD-L1 expression correlated with ROS levels in CD206+ TAMs as found in BMDMs. At 5 d posttreatment, we stained CD206+ TAMs for DCF-DA to measure intracellular ROS. Strikingly, we observed an increased positivity for DCF-DA in the PD-L1+ macrophages, validating the link between cellular redox status and PD-L1 levels found in vitro (Fig. 3C). It is reported that paclitaxel treatment is also able to induce PD-L1 expression in tumor cells, including the TNBC cell line MDA-MB-231 and a panel of ovarian cell lines (45–47). Therefore, we investigated PD-L1 levels in CD45−CD49f+ KBP mammary tumor cells as we did in TAMs. We did not observe any increased PD-L1 positivity within mammary tumor epithelial cells, either 24 h or 5 d posttreatment (Fig. 3D and SI Appendix, Fig. S4F). In vitro treatment of the same KBP cells with increasing doses of paclitaxel induced a very marginal increase in PD-L1 surface expression after 24 h (SI Appendix, Fig. S4G). Consistent with the in vitro results from the BMDM cytokine array, TAMs from paclitaxel-treated tumors produced higher levels of IL-10 and IL-17 and lower amount of IL-12 (Fig. 3 E–G and SI Appendix, Fig. S4H).
To investigate the involvement of NF-κB in PD-L1+ TAMs in vivo, we analyzed CD206+ PD-L1+ TAMs for the presence of phosphorylated p65 in both paclitaxel- and vehicle-treated KBP allografts. At the 5-d time point, when PD-L1 surface expression was high, these macrophages also showed an increase in p65 phosphorylation, suggesting activation of the NF-κB pathway in the same cellular compartment (Fig. 3H and SI Appendix, Fig. S4I). Overall, both in vitro and in vivo data elucidate a link between paclitaxel, ROS accumulation, and NF-κB activation in macrophages. We corroborated this signaling pathway by bioinformatics analysis of TCGA basal and HR-defective BC cohorts. In these datasets, we looked for correlation between expression signature of human BC-infiltrating TAMs (48, 49), our key genes of interest (i.e., PD-L1 and NFKB1/p65) and a comprehensive ROS-induced gene signature (50). We investigated gene correlations in the expression profiling of both M1 and M2 compartments identified in the two published studies (48, 49). These studies elucidated that macrophages express M1- and M2-type gene modules simultaneously and M1 and M2 genes positively correlate in macrophages, contrary to models supporting mutually exclusive M1 and M2 subsets (49). We found that in both basal and HR-defective BC cohorts, M1 and M2 signatures positively correlated with expression of PD-L1, NFKB1/p65, and activation of the ROS signaling pathway (Fig. 3I and SI Appendix, Fig. S4J).
PD-L1 Blockade Potentiates Antitumor Effects of Paclitaxel in Vivo.
Several ongoing clinical trials in TNBC patients are currently exploring the effectiveness of combining paclitaxel treatment with immune checkpoint inhibitors compared with single agent therapy (7, 51, 52). We hypothesized, based on our in vitro and in vivo observations, that inhibition of PD-L1 could revert the immune-suppressive and tumorigenic properties of TAMs to enhance the antitumor activity of paclitaxel. We first assessed if the use of anti–PD-L1 antibody (αPD-L1) could affect the viability of BMDMs whether alone or in combination with paclitaxel in vitro by SRB assay. Isotype-treated cells were included as control. We did not notice any difference in cell viability in both groups over a 5-d treatment (SI Appendix, Fig. S5A). In addition, we did not find any changes in the surface expression of CD206 and MHC-II, in both treated or control cells (SI Appendix, Fig. S5 B and C). αPD-L1 also did not affect paclitaxel-induced Pdl1 mRNA increase (SI Appendix, Fig. S5D).
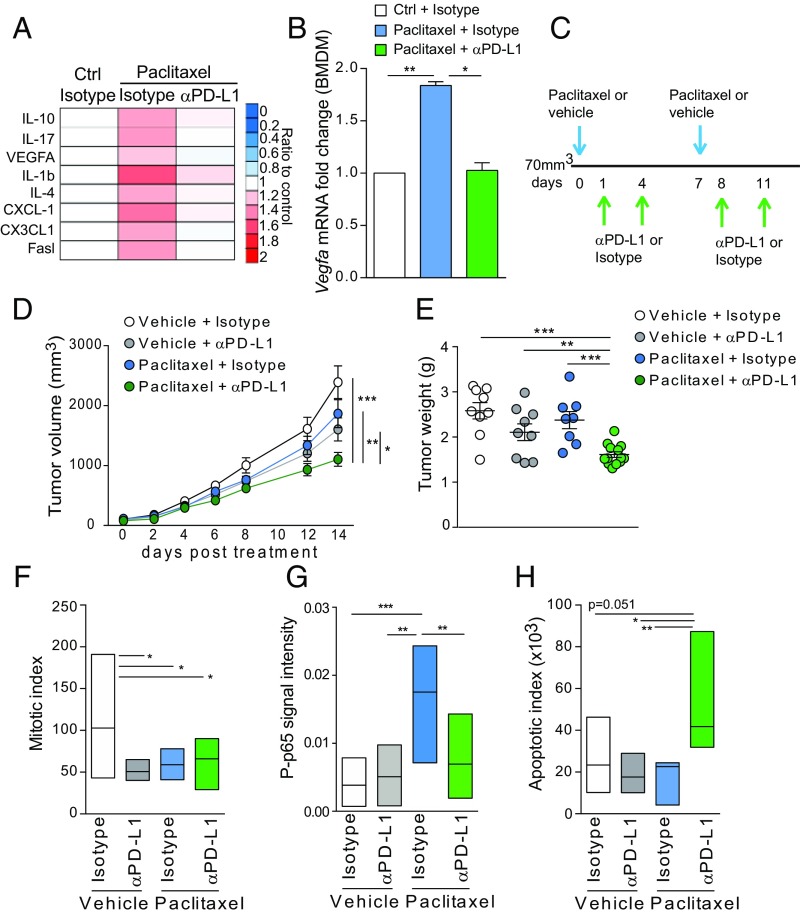
Then, we investigated if PD-L1 blockade could influence the production of cytokines in BMDMs. Interestingly, we found that PD-L1 inhibition restored all of the paclitaxel-induced cytokines to control levels (Fig. 4A). Vegfa mRNA levels were also negatively affected by αPD-L1/paclitaxel combination compared with isotype/paclitaxel (Fig. 4B).
Fig. 4.
PD-L1 blockade potentiates antitumor effects of paclitaxel in vivo. (A) Levels of indicated chemokines in BMDMs after treatment with vehicle, paclitaxel (100 nM) with αPD-L1 antibody (10 μg/mL), or isotype control (10 μg/mL) for 24 h. Values are the mean of biological duplicates and are represented as ratio to control cells treated with DMSO and isotype. (B) Vegfa mRNA levels in BMDMs treated as in A. (C) Schematic representation of paclitaxel and αPD-L1 treatment schedule for KBP tumor-bearing mice. Control group received vehicle and isotype. (D) Volume measurement of mammary tumors over time in mice transplated with KBP cells and treated according to the regimen described in C. (E) Weight of tumors isolated at humane endpoint posttreatment (day 14). n = 10–15 per group. (F) Mitotic index by direct counting of mitotic cells in H&E‐stained tumor sections. n = 5 per group. (G) Mean of P-p65 signal calculated as optical density by ImageJ assessed by immunohistochemistry of KBP tumors. n = 5 per group. (H) Quantification of cleaved caspase 3 staining performed by immunohistochemistry of KBP tumors. n = 10 per group. Data in B–H are presented as mean ± SEM of biological replicates. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
These in vitro data prompted us to investigate the antitumor effect of αPD-L1 in the KBP mouse model in combination with paclitaxel. We administrated paclitaxel (intravenously) and αPD-L1 (intraperitoneally) either in combination or as single agents. First, mice were treated with paclitaxel (to induce PD-L1) and then with αPD-L1 as summarized in Fig. 4C. Since paclitaxel was administrated once a week, we performed an additional injection of αPD-L1 to guarantee PD-L1 blockade. Both compounds were administrated when mammary tumors reached a palpable and measurable volume (70 mm3). Tumor-bearing mice were monitored until they reached a humane endpoint (tumor volume ≤2 cm3). Combinatorial treatment of KBP mice with paclitaxel and αPD-L1 significantly reduced both tumor volume and weight compared with control mice (Fig. 4 D and E). On the contrary, the administration of either paclitaxel or αPD-L1 as monotherapy did not show any effect (Fig. 4 D and E). Haemotoxylin and eosin (H&E) staining of KBP allografts showed that all treated tumors had a reduced mitotic index compared with tumors from the vehicle + isotype mouse group (Fig. 4F). We did not find any difference in blood vessel density based on immunohistochemical staining of the platelet endothelial cell adhesion molecule PECAM1/CD31 (SI Appendix, Fig. S5E). Compared with paclitaxel alone, cotreatment with PD-L1 blockade significantly impaired P-p65 signal and increased cleaved caspase 3 (Fig. 4 G and H and SI Appendix, Fig. S5 F and G).
Paclitaxel Combined with PD-L1 Blockade Leads to Antitumor Immune Activation.
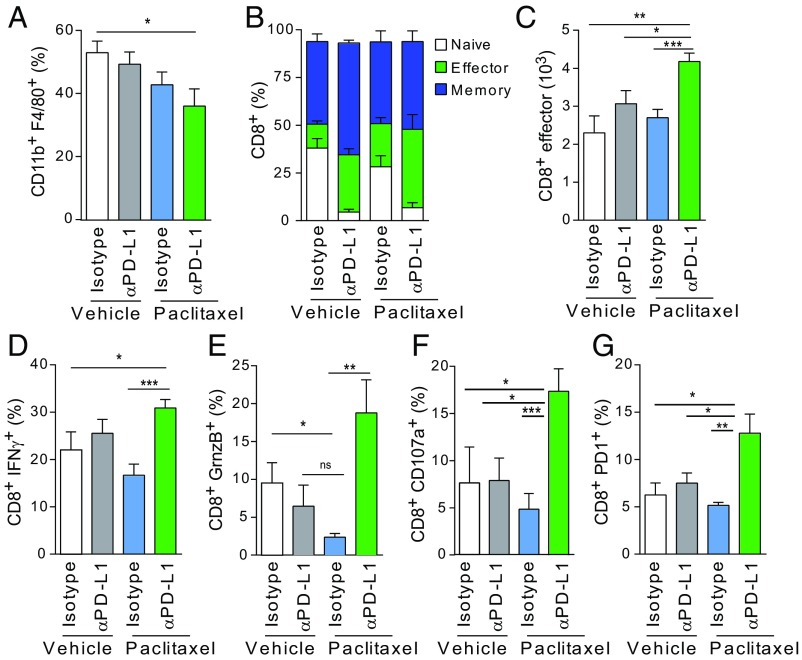
To evaluate the immune response in the KBP allografts, we collected and analyzed tumors from all mouse groups at endpoint for immune cell infiltrates. We observed a moderate reduction in the percentage of F4/80+CD11b+ TAMs in the αPD-L1/paclitaxel group, probably due to reduced infiltration in the TME rather than increased TAM cell death (Fig. 5A). Then, we looked at the impact of the αPD-L1/paclitaxel regimen on different T cell populations. αPD-L1/paclitaxel-treated tumors restored the number of CD4+ T cells that were reduced by paclitaxel alone (SI Appendix, Fig. S6 A and B). Within the CD4+ population, we found a reduced percentage of immune-suppressive CD4+ regulatory cells (as defined by FoxP3 and CD25 marker staining) in the tumors isolated from the αPD-L1/paclitaxel-treated group. CD4+FoxP3+ T cells were dramatically affected by αPD-L1 alone as previously published (53) (SI Appendix, Fig. S6C). While paclitaxel alone did not affect the CD8+ population, these cells were moderately reduced in tumors from the αPD-L1/paclitaxel cohort (SI Appendix, Fig. S6D). To better characterize the phenotype of CD8+ T cells, we stained them for CD44/CD62L markers. αPD-L1/paclitaxel-treated tumors contained a higher percentage of CD8 effector (CD8eff) cells than all other tumors (Fig. 5 B and C and SI Appendix, Fig. S6E). In the same tumors, CD8+ T cells presented the highest expression of IFN-gamma (IFN-γ), granzyme-B (GrnzB), CD107a and PD-1, and confirming their activation and cytotoxic activity in the TME (Fig. 5 D–G and SI Appendix, Fig. S6 F–I).
Fig. 5.
Paclitaxel and PD-L1 blockade combinatorial treatment promotes antitumor immune respose. (A) Percentage of CD11b+F4/80+ cells gated on live CD49f−CD45+ cells isolated from KBP tumors at endpoint. n = 10–15 per group. (B) Percentage of CD8+ T cells gated on CD49f−CD45+CD3+cells in control or treated cohorts (n = 10–15 per group). Cells were characterized based on the expression of CD44 and CD62L as follows: naïve (CD44−CD62L+), memory (CD44+CD62L+), or effector (CD44+CD62L−). Percentages are represented as stacked bar graphs. For the CD8+ effector cells, statistical significance was determined by Student’s t test: vehicle + isotype versus paclitaxel + αPD-L1 (*P = 0.01), vehicle + αPD-L1 versus paclitaxel + αPD-L1 (*P = 0.0152), paclitaxel + isotype versus paclitaxel/αPD-L1 (**P = 0.006). (C) Absolute numbers of CD8+ effector cells in control and treated cohorts (n = 10–15 per group). (D–G) Percentage of CD8+ T cells (gated on CD49f−CD45+CD3+) that were positive for the expression of IFN-γ, GrnzB, CD107a, and PD-1 as indicated in control or treated cohorts (n = 10–15 per group). Data in A–G are presented as mean ± SEM of biological replicates. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Discussion
The recent success of the PD-1/PD-L1 blockade has renewed interest in immunotherapies and in combining them with chemotherapy to achieve additive or synergistic clinical activity. Clinicians are currently exploring these combinatorial strategies for the treatment of TNBC, a very aggressive form of BC with poor prognosis. In TNBC, the expression of PD-L1 is almost undetectable in ductal carcinoma in situ (DCIS) tumor epithelial cells but increases to a higher extent in invasive ductal carcinoma (IDC) with the amplification of the CD274 locus encoding PD-L1 in about 30% of the cases (53). This important finding suggests that in TNBC, TME immune-suppressive functions progressively change during tumor evolution.
Together with anthracyclines, taxanes (including paclitaxel) are currently used as first-line therapy with variable success and frequent cases of relapse (1). The expression of PD-L1 in tumor and tumor-infiltrating cells in TNBC patients suggest that PD-L1 blockade may be a useful strategy to potentiate the antitumor effects of taxanes. Indeed, several clinical trials are currently exploring the combination of taxanes with PD-L1 inhibitors in TNBC (52). Very recently, the primary results of one of these trails, IMpassion130, a phase III trial of an anti–PD-L1 or anti–PD-1 antibody, have been reported in patients with metastatic TNBC (54). Although the study has not reached statistical significance yet, numerical increases in median overall survival were clearly observed in both the randomized population and in the subgroup where PD-L1 expression was assessed in tumor-infiltrating cells (54). One of the most important findings of this work is that PD-L1 expression levels in the tumor-infiltrating cells should be taken into consideration to guide treatment strategies in TNBC. Our data reporting the regulation of PD-L1 expression by paclitaxel in macrophages and TAMs align with this result. Although PD-L1 expression has been generally considered to be induced at the transcriptional level after exposure to IFN-γ released by T effector cells (55), novel ways of transcriptional regulation of PD-L1 are emerging in both immune and tumor cells (19, 46). Here we have shown that in macrophages, PD-L1 levels respond to intracellular redox imbalances caused by both metabolic alterations such as deprivation of antioxidants and chemotherapy such as taxane. Overall, these data suggest a scenario where any intracellular or extracellular stresses affecting TME redox status can influence the communication between tumor cells and the surrounding immune system. In these settings, TAMs respond to chemotherapy-induced ROS by up-regulating PD-L1, releasing VEGF-A to promote angiogenesis and suppressing T cell-mediated antitumor response. This suggests that administration of immunotherapy could potentiate paclitaxel efficacy by interfering with the immunosuppressive abilities of macrophages established by paclitaxel itself. Indeed, combinatorial αPD-L1 and paclitaxel therapy promotes the antitumoral properties of the TME by significantly increasing the percentage of tumor-infiltrating effector and cytotoxic CD8+ T cells. Given the broad expression of PD-L1 in the TME, PD-L1 blockade may affect a wide range of cells, including tumor cells, T and B cells, natural killer, dendritic cells, and macrophages (56). However, in our in vivo tumor model, ROS specifically increase PD-L1 in the macrophage compartment. Remarkably, it has been reported that TAMs interfere with the cytotoxic activity of paclitaxel and TAM depletion potentiates the antitumor effect of the paclitaxel (57–59). In conclusion, our work has revealed a unique scenario that further supports the combination of PD-L1 blockade with taxane for the treatment of TNBC patients.
Materials and Methods
Mice.
KBP mice were provided by J. Jonkers, Netherlands Cancer Institute, Amsterdam, The Netherlands, and were on the FVB background. KBP tumor cells were obtained and used for in vivo transplantation studies as previously described (44). For KBP transplantation studies, FVB recipient female mice were 8–10 wk old and purchased from The Jackson Laboratory. Mice were maintained and handled according to protocols approved by the Animal Care and Use Committee of the University Health Network, Toronto, Canada.
Preparation of Murine BMDMs.
Whole bone marrow was harvested from 10- to 12-wk-old female mice by flushing Hanks’ Balanced Salt Solution (HBSS) through femurs and tibias using a 27-gauge needle (BD Biosciences). Following red blood cell lysis, cells were cultured in 10% RPMI in 10-cm plates overnight. Nonadherent cells were collected and seeded in Petri dishes in medium containing 20 ng/mL M-CSF (PeproTech). After 3 d of culture, cell media were replenished with media containing 20 ng/mL M-CSF. Macrophages were harvested on day 4 and used for all of the in vitro assays described in the article. Polarization toward alternative-activated macrophages was obtained by culturing BMDMs with medium containing 20 ng/mL IL-4 (PeproTech) and 20 ng/mL M-CSF (PeproTech) for 24 h. For coculture experiments, BMDMs (1 × 106) were seeded in triplicate in six-well plates and incubated with or without KBP cells (2.5 × 105 cells per well). Cells were harvested 24 h later using enzyme-free cell dissociation medium (Millipore) and sorted on an Astrios FACS instrument (Beckman Coulter).
Human Samples and Preparation of Human Macrophages.
Healthy subjects were recruited at the Princess Margaret Cancer Centre’s blood donor center. The study was approved by the Research Ethics Board (REB) of the University Health Network (Toronto, Canada) and conducted after obtaining written consent from all participants. To generate human macrophages, CD14-positive monocytes were isolated from peripheral blood mononuclear cells using human CD14 microbeads (Miltenyi Biotech). Monocytes were cultured in DMEM plus 10% FBS (Gibco), and 100 units/mL penicillin and streptomycin (Pen-Strep) in the presence of recombinant human M-CSF (PeproTech) at 50 ng/mL. After 5–7 d, the differentiated macrophages were characterized by flow cytometry and used in all of the reported experiments.
KBP Mammary Tumor Induction and Treatment.
KBP cells (3 × 105) were transplanted into no. 4 mammary gland fat pads of syngeneic female FVB recipient mice (10 wk old). Diameters of developing tumors were measured in duplicate using digital calipers starting on day 14 posttransplantation. Tumor volume (mm3) was calculated as 1/2(width2 × height). Tumor diameters were measured, and volumes calculated as above, three times per week. For experiments with paclitaxel, anti–PD-L1 antibody (10F.9G2, BioXCell) or isotype control (LTF-2, BioXCell) antibodies, KBP transplanted female mice were monitored until tumors reached a volume of 70 mm3 and randomized. Mice were injected i.v. with paclitaxel (20 mg/kg; Medkoo) or vehicle (saline) once a week. When needed, the same mice were administrated with mouse anti–PD-L1 antibody (200 μg per mouse) or its isotype control (200 μg per mouse) twice a week intraperitoneally. Paclitaxel was purchased in a powder form and dissolved in a solution of ethanol/cremophor EL/1× PBS (1:1:18). Both anti–PD-L1 and isotype antibodies were diluted in appropriate dilution buffers that were provided by the manufacturer.
Mouse Mammary Tumor Dissociation.
Tumors were resected from no. 4 mammary fat pads of transplanted mice, cut into 2- to 3-mm2 pieces, and placed into a C-tube (Miltenyi Biotech) containing 5 mL Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% FBS, 2 mM l-glutamine, 100 units/mL penicillin, 100 μg/mL streptomycin, 0.05 mM β-mercaptoethanol, 0.26 units/mL Liberase TM (Sigma), and 20 units/mL DNase I (Sigma). Tumors were mechanically processed using a gentleMACS Octo dissociator with heaters (Miltenyi Biotech). Processed samples were filtered once through a 100-µm cell strainer (Falcon), and the corresponding C-tubes were rinsed with 5 mL cold IMDM and passed through the same strainer. Cells were filtered once using a 70-µm strainer (Falcon), followed by a 40-µm strainer (Falcon). Filtered samples were collected in 15-mL Falcon tubes and centrifuged at 278 × g for 8 min at 4 °C. Pellets were incubated with red blood cell lysis buffer for 7 min at room temperature (RT), and then centrifuged at 278 × g for 8 min at 4 °C before resuspension in 1× PBS−/− containing 1% BSA plus 2 mM EDTA. Cell suspensions were subjected to fluorescence-activated cell sorting (FACS)/flow cytometry as described below.
Flow Cytometry.
For mouse tumor-associated macrophages and BMDMs, cell surface marker staining for flow cytometry analysis was performed using the following antibodies (Abs), all from BioLegend unless indicated: anti-CD49f (AF488 GoH3; 1/200), anti-CD45.1 (AF700 A20; 1/400), anti-CD11b (Pacific Blue M1/70; 1/400), anti-F4/80-PE (BM8; 1/400), anti-F4/80-PerCP-Cy5.5 (BM8; 1/200), anti-MHC II (IA/IE)-PE (M5/114.15.2; 1/1600; Thermo Fisher Scientific), anti-CD206-AF647 (C068C2; 1/400), anti-PD-L1-Pe-Cy7 (10F.9G2; 1/400), anti-CD3-AF488 (145-2C11; 1/250), anti-CD8-APC-Cy7 (53-6.7; 1/200), anti-CD4-PerCP-Cy5.5 (RM4-5; 1/800), anti-CD25-PE (PC61; 1/200), anti-IFNγ-APC (XMG1.2; 1/200), anti-Granzyme B-PE (GB11; 1/200), anti-TNFα-PE-Cy7 (MP6-XT22; 1/800), anti-CD107a-BV421 (1D4B; 1/800), anti-CD62L-PE-Cy7 (MEL-14; 1/800; Thermo Fisher Scientific), anti-CD44-APC (IM7; 1/800) and anti-PD1-PE (RMP1-30; 1/200; Thermo Fisher Scientific).
For IFN-γ and Granzyme-B staining, cells were stimulated with phorbol myristate acetate (PMA) (20 ng/mL; Sigma) and ionomycin (1 μg/mL; Sigma) in presence of the intracellular protein transport inhibitor Brefeldin A (eBioscience). Cells were harvested 5 h later and stained for surface markers as follows: Cells were washed twice with 1× PBS−/−, fixed, and permeabilized on ice for 30 min with the Intracellular Permeabilization kit (Thermo Fisher Scientific). After washing with the permeabilization buffer, cells were stained for IFN-γ and Granzyme-B on ice for 30 min.
Mouse macrophages were identified as CD49flo/−CD45+CD11b+F4/80hi and mouse T cells as CD49flo/−CD45+CD3+. Mouse macrophages were treated with Mouse BD Fc Block (anti-CD16/CD32 2.4G2; eBioscience) at 1/100 dilution for at least 10 min prior to staining with appropriate Abs. For human macrophages, cell surface marker staining was performed with PDL1 BV421 (29E.2A3; 1/400) and CD11b BV510 (ICRF44; 1/400), both from BioLegend.
Human macrophages were treated with 1× PBS−/− containing 5% BSA and 2 mM EDTA for 30 min on ice prior to staining with Abs. For both mouse and human macrophages, Abs were prepared at the indicated dilutions in 1× PBS−/− containing 1% BSA and 2 mM EDTA for 30 min on ice. Dead cells were excluded by adding 5 μL of 7-AAD (BioLegend) during the last 10 min of staining with the Abs. Cells were then washed twice and further analyzed.
To quantify intracellular cytokine production, mouse mammary tumor cell suspensions were treated with PMA (5 ng/mL; Sigma) and ionomycin (500 ng/mL; Sigma) in presence of the intracellular protein transport inhibitor Brefeldin A (eBioscience) and Monensin (eBioscience), for 4 h before staining. The cells were fixed and permeabilized before immunostaining for CD45/CD11b/F4/80 (as described above) and IL-12 PE (C15.6; 1/400; BD), IL-17 (TC11-18H10.1; 1/400; BioLegend), and IL-10 (JES5-16E3; 1:400; BioLegend). All samples were analyzed with a Fortessa instrument (BD Biosciences) and data were processed with FlowJo (Tree Star, Inc.) and GraphPad software.
Analysis of Peripheral Blood Monocytes.
Peripheral blood (15 μL) was first collected from mouse tail vein into heparinized capillary tubes and then transferred into a 5-mL polystyrene tube containing 100 μL 1× PBS−/− plus 20 mM EDTA. After blocking with anti-CD16/CD32 Abs (1:100) for 10 min, samples were stained with anti-CD11b-Pacific Blue and anti-PD-L1-PE-Cy7 as described above. Samples were washed twice with 1× PBS−/− containing 1% BSA and 2 mM EDTA. Red blood cells were lysed at room temperature for 10 min with 1 mL of Fix/Lyse solution (Thermo Fisher Scientific). Cells were washed twice with 1× PBS−/− and analyzed with a Fortessa instrument (BD Biosciences). Data were processed with FlowJo (Tree Star, Inc.) and GraphPad software.
Phospho-p65 Staining by Flow Cytometry.
Tumors were dissociated according to a mouse mammary tumor dissociation method. The 106cells were suspended in 0.5 mL of 1× PBS and immediately fixed with 0.5 mL of 4% formaldehyde (final concentration 2%) at 37 °C for 10 min. Cells were washed by centrifugation with 1× PBS−/− containing 1% BSA and 2 mM EDTA prior to staining with anti-CD45.1 (AF700 A20; 1/400), anti-CD11b (Pacific Blue M1/70; 1/400), anti-F4/80 (PE BM8; 1/400), anti-CD206 (Fitc C068C2; 1/400) and anti-PD-L1 (Pe-Cy7 10F.9G2, 1/400) (see Flow Cytometry above for additional details). Cells were washed twice with 1× PBS−/− containing 1% BSA and 2 mM EDTA and permeabilized by slowly adding ice-cold Perm Buffer II (eBioscience) with gentle vortexing. Cells were incubated on ice for 30 min and washed twice. Cells were then suspended in 100 μL of primary P-p65 (Ser536) antibody (93H1, 1/1,600; Cell Signaling) or isotope control (rabbit IgG, 1/1,600; Thermo Fisher Scientific) and incubated on ice for 1 h. After two washes, cells were resuspended in 100 μL of secondary antibody (goat anti-rabbit APC conjugated; Thermo Fisher Scientific) diluted 1/500 and incubated on ice for 1 h. Cells were then washed twice and analyzed with a Fortessa instrument (BD Bioscience) and data were processed with FlowJo software (Tree Star, Inc.).
Phospho-p65 Staining by Immunofluorescence.
For analysis of p65 S536 phosphorylation, 2 × 105 BMDMs were seeded in a 12-well plate containing glass coverslips. The day after, cells were treated with BSO (200 μM) and paclitaxel (100 nM) in the presence or absence of SC514 (50 μM) for 3 h. Cell treatment with LPS at 1 μg/mL for 30 min was included as positive control of p65 S536 phosphorylation. After treatments, cells were fixed with 4% PFA for 15 min at 37 °C. Cells were then stained overnight at 4 °C with primary antibody (93H1, 1:1,600; Cell Signaling). The following day, cells were washed three times and subjected to FITC-conjugated secondary antibody (A-11008; Thermo Fisher Scientific) at 1:1,000 for 2 h at RT. Nuclei were counterstained with DAPI (P36962; Thermo Fisher Scientific). To quantify P-p65 nuclear fluorescence intensity, we randomly selected 100 nuclei for each sample and analyzed them with ImageJ software as follows: First, we applied an Otsu threshold to the DAPI channel to generate a mask marking the nuclear area. Then, with the tracing tool, we transposed each mask to the FITC-positive channel to calculate the mean intensity in the nuclear region.
Cell Lines and Treatments.
Mouse KBP cells were cultured in DMEM/F12 medium containing 10% FBS (Thermo Fisher Scientific), l-glutamine, 1 µg/mL hydrocortisone (Sigma), 5 µg/mL insulin (Sigma), 5 ng/mL epidermal growth factor (EGF) (Sigma) and Pen-Strep (Thermo Fisher Scientific). Human macrophages were cultured in DMEM supplemented with 10% FBS (Thermo Fisher Scientific), l-glutamine, and Pen-Strep (Thermo Fisher Scientific). The glutathione synthesis inhibitor, BSO (Sigma) was used at 200 µM (BMDMs) or 1 mM (human macrophages) for specific periods of time as indicated above. Chemotherapeutic drugs were administered in vitro at the following concentrations: 100 nM (paclitaxel, Medkoo), 0.5 μM (olaparib, Medkoo) and 2 μM (cisplatin, Medkoo). Anti–PD-L1 mouse or isotype control antibodies were both used at 10 μg/mL (BioXCell). For ROS scavenging, BSO- or paclitaxel-exposed cells were cotreated with 1 mM NAC (Sigma). The NF-κB antagonist SC514 (Sigma) was applied to cell cultures at 50 μM. The AhR inhibitor, CH-223191 (MedKoo) was used at 10 μM for 24 h.
SRB Colorimetric Assay.
BMDMs were seeded at 8,000 cells per well in triplicate. The day after, cells were treated with paclitaxel at 100 nM or DMSO for 24 h. The following day, anti–PD-L1 or isotype control antibodies were added to the cells at 10 μg/mL. Cells were stained with sulforhodamine B every 24 h and processed according to the manufacturer’s recommendations. Cell density was calculated using SoftMax Pro software (Molecular Devices).
Mouse Cytokine Array.
The Mouse Cytokine Antibody Array kit (Abcam) was used to measure chemokines, cytokines, and growth factors in the culture media of BMDMs and according to the manufacturer’s instructions. Membranes were developed for chemiluminescent detection and images were acquired with GelCapture software using MicroChemi 2.0/4.2 (FroggaBio).
RT-PCR.
RNA was isolated using the Nucleospin RNA Plus kit (Macherey-Nagel) and reverse transcribed using the iScript cDNA Synthesis kit (Bio-Rad) according to the manufacturers’ instructions. Quantitative RT-PCR was performed using SYBR Green primers (Applied Biosystems). Mouse and human ribosomal proteins S9 (rps9) were used as housekeeping genes to calculate relative mRNA expression. All mouse and human primer sequences are described in SI Appendix, Table S1.
Chromatin Immunoprecipitation.
ChIP assay was performed using the SimpleChip Plus Enzymatic Chromatin IP kit (9003, Cell Signaling Technology). The 8 × 106 BMDMs were left untreated or treated with 100 nM paclitaxel with or without 1 mM NAC for 1 h. Chromatin was prepared by enzymatic shearing according to the manufacturer’s instructions. Immunoprecipitation was performed at 4 °C overnight with 0.75 μg NF-κB p65 Ab (L8F6, Cell Signaling) and 1 μg mouse IgG as negative control. Following immunoprecipitation, samples were incubated with Chip-Grade Protein G Magnetic Beads from a kit at 4 °C for 2 h. The cross-linking was reversed by adding 5 M NaCl and proteinase K at 65 °C for 2 h. Real-time PCR was performed on DNA isolated from each ChIP reaction (n = 3) using Power SYBR Green PCR Master mix. Primers are indicated in SI Appendix, Table S1.
ROS Measurement.
To measure intracellular ROS, 2 × 105 BMDMs were incubated with 300 nM DCF-DA (C6827, Invitrogen) for 10 min at 37 °C. DCF-DA fluorescence was analyzed by flow cytometry using a FACS Fortessa (BD Biosciences) and data were processed with FlowJo software.
DNA Damage Measurement.
To measure intracellular DNA damage, 2 × 105 BMDMs were fixed and permeabilized with BD Cytofix/Cytoperm (BD Biosciences). Then, cells were stained with FITC-conjugated γ-H2AX (1/400; Millipore) for 1 h, washed twice, and analyzed by flow cytometry using a FACS Fortessa (BD Biosciences). Data were processed with FlowJo software.
Immunohistochemistry.
Tissue sections were cut from frozen tissue specimens in OCT 8–10 μm thick. Slides were dried overnight and immunohistochemistry was performed the next day. Slides were briefly washed in PBS followed by 15 min in 0.3% hydrogen peroxide-PBS to quench endogenous peroxidases. Slides were then washed in PBS before applying a histoblock solution (HB) (BSA, MgCl2, 0.2% Tween20) for 30 min. Slides were drained and anti-p65 antibody (93H1, 1:100; Cell Signaling) was applied and incubated overnight at 4 °C. The following day, slides were washed and secondary Abs applied for 30 min (goat anti-rabbit secondary, BA-1000; Vector Labs). ABC reagent (PK-6100, Vector Labs) was applied for 25 min following development with DAB (SK-4100, Vector Labs). Specimens were viewed with a brightfield microscope (Leica DM2500 equipped with a Micropublisher 3.3-QI imaging camera) using Q-Capture Pro software and processed with Adobe Photoshop CS5.
Cleaved Caspase 3 Staining.
Tissue specimens from treated mouse cohorts were processed for cleaved caspase 3 immunohistochemical staining as follows: Antigen retrieval was performed with microwave heating in 10 mM Na citrate (pH 6.0) treatment for 25 min. The primary Ab was prepared in histoblock solution (3% BSA, 20 mM MgCl2, 0.3% Tween 20 in 1× PBS with 5% goat serum with 0.2% Triton X-100 at a dilution of 1:1,000 overnight at 4 °C). Secondary Ab (goat anti-rabbit, BA-4001, Vector Laboratories) was incubated for 1 h at a dilution of 1:400. Signal was visualized with DAB peroxidase reagent (SK-4100 kit, Vector Laboratories). Slides were digitized using a Nanozoomer2.0 HT-Hamamatsu (Olympus). The analysis protocol package (APP) was developed using Visiopharm software to identify the cells that were positive for cleaved caspase 3. A ratio of positive cells to total cell number was then calculated for each slide.
Analysis of Mouse BMDM Databases.
Unsupervised clustering of Pdl1, Nfkb1/p65, and Rela/p50 gene expression profiles in mouse BMDMs exposed to LPS (data from GEO accession no. GSE27112) (41). The Spearman’s correlation coefficients (SCCs) of Cd274 and the two other genes are shown. Mouse genomic region including the Pdl1 gene and depicting the Nfkb1/p65 chromatin immunoprecipitation results in BMDMs untreated or exposed to LPS (two assays indicated in yellow and green; GEO accession no. GSE16723) (60). The positions of the Nfkb1/p65 enhancer described in Ghisletti et al. (42) and validated by ChIP-qPCR are also indicated in blue.
Analysis of Human Breast Cancer Datasets.
In TCGA database, Pam50 subtype calls annotated in clinical data were used to identify primary breast tumors of the basal subtype. The HR-deficient BC cohort was defined by using TCGA breast cancer RNAseq data and somatic mutations after being obtained following approval by the Data Access Committee (project no. 11689). The results published here are partly based upon data generated by TCGA managed by the National Cancer Institute and the National Human Genome Research Institute. Information about TCGA can be found at https://cancergenome.nih.gov/. Mutational signatures were defined using the R mutSignatures package (61). The expression signature scores were computed using the ssGSEA algorithm with standard parameters and using all genes included in each set (62). The Pearson Coefficient Correlation (PCC) and P values were computed in R, and immune cell types were inferred using the microenvironment cell populations-counter (MCP-counter) method (43). The “Chuang_oxidative_stress_response” gene set (ROS up-regulated genes only) was chosen because it includes a highly comprehensive gene expression signature derived from the cellular response to three different oxidants, including hydrogen peroxide, menadione, and t-butyl hydroperoxide (50).
Statistical Analyses.
Data are reported in bar graphs as the mean or median ± SEM, with P values calculated using Student’s t test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). The mean was calculated based on a minimum of n = 3 replicates in each experiment, and each experiment was performed at least three times. Data were analyzed either by Microsoft Excel or GraphPad Prism 7.
Supplementary Material
Acknowledgments
We thank Andrew Elia for assistance in immunohistochemical staining and Kelsie Thu, Christian Bassi, and Shawn P. Kubli for helpful discussions and technical advice. This study was supported by Susan G. Komen Career Catalyst Research Grant 410005437 (to C.G.); Foundation Canadian Institutes of Health Research Grant 143268 (to T.W.M.); Carlos III Institute of Health Grants PI15/00854 and PI18/01029, cofunded with Fondo Europeo de Desarrollo Regional (FEDER) (to M.A.P.); Generalitat de Catalunya Grant 2017-449 (to M.A.P.); Centres de Recerca de Catalunya (CERCA) program (M.A.P.); Italian Ministry of University and Research (P.C.); University of Turin-Progetti Ricerca Locale (P.C.); and National Cancer Institute/National Institutes of Health Grant CA190449 (to T.L.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819473116/-/DCSupplemental.
References
- 1.Senkus E, et al. ESMO Guidelines Committee Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26:v8–v30. doi: 10.1093/annonc/mdv298. [DOI] [PubMed] [Google Scholar]
- 2.Liedtke C, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 3.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savas P, et al. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat Rev Clin Oncol. 2016;13:228–241. doi: 10.1038/nrclinonc.2015.215. [DOI] [PubMed] [Google Scholar]
- 5.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emens LA, Middleton G. The interplay of immunotherapy and chemotherapy: Harnessing potential synergies. Cancer Immunol Res. 2015;3:436–443. doi: 10.1158/2326-6066.CIR-15-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solinas C, et al. Targeting immune checkpoints in breast cancer: An update of early results. ESMO Open. 2017;2:e000255. doi: 10.1136/esmoopen-2017-000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smyth MJ, Ngiow SF, Ribas A, Teng MW. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13:143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 9.Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res. 2015;75:2139–2145. doi: 10.1158/0008-5472.CAN-15-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowanetz M, et al. Differential regulation of PD-L1 expression by immune and tumor cells in NSCLC and the response to treatment with atezolizumab (anti-PD-L1) Proc Natl Acad Sci USA. 2018;115:E10119–E10126. doi: 10.1073/pnas.1802166115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conklin KA. Chemotherapy-associated oxidative stress: Impact on chemotherapeutic effectiveness. Integr Cancer Ther. 2004;3:294–300. doi: 10.1177/1534735404270335. [DOI] [PubMed] [Google Scholar]
- 12.Jezierska-Drutel A, Rosenzweig SA, Neumann CA. Role of oxidative stress and the microenvironment in breast cancer development and progression. Adv Cancer Res. 2013;119:107–125. doi: 10.1016/B978-0-12-407190-2.00003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassetta L, Kitamura T. Targeting tumor-associated macrophages as a potential strategy to enhance the response to immune checkpoint inhibitors. Front Cell Dev Biol. 2018;6:38. doi: 10.3389/fcell.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carron EC, et al. Macrophages promote the progression of premalignant mammary lesions to invasive cancer. Oncotarget. 2017;8:50731–50746. doi: 10.18632/oncotarget.14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartley G, Regan D, Guth A, Dow S. Regulation of PD-L1 expression on murine tumor-associated monocytes and macrophages by locally produced TNF-α. Cancer Immunol Immunother. 2017;66:523–535. doi: 10.1007/s00262-017-1955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffith OW, Meister A. Glutathione: Interorgan translocation, turnover, and metabolism. Proc Natl Acad Sci USA. 1979;76:5606–5610. doi: 10.1073/pnas.76.11.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray PJ. Macrophage polarization. Annu Rev Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 18.Mantovani A, et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Prima V, Kaliberova LN, Kaliberov S, Curiel DT, Kusmartsev S. COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc Natl Acad Sci USA. 2017;114:1117–1122. doi: 10.1073/pnas.1612920114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruffell B, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26:623–637. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makita N, Hizukuri Y, Yamashiro K, Murakawa M, Hayashi Y. IL-10 enhances the phenotype of M2 macrophages induced by IL-4 and confers the ability to increase eosinophil migration. Int Immunol. 2015;27:131–141. doi: 10.1093/intimm/dxu090. [DOI] [PubMed] [Google Scholar]
- 22.Aras S, Zaidi MR. TAMeless traitors: Macrophages in cancer progression and metastasis. Br J Cancer. 2017;117:1583–1591. doi: 10.1038/bjc.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang N, et al. CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-κB/SOX4 signaling. Cell Death Dis. 2018;9:880. doi: 10.1038/s41419-018-0876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holen I, et al. IL-1 drives breast cancer growth and bone metastasis in vivo. Oncotarget. 2016;7:75571–75584. doi: 10.18632/oncotarget.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spiegel A, et al. Neutrophils suppress intraluminal NK cell-mediated tumor cell clearance and enhance extravasation of disseminated carcinoma cells. Cancer Discov. 2016;6:630–649. doi: 10.1158/2159-8290.CD-15-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatano Y, Shimizu T, Tomioka H. Unique macrophages different from M1/M2 macrophages inhibit T cell mitogenesis while upregulating Th17 polarization. Sci Rep. 2014;4:4146. doi: 10.1038/srep04146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elgueta R, et al. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229:152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grohmann U, et al. Positive regulatory role of IL-12 in macrophages and modulation by IFN-gamma. J Immunol. 2001;167:221–227. doi: 10.4049/jimmunol.167.1.221. [DOI] [PubMed] [Google Scholar]
- 29.Ma X, et al. Regulation of IL-10 and IL-12 production and function in macrophages and dendritic cells. F1000 Res. 2015;4:1465. doi: 10.12688/f1000research.7010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noy R, Pollard JW. Tumor-associated macrophages: From mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 32.Brown SB, Savill J. Phagocytosis triggers macrophage release of Fas ligand and induces apoptosis of bystander leukocytes. J Immunol. 1999;162:480–485. [PubMed] [Google Scholar]
- 33.Tardáguila M, et al. CX3CL1 promotes breast cancer via transactivation of the EGF pathway. Cancer Res. 2013;73:4461–4473. doi: 10.1158/0008-5472.CAN-12-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011;21:103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Natoli G, Chiocca S. Nuclear ubiquitin ligases, NF-kappaB degradation, and the control of inflammation. Sci Signal. 2008;1:pe1. doi: 10.1126/stke.11pe1. [DOI] [PubMed] [Google Scholar]
- 36.Kishore N, et al. A selective IKK-2 inhibitor blocks NF-kappa B-dependent gene expression in interleukin-1 beta-stimulated synovial fibroblasts. J Biol Chem. 2003;278:32861–32871. doi: 10.1074/jbc.M211439200. [DOI] [PubMed] [Google Scholar]
- 37.Bristow CL, et al. NF-kappaB signaling, elastase localization, and phagocytosis differ in HIV-1 permissive and nonpermissive U937 clones. J Immunol. 2008;180:492–499. doi: 10.4049/jimmunol.180.1.492. [DOI] [PubMed] [Google Scholar]
- 38.Vogel CF, et al. Cross-talk between aryl hydrocarbon receptor and the inflammatory response: A role for nuclear factor-κB. J Biol Chem. 2014;289:1866–1875. doi: 10.1074/jbc.M113.505578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goode G, Pratap S, Eltom SE. Depletion of the aryl hydrocarbon receptor in MDA-MB-231 human breast cancer cells altered the expression of genes in key regulatory pathways of cancer. PLoS One. 2014;9:e100103. doi: 10.1371/journal.pone.0100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terashima J, Tachikawa C, Kudo K, Habano W, Ozawa S. An aryl hydrocarbon receptor induces VEGF expression through ATF4 under glucose deprivation in HepG2. BMC Mol Biol. 2013;14:27. doi: 10.1186/1471-2199-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng CS, et al. The specificity of innate immune responses is enforced by repression of interferon response elements by NF-κB p50. Sci Signal. 2011;4:ra11. doi: 10.1126/scisignal.2001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Becht E, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorrini C, et al. Estrogen controls the survival of BRCA1-deficient cells via a PI3K-NRF2-regulated pathway. Proc Natl Acad Sci USA. 2014;111:4472–4477. doi: 10.1073/pnas.1324136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rom-Jurek EM, et al. Regulation of programmed death ligand 1 (PD-L1) expression in breast cancer cell lines in vitro and in immunodeficient and humanized tumor mice. Int J Mol Sci. 2018;19:E563. doi: 10.3390/ijms19020563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samanta D, et al. Chemotherapy induces enrichment of CD47+/CD73+/PDL1+ immune evasive triple-negative breast cancer cells. Proc Natl Acad Sci USA. 2018;115:E1239–E1248. doi: 10.1073/pnas.1718197115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng J, et al. Chemotherapy induces programmed cell death-ligand 1 overexpression via the nuclear Factor-κB to foster an immunosuppressive tumor microenvironment in ovarian cancer. Cancer Res. 2015;75:5034–5045. doi: 10.1158/0008-5472.CAN-14-3098. [DOI] [PubMed] [Google Scholar]
- 48.Chung W, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308 e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuang YY, et al. Gene expression after treatment with hydrogen peroxide, menadione, or t-butyl hydroperoxide in breast cancer cells. Cancer Res. 2002;62:6246–6254. [PubMed] [Google Scholar]
- 51.Kwa MJ, Adams S. Checkpoint inhibitors in triple-negative breast cancer (TNBC): Where to go from here. Cancer. 2018;124:2086–2103. doi: 10.1002/cncr.31272. [DOI] [PubMed] [Google Scholar]
- 52.Soliman HH. nab-Paclitaxel as a potential partner with checkpoint inhibitors in solid tumors. OncoTargets Ther. 2016;10:101–112. doi: 10.2147/OTT.S122974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gil Del Alcazar CR, et al. Immune escape in breast cancer during in situ to invasive carcinoma transition. Cancer Discov. 2017;7:1098–1115. doi: 10.1158/2159-8290.CD-17-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmid P, et al. IMpassion130 Trial Investigators Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 55.Yamazaki T, et al. Expression of programmed death 1 ligands by murine T cells and APC. J Immunol. 2002;169:5538–5545. doi: 10.4049/jimmunol.169.10.5538. [DOI] [PubMed] [Google Scholar]
- 56.Gibbons Johnson RM, Dong H. Functional expression of programmed death-ligand 1 (B7-H1) by immune cells and tumor cells. Front Immunol. 2017;8:961. doi: 10.3389/fimmu.2017.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olson OC, Kim H, Quail DF, Foley EA, Joyce JA. Tumor-associated macrophages suppress the cytotoxic activity of antimitotic agents. Cell Rep. 2017;19:101–113. doi: 10.1016/j.celrep.2017.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeNardo DG, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeNardo DG, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barish GD, et al. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.