Activating mutations in RAS genes (KRAS, HRAS, and NRAS) are oncogenic drivers arising in about one-third of human malignancies (1, 2). Cancers with oncogenic RAS mutations are among those with the poorest prognosis, the most notorious example being pancreatic cancer with a 95% mutation frequency in KRAS and a 7% survival rate beyond 5 y of diagnosis. As such, targeting oncogenic RAS proteins or their functional output has been a longstanding priority for development of effective cancer therapies. Unfortunately, therapeutic targeting of oncogenic RAS proteins directly or of their individual downstream effector pathways has not been successful for the treatment of the vast majority of human cancers, suggesting that functional redundancies provide workarounds that sustain oncogenic activity. In PNAS, Lee et al. (3) use a novel combinatorial knockdown screening approach to identify essential RAS signaling and stress adaptation programs that, when cotargeted, compromise RAS-mediated cancer cell survival.
RAS proteins are small GTPases that transduce signals from upstream growth factor receptors to downstream signaling pathways to stimulate growth, proliferation, and survival. In cancers, oncogenic mutations in RAS proteins such as KRAS G12V render them in the constitutively “on” position, decoupling regulatory growth signals from effector mechanisms. These unregulated growth signals drive the cancer phenotype through constitutive activation of the downstream RAF, RalGDS, and PI3K pathways (Fig. 1) (1, 2).
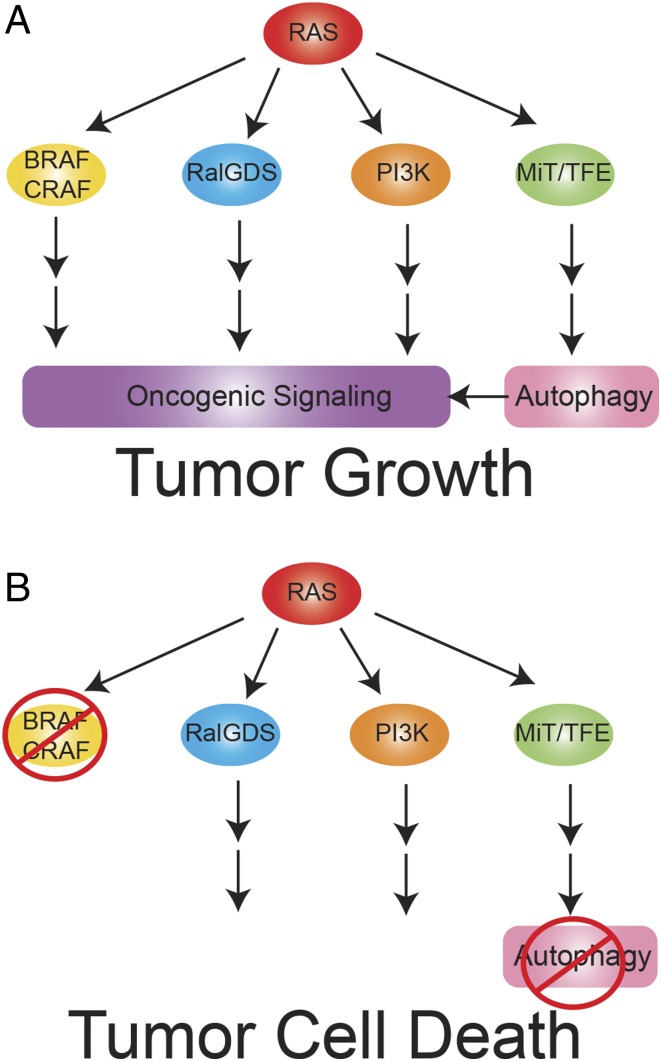
Fig. 1.
Essential codependency of RAS-driven cancers on BRAF, CRAF, and autophagy. BRAF and CRAF provide key functional oncogenic signaling downstream of RAS that requires autophagy mediated by ATG7 to sustain survival. Coordinate blockade of BRAF, CRAF, and ATG7 provides the one-two punch and lethal blow to Ras-driven cancer cells.
Targeting oncogenic RAS proteins directly has proved difficult, with the possible exception of the KRAS V12C mutation in a small subset of human cancers in which the cysteine residue renders RAS vulnerable to inactivation (4). To stimulate the quest to target RAS and the downstream RAS effectors, the RAS Initiative at the National Cancer Institute (https://www.cancer.gov/research/key-initiatives/ras) was formed to provide a large, coordinated effort. Targeting single RAS downstream effector pathways, such as the RAF/MEK/ERK MAPK pathway using inhibitors of its components, has activity in preclinical models but generally fails to produce durable responses in patients (4). Multiple redundantly functioning paralogs of each signaling component and the retention of signaling activity through multiple effector pathways are thought to limit this type of approach by providing inhibitor bypass mechanisms. Combining inhibition of multiple effector arms of RAS downstream signaling has also proved to be toxic to normal cells, as has deep inhibition of multiple paralogs in a single arm. Thus, standard approaches to find a therapeutic window for oncogenic RAS signaling inhibition has proved elusive. Numerous unbiased synthetic lethal screens to identify novel single vulnerabilities of RAS-driven cancer cells have also yet to bring forth superior targets to effectively block oncogenic signaling by RAS sufficient for therapeutic efficacy. These findings suggest that multiple genes downstream of RAS may have to be cotargeted to overcome paralog redundancy and pathway cooperativity to block the oncogenic activity of RAS, but which ones? Also, while doing so, is it possible to reduce toxicity to normal cells sufficiently for a therapeutic window?
To address redundant effector pathways and paralog function downstream of RAS, Lee et al. (3) develop a combinatorial siRNA approach to simultaneously target multiple genes in KRAS-driven cells in comparison with KRAS wild-type human cancer cell lines and normal cells. They focus on cotargeting known downstream RAS effectors with stress response pathways using 73 genes in 29 gene nodes, looking for selective loss of viability in RAS mutant cells (and not in RAS wild-type cancer cells and normal cells). Among the RAS effector nodes, only knockdown of the RAF node (particularly BRAF and CRAF) most closely replicated RAS dependency in colorectal and pancreatic cancer cell lines identifying the BRAF/CRAF axis as a superior target to the MEK and ERK nodes (Fig. 1).
Lee et al. (3) assess RAS-specific toxicity and the efficacy of targeting node combinations by evaluating the knockdown of 378 node-pair combinations across RAS mutant and wild-type cancer cell lines and normal cells. Specific combinations were superior to targeting the RAF node alone, including targeting RAF in combination with the RAC, RAL, ROCK, and ATG (autophagy) nodes. To augment targeting of the RAF node alone, it was combined with knockdown of RAC, RAL, and ATG nodes, followed by deconvolution of the paralogs within the nodes. Toxicity of the combinations to RAS wild-type cancer cell lines and normal cells distinguished general toxicity from RAS-specific addiction to the pathway. Targeting BRAF, CRAF, and the essential autophagy gene ATG7 in combination provided the best discrimination between KRAS-mutant cells and normal, untransformed cells (Fig. 1). Preserving ARAF increased selectivity to oncogenic RAS signaling by decreasing toxicity to normal cells. Although knockdown of ATG7 alone was not toxic to normal cells in these nutrient-replete conditions, it importantly converted a cytostatic response of BRAF and CRAF knockdown to one of cell death in RAS-transformed cancer cell lines. Thus, the authors (3) identify that targeting BRAF and CRAF in combination with ATG7 is a promising therapeutic approach for RAS-driven cancers; however, why is this the case?
Autophagy is a catabolic process whereby intracellular components are captured, degraded, and recycled in lysosomes to sustain metabolism during interruptions in nutrient availability (5). Basal autophagy functions at a low level in normal cells but is activated by stress and particularly starvation, where it is essential for adult mice to maintain circulating glucose levels to survive fasting (6). In contrast to normal cells, RAS-driven cancer cells surprisingly up-regulate basal autophagy (7–12) by activating the MiT/TFE transcription program (13). In RAS-driven cancer cells, autophagy scavenges intracellular nutrients, which are recycled into central carbon metabolism to promote survival (5, 14). Without autophagy, Ras-driven lung and pancreatic cancers cells are sensitized to nutrient deprivation, and tumors have impaired survival and growth and fail to efficiently progress to malignancy, prompting efforts to target autophagy and downstream components including lysosomes in RAS-driven cancers (5, 15).
The next issue that Lee et al. (3) address is the mechanism by which autophagy is important for RAS-driven cancers. Metabolic analysis of RAS-driven tumor cells with and without the essential autophagy gene ATG7 revealed that autophagy recycles macromolecules into central carbon metabolism. By sustaining the supply
In PNAS, Lee et al. use a novel combinatorial knockdown screening approach to identify essential RAS signaling and stress adaptation programs that, when cotargeted, compromise RAS-mediated cancer cell survival.
of metabolic substrates during nutrient deprivation, autophagy mitigates energy crisis and death from the depletion of nucleotide pools, enabling the survival of RAS-driven tumor cells (14). Indeed, RAS-driven cancers are more sensitive to acute loss of autophagy than most normal tissues, indicating a therapeutic window for targeting autophagy (6, 16). Recent evidence suggests that RAS-driven lung cancers with loss of LKB1 and thereby nutrient stress adaptation are particularly autophagy dependent (17). Host as well as tumor cell-autonomous autophagy also promotes tumor growth by sustaining microenvironmental and circulating nutrients critical for tumor growth, underscoring the importance of metabolic maintenance in cancer (18, 19). Whereas the findings of Lee et al. (3) improve upon our understanding the functional dependency of RAS-driven cancers on autophagy, they raise important points about how to move forward both preclinically and clinically.
The majority of the work identifying the important role for autophagy in RAS-driven and other cancers has been performed using genetic inactivation of essential autophagy genes in genetically engineered mouse models for cancer (5). Development of specific and potent autophagy inhibitors that work in vivo has been limited thus far. Lee et al. (3) point to the therapeutic importance of targeting the E1-like enzyme ATG7, but it is yet unknown whether targeting other autophagy pathway components upstream (e.g., ULK1 or VPS34) or downstream (ATG4 or lysosome function) of ATG7 would be similarly active with coordinate BRAF/CRAF inhibition. Current therapeutic efforts to target autophagy in cancer use hydroxychloroquine (HCQ) or its analogs that disrupt lysosome function (15). Whether this approach can be improved by further mechanistic studies, drug combinations, more potent analogs, or a defined patient population is under scrutiny. Most of the genetic functional studies identifying the role for autophagy in RAS-driven cancers have been performed in mice in vivo, whereby knockout of a single essential autophagy gene has antitumor activity. Because the study of Lee et al. (3) is limited to functional assessment of RAS effectors in vitro in nutrient-replete conditions where autophagy is less important, coordinate BRAF/CRAF and ATG7 inhibition should be examined in vivo, where nutrients are limited and autophagy is more important. Since autophagy dependence of RAS-driven cancer cells in vitro may be mitigated by nutrient-replete conditions, perhaps less RAF signaling in vivo in tumors increases autophagy addiction. Moving forward, sparing ARAF by inhibiting BRAF/CRAF dimerization with coordinate autophagy pathway inhibition is a promising strategy. Because BRAF-driven cancers are also autophagy dependent, this approach may have broad utility beyond RAS-driven cancers. Indeed, BRAF-driven cancers are sensitive to coordinate BRAF and autophagy inhibition with HCQ, and genetic loss of autophagy enhances antitumor activity of MAPK pathway inhibitors (20, 21).
Acknowledgments
E.W.’s research is supported by the National Institutes of Health (Grants R01 CA163591 and R01 CA193970) and by the NIH Grant P30 CA072720 (to Rutgers Cancer Institute of New Jersey).
Footnotes
Conflict of interest statement: The author is a founder of Vescor Therapeutics, LLC and a stockholder in Forma Therapeutics.
See companion article on page 4508.
References
- 1.Li S, Balmain A, Counter CM. A model for RAS mutation patterns in cancers: Finding the sweet spot. Nat Rev Cancer. 2018;18:767–777. doi: 10.1038/s41568-018-0076-6. [DOI] [PubMed] [Google Scholar]
- 2.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee C-S, et al. MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc Natl Acad Sci USA. 2019;116:4508–4517. doi: 10.1073/pnas.1817494116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spiegel J, Cromm PM, Zimmermann G, Grossmann TN, Waldmann H. Small-molecule modulation of Ras signaling. Nat Chem Biol. 2014;10:613–622. doi: 10.1038/nchembio.1560. [DOI] [PubMed] [Google Scholar]
- 5.Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karsli-Uzunbas G, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JY, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lock R, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenfeldt MT, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 11.Yang A, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perera RM, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo JY, et al. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 2016;30:1704–1717. doi: 10.1101/gad.283416.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307–3318. doi: 10.1002/cncr.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang A, et al. Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 2018;8:276–287. doi: 10.1158/2159-8290.CD-17-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhatt V, et al. Autophagy modulates lipid metabolism to maintain metabolic flexibility for Lkb1-deficient Kras-driven lung tumorigenesis. Genes Dev. January 28, 2019 doi: 10.1101/gad.320481.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poillet-Perez L, et al. Autophagy maintains tumour growth through circulating arginine. Nature. 2018;563:569–573. doi: 10.1038/s41586-018-0697-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sousa CM, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy JM, et al. Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 2014;4:773–780. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie X, Koh JY, Price S, White E, Mehnert JM. Atg7 overcomes senescence and promotes growth of BrafV600E-driven melanoma. Cancer Discov. 2015;5:410–423. doi: 10.1158/2159-8290.CD-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]