Significance
Amyotrophic lateral sclerosis (ALS) is a devastating, motor neuron degenerative disease without any cure to date. About 95% of ALS patients feature abnormalities in the RNA/DNA binding protein TDP-43, involving its nucleus-cytoplasmic mislocalization in spinal motor neurons. How TDP-43 pathology triggers neuronal apoptosis remains unclear. Here, we report that TDP-43 participates in the DNA damage response and its nuclear clearance in motor neurons causes DNA double-strand break repair defects in ALS. Our findings uncover a link between TDP-43 pathology and impaired DNA repair, and suggest potential avenues for DNA repair-targeted therapies for TDP-43–ALS.
Keywords: TDP-43, DNA damage response, DNA double-strand break repair, amyotrophic lateral sclerosis, neurodegeneration
Abstract
Genome damage and their defective repair have been etiologically linked to degenerating neurons in many subtypes of amyotrophic lateral sclerosis (ALS) patients; however, the specific mechanisms remain enigmatic. The majority of sporadic ALS patients feature abnormalities in the transactivation response DNA-binding protein of 43 kDa (TDP-43), whose nucleo-cytoplasmic mislocalization is characteristically observed in spinal motor neurons. While emerging evidence suggests involvement of other RNA/DNA binding proteins, like FUS in DNA damage response (DDR), the role of TDP-43 in DDR has not been investigated. Here, we report that TDP-43 is a critical component of the nonhomologous end joining (NHEJ)-mediated DNA double-strand break (DSB) repair pathway. TDP-43 is rapidly recruited at DSB sites to stably interact with DDR and NHEJ factors, specifically acting as a scaffold for the recruitment of break-sealing XRCC4-DNA ligase 4 complex at DSB sites in induced pluripotent stem cell-derived motor neurons. shRNA or CRISPR/Cas9-mediated conditional depletion of TDP-43 markedly increases accumulation of genomic DSBs by impairing NHEJ repair, and thereby, sensitizing neurons to DSB stress. Finally, TDP-43 pathology strongly correlates with DSB repair defects, and damage accumulation in the neuronal genomes of sporadic ALS patients and in Caenorhabditis elegans mutant with TDP-1 loss-of-function. Our findings thus link TDP-43 pathology to impaired DSB repair and persistent DDR signaling in motor neuron disease, and suggest that DSB repair-targeted therapies may ameliorate TDP-43 toxicity-induced genome instability in motor neuron disease.
Amyotrophic lateral sclerosis (ALS) is a rapidly progressive, fatal degenerative disease of motor neurons without an effective treatment. ALS affects neurons in the motor cortex, brainstem, and upper and lower spinal cord, gradually inducing muscle atrophy, denervation, and severe motor dysfunction. The ALS group of motor neuron diseases is highly complex, involving more than a dozen genes (reviewed in ref. 1). The transactivation response DNA-binding protein (TARDBP) of 43 kDa (TDP-43), has nuclear clearance, cytosolic sequestration/aggregation, and fragmentation in motor neurons characteristically observed in nearly 95% of sporadic ALS patients (2, 3). ALS can develop both from familial (∼10% incidences) and sporadic causes (∼90% cases). Furthermore, a number of frontotemporal lobar degeneration (FTLD) patients develop ALS-like tau-negative, ubiquitin-positive inclusions of TDP-43 in cortical neurons, a subtype of motor neuron disease named FTLD-TDP (4).
TDP-43 protein, encoded by the TARDBP gene located on chromosome 1, is an RNA/DNA-binding protein of the heterogeneous ribonucleoprotein family (5). Structurally, TDP-43 is composed of N-terminal domains comprising a bipartite nuclear localization sequence (NLS), two distinct RNA recognition motifs (RRM), namely RRM1 and RRM2, and a bipartite nuclear export sequence (NES). The C-terminal disordered domain comprising a prion-like motif is the primary contributor to its aggregation propensity (6). Since its first implication in ALS and FTLD (3, 7–9), the involvement of TDP-43 in mRNA processing and microRNA biogenesis has been well documented (reviewed in ref. 1). In addition, TDP-43 may act as a structural component in stress granule formation and in regulation of neurite growth (10). Studies have linked TDP-43 toxicity to other cellular pathways, including autophagy, loss of synaptic transmission, inflammation, and microglia infiltration, and their involvement in motor neuron death in both familial and sporadic forms of ALS (11, 12). However, none of these processes exclusively drive the motor neurons to death, nor is their intervention sufficient to rescue degenerating neurons. Hence, further investigation is warranted to identify other functions of TDP-43 responsible for survival of motor neurons.
In addition to its RNA-binding activity, TDP-43 also binds to DNA (13, 14); however, its possible role in DNA transactions have not been investigated. Furthermore, significant accumulation of genomic damage is consistently observed in multiple neurodegenerative diseases and a previous proteomic study identified a key DNA repair protein “Ku” in the TDP-43 immunoprecipitation (IP) complex from human cells (15). This raised the unexplored possibility of TDP-43’s involvement in DNA damage response (DDR).
Here, we have documented TDP-43’s involvement in DDR as a key component of nonhomologous end joining (NHEJ), the major pathway for repair of DNA double-strand breaks (DSBs) in the postmitotic neurons. Our results showed significant DSB accumulation and reduced NHEJ levels in TDP-43–depleted human neural stem cell-derived motor neurons, as well as in sporadic ALS patients’ spinal cord specimens with TDP-43 pathology. The NHEJ defects were due to reduced recruitment of X-ray repair cross-complementing protein 4 (XRCC4)-like factor (XLF)-DNA ligase 4 (Lig4) complex at DSB sites, which is critical for break end ligation for NHEJ. Consistently, loss of TDP-43 correlated with reduced Lig4 activity. These observations, together with enhanced activation of the DDR factors, are consistent with the role of TDP-43 in DDR. Defective genome repair, the resulting persistent accumulation of unrepaired DSBs and sustained DDR activation, thus contribute to neuronal death in ALS and other TDP-43–associated neurodegenerative diseases. Our study thus provides a paradigm about the mechanism of TDP-43 toxicity, which may help develop DNA repair-targeted therapeutic approaches for ameliorating a broad range of motor neuron diseases involving TDP-43 pathology.
Results
TDP-43 Is a Component of the DDR Signaling for DSB Repair.
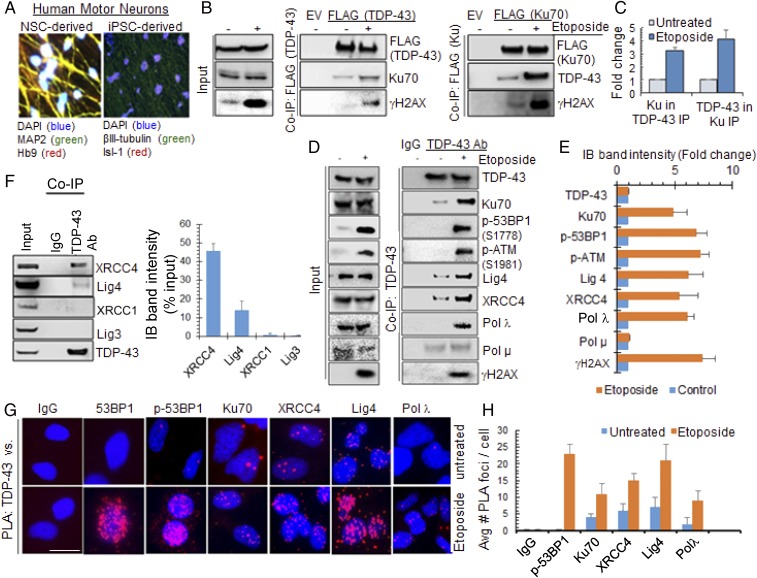
Following up on a proteomic analysis by Taylor and coworkers (15), which showed the NHEJ-initiating DSB sensor protein Ku70 as an interacting partner of TDP-43 in mammalian cells, we confirmed the in cell association of TDP-43 with Ku70 by co-IP and in situ proximity ligation assay (PLA), using both endogenous TDP-43 and ectopic FLAG–TDP-43. These in cell studies were performed in three neuronal lines, namely differentiated neuroblastoma SH-SY5Y cell, human induced pluripotent stem cell (iPSC)- or fetal stem cell-derived neural progenitor (NP) cells (NPCs), as well as differentiated motor neurons. Human motor neurons were generated from a genetically unmodified human fetal neural stem cell (hNSC, K048) line (Fig. 1A and SI Appendix, Fig. S1B) (16) and from an iPSC (KYOU-DXR0109B) line (SI Appendix, Fig. S1 C–F) (17). Culture conditions and growth factors were optimized for generating up to ∼80% efficiency of differentiation into motor neurons. Motor neurons were used in most experiments that required <10,000 cells, including in cell studies by microscopy, comet, or PCR-based genome damage analyses. However, iPSC-derived NP cells (18) were used for co-IP studies, unless otherwise indicated. In view of Ku’s high affinity for DNA, we eliminated the possibility of DNA-mediated protein–protein interactions by pretreating cell extracts with benzonase (DNase+RNase) for 30 min at 37 °C, before performing co-IP assays. FLAG co-IP from total cell extracts of NP or differentiated SH-SY5Y cells (19, 20) (SI Appendix, Fig. S1 A–E) ectopically expressing FLAG–TDP-43, FLAG-Ku70, or empty FLAG-tag vector after treating with DSB-inducing topoisomerase II inhibitor etoposide or DMSO control, revealed strong interaction between TDP-43 and Ku70 (Fig. 1 B and C). Induction of DNA damage significantly (>threefold) enhanced the interaction. The presence of phospho-(S139)-histone H2AX (γH2AX) in the co-IP complex of TDP-43 and Ku70 suggests the interaction of TDP-43 with Ku70 at the chromatin level. DNA-dependent protein kinase catalytic subunit (DNA-PKcs) forms the DNA-PK holoenzyme after binding to Ku70/80 heterodimer, a key early response factor in NHEJ (21, 22). PLA analysis in irradiated (IR, 3 Gy) NPCs, fixed at 30-min post-IR, showed strong interaction foci between TDP-43 and Ku70 or DNA-PKcs, compared with the control cells (SI Appendix, Fig. S2 A and B). TDP-43 similarly showed enhanced interaction with p53 binding protein 1 (53BP1), via its DDR-linked phospho-(S1778)-53BP1 (23), in etoposide-treated cells as showed by endogenous 53BP1 co-IP using anti-53BP1 antibody (Ab) or normal rabbit IgG (SI Appendix, Fig. S2 C and D). These data suggest the involvement of TDP-43 in early DSB response mechanisms.
Fig. 1.
TDP-43 complexes with DNA DSB repair and DDR proteins in a damage-dependent fashion. (A) Representative images of human motor neurons derived from hNSC and iPSC line (images acquired at 40× magnification). Immunofluorescence with neuronal marker MAP2 or βIII-tubulin and motor neuron marker Hb9 or Isl-1. See also SI Appendix, Fig. S1. (B and C) TDP-43 (Left) or Ku70 (Right) co-IP from NPCs, transfected with FLAG-tagged plasmids and treated with etoposide to induce DNA damage. The IPs with anti-FLAG Ab were probed for anti–TDP-43, anti-Ku70, and anti-γH2AX Abs. (C) Quantitation of the level of Ku70 in TDP-43 IP and vice versa. (D and E) TDP-43 IP using anti-TDP-43 Ab from etoposide-treated NPC nuclear extracts. Histogram shows quantitation of IB band intensity. (F) IB of TDP-43 IP from differentiated SH-SY5Y cells showing specific association with NHEJ-related XRCC4/Lig4 but not with XRCC1/Lig3. (G and H) PLA in etoposide- (5 μM, 4 h) treated motor neurons. PLA of anti-TDP-43 vs. IgG, 53BP1, p53BP1, Ku70, XRCC4, Lig4, or Polλ Abs. (Scale bars, 10 μm.) Average number of PLA foci from >25 cells were quantified. Nuclei were stained with DAPI.
We next examined the association of TDP-43 with key components of DDR signaling and the NHEJ pathway. Endogenous TDP-43 co-IP from nuclear extracts of the iPSC-derived NPC line (Fig. 1 D and E) treated with etoposide contained Ku70, p-(S1778)-53BP1, XRCC4/Lig4 complex, and DNA polymerase (Pol) λ, at ∼6- to 10-fold higher levels compared with that of control cells. Notably, the XRCC1/Lig3 complex, primarily involved in DNA single-strand break (SSB) repair (24), was not detected in the TDP-43 co-IP (Fig. 1F and SI Appendix, Fig. S2E), suggesting specific involvement of TDP-43 in NHEJ-mediated DSB repair. Furthermore, NHEJ-associated X family DNA Pols (25, 26), Polμ and Polλ, were present in the TDP-43 co-IP complex at a basal level; however, DNA damage-dependent enhanced association with TDP-43 was observed only for Polλ. Similar interaction patterns were observed in TDP-43 co-IP from chromatin extracts of differentiated SH-SY5Y cells (SI Appendix, Fig. S2 F and G). In parallel, these interactions were confirmed by PLA in hNSC-derived motor neurons treated with etoposide or DMSO control (Fig. 1 G and H).
To evaluate DNA damage dose-dependent enhancement of TDP-43’s association with NHEJ and DDR factors, we isolated endogenous TDP-43 IP from nuclear extracts of NP cells after treatment with varying doses of etoposide (0.5–10 μM) (SI Appendix, Fig. S2 H and I). The immunoblot (IB) analysis showed etoposide dose-dependent increase in the levels of repair factors Ku70, XRCC4, Lig4, and Polλ, together with DDR factors γH2AX, p53BP1, and p-(S1981)-ataxia-telangiectasia mutated (ATM). Moreover, another DSB-inducing radiomimetic drug bleomycin also induced TDP-43’s association with γH2AX (SI Appendix, Fig. S5), indicating the ubiquitous nature of the interaction in response to DNA damage.
The kinetics of association and dissociation of TDP-43 with DDR factors further confirm its early recruitment and sustained retention/association as part of the DSB repair machinery, and its dissociation closely follows the time course of repair completion. SI Appendix, Fig. S3 H and I show the PLA of TDP-43 vs. Ku70 or p53BP1 at the indicated time intervals following etoposide treatment in NSC-derived motor neurons. The quantitation of average PLA signals showed that the association of TDP-43 with Ku70 and p53BP1 peaked at 5 min after etoposide treatment, before gradually dissociating by 1 h (SI Appendix, Fig. S3I). Furthermore, IB analysis of endogenous TDP-43 co-IP from IR (6 Gy) SH-SY5Y cells showed similar association/dissociation patterns. IB of the input nuclear extracts confirmed activation of γH2AX and p53BP1 post-IR, peaking at 1–5 min before gradually disappearing with time (SI Appendix, Fig. S3A). The level of Ku70 or TDP-43 was unaffected in the input, as expected. IB of co-IP eluates demonstrated a similar dynamic association of Ku70, p53BP1, and γH2AX with TDP-43 (SI Appendix, Fig. S3 B–D). Notably, TDP-43’s interaction with Ku70 and p53BP1 increased significantly, immediately (∼1 min) after DSB induction, followed by gradual dissociation to almost basal level of interaction at ∼2 h (SI Appendix, Fig. S3C). The DDR and NHEJ factors identified in this study that interact with TDP-43 are schematically listed in SI Appendix, Fig. S3G.
TDP-43 Is Rapidly Recruited at DSB Sites and Retained Until Completion of Repair.
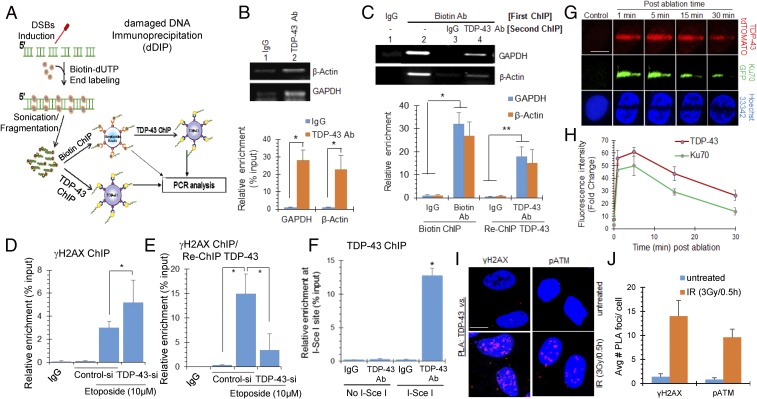
We next evaluated binding of TDP-43 to DSB ends in chromatin by the damaged DNA IP assay (dDIP) (Fig. 2 A–C). The bleomycin-treated cells were fixed with 4% paraformaldehyde and subjected to chromatin IP (ChIP) (25) with either anti–TDP-43 or anti-biotin Ab and control IgG. Binding of TDP-43 to chromatin was first confirmed by fold enrichment of two randomly selected housekeeping genes, GAPDH and β-actin, in control vs. treated TDP-43 ChIP eluates (Fig. 2B). First, ChIP analysis using anti-biotin Ab, followed by a re-ChIP with anti–TDP-43 Ab showed enrichment of the same gene segments (Fig. 2C), suggesting TDP-43’s binding to the DSB ends. A similar dDIP analysis was performed in unstressed cells as control, where we did not observe a significant association of anti-biotin or anti–TDP-43 Ab with the selected gene segments in control cells (SI Appendix, Fig. S4A).
Fig. 2.
TDP-43 is recruited at DSB sites in neuronal genome. (A–C) dDIP for identifying enrichment of TDP-43 at DSB sites. Differentiated neuroblastoma cells were treated with bleomycin (3 μg/mL) for 1 h, and subjected to biotin-conjugation at DSB sites, followed by ChIP analysis. (A) Scheme of dDIP assay. (B) ChIP using control IgG or anti-TDP-43 Ab followed by qRT-PCR with primers targeting randomly selected regions of GAPDH and β-actin genes. Agarose gel separation of PCR products (Upper) and real-time PCR-based quantitation (Lower). (C) ChIP/re-ChIP assay. Agarose gel separation of PCR products (Upper) and relative enrichment of biotin/TDP-43 at DSB sites by real time PCR based quantitation (Lower). (D and E) ChIP/re-ChIP assay to show association of TDP-43 with γH2AX-enriched genomic sequences. NPCs were transfected with control or TDP-43 siRNA. After 72 h, cells were treated with etoposide (10 μM, 4 h) and subjected to first ChIP with anti-γH2AX Ab (D), followed by re-ChIP with anti-TDP-43 Ab (E). Increase in γH2AX-bearing genomic sequences in TDP-43 ChIP shows TDP-43 binding to DSB sites overlaps with γH2AX. TDP-43 ChIP in TDP-43 siRNA treated cells confirms Ab specificity. (F) Association of TDP-43 with I-SceI–induced defined DSB sites. SH-SY5Y cells were transfected with either I-SceI expression plasmid pCBASceI or combination of the NHEJ reporter construct pimEJ5GFP and pCBASceI. Six hours posttransfection, cells were harvested for ChIP assay with control IgG or anti-TDP-43 Ab, then ChIP eluates were subjected to qRT-PCR quantitation of defined sequences adjacent to the I-SceI cleavage site. (G and H) Live cell imaging after laser ablation at 405 nm to induce DSB-rich damage track. NPCs ectopically expressing Ku70-GFP or TDP-43-tdTOMATO were laser-ablated and recruitment of Ku70 or TDP-43 at the laser track was monitored (single-channel) at indicated time-points. Nuclei were stained with Hoechst33342 dye (G). Quantitation of fluorescence intensity at laser tracks from 5 to 10 cells (H). (Scale bars, 10 μm.) (I and J) iPSC-derived motor neurons were treated with 3 Gy of IR and allowed to recover for 30 min, then subjected to PLA between anti–TDP-43 and DDR markers anti-γH2AX or anti-pATM Ab (I). Quantitation of average number of PLA foci from >25 randomly selected cells (J). (Scale bars, 10 μm.) All values are from three independent experiments ±SD. *P < 0.01; **P < 0.05.
To analyze the specificity and global nature of TDP-43’s recruitment at the DSB sites generated either by genome-wide DSB induction with etoposide or at defined I-SceI endonuclease cleavage sites, we performed ChIP with anti-γH2AX Ab or mouse IgG, followed by re-ChIP with anti–TDP-43 Ab (Fig. 2 D–F). For the ChIP assay, chromatin fragmentation was optimized to obtain 250–650 bp in sizes by sonication (SI Appendix, Fig. S4B). For the first experiment, to test TDP-43’s enrichment at etoposide-induced DSBs, cells were transfected with control or TDP-43 siRNA, 72 h before damage induction with etoposide (Fig. 2 D and E). The first ChIP with anti-γH2AX Ab showed ∼threefold enrichment at a randomly selected HPRT gene segment, over corresponding IgG, which further increased by ∼twofold in TDP-43 knockdown (KD) cells (Fig. 2D). The re-ChIP with anti–TDP-43 Ab showed a marked increase (∼15-fold) in enrichment at the amplified HPRT gene segment, indicating TDP-43’s association at etoposide-induced DSB sites. Significantly reduced amplification in TDP-43 KD cells confirmed the specificity of anti–TDP-43 Ab (Fig. 2E). For the second experiment to test TDP-43’s enrichment at defined DSBs, we used SH-SY5Y cells stably transfected with an I-SceI recognition sequence containing vector (pimEJ5GFP). ChIP was performed at 6 h posttransfection with or without the I-SceI expression vector (pCBASceI), with anti-TDP-43 Ab or control IgG as before. Amplification of DNA sequence adjacent to the I-SceI site demonstrated ∼10-fold enrichment of TDP-43 (Fig. 2F). Together, these results suggest that TDP-43 is enriched in proximity of DNA DSBs following damage induction in the chromatin.
To gain further insight into the recruitment of TDP-43 to damaged chromatin, we analyzed repair kinetics after laser micro-IR (26) in NPCs transfected with either tdTomato reporter-tagged TDP-43 or EGFP reporter-tagged Ku70 vector constructs (Fig. 2 G and H). Given that Ku70, the key component of DNA-PK holoenzyme, is recruited at the DSB site very early, Ku70 recruitment at micro-IR–mediated chromatin damage served as a positive control (27). GFP-Ku70 was recruited at substantially high intensity within a minute at the laser track, followed by its gradual dissociation. Similarly, TDP-43–tdTomato was recruited at the laser track within 1 min (the earliest time point measured in this assay) with strong intensity, which was sustained for ∼15 min before a gradual decline. Furthermore, PLA of anti–TDP-43 vs. early DDR marker anti-γH2AX or anti-pATM Ab after IR (3 Gy) in iPSC-derived motor neurons was consistent with this scenario (Fig. 2 I and J). The PLA foci analysis show the number of foci peaked within 1–5 min before gradually disappearing by 3 h (SI Appendix, Fig. S5B).
To further evaluate the ability of TDP-43 to bind directly to the DSB ends, we performed an in vitro biotin-affinity pull-down assay (28) using purified recombinant TDP-43 (SI Appendix, Fig. S4C) with three distinct DNA oligos mimicking a DSB, SSB, or intact duplex. The oligos were either 5′-biotinylated with one open blunt-end terminus (DSB) or both 5′- and 3′-biotinylated (intact duplex) or both ends biotinylated with an internal SSB nick. IB analysis of the biotin-affinity coeluated products against anti–TDP-43 Ab showed that TDP-43 binding required an unblocked DSB-like blunt terminus in DNA (SI Appendix, Fig. S4C). Collectively, these results suggest TDP-43’s direct binding to DSB ends, both in vitro and in chromatin. Moreover, the early recruitment of TDP-43 at DSB sites and its sustained presence until the repair completion suggest a vital role of TDP-43 in DSB repair/DDR signaling.
Loss of TDP-43 Causes Accumulation of DSBs in Neuronal Genome.
Based on TDP-43’s cross-talk with DDR and NHEJ factors, we hypothesized that nuclear loss or functional inactivation of TDP-43 could cause unrepaired DSB accumulation. To test this, we first optimized TDP-43 KD in iPSC-derived NP cells using GFP-tagged lentiviral shRNA transduction, which showed 80–90% uptake in neurons and ∼80% depletion of TDP-43 (SI Appendix, Fig. S6A). Alkaline and neutral comet analyses of TDP-43 KD cells, 96 h post-shRNA transduction, showed ∼20-fold increase in DNA strand breaks compared with the control cells (SI Appendix, Fig. S6B). Neutral comet analysis reflects DSBs exclusively, whereas alkaline comet analysis could be used to quantitate DSBs, SSBs, and other alkali-labile apurinic/apyrimidinic (AP) sites (29, 30). Comparable increase in the mean alkaline vs. neutral comet tail moment in TDP-43 KD cells suggests that a majority of these breaks are DSBs. Furthermore, significant increase in the number of foci of γH2AX, p53BP1 and pATM at 96 h after TDP-43 KD, confirmed the accumulation of endogenous DSBs and DDR activation in neurons (SI Appendix, Fig. S6C). The TUNEL assay at indicated time-points also suggested accumulation of DNA strand breaks (SI Appendix, Fig. S6D). Consistent with our results in neurons, we observed similar accumulation of DSBs in TDP-43 siRNA-treated differentiated SH-SY5Y cells, as analyzed by the comet assay (SI Appendix, Fig. S6 E–G), long-amplicon (LA) PCR-based amplification of isolated genomic DNA (SI Appendix, Fig. S6H), and γH2AX foci accumulation (SI Appendix, Fig. S6I).
DNA Damage and Activation of DDR Signaling After Conditional TDP-43 Knockout by Inducible CRISPR/Cas9 Technique.
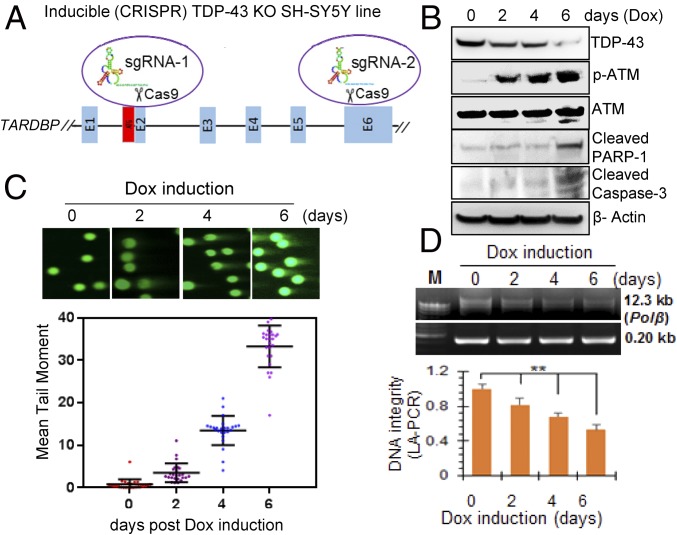
Given that TARDBP gene knockout (KO) is lethal to the neurons (31), we developed doxycycline (Dox)-inducible CRISPR/Cas9 (32) -based TDP-43 KO in an SH-SY5Y line (iCRISPR-TDP-43-KO-SH-SY5Y) to investigate the effect of dose-dependent depletion of TDP-43 in neurons. The iCRISPR/Cas9 strategy is schematically shown in Fig. 3A. iCRISPR-TDP-43-KO-SH-SY5Y cells were first differentiated with retinoic acid (10 μM) for 4 d (SI Appendix, Fig. S1A) and then induced with Dox (5 μg/mL). The TDP-43 level was measured at 0, 2, 4, 6, and 8 d of induction. Complete TDP-43 KO was observed in 8 d (SI Appendix, Fig. S7 A and B); however, the cells were detached from the petri dish and appeared nonviable by then. This was perhaps expected, because TDP-43 is essential for survival. On the other hand, ∼30%, 50%, and 75% depletion of TDP-43 was observed in viable cells after 2, 4, and 6 d of Dox induction, respectively (SI Appendix, Fig. S7 D and E). IB of total extracts of iCRISPR-TDP-43-KO-SH-SY5Y cells showed activation of ATM as reflected in pATM level, as well as the formation of apoptosis markers cleaved poly-ADP ribose polymerase (PARP) 1 and cleaved caspase-3 (Fig. 3B). Gradual increase in apoptotic cells after TDP-43 depletion was confirmed by FACS analysis using Annexin V/propidium iodide (PI) staining (SI Appendix, Fig. S7 D and E) and by MTT assay (SI Appendix, Fig. S7F). Neutral comet analysis showed presence of DSBs, the extent of which correlated with reduction in the TDP-43 level (Fig. 3C). Similarly, LA-PCR analysis (33) showed a decrease in DNA integrity proportionate to TDP-43 level (Fig. 3D). Notably, genome damage data on day 2 of Dox induction showed a marked increase in DSBs together with ATM activation (Fig. 3B and SI Appendix, Fig. S7C), without a significant increase in apoptotic cell population, suggesting that DSB damage likely precedes cell death. Collectively, these studies demonstrate that conditional TDP-43 depletion is strongly correlated with unrepaired DSB accumulation, sustained DDR activation, and subsequently apoptotic cell death in neurons and, thus, underscores the essential role of TDP-43 in maintaining their genomic integrity.
Fig. 3.
TDP-43 depletion correlates to DSB accumulation in the neuronal genome. CRISPR/Cas9-mediated conditional TDP-43 KO in SH-SY5Y cells. (A) Scheme of inducible CRISPR/Cas9-mediated KO of TDP-43 indicating sgRNA targeting location on the TARDBP gene. (B) Dox (5 μg/mL) induction resulted in progressive depletion of TDP-43 from 2 to 6 d. IB with TDP-43, pATM, ATM, cleaved caspase-3, cleaved PARP1, and β-actin antibodies shows DDR activation and apoptosis induction after TDP-43 KO. (C) Neutral comet analysis and quantitation of mean comet tail moment of at least 25 randomly selected cells shows the presence of DSBs in TDP-43 KO cells. Images were acquired at 20× magnification. (D) DNA integrity in TDP-43 KO cells measured by LA-PCR analysis. A 0.8% agarose gel image of PCR products, quantitation of PCR products by picogreen-based DNA quantitation from triplicate experiments. **P < 0.05. Data are presented as means ± SD. P values are based on two-way ANOVA.
TDP-43 Is Required for Optimal DSB Repair via NHEJ in Neurons.
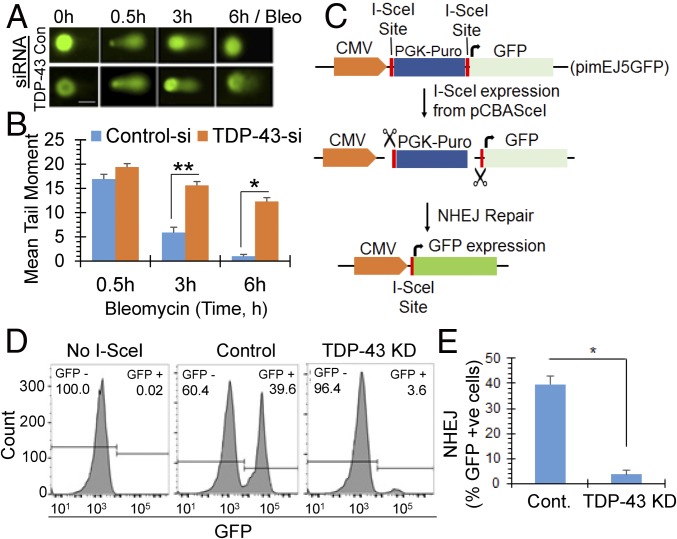
We next investigated whether TDP-43 depletion affected DSB repair via the NHEJ pathway in neurons. Comet analysis to monitor DSB repair kinetics showed significantly delayed repair in TDP-43 siRNA-transfected NP cells compared with control siRNA-treated cells, after exposure to bleomycin (Fig. 4 A and B). While control cells showed nearly complete recovery from the exogenous genome damage by 6 h, ∼10-fold higher mean comet tail moment persisted in TDP-43 KD cells, indicating substantial reduction in DSB repair efficiency due to loss of TDP-43. Similar delay in DSB repair was reflected in the slower disappearance of 53BP1 foci in TDP-43 KD cells relative to control after bleomycin treatment (SI Appendix, Fig. S8A). Finally, cell viability analysis by MTT or clonogenic survival assay indicated that TDP-43 depletion acted synergistically with IR or bleomycin to reduce cell viability (≥sixfold) (SI Appendix, Fig. S8 B and F).
Fig. 4.
TDP-43 is required for DSB repair via the NHEJ pathway in neuronal genomes. (A and B) Loss of TDP-43 affects DSB repair kinetics. NPCs were first transfected with control or TDP-43 siRNA, and treated with bleomycin (10 μM, 30 min) 48 h posttransfection. Neutral comet assay (A) at various time points after DSB induction and quantitation of mean comet tail moment in at least 25 randomly selected cells (B) show delayed repair in TDP-43 KD cells. (Scale bar, 10 μm.) (B) *P < 0.01; **P < 0.05. (C–E) I-SceI–based NHEJ reporter assay. (C) Schematic of in cell NHEJ assay using an I-SceI–based reporter pimEJ5GFP vector. (D) Percentage of GFP+ cells measured by flow cytometry in control vs. TDP-43 down-regulated cells with siRNA treatment. (E) Quantitation histogram of GFP+ (NHEJ) cells. *P < 0.01. Data are presented as mean ± SD. P values are based on two-way ANOVA.
The delayed DSB repair of both intrinsic and induced DSBs in TDP-43 KD cells, together with the association of TDP-43 with NHEJ factors, suggest a function of TDP-43 in NHEJ, which we explored using an I-SceI plasmid-based NHEJ reporter assay (34). In this approach, neuronal cells were stably transfected with NHEJ reporter plasmid pimEJ5GFP harboring PGK-Puromycin cassette within the two I-SceI recognition sites. I-SceI sites are located between the promoter and GFP reporter coding sequence (schematically shown in Fig. 4C) (35). Puromycin-resistant cells were sequentially treated with either control or TDP-43 siRNA for 72 h followed by pCBASceI transfection to introduce DSB (36). NHEJ-mediated error-free repair of I-SceI sites after break induction would result in expression of GFP, and therefore the percentage of GFP+ cells would provide a relative measure of NHEJ (37). Notably, TDP-43 KD (∼90%) significantly reduced the percentage of GFP-expressing cells compared with control cells (Fig. 4 D and E). A similar experiment in cycling HEK293 cells showed significantly reduced level of GFP expression in TDP-43 KD cells compared with that of control by FACS analysis as well as immunfluoresence (SI Appendix, Fig. S8 C and D). DNA-PKcs inhibitor NU7441 served as positive control for NHEJ inhibition. Taken together, these results suggest that TDP-43 depletion causes reduction in NHEJ-mediated DSB repair proficiency. Furthermore, as described in SI Appendix, SI Results and Fig. S8 G–J, TDP-43 KD induced DSB repair defects were confirmed using a second shuttle vector-based approach (38).
Caenorhabditis elegans Mutant Lacking TDP-1 Is Sensitive to DNA Damage due to Defective NHEJ.
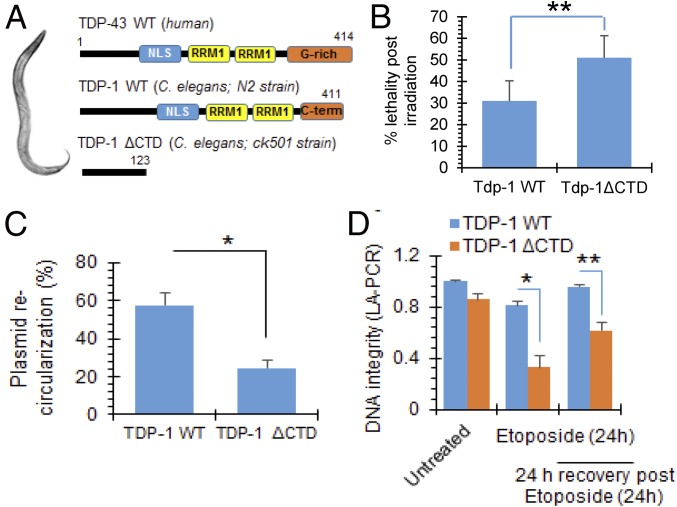
To establish that involvement of TDP-43 in maintenance of genomic integrity is universal among all metazoans, we investigated whether TDP-1, the TDP-43 homolog in C. elegans, participated in DSB repair. Human TDP-43 and nematode TDP-1 are functional orthologs with similar RNA binding activities and have a conserved N terminal, NLS, and RRMs (Fig. 5A) (39, 40). We utilized standard laboratory strains N2 (Bristol) as WT and an endogenous knockin Tdp-1(ok803) loss-of-function strain (TDP-1ΔCTD; CK501) with deletion of C-terminal 299-aa residues lacking the NLS and the two RRMs (41). Sensitivities of WT or TDP-1ΔCTD C. elegans to genotoxic DSBs were assayed by evaluating numbers of viable embryos following ionizing radiation exposure. Stage-matched day 1 adult worms were irradiated for 10 min at 390 rad/min (total exposure of 40 Gy). IR C. elegans were allowed to lay eggs for 4 h, and the embryos were scored 24 h later for survival analysis by counting live worms vs. dead eggs. Data showed ∼25% increased lethality in CK501 compared with N2 after IR (Fig. 5B). Total worm extracts (42, 43) were then incubated with linearized plasmid substrate to analyze the plasmid recircularization efficiency via a colony formation assay (44). Mutant extract showed significantly reduced recircularization efficiency compared with N2 worms (Fig. 5C), indicating impaired DSB repair. To further compare genome integrity in N2 vs. CK501 worms, genomic DNA was isolated from control or etoposide-treated worms and separated in 1% agarose gel electrophoresis and band intensity of intact DNA was quantified (SI Appendix, Fig. S9 A and B). For this assay, etoposide treatment was continued for 24 h, and worms were either harvested immediately as 0-h recovery or allowed to recover for 24 h. Untreated mutant worms showed increased smear compared with WT, indicating the presence of higher basal level DNA damage. While the WT worms recovered from etoposide-induced DNA damage, CK501 worms showed persistent genome damage (indicated by smear and reduction in intact band intensity). Similarly, LA-PCR analysis from these C. elegans genomic DNA templates (45) consistently showed failure of mutant C. elegans to recover from etoposide-induced DNA damage, as measured by picogreen-based quantitation of PCR amplified products, suggesting defective DSB repair (Fig. 5D and SI Appendix, Fig. S9C). Together, these studies establish the direct linkage between loss of TDP-1 function and the DSB repair defect in C. elegans.
Fig. 5.
Loss of TDP-1 (TDP-43 homolog) induces genomic instability in C. elegans. See also SI Appendix, Fig. S9. Analysis of DNA damage and repair defect in mutant C. elegans strain expressing truncated TDP-1 with loss-of-function. (A) Scheme showing human TDP-43, WT TDP-1 (N2 strain), and mutant TDP-1 (CK501 strain) with C-terminal 299-aa deletion. (B) Comparison of percent lethality of N2 and CK501 worm embryos after exposure to ionizing radiation (40 Gy) to induce DSB damage. The data represent two independent experiments (n = 148–517 worms scored per genotype per experiment). **P < 0.05. (C) Plasmid recircularization assay. DSB-containing plasmid was incubated with either N2 or CK501 total protein extracts at 30 °C for 1 h followed by addition of EDTA to stop the reaction, then 10 μL of the reaction mix were transformed in Escherichia coli for colony formation assay. *P < 0.01. (D) LA-PCR analysis. A 13.7-kb region of polβ gene was amplified by LA-PCR from the genomic DNA isolated from worms and quantitated by picogreen-based quantitation of PCR products. *P < 0.01; **P < 0.05. Data are presented as mean ± SD. P values are based on two-way ANOVA.
TDP-43 Pathology Strongly Correlates with DNA Damage, DDR Activation, and Neurodegeneration in Sporadic ALS Patients.
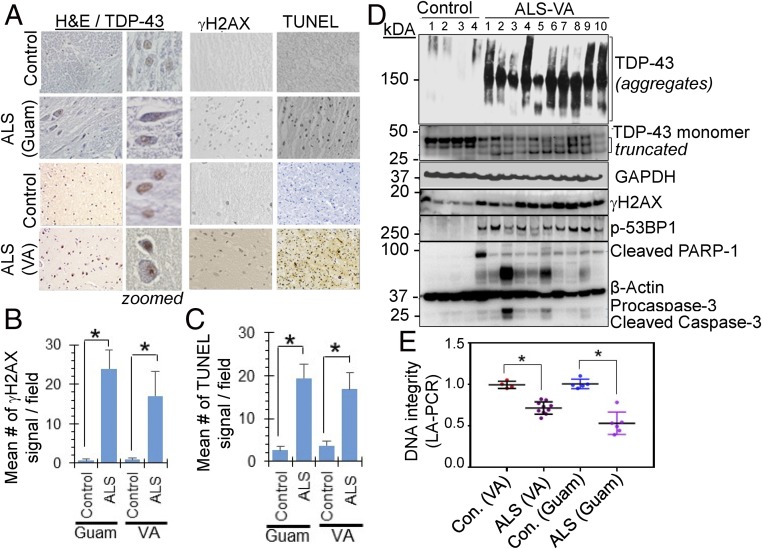
We next examined if loss of nuclear TDP-43 observed in ALS patients could be correlated with genome damage in affected spinal cord tissue. We obtained sporadic ALS-affected spinal cord tissues from the Department of Veterans Affairs (VA) Brain Biorepository (ALS-VA) and Guamanian ALS (ALS-Guam) tissues from the Binghamton Biospecimen Archive. Clinical characteristics of the patients are shown in SI Appendix, Tables S1 and S2. As shown in representative immunohistochemistry (IHC) images, the typical pathology of increased extranuclear TDP-43 was confirmed in spinal cord sections (cervical region) of both ALS-Guam and ALS-VA cases, and this was absent in appropriately matched controls (Fig. 6A, Left). Importantly, ALS spinal cord showed significantly higher staining for γH2AX and TUNEL compared with matched control spinal cord sections (Fig. 6A, Right). Fig. 6 B and C show the quantitation of γH2AX and TUNEL IHC signals per field from 10 sporadic ALS-VA and 5 ALS-Guam cases in comparison with matched controls. The ALS spinal cord also showed strong Thioflavin S+ reactivity, indicating the presence of protein aggregates (SI Appendix, Fig. S10A). Furthermore, genomic DNA isolated from both ALS-Guam and ALS-VA spinal cord tissue showed significantly reduced integrity due to the presence of strand breaks, as analyzed by LA-PCR (Fig. 6E and SI Appendix, Fig. S10 B–D).
Fig. 6.
TDP-43 nuclear clearance correlates with DNA strand breaks and DDR activation in the spinal cord of sporadic ALS patients. (A–C) IHC of spinal cord from sporadic ALS patients for anti–TDP-43, anti-γH2AX Ab, and TUNEL staining. Representative images acquired at 20× magnification from ALS-Guam and ALS tissue obtained from the Department of Veterans Affairs Brain Biorepository (ALS-VA) (A). (B) Quantitation of mean γH2AX signal, and mean TUNEL signal (C) per field in 5 ALS-Guam and 10 ALS-VA spinal cord specimens together with matched controls. *P < 0.01. See SI Appendix, Tables S1 and S2 for patient details. (D) Immunoblotting of total tissue extracts from control and ALS spinal cord-cervical with anti–TDP-43 Ab detecting monomeric, oligomeric and truncated forms, anti-γH2AX, anti-p53BP1, and the apoptotic markers cleaved PARP-1 and cleaved caspase-3 Ab. (E) LA-PCR analysis of DNA strand breaks in genomic DNA isolated from control and ALS spinal cord. *P < 0.01. Data are presented as means ± SD. P values are based on two-way ANOVA.
Given that sporadic ALS–TDP-43 proteinopathy may involve both TDP-43 aggregation and fragmentation, the status of the TDP-43 protein was analyzed by IB in the ALS-VA spinal cord (cervical) and matched controls. Notably, all of 10 ALS samples showed reduced TDP-43 monomer levels compared with controls (Fig. 6D and SI Appendix, Fig. S10H) as well as significant increase in ubiquitinated proteins in sporadic ALS cases compared with the controls (SI Appendix, Fig. S11), consistent with the previous observation (3). In addition, the ALS samples also showed characteristic IB patterns representing TDP-43 aggregates/oligomeric forms and fragmentation, which were present at a negligible level in the controls. Furthermore, the presence of TDP-43 fragmentation and its aggregation in ALS samples were strongly correlated with higher levels of γH2AX and p53BP1 accumulation, together with the presence of cleaved PARP-1 and cleaved caspase-3, compared with their negligible level in controls (Fig. 6D; quantitation of relative IB band intensity in SI Appendix, Fig. S10 E–G), suggesting a link between accumulated DNA damage and the apoptotic death of affected spinal cord neurons.
To assess the linkage of TDP-43 pathology and genome damage accumulation in ALS spinal cord with DSB repair defects, we performed a plasmid recircularization assay, as an in vitro surrogate NHEJ assay with extracts from control and ALS spinal cord tissue. Four controls and 10 ALS-VA samples (SI Appendix, Table S1) were grouped into two sets each: control-I (containing control #1 and #2) and control-II (containing control #3 and #4); ALS-I (containing ALS#1–5) and ALS-II (containing ALS# 6–10). The grouped extracts were homogenously mixed with same amount of total protein from each sample. Both the ALS groups showed significant loss of monomeric TDP-43; as shown in Fig. 6D, the plasmid recircularization efficiency reduced about 50% in ALS tissue, suggesting defective DSB ligation (SI Appendix, Fig. S10I). These data provide strong correlation of TDP-43 pathology, NHEJ defects, DSB damage, and neurodegeneration in ALS pathology.
TDP-43 Acts as a Scaffold in Recruiting XRCC4-Lig4 Complex at DSB for Efficient DNA Ligation in NHEJ.
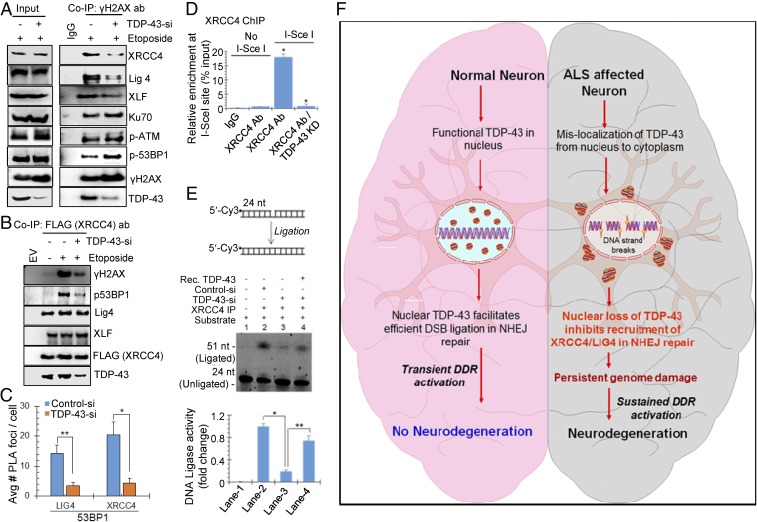
To gain molecular insights into role of TDP-43 in DSB repair, we examined recruitment of key NHEJ factors at damage sites, by testing their presence in γH2AX or 53BP1 co-IPs from nuclear extracts of NPCs first transfected with control or TDP-43 siRNA, following DSB induction with etoposide. IB of endogenous co-IPs showed substantially reduced association of both γH2AX (Fig. 7A and SI Appendix, Fig. S12A) and 53BP1 (SI Appendix, Fig. S12 C and D) with XRCC4, Lig4, as well as XLF, key factors in the NHEJ ligation complex after TDP-43 KD. It is important to note that unlike the XRCC4-Lig4-XLF complex, the association of Ku, pATM, and p53BP1 increased with γH2AX formation in absence of TDP-43, suggesting that TDP-43 primarily works downstream of 53BP1 in the NHEJ pathway. To further confirm reduced association of XRCC4 with DSB markers, we similarly performed FLAG-XRCC4 co-IP, which showed significantly reduced interactions with γH2AX and p53BP1 due to TDP-43 depletion (Fig. 7B and SI Appendix, Fig. S12B). Levels of XLF and Lig4 in FLAG-XRCC4 co-IP remained unaffected, indicating TDP-43’s role in recruitment of the NHEJ ligation complex, but may not on the complex formation itself. Defective in cell association of XRCC4 and Lig4 at DSB sites was further confirmed by a complementary PLA experiment between anti-53BP1 and anti-Lig4 or anti-XRCC4 Ab in NPCs treated with control or TDP-43 siRNA for 72 h (Fig. 7C and SI Appendix, Fig. S12G), revealing ∼10-fold reduction in PLA signal due to TDP-43 down-regulation.
Fig. 7.
TDP-43 facilitates recruitment and activity of the NHEJ ligation complex (XLF/XRCC4/Lig4) at DSB sites. (A and B) NPCs were transfected with control or TDP-43 siRNA, followed by treatment with etoposide (10 μM) for 4 h. Total nuclear extracts were subjected to IP with control IgG or anti-γH2AX Ab, then co-IP eluates were probed with indicated antibodies. The level of XRCC4, Lig4, and XLF were reduced in γH2AX after TDP-43 KD. Quantitation of IB band intensity shown in SI Appendix, Fig. S12A. (B) NPCs were cotransfected with TDP-43 siRNA and FLAG-XRCC4 or empty vector, and then treated with etoposide as before. FLAG co-IP from the total cell lysates were probed with indicated Ab showing TDP-43–dependent interaction of XRCC4 with γH2AX and p53BP1. Quantitation of IB band intensity shown in SI Appendix, Fig. S12B. (C) PLA of XRCC4 or Lig4 vs. p53BP1 in NPCs show reduced association after TDP-43 KD by siRNA. Quantitation of average number of PLA foci per cell from at least 25 cells. Representative PLA images in SI Appendix, Fig. S12G. (D) ChIP analysis with control IgG or anti-XRCC4 Ab revealed reduced enrichment at the I-SceI–defined DSB site in TDP-43 KD cells. (E) DNA ligation analysis. Total lysates from TDP-43 KD NPC, treated with etoposide, were subjected to IP with anti-XRCC4 Ab. The co-IP eluates were incubated with a Cy-3–labeled nicked duplex oligonucleotide substrate (Top). Reduced ligation in TDP-43 KD cells was rescued by addition of recombinant TDP-43. Quantitation of ligated products shown in histogram. Data are presented as means ± SD. *P < 0.01; **P < 0.05. (F) A model showing genome damage followed by persistent DDR signaling in ALS spinal cord neurons caused by TDP-43 pathology. TDP-43 is a critical component of NHEJ-mediated DSB repair in healthy neurons. TDP-43’s nucleo-cytoplasmic mislocalization in motor neurons inhibits DSB repair and contributes to neurodegeneration in ALS. Our findings uncover a link between TDP-43 pathology and impaired DSB repair and suggest potential avenues for DNA repair-targeted therapies for TDP-43–ALS.
We next investigated whether reduced in cell interaction of DSB markers with the XRCC4/Lig4 complex is correlated in their actual recruitment at DSB sites, by ChIP/re-ChIP analyses. A first ChIP with control IgG and anti-γH2AX Ab (as in Fig. 2D) followed by re-ChIP with anti-XRCC4 Ab (SI Appendix, Fig. S12E) or anti-53BP1 Ab (SI Appendix, Fig. S12F) and respective control IgG, was performed in etoposide-treated NPC with or without TDP-43 KD. Quantitative PCR amplification of HPRT gene segment from ChIP products demonstrated a significantly reduced enrichment of XRCC4 after TDP-43 depletion. The enrichment of 53BP1 was increased after etoposide treatment, as expected, with an additional ∼fivefold increase after TDP-43 KD, consistent with co-IP results, suggesting enhanced accumulation of unrepaired DSBs. A similar re-ChIP analysis at defined I-SceI cleavage sites also confirmed defective recruitment of XRCC4 in the proximity of DSBs due to loss of TDP-43 (Fig. 7D). Taken together, the ChIP data demonstrate that TDP-43 is required for optimal recruitment of the XRCC4-Lig4 complex at both genome-wide DSBs and I-SceI–induced defined break sites.
We next tested whether the reduced recruitment of XRCC4-Lig4 at DSBs in the absence of TDP-43 resulted in impaired DNA DSB ligation activity. For this experiment, endogenous XRCC4 IP complexes were isolated from etoposide-treated NPCs with or without TDP-43 KD by siRNA. IP eluates were used to test DNA ligation activity by two approaches: either using a 5′Cy3-labeled nicked duplex oligonucleotide substrate (Fig. 7E) or a plasmid recircularization assay (SI Appendix, Fig. S12H). Both assays showed significantly (∼four- to sixfold) reduced DNA ligation activity with the XRCC4 IP complex after TDP-43 KD, which was mostly rescued by the addition of recombinant TDP-43 to reaction mix. Absence of other DNA ligases, namely DNA Ligase1 (Lig1) and DNA Lig3, in XRCC4 co-IP complexes was confirmed by IB with respective Ab (SI Appendix, Fig. S13B). Consistently, recombinant TDP-43 showed direct interaction with purified XRCC4-Lig4 complex in the absence of DNA (SI Appendix, Fig. S13A). These data not only reveal a scaffolding activity of TDP-43 for recruiting the NHEJ ligation complex (schematically represented in SI Appendix, Fig. S12I), but also provided a direct linkage of TDP-43 depletion and DSB ligation defects in the NHEJ pathway.
In summary, our data reveal that TDP-43 is a critical component of genomic DSB repair and the DDR signaling. Thus, TDP-43 pathology leading to its loss of functions in the ALS spinal cord neurons, impairs DSB repair by inhibiting NHEJ, leading to persistent accumulation of damage and sustained activation of DDR signaling, which contribute to neuronal cell death by apoptosis (Fig. 7F).
Discussion
Since the discovery of TDP-43 toxicity in ALS in 2006 (3, 7), research has mainly focused on the cytoplasmic aggregation of TDP-43 and disruption of its RNA-binding functions (46–51). TDP-43, whose nuclear clearance is a hallmark of degenerating motor neurons in ALS, has also been shown to have the ability to bind to DNA (13, 14). This makes TDP-43 a member of the distinct subgroup of RNA-binding proteins with functional DNA-binding activity, which we name RNA/DNA-binding proteins (RDBPs). However, the physiological functions/relevance of the DNA-binding activity of TDP-43, and the eventual loss of this function in disease-affected neurons, have not been previously investigated. Evidence for the presence of DNA damage in TDP-43 toxicity-linked neurodegenerative diseases (52, 53), and identification of the DNA repair protein Ku in the TDP-43 interactome in human cells (15), provided the premise to investigate the role of TDP-43 in DDR in this study.
We have identified TDP-43 as an integral component of early DDR, which is activated for repair of DNA DSBs via NHEJ. TDP-43 showed strong in cell association with key DDR marker proteins γH2AX, pATM, and p53BP1 in a DNA damage dose- and time-dependent manner. We induced DSBs using IR, radiomimetic bleomycin, or a DNA topoisomerase II inhibitor etoposide, all of which consistently showed TDP-43’s association with DSB repair proteins and its recruitment at damage sites. Furthermore, laser ablation/live cell imaging revealed that TDP-43 was recruited at DSB sites as early as 1 min, together with Ku70, after damage induction and persisted at DSB until completion of repair. We confirmed recruitment of TDP-43 at DSBs using multiple complementary approaches including dDIP, ChIP/re-ChIP, and PLA with DSB markers, all of which consistently showed strong association of TDP-43 at DSBs. Importantly, TDP-43 was able to directly bind DNA oligonucleotide containing a DSB-mimicking blunt end in vitro.
The DDR in mammalian cells is a complex and highly orchestrated signaling process. While there are minor variations in the proposed early events following DNA DSB damage, ATM together with the Mre11/Rad50/NBS1 complex is the first responder. Activated ATM (autophosphorylation at serine 1981) phosphorylates the H2AX bound to the DSB site, which then facilitates recruitment of other DDR proteins, including 53BP1, to the vicinity of the DNA break (54). Repair of DSBs can occur via one of three subpathways, two of which are homologous recombination in S/G2 cells, and NHEJ in all cells including postmitotic neurons. The third subpathway is via error-prone microhomology-dependent alternative end-joining and primarily detectable in immune cells for Ab diversification and in some drug-resistant tumors, and has not been shown to significantly contribute to DSB repair in neurons. NHEJ involves the DNA-PK holoenzyme (DNA-PKcs/Ku70/Ku80), a DNA polymerase (μ or λ), and XRCC4/DNA Lig4 (55, 56).
Consistent with TDP-43’s recruitment at DSBs and its stable in cell association with NHEJ factors, si/shRNA, or CRISPR/Cas9-mediated loss of TDP-43 in multiple neuronal cell models, including human iPSC or NSC-derived motor neurons and differentiated SH-SY5Y and HEK293 cells, resulted in a marked increase in levels of unrepaired DSBs in the genome and activation of DDR signaling, even in unstressed conditions. It is important to mention that motor neuron differentiation from hNSCs was optimized to obtain up to ∼80% efficiency (SI Appendix, Fig. S1 C–F) (17, 18).
To gain molecular insight into TDP-43’s role in specific DSB repair reactions, we tested the recruitment of key NHEJ factors by co-IP, ChIP, and PLA analyses. The data clearly demonstrated that loss of TDP-43 causes a strong inhibition in recruitment of XRCC4, Lig4, and XLF, three key components for DSB ligation in the NHEJ pathway. Because levels of these proteins were unchanged after TDP-43 KD, our findings suggested the involvement of defective protein–protein or protein–DNA interactions during NHEJ repair. The recruitment of Ku or 53BP1 was not inhibited in the absence of TDP-43, which suggested that TDP-43 might work as downstream factor of 53BP1 in NHEJ pathway, likely acting as a scaffold for recruiting/stabilizing the ligation complex. Furthermore, substantial impairment of DSB ligation activity in the XRCC4 immunocomplex isolated from neuronal cells lacking TDP-43, showed that reduced recruitment of the NHEJ ligation complex in the absence of TDP-43 indeed resulted in DNA end-break ligation defect. As shown in SI Appendix, Fig. S13B, the XRCC4 co-IP contained Lig4 but not other nuclear DNA ligases (Lig3 or Lig1), thus reduced ligation activity of XRCC4 co-IP after TDP-43 KD could be directly attributed to Lig4 defect, the only DNA ligase involved in NHEJ. It is important to note that the level of XRCC4 and Lig4 were not affected in TDP-43–depleted cells, suggesting a direct role of TDP-43 in regulating the Lig4 function rather than via RNA processing. Our observation of TDP-43’s pairwise interaction with the XRCC4/Lig4 complex in vitro supports this scenario. We recently reported the role of FUS in regulating recruitment and functions of the XRCC1/Lig3 complex in DNA SSB repair (57). Together these studies suggest a scaffolding role of RDBPs in DNA break sealing, likely critical for stabilizing the DNA termini at break sites.
The data in cultured motor neurons linking loss of TDP-43 to genomic DSB accumulation were further recapitulated in two in vivo model systems, including C. elegans lacking functional TDP-1 and human ALS patients’ spinal cord tissue. Unlike in human neurons, though, hemizygous expression of functionally dead, truncated TDP-1 causes DNA damage accumulation but does not show motor phenotype in worms; however, it does make them highly sensitive to exogenously induced DSBs (γ-IR or etoposide treatment). Reduced plasmid recircularization in extracts of C. elegans lacking TDP-1, together with their increased lethality in response to DSB stress, provide strong in vivo evidence for the involvement of TDP-43 in DSB repair.
Here, we have provided evidence for the presence of unrepaired DSB damage and activation of DDR factors associated with clearance of nuclear TDP-43 in the spinal cord tissue of sporadic ALS patients. In addition, plasmid recircularization assay using spinal cord tissue extracts showed ∼70% reduction in DSB ligation in ALS compared with controls. Higher ALS predisposition has been reported in military veterans, perhaps due to potential exposure to lead metal complexes and other toxicants used in warfare combined with stress and other unknown factors; on the other hand, a significantly higher incidence of ALS in Guam residents was attributed to the consumption of unnatural amino acid β-methylamino-l-alanine through cycad products in the diet together with metal toxicity (58–60). TDP-43 pathology has also previously been reported in a majority of ALS-Guam patients (61, 62), and this was confirmed in this study. Remarkably, the extent of genome damage in the ALS spinal cord also broadly correlated with the activation of apoptotic markers. Notably, we observed the presence of both truncated and aggregated TDP-43 species in the spinal cord of sporadic ALS patients (consistent with previous reports); however, both exhibited a comparable level of genome damage. This suggested the correlation of loss of monomeric TDP-43 to genomic instability in ALS patients.
Our studies thus report a key role of TDP-43 in DDR and DSB repair and suggest potential DNA repair based therapies for TDP-43-associated neurodegeneration.
Materials and Methods
For detailed and additional methods, see SI Appendix.
hNSC Culture and Differentiation.
The hNSC (63) line (K048) was cultured as neurospheres in complete medium containing appropriate growth factors (16). The normal iPSC line (ATCC #KYOU-DXR0109B) was grown in CellMatrix basement membrane gel (Gibco) and Pluripotent Stem Cell SFM XF/FF media (Gibco) at 37 °C and 5% CO2. For differentiation into motor neurons, see details in SI Appendix.
C. elegans.
C. elegans strains N2 (Bristol) and RB929 tdp-1 (ok803) were provided by the Caenorhabditis Genetic Center (Minneapolis, MN), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and from the National Bioresource Project (Japan).
Human ALS Tissue Specimens.
Human postmortem spinal cord tissue specimens from sporadic ALS patients and age-matched controls were obtained from two bio-repositories, namely the Department of Veterans Affairs Brain Biorepository (ALS-VA) and Binghampton Biorepository Archive (ALS-Guam) (SI Appendix, Tables S1 and S2).
CRISPR/Cas9-Mediated TDP-43 KO.
Two single-guide RNAs were designed based on the exact DNA sequence of TDP-43 coding DNA region in SH-SY5Y cells. An inducible humanized Cas9 system was used for conditional knockout of TDP-43.
PLA.
In situ protein–protein association was analyzed using PLA (Duolink; Sigma) as per the manufacturer’s instructions (18, 64).
Live-Cell Imaging of Micro-IR Based DSB Repair.
For live cell imaging of DSB repair, cells were transfected with TDP-43-tdTOMATO or Ku70-EGFP reporter plasmids. DSBs were generated in a laser ablation track in the region of interest by a 405-nm diode laser, set to 100% transmission output for 150 iterations. Following IR, cells were maintained for indicated time periods scanning the fluorescence intensity of the laser-track for 30 min with 1 min interval. Data were collected from 5 to 15 cells and measured by ImageJ and normalized to the signal of the whole nucleus (26, 65).
NHEJ Assay for the Repair of DSBs.
We employed two plasmid based approaches to measure NHEJ-mediated DSB repair proficiency, namely the standard I-SceI based GFP reporter assay (35) and a shuttle plasmid containing unligatable DSB termini (38).
Supplementary Material
Acknowledgments
The authors thank other members of M.L.H. laboratory, S. Rangaswamy, P. Basu, and V. Bora for assistance. Control and sporadic ALS spinal cord tissue specimens were provided by the Department of Veterans Affairs Biorepository. The Guamanian ALS specimens were obtained from Binghamton Biospecimen Archive. This research was primarily supported by National Institute of Neurological Disorders and Stroke-NIH Grant R01 NS088645; Muscular Dystrophy Association Grant MDA 294842; and the Houston Methodist Research Institute (M.L.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818415116/-/DCSupplemental.
References
- 1.Guerrero EN, et al. TDP-43/FUS in motor neuron disease: Complexity and challenges. Prog Neurobiol. 2016;145–146:78–97. doi: 10.1016/j.pneurobio.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mackenzie IR, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 3.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 4.Mackenzie IR, et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–113. doi: 10.1007/s00401-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ou SH, Wu F, Harrich D, García-Martínez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang C, et al. The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PLoS One. 2010;5:e15878. doi: 10.1371/journal.pone.0015878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 8.Cairns NJ, et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol. 2007;171:227–240. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson Y, et al. Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol. 2007;113:521–533. doi: 10.1007/s00401-006-0189-y. [DOI] [PubMed] [Google Scholar]
- 10.Alami NH, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bose JK, Huang CC, Shen CK. Regulation of autophagy by neuropathological protein TDP-43. J Biol Chem. 2011;286:44441–44448. doi: 10.1074/jbc.M111.237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swarup V, et al. Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor κB-mediated pathogenic pathways. J Exp Med. 2011;208:2429–2447. doi: 10.1084/jem.20111313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo PH, Chiang CH, Wang YT, Doudeva LG, Yuan HS. The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids. Nucleic Acids Res. 2014;42:4712–4722. doi: 10.1093/nar/gkt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalmansingh AS, Urekar CJ, Reddi PP. TDP-43 is a transcriptional repressor: The testis-specific mouse acrv1 gene is a TDP-43 target in vivo. J Biol Chem. 2011;286:10970–10982. doi: 10.1074/jbc.M110.166587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan PM, et al. Generation of spinal motor neurons from human fetal brain-derived neural stem cells: Role of basic fibroblast growth factor. J Neurosci Res. 2009;87:318–332. doi: 10.1002/jnr.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du ZW, et al. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat Commun. 2015;6:6626. doi: 10.1038/ncomms7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasquez V, et al. Chromatin-bound oxidized α-synuclein causes strand breaks in neuronal genomes in in vitro models of Parkinson’s disease. J Alzheimers Dis. 2017;60:S133–S150. doi: 10.3233/JAD-170342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Encinas M, et al. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem. 2000;75:991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- 20.Hegde ML, et al. Specific inhibition of NEIL-initiated repair of oxidized base damage in human genome by copper and iron: Potential etiological linkage to neurodegenerative diseases. J Biol Chem. 2010;285:28812–28825. doi: 10.1074/jbc.M110.126664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang HAS, Butler EB, Pandita TK, Mitra S, Hegde ML. A perspective on chromosomal double strand break markers in mammalian cells. Jacobs J Radiat Oncl. 2014;1:003. [PMC free article] [PubMed] [Google Scholar]
- 22.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Lee JH, Cheong HM, Kang MY, Kim SY, Kang Y. Ser1778 of 53BP1 plays a role in DNA double-strand break repairs. Korean J Physiol Pharmacol. 2009;13:343–348. doi: 10.4196/kjpp.2009.13.5.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leduc F, et al. Genome-wide mapping of DNA strand breaks. PLoS One. 2011;6:e17353. doi: 10.1371/journal.pone.0017353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei L, et al. Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Mol Cell Biol. 2008;28:7380–7393. doi: 10.1128/MCB.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mari PO, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegde ML, et al. Enhancement of NEIL1 protein-initiated oxidized DNA base excision repair by heterogeneous nuclear ribonucleoprotein U (hnRNP-U) via direct interaction. J Biol Chem. 2012;287:34202–34211. doi: 10.1074/jbc.M112.384032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins AR. The comet assay for DNA damage and repair: Principles, applications, and limitations. Mol Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 30.Olive PL, Banáth JP. The comet assay: A method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 31.Wu LS, et al. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 32.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovalenko OA, Santos JH. Analysis of oxidative damage by gene-specific quantitative PCR. Curr Protoc Hum Genet. 2009;Chapter 19:Unit 19.1. doi: 10.1002/0471142905.hg1901s62. [DOI] [PubMed] [Google Scholar]
- 34.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: Suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, et al. Protein phosphatase PP4 is involved in NHEJ-mediated repair of DNA double-strand breaks. Cell Cycle. 2012;11:2643–2649. doi: 10.4161/cc.20957. [DOI] [PubMed] [Google Scholar]
- 38.Dutta A, et al. Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res. 2017;45:2585–2599. doi: 10.1093/nar/gkw1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang T, Hwang HY, Hao H, Talbot C, Jr, Wang J. Caenorhabditis elegans RNA-processing protein TDP-1 regulates protein homeostasis and life span. J Biol Chem. 2012;287:8371–8382. doi: 10.1074/jbc.M111.311977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayala YM, et al. Human, Drosophila, and C. elegans TDP43: Nucleic acid binding properties and splicing regulatory function. J Mol Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 41.Liachko NF, Guthrie CR, Kraemer BC. Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J Neurosci. 2010;30:16208–16219. doi: 10.1523/JNEUROSCI.2911-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emmons SW, Klass MR, Hirsh D. Analysis of the constancy of DNA sequences during development and evolution of the nematode Caenorhabditis elegans. Proc Natl Acad Sci USA. 1979;76:1333–1337. doi: 10.1073/pnas.76.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asagoshi K, et al. Single-nucleotide base excision repair DNA polymerase activity in C. elegans in the absence of DNA polymerase β. Nucleic Acids Res. 2012;40:670–681. doi: 10.1093/nar/gkr727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craig AL, Moser SC, Bailly AP, Gartner A. Methods for studying the DNA damage response in the Caenorhabdatis elegans germ line. Methods Cell Biol. 2012;107:321–352. doi: 10.1016/B978-0-12-394620-1.00011-4. [DOI] [PubMed] [Google Scholar]
- 45.Hunter SE, Jung D, Di Giulio RT, Meyer JN. The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods. 2010;51:444–451. doi: 10.1016/j.ymeth.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Conti L, et al. TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic and mitotic cellular pathways. Nucleic Acids Res. 2015;43:8990–9005. doi: 10.1093/nar/gkv814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costessi L, Porro F, Iaconcig A, Muro AF. TDP-43 regulates β-adducin (Add2) transcript stability. RNA Biol. 2014;11:1280–1290. doi: 10.1080/15476286.2014.996081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishiguro A, Kimura N, Watanabe Y, Watanabe S, Ishihama A. TDP-43 binds and transports G-quadruplex-containing mRNAs into neurites for local translation. Genes Cells. 2016;21:466–481. doi: 10.1111/gtc.12352. [DOI] [PubMed] [Google Scholar]
- 51.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci USA. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu Z, et al. Neurodegeneration-associated TDP-43 interacts with fragile X mental retardation protein (FMRP)/Staufen (STAU1) and regulates SIRT1 expression in neuronal cells. J Biol Chem. 2012;287:22560–22572. doi: 10.1074/jbc.M112.357582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill SJ, et al. Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc Natl Acad Sci USA. 2016;113:E7701–E7709. doi: 10.1073/pnas.1611673113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiloh Y, Ziv Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 55.Barnes DE. Non-homologous end joining as a mechanism of DNA repair. Curr Biol. 2001;11:R455–R457. doi: 10.1016/s0960-9822(01)00279-2. [DOI] [PubMed] [Google Scholar]
- 56.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H, et al. Mutant FUS causes DNA ligation defects to inhibit oxidative damage repair in amyotrophic lateral sclerosis. Nat Commun. 2018;9:3683. doi: 10.1038/s41467-018-06111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cox PA, Sacks OW. Cycad neurotoxins, consumption of flying foxes, and ALS-PDC disease in Guam. Neurology. 2002;58:956–959. doi: 10.1212/wnl.58.6.956. [DOI] [PubMed] [Google Scholar]
- 59.Garruto RM. Pacific paradigms of environmentally-induced neurological disorders: Clinical, epidemiological and molecular perspectives. Neurotoxicology. 1991;12:347–377. [PubMed] [Google Scholar]
- 60.Arif M, Kazim SF, Grundke-Iqbal I, Garruto RM, Iqbal K. Tau pathology involves protein phosphatase 2A in parkinsonism-dementia of Guam. Proc Natl Acad Sci USA. 2014;111:1144–1149. doi: 10.1073/pnas.1322614111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maekawa S, et al. TDP-43 is consistently co-localized with ubiquitinated inclusions in sporadic and Guam amyotrophic lateral sclerosis but not in familial amyotrophic lateral sclerosis with and without SOD1 mutations. Neuropathology. 2009;29:672–683. doi: 10.1111/j.1440-1789.2009.01029.x. [DOI] [PubMed] [Google Scholar]
- 62.Geser F, et al. Pathological TDP-43 in parkinsonism-dementia complex and amyotrophic lateral sclerosis of Guam. Acta Neuropathol. 2008;115:133–145. doi: 10.1007/s00401-007-0257-y. [DOI] [PubMed] [Google Scholar]
- 63.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921, and erratum (2001) 412:565. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 64.Hegde ML, et al. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc Natl Acad Sci USA. 2013;110:E3090–E3099. doi: 10.1073/pnas.1304231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang WY, et al. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci. 2013;16:1383–1391. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.