Significance
The propagation of most flowering plant species is determined by the success of seed germination, which is of both economic and ecologic importance. Mitochondria are the energy resource and crucial organelles for plant seed germination. Studying the underlying mechanism is important for us to understand the basic principles of plant development and improve crop yields. Here we identify HSP24.7 as a central activator for temperature-dependent seed germination. HSP24.7 modulates cytochrome C/C1 production in the mitochondrial electron transport chain and induces the generation of reactive oxygen species, which accelerates seed germination. Our work provides a comprehensive framework of how mitochondria regulate seed germination in response to the dynamics of environmental temperature.
Keywords: seed germination, mitochondria, ROS, temperature, HSP
Abstract
Seed germination is an energy demanding process that requires functional mitochondria upon imbibition. However, how mitochondria fine tune seed germination, especially in response to the dynamics of environmental temperature, remains largely unknown at the molecular level. Here, we report a mitochondrial matrix-localized heat shock protein GhHSP24.7, that regulates seed germination in a temperature-dependent manner. Suppression of GhHSP24.7 renders the seed insensitive to temperature changes and delays germination. We show that GhHSP24.7 competes with GhCCMH to bind to the maturation subunit protein GhCcmFc to form cytochrome C/C1 (CytC/C1) in the mitochondrial electron transport chain. GhHSP24.7 modulates CytC/C1 production to induce reactive oxygen species (ROS) generation, which consequently accelerates endosperm rupture and promotes seed germination. Overexpression of GhHSP24.7’s homologous genes can accelerate seed germination in Arabidopsis and tomato, indicating its conserved function across plant species. Therefore, HSP24.7 is a critical factor that positively controls seed germination via temperature-dependent ROS generation.
Rapid and uniform seed germination is critical to maximize crop yield potential in modern agricultural cultivation practice. Seed germination is a complex process that begins with water uptake and ends up with the radicle emergence from the surrounding seed tissues (1). In cotton seeds, the embryo is enclosed by a thin living cell layer with endosperm origin and a dead outer layer called testa (2). From a mechanical point of view, the germination process is also controlled by an interplay between two opposing forces: the growth potential of the radicle and the resistance of the seed-covering layers (3).
Temperature is an environment signal, activating or repressing Arabidopsis seed germination during seasonal changes (4). Studies on molecular networks controlling temperature-dependent Arabidopsis seed germination have indicated a role for DOG1 in endosperm weakening and in determining temperature response for germination (5). However, little is known about how the temperature information is sensed and replayed to elicit germination.
Mitochondria play a crucial role in seed germination. One of the earliest events of seed germination is the progressive transition of metabolically quiescent promitochondria into metabolically and energetically active mature mitochondria (6). This transition is a tightly regulated process. Delayed germination will occur if it is interrupted (7). In pea seeds, mitochondria played a central role in allowing plants to adapt to extreme temperatures (8). A rapid resumption of mitochondrial energy metabolism is required during early seed imbibition to fuel the high cellular energy demand. However, how seed mitochondria regulate germination after they exit metabolic stasis remains less understood. Available reports show that the mitochondrial electron transport chain (mETC) is considered as the major source of reactive oxygen species (ROS) (9). However, at the crossroad of seed dormancy and germination, whether and how mitochondria can modulate ROS production by adjusting their electron transport activity in an environment-dependent manner remains unclear.
Here, we report that a mitochondria-localized small heat shock protein (GhHSP24.7) can bind to its client protein GhCcmFc to modulate cytochrome C/C1 (CytC/C1) production in the mETC and induce ROS generation, thereby activating seed germination in response to increased temperature. Our work reveals a molecular mechanism of a plant life cycle transition from seed to seedling in response to temperature changes and provides a potential target for fast and uniform seed germination that might eventually be useful in breeding crops with improved yield.
Results
GhHSP24.7 Regulates Seed Germination in a Temperature-Dependent Pathway.
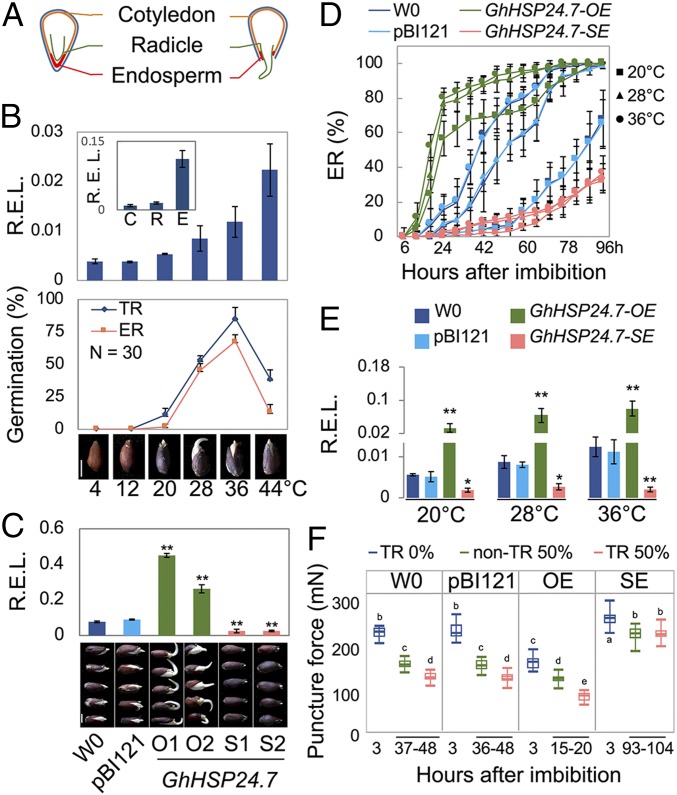
The mature cotton seed we used contained a fully developed embryo with differentiated meristems, radicle, and cotyledons. The seed-covering layers included testa and endosperm. The embryo was enclosed by a thin layer of endosperm, typically consisting of two layers of cells on the edge. The micropylar endosperm, which formed a cap-like structure covering the radicle tip, had approximately 10 cell layers (Fig. 1A and SI Appendix, Fig. S1A). Testa rupture (TR) and endosperm rupture (ER) are temporally independent events during seed germination (SI Appendix, Fig. S1B). Cotton seed germination is sensitive to temperature changes. Either low or high temperature delays seed germination. The germination assay at temperatures from 4 °C to 44 °C showed that the conditions below 12 °C and above 44 °C were suppressive to germination (Fig. 1B). The germination efficiency was positively related to temperature in the warm condition at a narrow window between 20 °C and 36 °C, indicating a conserved endogenous thermal system modulating seed germination. We, therefore, attempted to identify the gene(s) response to it. A group of mitochondria I subfamily small heat shock proteins (msHSPs) were proactively expressed during seed germination (10, 11) (SI Appendix, Fig. S1C). Among them, GhHSP24.7 was expressed predominantly in the endosperm after seed imbibition (Fig. 1B and SI Appendix, Fig. S1D). The GhHSP24.7 showed high expression correlation (R = 0.99) with temperature changes from 4 °C to 36 °C in the seed germination assay (Fig. 1B). These observations suggest that GhHSP24.7 may play an important role in seed germination in response to temperature increase.
Fig. 1.
GhHSP24.7 regulated seed germination in a temperature-dependent manner. (A) Illustration of the mature cotton seed. (B, Upper) Relative expression level (R.E.L.) of GhHSP24.7 in 3-h imbibed seeds at the indicated temperature. The Inset histogram shows the R.E.L. of GhHSP24.7 in cotyledon (C), radicle (R), and endosperm (E) of the same dissected seed. Middle and Lower show 48-h germination seeds at indicated temperature. (Scale bar, 4 mm.) (C, Upper) R.E.L. of GhHSP24.7 in the 3-h imbibed seeds at 28 °C. Lower shows photographs of 2-d-old germinated seeds at 28 °C. Two lines of overexpression (GhHSP24.7-OE for O1 and O2) and suppression (GhHSP24.7-SE for S1 and S2) of transgenic plants are selected. The controls are W0 and pBI121. (Scale bar, 4 mm.) (D) Line plot shows ER percentage at each time point at 20 °C, 28 °C, and 36 °C. (E) Histogram showing the GhHSP24.7 R.E.L. in the 3-h imbibed seeds at indicated temperature. (F) Box plots showing the puncture force of endosperm cell layer (PFE) of imbibed seeds with or without TR at 28 °C at the time indicated (n = 15). TR percentage of the seed population at each time point is indicated. OE, GhHSP24.7-OE and SE, GhHSP24.7-SE. Letters represent data that are significantly different according to Student’s t test, P < 0.05. For all panels, data are means ± SEM, n = 3; *P < 0.05, **P < 0.01, Student’s t test.
To investigate the roles of GhHSP24.7, we developed GhHSP24.7 overexpression (GhHSP24.7-OE) and suppression (GhHSP24.7-SE using the antisense strategy) transgenic cotton lines driven by a 35S promoter. Two independent lines of each were selected for detailed analysis (Fig. 1C and SI Appendix, Fig. S1 E and F). A transgenic line with an empty vector (pBI121) and untransformed cotton (accession W0) were used as controls. GhHSP24.7-SE seeds with reduced expression of GhHSP24.7 evidently delayed germination compared with the control groups at 28 °C (Fig. 1C). In contrast, GhHSP24.7-OE seeds germinated more quickly, revealing that GhHSP24.7 plays a positive role in seed germination.
To test if the GhHSP24.7-modulated impacts on seed germination are temperature dependent, a germination assay was performed at 20 °C, 28 °C, and 36 °C (Fig. 1D and SI Appendix, Fig. S2). It took about 86 h to reach 50% seed germination (ER) at 20 °C for the control groups, but only about 40 h at 36 °C. For the GhHSP24.7-SE seeds, the delayed germination was not influenced by the temperature changes from 20 °C to 36 °C, suggesting that temperature-dependent germination was disturbed due to the suppression of GhHSP24.7 (Fig. 1E). By contrast, the GhHSP24.7-OE seeds showed a fast-germination phenotype, which resembled the fast germination of the control groups at high temperature (Fig. 1D). The above data indicated the GhHSP24.7 regulates seed germination in a temperature-dependent manner. Here, the behavior of GhHSP24.7-OE seeds in germination mimicked that of the control seeds under warm conditions, while the behavior of GhHSP24.7-SE seeds represented that of the control seeds under cold conditions.
Abscisic acid (ABA) and gibberellins (GAs) are known to be major regulators of seed dormancy and germination. To examine whether the function of GhHSP24.7 is dependent on the hormones, we applied ABA, fluridone (ABA biosynthesis inhibitor), and GAs to the germinating seeds, respectively (SI Appendix, Fig. S3A). The fast-germination phenotype of GhHSP24.7-OE seeds was not suppressed by ABA. Neither was the delayed germination of the GhHSP24.7-SE seeds recovered by fluridone. All of the transgenic and control seeds exhibited similar responses to GAs. In addition, there was no significant difference in the endogenous ABA and GA contents between these transgenic and control seeds (SI Appendix, Fig. S3B). These data suggest that GhHSP24.7 regulates seed germination via a pathway which might be independent of the ABA and GA signaling.
GhHSP24.7 Induces Endosperm Weakening.
Seed dormancy and germination are balanced between the resistance of the seed-covering layers (testa and endosperm) and the embryo growth potential (12). The latter determines embryo growth by water uptake and can be quantified using solutions with different concentrations of polyethylene glycol (PEG) (5). GhHSP24.7 did not affect the embryo growth potential at any tested ambient water potential (SI Appendix, Fig. S3C). The resistance of the seed-covering layers was further tested by measuring the puncture force of endosperm (PFE). A high PFE represents strong resistance of the seed-covering layers, which is a suppressive force for seed germination, or vice versa (13). We found that the PFE decreased along with the progress of ER. The GhHSP24.7-SE seeds, which exhibited delayed germination, showed consistently high PFE (Fig. 1F). The PFE of the controls dropped quickly from 20 °C to 36 °C (SI Appendix, Fig. S2E), which was in agreement with the fast germinating dynamics under relatively warm conditions. However, the GhHSP24.7-SE seeds still exhibited a high PFE even at 36 °C (SI Appendix, Fig. S2E). Taken together, these results suggest that GhHSP24.7 plays a unique role in the decay of the endosperm during seed germination.
GhCcmFc Is the Client of GhHSP24.7.
HSPs are known chaperones to protect their client proteins against stress conditions. To determine the client of GhHSP24.7, a yeast two-hybrid (Y2H) screen was performed using a cotton leaf cDNA library. A CytC maturation protein Fc (CcmFc) was identified as an interactor of GhHSP24.7 (Fig. 2A). CytC is one of the essential components of the mETC (14), and transfers electrons from complex III to IV (15). The maturation of CytC is activated and stabilized by binding with heme, which is synthesized from different modules in plant, namely CytC maturation A-I (CcmABCDEFGH) (16). CcmF delivers heme to apocytochrome C with the assistance of CCMH. GhHSP24.7::GFP fusion signals were detected in punctuate spots that overlapped with the mitochondria marker AtPGN::RPF (17) (Fig. 2B). Similarly, GhCcmFc was also localized on mitochondria (Fig. 2B). The interaction between GhHSP24.7 and GhCcmFc was confirmed by bimolecular fluorescence complementation (BiFC) (Fig. 2C) and in vitro protein coimmunoprecipitation (Co-IP) assays (Fig. 2D).
Fig. 2.
GhHSP24.7 competed with GhCCMH to bind with GhCcmFc in mitochondria. (A) Y2H validated that GhHSP24.7 interacted with GhCcmFc. For GhCcmFc, the N-terminal membrane signal peptide with 40 amino acids was deleted. (B) GhHSP24.7::GFP (G), GhCcmFc::GFP (G) and GhCCMH::GFP (G) signals are localized in mitochondria. AtPGN::RFP (R) is a mitochondrial marker. (C) BiFC assay shows GhHSP24.7 and GhCcmFc, GhCcmFc and GhCCMH colocalized in the mitochondria. (D and E) GST and His pull-down analysis reveals an interaction between GhHSP24.7, GhCcmFc, and GhCCMH in vitro. (F) Y2H assay to determine interactions between GhCcmFc, GhCCMH, and GhHSP24.7. (G) Competitive pull-down of GhCcmFc with GhHSP24.7 and GhCCMH in vitro. Beads containing His-fused GhCcmFc were assayed for their ability to bind a soluble TST-fused GhCCMH with increasing doses of GST-fused GhHSP24.7. Abbreviations: 488, 488-nm excitation laser; 561, 561-nm excitation laser; B.F., bright field; M, merged.
The mitochondrial gene coded CcmFc protein is conserved in plant (SI Appendix, Fig. S4 A and C). CcmFc, CCMH, and CcmI form a complex to synthesize holocytochrome, which transfers electrons from complex III to complex IV on the mETC (14). The direct interaction between CcmF and CCMH widely exists in prokaryotic cells such as Rhodobacter capsulatus (18). We employed BiFC, Co-IP, and Y2H (Fig. 2 C, E, and F) assays to confirm their interaction in vitro. On the other hand, the expression of both GhCCMH and GhCcmFc (SI Appendix, Fig. S4 E and F) was synchronized with the expression of GhHSP240.7 (SI Appendix, Fig. S4G). We did not detect any direct interaction between GhHSP24.7 and GhCCMH (Fig. 2F and SI Appendix, Fig. S5D). A competitive protein pull-down assay showed that the binding efficiency of GhCCMH and GhCcmFc was reduced with increasing amounts of GhHSP24.7, indicating that GhHSP24.7 and GhCCMH antagonistically interact with GhCcmFc (Fig. 2G). These data demonstrate that GhCcmFc is the client of GhHSP24.7 in mitochondria. The GhHSP24.7 competes with GhCCMH to form the GhCcmFc–GhHSP24.7 complex, preventing the GhCcmFc–GhCCMH interaction.
To study the membrane association and topology of GhHSP24.7, GhCcmFc, and GhCCMH in the mitochondria, the soluble and nonsoluble fractions of mitochondrial extraction were isolated (19) (SI Appendix, Fig. S6A). As shown in SI Appendix, Fig. S6B, GhHSP24.7::GFP was detected only in the soluble fractions after each treatment, indicating that GhHSP24.7 is not a mitochondrial membrane protein. In contrast, GhCcmFc::GFP and GhCCMH::GFP were detected only in the insoluble fractions after each treatment (SI Appendix, Fig. S6B), indicating that GhCcmFc and GhCCMH are integral proteins of the mitochondrial membrane. We further dissected the localization and topology of GhHSP24.7::GFP, GhCcmFc::GFP, and GhCCMH::GFP by a set of protease protection experiments with thermolysin, which degrades proteins on the surface of the organelles, and trypsin, which can access the intermembrane space (19). The matrix protein GhHSP24.7::GFP was protected from both enzymes (SI Appendix, Fig. S6 C and D). However, GhCcmFc::GFP and GhCCMH::GFP were resistant to thermolysin but disrupted by trypsin (SI Appendix, Fig. S6 C and D), indicative of a topology in which GhCcmFc and GhCCMH are anchored to the mitochondrial inner membrane with their C terminals facing the intermembrane space (SI Appendix, Fig. S6A), as occurs with CCMH in Arabidopsis (20).
To identify the domains in GhCcmFc and GhCCMH responsible for interaction, a series of truncated constructs were generated for Y2H assays. GhCcmFc and GhCCMH were divided into five and two sections according to the transmembrane domain, respectively (SI Appendix, Fig. S6E). GhHSP24.7 does not contain transmembrane domains, consistent with its location in the mitochondrial matrix. As shown in SI Appendix, Fig. S6F, only subsection GhCcmFc-4 was able to interact with GhHSP24.7 and GhCCMH-1 but not with GhCCMH-2, suggesting that the GhCcmFc-4 is necessary and sufficient for the interaction of GhHSP24.7 and GhCCMH. Together, these results indicate GhHSP24.7 binds with the C terminal of GhCcmFc in competition with GhCCMH in the matrix site of the mitochondrial inner membrane.
The Elevated GhHSP24.7 Reduces the Production of CytC/C1.
Both CcmFc and CCMH are required for holocytochrome assembly of CytC/C1 in mitochondria (16) (Fig. 3A). So, the destabilization of the CcmFc and CCMH complex might impair CytC/C1 maturation and substantially influence the mETC components. Given GhHSP24.7 is a competitor of GhCCMH for GhCcmFc binding (Fig. 2G), we wondered whether the elevated expression of GhHSP24.7 during seed germination could affect CytC/C1 in mETC. Western blotting analysis revealed that the CytC/C1 levels were significantly lower in the GhHSP24.7-OE lines than in the controls constantly after imbibition (Fig. 3B). Interestingly, the AOX level was similar in both the transgenic lines and the controls at 3 h postimbibition, but was higher in the GhHSP24.7-OE lines than in the controls at 9, 15, and 21 h postimbibition (Fig. 3B). However, the levels of Nad9 (complex I), Cox2 (complex IV), and α-ATPase (complex V) were similar in both the transgenic lines and the controls (Fig. 3B).
Fig. 3.
GhHSP24.7 altered CytC/C1 activity in the mETC. (A) Schematic graph illustrates the suppressors of the mETC applied in this assay. (B) Western blots show major components of the mETC during the seed germination over a time course at 3, 9, 15, and 21 h postimbibition at 28 °C. (C) Histograms show the oxygen uptake rate in the 3-, 9-, 15-, and 21-h imbibed seeds at 28 °C. NaN3 is an inhibitor of COX. SHAM is an inhibitor of the AOX. Data are means ± SEM of n = 3 (*P < 0.05; **P < 0.01 compared with the control by Student’s t test).
One of the initial changes during the early stages of seed germination is the resumption of respiratory activity. The dynamics of the respiration, represented by the oxygen uptake at 28 °C, displayed an S-shaped curve for increase in respiration during cotton seed germination (SI Appendix, Fig. S8). GhHSP24.7-SE exhibited a similar total respiration level to the control groups in terms of oxygen uptake (Fig. 3C). However, the total respiration level was lower in the GhHSP24.7-OE lines than the controls (Fig. 3C). In plants, the respiration is usually regulated by two pathways in mitochondria, the cytochrome oxidase pathway (COX) and the alternative oxidase pathway (AOX) (21) (Fig. 3A). To determine which pathway GhHSP24.7 is involved in, the oxygen consumption in germinating seeds was measured in the presence of one of the two specific respiration inhibitors, NaN3 (inhibitor of the COX pathway) and salicylhydroxamic acid (SHAM) (inhibitor of the AOX pathway) (21) (Fig. 3A). The oxygen consumption of the COX pathway decreased remarkably in the GhHSP24.7-OE lines over time as shown by measurements taken at 3, 9, 15, and 21 h after imbibition (Fig. 3C). Interestingly, the oxygen consumption of the AOX respiration pathway was similar in the transgenic lines and the controls at 3 h postimbibition, but was higher in the GhHSP24.7-OE lines than in the controls at 9, 15, and 21 h postimbibition, which was consistent with the AOX levels (Fig. 3B). These results suggest that the elevated GhHSP24.7 level impaired the maturation of CytC/C1, which led to the inhibition of the COX pathway. Consequently, the AOX pathway was activated.
GhHSP24.7 Mediates ROS Release in Seed Germination.
Complex III in the ETC produces ROS upon the leakage of electrons caused by reduced production of mature CytC (22). ROS play a positive role in seed germination (23). To test whether GhHSP24.7 modulates CytC/C1 production to induce ROS production, we examined the ROS content in the seed radicle by 5-(and-6)-chloromethyl-2.7-dichlorofluorescein diacetate (DCFH-DA) staining (24). The GhHSP24.7-SE lines showed low ROS levels, but the GhHSP24.7-OE lines showed the opposite trend (Fig. 4 A and B and SI Appendix, Fig. S9). These differences were also observed in the measurements of H2O2 and O2− concentrations in the transgenic seeds (Fig. 4C and SI Appendix, Fig. S9C). This trend was in agreement with the CytC/C1 content of the transgenic lines (Fig. 3B). Both H2O2 and O2− concentrations showed high correlation (R = 0.97 for H2O2 and R = 0.99 for O2−) with temperature changes in the germination assay (Fig. 4C and SI Appendix, Fig. S9C). The GhHSP24.7-SE seeds had constantly low H2O2 and O2− levels at various temperatures. Treated with the ROS-generating compound methylviologen (MV), the delayed germination in the GhHSP24.7-SE seeds was rescued to the control level (Fig. 4 E and F and SI Appendix, Fig. S10). In contrast, the treatment of the H2O2 suppressor, N-acetyl-l-cysteine (NAC) delayed the seed germination both in GhHSP24.7-OE and the controls (Fig. 4 E and F and SI Appendix, Fig. S10). All these findings suggest that GhHSP24.7 positively regulates the endogenous ROS generated from mitochondria during seed germination.
Fig. 4.
GhHSF24.7 regulated seed germination by fine tuning ROS release. (A) DCFH-DA staining on vertical dissections of radicles of the 24-h imbibed seeds at 28 °C. Abbreviations are the same as in Fig. 2. (B) Quantified florescence intensity of A. (C and D) Upper shows H2O2 concentration in the seeds and mitochondria of transgenic lines over time after imbibition at 28 °C. Lower shows H2O2 concentration in the 3-h imbibed seeds and mitochondria at the indicated temperature. (E) Photographs show the germination status of seeds treated with H2O2, MV, and NAC 48 h after imbibition at 28 °C. (F) ER percentage of seeds treated with H2O2, MV, or NAC 48 h after imbibition at 28 °C. For all panels, data are means ± SEM of n = 3 (*P < 0.05; **P < 0.01 compared with the control by Student’s t test).
We further examined how altered GhHSP24.7 expression affected the cellular structure of the germinating seed. After 12 h imbibition at 28 °C, the endosperm and micropylar endosperm maintained a fine cell structure (Fig. 5A). The two layers of cells in the endosperm began to separate, representing the decay of the cell structure of micropylar endosperm. However, in the GhHSP24.7-OE seeds the endosperm layer was distinctively ruptured with deformed cells in micropylar endosperm (Fig. 5A). The PFE was examined following treatment with H2O2 and NAC. H2O2 decreased the PFE of the controls to the level of the GhHSP24.7-OE lines (Fig. 5B), while NAC rescued the PFE of the GhHSP24.7-OE seeds to the level of GhHSP24.7-SE seeds. Therefore, the endogenous ROS play a positive role in endosperm weakening during seed germination.
Fig. 5.
GhHSP24.7 induced endosperm weakening during seed germination. (A, Left) Dissected cotton seed. Photographs (Upper Right) show an enlarged view of the endosperm rupture on the side. Arrows show the ruptured cell layer of the endosperm. Photographs (Lower Right) show a section of micropylar endosperm. The imbibition hours at 28 °C are shown on the Right. (B) PFE for endosperm cell layer of seeds treated with H2O2 or NAC to TR or non-TR at 28 °C. TR percentage is indicated. Data are means ± SEM of n = 15. Different letters indicate that data are of significant difference (Student’s t test, P < 0.05).
Endosperm decay starts with the components’ destruction in cell wall, while, the completion of seed germination requires an increase in cell wall extensibility in the endosperm tissues surrounding the radicle (25). The cell wall components of the GhHSP24.7 transgenic lines and the controls were compared using fluorescence in situ hybridization, with antibodies against the major cell wall components and extension in seed sections. The fluorescence signals of cellulose, homogalacturonan, and galactosylated xyloglucan were reduced in the endosperm of the GhHSP24.7-OE lines, but became more abundant in the GhHSP24.7-SE lines (SI Appendix, Fig. S11 A and B). Meanwhile, the extent of cell wall loosening appeared higher in the GhHSP24.7-OE lines than in other groups (SI Appendix, Fig. S11 A and B). Therefore, the cell wall matrix has become more fragile in the endosperm of the GhHSP24.7-OE lines. In contrast, in the GhHSP24.7-SE seeds, the cell wall in the endosperm tended to have a structural characteristic that prevented rupture.
The Function of HSP24.7 Is Conserved for Seed Germination.
To determine whether the function of HSP24.7 in seed germination is conserved in plants, we examined its functional orthologs in Arabidopsis and tomato. Based on sequence similarity, two mitochondrial sHSP (AtHSP23.5 and AtHSP23.6) in Arabidopsis and two mitochondrial sHSP (SlHSP23.8B and SlHSP21.5B) in tomato were identified in one clustered subgroup (Fig. 6A). To elucidate the role of these homologs in regulating seed germination, we obtained transgenic Arabidopsis and tomato lines to overexpress these genes, respectively (Fig. 6C). Similar to the phenotypes observed in GhHSP24.7 transgenic cottons, AtHSP23.6 and SlHSP23.8B overexpression seeds exhibited fast germination phenotypes under favorable temperatures (Fig. 6B). These data suggest that the function of HSP24.7 in seed germination is conserved in plants.
Fig. 6.
The homologs of GhHSP24.7 played a positive role in seed germination. (A) Phylogenic tree of mitochondria I subfamily sHSP from Gossypium hirsutum, Arabidopsis, and tomato was constructed using MEGA 5.0 by the neighbor-joining method. (B and C) Germination assay of AtHSP23.6/23.5 and SlHSP23.8B/21.5B overexpression seeds. Upper shows photographs of 30-h imbibed Arabidopsis (Left) at 22 °C and 48-h imbibed tomato (Right) seeds at 28 °C. Lower shows the germination percentage of transgenic seeds on 1/2 MS medium. Germination of seeds was determined based on radicle extrusion. Data are means ± SEM of n = 3. (D) qRT-PCR analysis of transgenic lines used in B and C. Data are means ± SEM of n = 3 (**P < 0.01 compared with the control by Student’s t test).
Discussion
Fine Tuning of mETC Activity During Seed Germination.
Here, we discovered that a mitochondrion-localized plant sHSP, GhHSP24.7, which acts in cooperation with its in vivo client GhCcmFc, to modulate proper ROS production from the mETC for seed germination (Fig. 7). The CytC/C1 maturation subunit protein GhCcmFc is a client of GhHSP24.7. It is known that cytochrome bc1 (Cytbc1) complex is bifunctional in plants, as it also contains the mitochondrial processing peptidase required to remove the targeting signals from proteins imported into mitochondria (26). Hence, Cytbc1 has an important role in mitochondrial biogenesis. Mitochondrial biogenesis is also involved in seed germination (27). Therefore, the peptidase activity of Cytbc1 may also contribute to seed germination. The importance of Cytbc1 in seed germination and how each of its functions contributes to seed germination are very interesting topics in future studies. GhHSP24.7 is identified as an inhibitor for the formation of the GhCcmFc and GhCCMH complex instead of protecting this client protein. The GhHSP24.7 level is within a gentle and comfortable dynamic during the seed germination. This finding expands our understanding of sHSP’s functions in general and sheds a light on the mechanism by which sHSPs regulate seed germination under favorable conditions.
Fig. 7.
Schematic chart shows HSP24.7 modulated seed germination by fine tuning CytC/C1 activity and ROS generation from the mETC during seed germination. At the low-temperature condition, the seed remained dormant. When the temperature went up, the mitochondria were activated to accelerate the respiration rate. In addition, HSP24.7 occupied CcmF to deactivate CytC/C1, leading the inhibition of the COX pathway and induction of ROS. As a result, the AOX pathway was activated, which had a decreasing effect on ROS levels to some extent. With the ROS release to weaken the endosperm in the seed, the radicle reached out to accomplish the germination.
HSP24.7 Mediates ROS-Regulating Seed Germination.
In recent decades, major advances have been made in our understanding of the role of ROS in seed germination. ROS is a dominant factor to stimulate the transition from a quiescent seed to a metabolically active organism. Production of ROS has been demonstrated during the early imbibition period in major crops, such as wheat (28). In germinating seeds, ROS can be generated from all metabolically active compartments, such as endogenously ROS produced in chloroplasts and mitochondria, apoplastic ROS produced by NADPH oxidase (29). The domination and continuation of respiration in mitochondria in imbibed seeds can lead to electron leakage. Because the electrons of the mETC have sufficient free energy to directly reduce O2, the electron leakage from the mETC is an unavoidable source of increased ROS production in mitochondria (30). Nevertheless, the mechanism of ROS release during the respiration-dominant stage of germination, especially in response to the temperature changes is not clear. Based on the data obtained from this study, we conclude that GhHSP24.7 abundance is sensitive to the dynamic of temperature during seed germination. In response to warm conditions, GhHSP24.7 level is elevated to inhibit the COX pathway and block the electron transport from complex III in mitochondria, leading to the release of additional ROS, which promotes seed germination (Fig. 7). When the COX pathway is reduced, the AOX pathway is activated, which has a decreasing effect on ROS levels to some extent, but the effect is not enough to suppress ROS levels in the GhHSP24.7-OE lines to match levels in the controls. Hence, the higher ROS levels in the GhHSP24.7-OE lines accelerate endosperm rupture to promote seed germination. Moreover, the elevated ROS level may also affect other mechanisms sensitive to ROS in the mitochondria, such as iron–sulfur assembly (31), which likely relate to seed germination. The function of GhHSP24.7 in seed germination control is widely conserved in the plant kingdom. GhHSP24.7 therefore can fine tune seed germination by manipulating ROS release. Our study led to the discovery of a mechanism to regulate ROS production in mitochondria during seed germination.
Materials and Methods
Details about plant materials and growth conditions are described in SI Appendix, Materials and Methods. Seed germination, puncture force measurement, gene expression, plant transformation, yeast two-hybrid system, BiFC assay, pull-down assay, ROS detection, oxygen consumption measurement, and mitochondrial assays are also described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Jiawei Wang (Shanghai Institute of Plant Physiology and Ecology) and Prof. Yuxian Zhu (Wuhan University) for their critical comments; and Prof. Baocai Tan (Shandong University) for providing the antibodies of mitochondrial complexes. This work was financially supported in part by grants from the National Natural Science Foundation of China (U1503284), the National Key Research and Development Program (2016YFD0100605), the Distinguished Discipline Support Program of Zhejiang University, and the Australian Research Council (DP180103834).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815790116/-/DCSupplemental.
References
- 1.Weitbrecht K, Müller K, Leubner-Metzger G. First off the mark: Early seed germination. J Exp Bot. 2011;62:3289–3309. doi: 10.1093/jxb/err030. [DOI] [PubMed] [Google Scholar]
- 2.Reeves RG, Valle CC. Anatomy and microchemistry of the cotton seed. Bot Gaz. 1932;93:259–277. [Google Scholar]
- 3.Steinbrecher T, Leubner-Metzger G. The biomechanics of seed germination. J Exp Bot. 2017;68:765–783. doi: 10.1093/jxb/erw428. [DOI] [PubMed] [Google Scholar]
- 4.Donohue K. Niche construction through phenological plasticity: Life history dynamics and ecological consequences. New Phytol. 2005;166:83–92. doi: 10.1111/j.1469-8137.2005.01357.x. [DOI] [PubMed] [Google Scholar]
- 5.Graeber K, et al. DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc Natl Acad Sci USA. 2014;111:E3571–E3580. doi: 10.1073/pnas.1403851111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrie C, et al. How do plants make mitochondria? Planta. 2013;237:429–439. doi: 10.1007/s00425-012-1762-3. [DOI] [PubMed] [Google Scholar]
- 7.Giegé P, Sweetlove LJ, Cognat V, Leaver CJ. Coordination of nuclear and mitochondrial genome expression during mitochondrial biogenesis in Arabidopsis. Plant Cell. 2005;17:1497–1512. doi: 10.1105/tpc.104.030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stupnikova I, et al. Pea seed mitochondria are endowed with a remarkable tolerance to extreme physiological temperatures. Plant Physiol. 2006;140:326–335. doi: 10.1104/pp.105.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noctor G, De Paepe R, Foyer CH. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 2007;12:125–134. doi: 10.1016/j.tplants.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Zhang T, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 11.Ma W, et al. Identification and characterization of the GhHsp20 gene family in Gossypium hirsutum. Sci Rep. 2016;6:32517. doi: 10.1038/srep32517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 13.Müller K, Tintelnot S, Leubner-Metzger G. Endosperm-limited Brassicaceae seed germination: Abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol. 2006;47:864–877. doi: 10.1093/pcp/pcj059. [DOI] [PubMed] [Google Scholar]
- 14.Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. Cytochrome c biogenesis: Mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev. 2009;73:510–528. doi: 10.1128/MMBR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margoliash E, Barlow GH, Byers V. Differential binding properties of cytochrome c: Possible relevance for mitochondrial ion transport. Nature. 1970;228:723–726. doi: 10.1038/228723a0. [DOI] [PubMed] [Google Scholar]
- 16.Sanders C, Turkarslan S, Lee DW, Daldal F. Cytochrome c biogenesis: The Ccm system. Trends Microbiol. 2010;18:266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laluk K, Abuqamar S, Mengiste T. The Arabidopsis mitochondria-localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol. 2011;156:2053–2068. doi: 10.1104/pp.111.177501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders C, et al. The cytochrome c maturation components CcmF, CcmH, and CcmI form a membrane-integral multisubunit heme ligation complex. J Biol Chem. 2008;283:29715–29722. doi: 10.1074/jbc.M805413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan R, Jones AD, Hu J. Cardiolipin-mediated mitochondrial dynamics and stress response in Arabidopsis. Plant Cell. 2014;26:391–409. doi: 10.1105/tpc.113.121095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer EH, et al. AtCCMH, an essential component of the c-type cytochrome maturation pathway in Arabidopsis mitochondria, interacts with apocytochrome c. Proc Natl Acad Sci USA. 2005;102:16113–16118. doi: 10.1073/pnas.0503473102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang XY, et al. Arabidopsis kinesin KP1 specifically interacts with VDAC3, a mitochondrial protein, and regulates respiration during seed germination at low temperature. Plant Cell. 2011;23:1093–1106. doi: 10.1105/tpc.110.082420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: Central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 23.Oracz K, et al. The mechanisms involved in seed dormancy alleviation by hydrogen cyanide unravel the role of reactive oxygen species as key factors of cellular signaling during germination. Plant Physiol. 2009;150:494–505. doi: 10.1104/pp.109.138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schopfer P, Plachy C, Frahry G. Release of reactive oxygen intermediates (superoxide radicals, hydrogen peroxide, and hydroxyl radicals) and peroxidase in germinating radish seeds controlled by light, gibberellin, and abscisic acid. Plant Physiol. 2001;125:1591–1602. doi: 10.1104/pp.125.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penfield S, Li Y, Gilday AD, Graham S, Graham IA. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell. 2006;18:1887–1899. doi: 10.1105/tpc.106.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kmiec B, Teixeira PF, Glaser E. Shredding the signal: Targeting peptide degradation in mitochondria and chloroplasts. Trends Plant Sci. 2014;19:771–778. doi: 10.1016/j.tplants.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 27.Law SR, Narsai R, Whelan J. Mitochondrial biogenesis in plants during seed germination. Mitochondrion. 2014;19:214–221. doi: 10.1016/j.mito.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Caliskan M, Cuming AC. Spatial specificity of H2O2-generating oxalate oxidase gene expression during wheat embryo germination. Plant J. 1998;15:165–171. doi: 10.1046/j.1365-313x.1998.00191.x. [DOI] [PubMed] [Google Scholar]
- 29.Bailly C. Active oxygen species and antioxidants in seed biology. Seed Sci Res. 2004;14:93–107. [Google Scholar]
- 30.El-Maarouf-Bouteau H, Bailly C. Oxidative signaling in seed germination and dormancy. Plant Signal Behav. 2008;3:175–182. doi: 10.4161/psb.3.3.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remes B, Berghoff BA, Förstner KU, Klug G. Role of oxygen and the OxyR protein in the response to iron limitation in Rhodobacter sphaeroides. BMC Genomics. 2014;15:794. doi: 10.1186/1471-2164-15-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.