Significance
In malignant melanoma, one of the most aggressive cancers, the components of the WNT pathway are frequently deregulated and have been shown to drive self-renewal and metastatic progression of melanoma cells. Identifying tumorigenic properties of key members of the WNT signaling network in the pathogenesis of melanoma represents a pivotal task in the field and underlies successful design of targeted therapeutic approaches. In the present study, we characterized Frizzled 3 receptor (FZD3) as a critical factor underlying tumorigenic properties of aggressive human melanoma. Using in vivo and in vitro tumorigenic assays, combined with functional transcriptomics analysis of FZD3 knockdown in human patient-derived cells, we determined key signaling nodes regulated by FZD3 activity during malignant transformation.
Keywords: frizzled, melanoma, MAPK, systems biology, gene expression
Abstract
Frizzled 3 receptor (FZD3) plays an important role in the homeostasis of the neural crest and its derivatives, which give rise to pigment-synthesizing cells, melanocytes. While the role for FZD3 in specification of the melanocytic lineage from neural crest is well established, its significance in the formation of melanoma, its associated malignancy, is less understood. In this study we identified FZD3 as a critical regulator of human melanoma tumorigenesis. Down-regulation of FZD3 abrogated growth, colony-forming potential, and invasive capacity of patient-derived melanoma cells. Xenotransplantation of tumor cells with down-regulated FZD3 levels originating from melanomas carrying the BRAF(V600) mutation uniformly suppressed their capacity for tumor and metastasis formation. FZD3 knockdown leads to the down-regulation of the core cell cycle protein components (cyclins D1, E2, B1, and CDKs 1, 2, and 4) in melanomas with a hyperactive BRAF oncogene, indicating a dominant role of this receptor during melanoma pathogenesis. Enriched pathway analysis revealed that FZD3 inhibits transcriptional networks controlled by CREB5, FOXD1, and ATF3, which suppress the activity of MAPK-mediated signaling. Thus, FZD3 establishes a positive-feedback mechanism that activates MAPK signal transduction network, critical to melanoma carcinogenesis. Importantly, high levels of FZD3 mRNA were found to be correlated with melanoma advancement to metastatic stages and limited patient survival. Changes in gene-expression patterns mediated by FZD3 activity occur in the absence of nuclear β-catenin function, thus representing an important therapeutic target for the melanoma patients whose disease progresses independent of canonical WNT signaling.
Melanomas arise from the malignant transformation of the melanocytic cell lineage and have high metastatic potential. Multiple studies have indicated that this aggressive skin cancer partially recapitulates a developmental program of normal melanocytes, but in an uncontrolled and disorganized manner (1–4). Consistent with these findings is the notion that the genes controlling phenotypic properties of melanocyte precursors also play an important role in melanoma pathogenesis. In vertebrates, melanocytes are derived from the neural crest cell population, which appears at the dorsal side of the neural tube during the final, closing stages of its formation in late embryogenesis (5). Upon dorsal–ventral migration melanocytic neural crest derivatives give rise to highly migratory precursors, melanoblasts, which undergo multiple steps of differentiation to become pigment-synthesizing cells, melanocytes (6). The WNT signaling pathway plays a critical role during melanocyte specification from the neural crest. In humans, the highly conserved WNT molecular network encompasses 19 closely related WNT ligands and 10 Frizzled receptors (FZD1–10) that direct self-renewal and regeneration of many tissues during their development (7, 8). FZDs are G protein-coupled receptors characterized by seven transmembrane-spanning domains, a cysteine rich N-terminal ligand binding domain, and a C-terminal intracellular activation domain. Depending on their activation, ligands, and intracellular binding adapter proteins, FZDs are capable of transmitting extracellular signals into diverse transcriptional program outputs that determine cell fate during normal and pathogenic development (9–11).
Previous studies using Xenopus and mouse models have delineated FZD3 as one of the few FZD family members that are predominantly expressed at the dorsal site of the neural tube, coinciding with neural crest appearance (12, 13). Subsequently, it was shown that the injection of FZD3 mRNA can induce formation of the neural crest in embryos and explants, while inhibition of FZD3 receptor action blocks endogenous neural crest formation, demonstrating a critical role for this receptor in neural crest biogenesis (13, 14). Using mouse knockout approaches, it was demonstrated that FZD3 is also required for axonal development in the forebrain and CNS (15, 16). In humans, FZD3 expression underlies proliferation and specification of the human neural crest and its melanocytic derivatives in vitro (17). While the above experimental evidence points to a major role for FZD3 in melanocyte biology, little is known about the functional significance of this receptor’s activity in melanoma initiation and progression. Interestingly, a recent study reported that FZD3 is overexpressed in 20% of melanoma patients whose tumors were devoid of infiltrating T cells, pointing to the importance of this receptor in the immune-evasive properties of melanoma (18).
FZD3 is distinct from most other FZD receptor family members in that it is not strongly linked to the canonical, β-catenin–dependent, signal transduction pathway. Instead, FZD3 is mostly associated with noncanonical, β-catenin–independent, signaling. This fact bears special significance when trying to understand the role of the WNT/FZD signaling axis in melanoma pathogenesis that remains the subject of heated debate (12, 19–21). In contrast to other cancers where activation of the canonical, β-catenin–dependent, pathway was shown to be a driving force behind tumor initiation and progression, human melanoma represents a type of tumor where nuclear and transcriptionally active β-catenin has been reported to correlate with a more favorable prognosis and a less-aggressive disease (22, 23). Other studies however, had clearly shown that the stabilization of β-catenin and its accumulation in the cell leads to an increased melanoma metastasis, both in vitro and in vivo (24, 25). These seemingly contradictory outcomes may reflect a different spectrum of driver mutations and species-related variability (human vs. mouse) in the model systems that are being used in these studies (26).
Due to the high significance of FZD3 in the homeostasis of the neural crest and the arising melanocytic cell lineage, we hypothesized that FZD3 may exert important influences on melanoma pathogenesis. In this study using patient-derived cells and xenograft assays, we demonstrate that indeed, FZD3 plays a critical role in the regulation of proliferation and metastatic progression of human melanomas, and it does so independent of β-catenin nuclear activity. Global gene-expression analyses reveal a pleotropic function for this receptor in the control of cell cycle progression and invasion. Moreover, using clinical datasets we demonstrate that the high levels of FZD3 expression correlate with the disease progression and diminished survival of advanced melanoma patients, revealing its significance as a therapeutic target.
Results
FZD3 Down-Regulation Suppresses Proliferation and Colony-Forming Capacity of Melanoma Patient-Derived Cells.
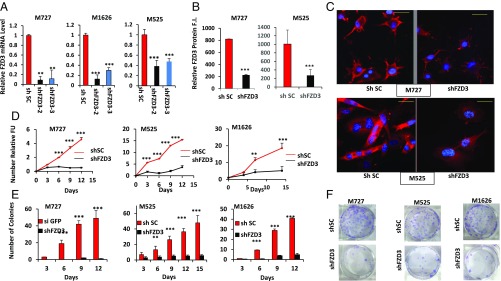
Based on the critical involvement of FZD3 in the homeostasis of melanocytic cell lineage, including neural crest stem cells, we hypothesized that this receptor can also play a critical role in the regulation of melanoma pathogenesis in human patients. To test this hypothesis, we employed lentiviral-based short-hairpin RNAs (shRNAs) targeting FZD3 mRNA expression in melanoma patient-derived cells. Using two independent shRNA sequences targeting different regions of FZD3 mRNA, and three independently derived cell cultures (M727, M1626, and M525), we were able to achieve significant levels of FZD3 down-regulation at the mRNA and protein levels (Fig. 1 A–C). qRT-PCR revealed that shFZD3 shRNA clone-2 (referred as shFZD3 hereafter) displayed the most potent knockdown efficiency among tested shRNAs, inducing ∼65% to ∼90% FZD3 mRNA down-regulation in all three independently derived patient melanoma cell cultures (Fig. 1A). FZD3 down-regulation was also confirmed at the protein level using an immunohistochemical approach and FZD3-specific antibody. Our results indicate that shFZD3 causes a significant decrease in FZD3 cell surface protein levels (Fig. 1 B and C).
Fig. 1.
Down-regulation of FZD3 inhibits melanoma cell proliferation and colony-forming capacity. (A) Expression of FZD3 was analyzed by qRT-PCR following transduction of melanoma M727, M1626, and M525 cells with the control shRNA (shSC) or FZD3-specific shRNA (shFZD3-2 and shFZD3-3). Error bars represent SEM of duplicate experiments with three replicates each. (B and C) Immunofluorescence analysis of FZD3 protein expression in the indicated melanoma cells transduced with control shRNA (shSC) or shFZD3. The y axis indicates relative FZD3 protein fluorescence intensity. Red color indicates positive FZD3 staining. (Scale bars, 50 μm.) (D) Proliferation kinetics of shFZD3 and shSC-transduced melanoma cells, M727, M525, and M1626 quantified using Resazurin assay. Fluorescence values were normalized to the background readings collected from the wells that had no growing cells but contained growth culture media and Resazurin reagent. (E) Numbers of colonies formed by M727, M525, and M1626 melanoma cells following their transduction with the control shRNA (shSC) or shFZD3. Error bars represent SEM of duplicate experiments with two replicates each. *P < 0.05, **P < 0.005, ***P < 0.0005. (F) Representative images of Crystal violet-stained plates demonstrating colony growth potential of indicated melanoma cell lines transduced with the control shRNA (shSC) or shFZD3.
To evaluate the effects of FZD3 down-regulation on the proliferative capacity of melanoma, we measured growth kinetics of the cells transduced with shFZD3 or shScramble (shSC) vectors using a Resauzarin assay. The resulting growth curves of M525, M727, and M1626 cells indicated a significant reduction in the proliferative rate of shFZD3-transduced melanoma cells compared with the control (shSC) transduced counterparts (Fig. 1D). These results were also supported by the overall decreased cell density in response to FZD3 down-regulation recorded by phase-contrast microscopy at the end of the monitoring period (SI Appendix, Fig. S1A). We next examined clonogenic properties of melanoma following FZD3 knockdown by seeding the transduced cells at the rare density of 103 cells per well in six-well plates, and periodically (every 3 d) counting the number of arising colonies. A more than 90% reduction in the number of colonies formed was observed following FZD3 knockdown in M525, M727, and M1626 cells compared with the shSC controls (Fig. 1 E and F). In addition to the overall reduced colony number, the colonies that arose from the shFZD3-transduced cells were much smaller in size (SI Appendix, Fig. S1B), indicating a significant decrease in colony-forming capacity of the cells lacking FZD3. In summary, the above experiments revealed that FZD3 expression is required for melanoma cell proliferation and colony formation.
To demonstrate that the observed changes in the tumorigenic phenotype of melanoma cells were specific to FZD3 down-regulation and not caused by the shRNA-mediated off-target effects, we performed FZD3 rescue experiments. In these assays we overexpressed FZD3 cDNA using lentiviral vector pFZD3_FL. To generate stably expressing FZD3_FL cells and the matched controls, we transduced melanoma M727 cells with either pFZD3_FL or pLM lentiviral particles and subjected them to puromycin selection at (1 μg/mL). Using puromycin-selected cells, we transduced them with either shFZD3- or shSC-expressing vectors as described above. Introduction of shFZD3 into pLM M727 cells caused ∼75% reduction of FZD3 mRNA level; however, introduction of shFZD3 into pFZD3_FL M727 cells had no significant effect on FZD3 mRNA levels, which remained virtually unchanged (SI Appendix, Fig. S2A). We also confirmed rescue of FZD3 at the protein level by performing immunohistochemical analysis using anti-FZD3 mAb on the melanoma cells transduced with the pLM+shSC, pLM+shFZD3, and pFZD3_FL+shFZD3 (SI Appendix, Fig. S2 B and C). Importantly, while pLM-transduced cells ceased to proliferate in response to shFZD3 expression, FZD3_FL-transduced cells continued to grow in the presence of shFZD3 at the similar rate as control shSC-transduced counterparts (SI Appendix, Fig. S2D). To further validate FZD3 rescue phenotype, we performed a colony assay by seeding the above generated cell cultures at the density of 103 cells per well in six-well plates, and periodically (every 3 d) counting the number of colonies for the period of 15 d. At the end of the experiment, we observed ∼90% reduction in the number of colonies formed by pLM_shFZD3 cells compared with the pLM_shSC control cells (SI Appendix, Fig. S2E). However, cells expressing pFZD3_FL in the presence of shFZD3 retained their colony-forming capacity, and their colony number closely matched the one produced by control, pLM_shSC-transduced cells (SI Appendix, Fig. S2E). These results confirmed that the suppression of melanoma cell proliferation, and colony-forming capacity is specific to FZD3 down-regulation and is not due to the off-target effects caused by shRNA transduction.
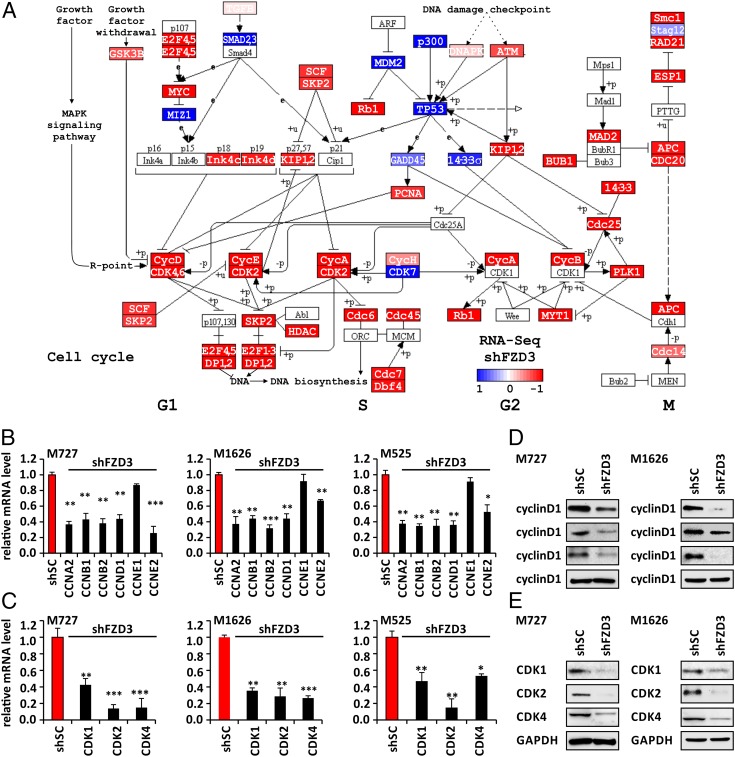
Global Gene-Expression Analysis Reveals FZD3 as a Critical Regulator of Cell Cycle Progression in Melanoma.
To gain molecular insights into the regulatory function of FZD3 in melanoma, we analyzed global gene-expression changes in the independently patient derived cell cultures (M727, M525, and M1626) in response to FZD3 down-regulation. For each patient-derived cell culture, the RNA was isolated from two groups of cells, shFZD3 and shSC, in multiple biological replicates, and the RNA-sequencing (RNA-Seq) library was created as previously described (27). Upon completion of RNA sequencing, we derived gene-expression profiles of M727, M525, M1626 cells that had 25,548 unique transcripts in common, including long-noncoding RNAs. After exclusion of lowly expressed genes, the dataset comprised 10,693 transcripts. We then identified 519 differentially expressed genes with log-fold changes greater than 1.0 and P values below 0.05. It is important to mention that these datasets included both slowly growing and highly aggressive metastasis-forming melanomas (SI Appendix, Table S1). Despite these differences, the transcriptional response among patient-derived cells with FZD3 knockdown was uniform and showed separation of clusters in the principal component analysis (PCA) (Fig. 2A). Hierarchical cluster analysis of the melanoma specimens in combination with knockdown of FZD3 showed that the majority of transcripts have a positive association with FZD3 activity, as 79.1% of differentially expressed transcripts were down-regulated following knockdown of FZD3 (Fig. 2B). Using these data, we defined the group of 373 genes as positively regulated by FZD3 activity (significantly down in the melanoma specimens with shRNA knockdown of FZD3), and 146 genes as negatively regulated by FZD3 activity (Fig. 2B).
Fig. 2.
Transcriptomic profiling of FZD3 knockdown in human melanoma reveals its pleotropic control of tumorigenic properties. RNA-Seq data using two biological replicates for each shFZD3 and shSC generated melanoma cell lines. (A) PCA of FZD3 knockdown (blue) vs. control specimens (red) shows uniform shift of transcriptional profiles. (B) Hierarchical cluster analysis using correlation distance of log-transformed, normalized FPKM RNA-Seq values of melanoma specimens in combination with knockdown of FZD3. Transcriptomic impact of FZD3 relates to 71.9% of gene activation (down in the melanoma specimens with shRNA knockdown of FZD3, red cluster), and 28.1% of transcriptional down-regulation (blue cluster). Both fractions are equally important, contributing to rewiring of signaling pathways in FZD3-activated melanoma. (C) Pathway enrichment analysis identifies major networks of cell cycle (red), transcription factor networks (purple), MAPK signaling (cyan), and cytokine interactions (blue). (D) Upstream regulator analysis shows hierarchical structure of regulatory network of FZD3. Transcription factor networks of FOXD1, CREB5, and ATF3 (purple) control activity of MAPK effectors (cyan). E2F, MYC, and AP1 targets, and cell cycle (red) are strongly engaged as downstream effectors resulting in a robust proliferative phenotype.
Signaling pathway analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) revealed major cellular networks of cell cycle, transcriptional regulators, MAPK signaling, and cytokine interactions to be significantly enriched with P values below 0.05 and q values below 0.2 (Fig. 2C). Further on, upstream-regulator analysis discovered a hierarchical structure of regulatory events that act in concert with the above identified pathways under the control of FZD3 (Fig. 2D). Thus, following FZD3 knockdown, transcriptional networks controlled by FOXD1, CREB5, and ATF3 become up-regulated and suppressed the activity of MAPK signaling, which affected downstream cell cycle regulatory pathways. This is reflected by strongly enriched networks of E2F, MYC, and AP1 transcription factors highly ranking in the pathway analysis with a normalized enrichment score above 2.0 and P values and q values below 10e−20 (Fig. 2D). Further downstream, in the cell cycle category, various cyclins (CCNs), cell division cycle homologs (CDCs), and cyclin dependent kinases (CDKs) (CCND1, CCNB1, CCNE2, CCNA2, CDCA2, CDCA5, CDCA8, CDC25C, CDC45, CDC20, CDC6, CDCA7L, CDK2, CDKN3) were found to be significantly up-regulated in the absence of FZD3 knockdown (Fig. 3A).
Fig. 3.
FZD3 knockdown leads to the inhibition of the cell cycle regulators in BRAFV600D/E mutant human melanoma cells. (A) KEGG pathway map of cell cycle, red marked genes are significantly down-regulated in FZD3 knock-down melanoma cells, as indicated in the heatmap (log2 fold-change > 1, P < 0.05). Quantification of cyclins (B) and CDKs (C) mRNA levels in control shRNA (shSC) and shFZD3-transduced melanoma cells (M727, M525, and M1626) using qRT-PCR and gene-specific primers. Error bars represent SEM of duplicate experiments with three replicates each. *P < 0.05, **P < 0.005, ***P < 0.0005. (D and E) Western blot analysis of cyclinD1, cyclinE2, cyclinB1 (D) and CDK1, 2, 4 (E) in M727 and M1626 melanoma cells after they were transduced with control shRNA (shSC) and shFZD3.
To validate changes in the critical cell cycle regulators, as revealed by our global transcriptome analysis of the FZD3 network, we examined their expression at the mRNA and protein levels in response to FZD3 down-regulation. Using M727, M525, and M1626 patient-derived cells with FZD3 knockdown and matching controls, shSC cells, we examined mRNA levels of cyclinA2, cyclinB1, cyclinB2, cyclinD1, cyclinE1, and cyclinE2 using qRT-PCR and corresponding primers (SI Appendix, Table S2). Our data demonstrate that FZD3 down-regulation in multiple independently derived melanoma cells significantly suppressed mRNA expression of cyclinA2, cyclinB1, cyclinB2, cyclinD1, and cyclinE2, but not cyclinE1 (Fig. 3B). Cyclins represent an integral part of cell cycle regulatory complex when bound to corresponding CDKs. Interestingly enough, analysis of the global transcriptional network controlled by FZD3 predicted changes in the genes encoding not only the cyclins but also multiple CDK subunits. Using corresponding primers (SI Appendix, Table S2) and qRT-PCR, we confirmed that indeed FZD3 knockdown leads to the significant reduction in the mRNA levels for CDK1, CDK2, and CDK4 (Fig. 3C). Moreover, protein analysis of cyclins and CDKs further confirmed significant loss of their expression in shFZD3-transduced cells compared with shSC controls (Fig. 3D).
The above findings revealed that FZD3 cell surface expression was critical in modulating the activity of the MAPK pathway and its downstream targets associated with proliferation of melanoma cells. The most common genetic alteration augmenting MAPK signaling network activity in melanoma resides within the BRAF kinase, which is mutated in ∼65% of the patients. Mutations in this protein at the V600 amino acid position enable cell proliferation independent of the upstream regulatory signals. We therefore decided to determine the status of BRAF in the patient-derived cells that were sensitive to the FZD3 inhibition. Analysis of the BRAF mutation hotspot sequence region revealed that M727 cells carried a BRAFV600E mutation, M1626 had a BRAFV600D mutation, and M525 had no alterations in the BRAF sequence (SI Appendix, Fig. S3). This indicated that FZD3 has the capability to exert significance influence on the MAPK signaling output even in the presence of the oncogenic BRAF mutations.
In summary, our results demonstrated that activity of the FZD3 receptor positively regulates proliferation of melanoma cells by engaging critical transcriptional networks that lead to the strong activation of cell cycle machinery.
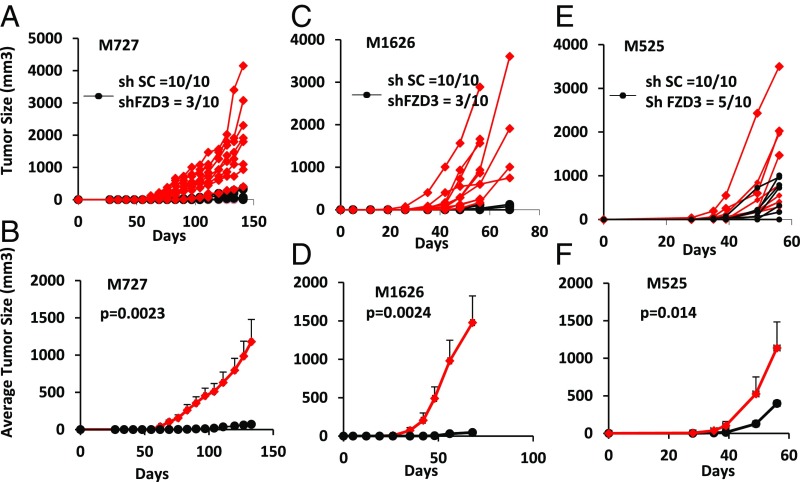
Inhibition of FZD3 Expression Suppresses Melanoma Initiation and Growth in Vivo.
To examine the regulatory effects of FZD3 on melanoma pathogenesis in vivo, we tested ability of patient-derived melanoma cells to initiate and maintain tumor growth following stable FZD3 knockdown. M727, M525, and M1626 cells were transduced with shFZD3 and shSC lentiviral vectors, as described above, and intradermally injected into two dorsal flanks of immunocompromised NSG mice (n = 10 for each shFZD3 or shSC group). Mice were monitored on a weekly basis for the time of tumor initiation following by regular measurements of the tumor size in each group. Our results demonstrate that FZD3 down-regulation suppressed tumor-forming capacity of multiple independent patient-derived cells carrying a BRAFV600D/E mutation, which either failed to initiate tumors at the site of an injection or were giving rise to many smaller tumors at a significantly later time period, compared with the mock-transduced counterparts (Fig. 4). For M727 (BRAFV600E) melanoma, tumor initiation rate was decreased by 70% (3 of 10) in shFZD3 group compared with 100% (10 of 10) for the control, shSC-transduced cells (Fig. 4A). The overall tumor volume among shFZD3 M727 was decreased by a mean of 94.9% (P = 0.0023) in size compared with the tumors derived from shSC-transduced cells (Fig. 4B). Similarly dramatic results were obtained with M1626 melanoma (BRAFV600D) cells where tumor initiation rate was decreased by 70% (3 of 10) in the shFZD3 group compared with 100% (10 of 10) for the control, shSC-transduced cells (Fig. 4C). The overall tumor volume in shFZD3 M1626 was decreased by a mean of 97% (P = 0.0024) in size compared with the tumors derived from shSC-transduced cells (Fig. 4D). Finally, in M525 (BRAFWT) melanoma the tumor initiation rate was decreased by 50% (5 of 10) among the cells transduced with shFZD3, compared with 100% (10 of 10) by those transduced with the control vector (Fig. 4E). The overall tumor volume in shFZD3 M525 was decreased by a mean of 65% (P = 0.014) compared with the tumors derived from shSC-transduced cells (Fig. 4F). These results demonstrated that the FZD3 receptor is a critical signaling component required for melanoma initiation and growth in vivo. Importantly, knockdown of FZD3 resulted in the very effective suppression melanoma tumors that carried a BRAF mutation, considered to be a driving oncogenic event in the pathogenesis of this disease.
Fig. 4.
Down-regulation of FZD3 inhibits melanoma initiation and growth in the presence of BRAFV600D/E mutation. Indicated patient-derived melanoma cells were transduced with either shFZD3 or shSC lentivectors and intradermally xenotransplanted into two dorsal flanks of NSG mice (100 cells per injection, n = 10 for each shFZD3 or shSC group). Tumor growth was monitored and measured on the weekly basis. Individual tumor growth curves for M727 (A), M1626 (C), and M525 (E) are plotted; the average tumor growth for M727 (B), M1626 (D), and M525 (F) is summarized, with indicated P values for each tumor sample.
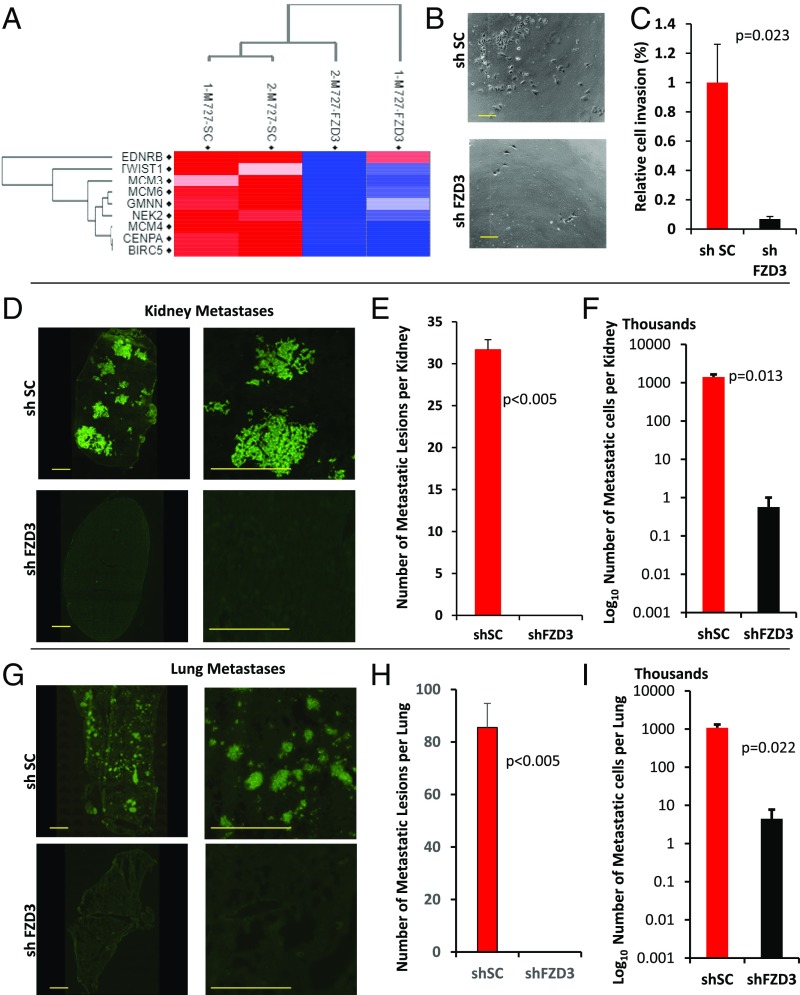
FZD3 Expression Mediates Melanoma Invasiveness and Metastasis.
Severe lethality of advanced melanomas is attributed to their aggressive invasive properties, underlying ability of these cells to colonize distant organs, which eventually fail due to a widespread disease. Further analysis of the global gene-expression profile in M727 cells capable of forming spontaneous metastases in mice revealed that FZD3 knockdown leads to the significant down-regulation of EDNRB, TWIST1, MCM3, MCM4, MCM6, BIRC5, and other proteins previously associated with melanoma invasiveness and progression signatures in human patients (28) (Fig. 5A). Therefore, we set out to determine how FZD3 down-regulation affects metastatic properties of melanoma. First, we performed an in vitro assay using BD invasion chamber wells: M727_shFZD3 and M727_shSC cells were plated into the top wells (103 cells per well) containing serum-free media and separated by the Matrigel-coated membrane from the bottom chambers containing media supplemented with 10% FBS serum. Cells were maintained in a TC incubator for 24 h and the number of invading cells reaching the bottom chamber was counted under the light microscope (Fig. 5B). The resulting data demonstrate that FZD3 down-regulation leads to the significant reduction (∼80%) of melanoma cells capable of invading the bottom chamber by migrating through the Matrigel-coated membrane (Fig. 5C).
Fig. 5.
FZD3 mediates invasive and metastatic properties of melanoma. (A) FZD3 knockdown leads to the significant down-regulation of the genes associated with melanoma progression and metastasis. Heatmap analysis of RNA-Seq data using two biological replicate experiments analyzing M727 cells transduced with shSC and shFZD3. (B and C) Matrigel cell invasion assay of M727 cells transduced with shSC or shFZD3 lentivectors. (Scale bars, 200 μm.) Error bars represent SEM of duplicate experiments with two replicates each. (D) Representative images of human melanoma Ku-80 nuclear antigen immune-fluorescence staining of kidneys and (G) lungs tissue sections from mice transplanted with indicated M727 cells. Green signal indicates positive Ku-80 signal and marks the presence of human melanoma cells throughout the mouse tissue. (Scale bars, 1,000 μm.) Average number of metastatic lesions per (E) kidney and (H) lungs (n = 5 mice). Estimated number of metastatic melanoma cells found per (F) kidney and (I) lung of transplanted mice using human Alu real-time PCR approach. Error bars represent SEM of triplicate experiments with two replicates each.
Next, we analyzed the ability of shFZD3 and shSC M727 cells to form spontaneous metastases in vivo upon their intradermal injection into the skin of NSG mice. At the end of the experiment we performed pathological examination of the mouse organ tissues that were sectioned and stained using an H&E procedure, as well as immunofluorescence staining using the human melanoma-specific nuclear antigen, Ku-80. Microscopic analysis revealed multiple metastatic lesions in the visceral organs of all animals (five of five) transplanted with shSC M727 cells, while mice transplanted with shFZD3 M727 cells were virtually free of metastases (Fig. 5 D–F and SI Appendix, Fig. S4A). Detailed examination of the kidneys at the end point of the monitoring period (141 d) uncovered metastatic lesions in 100% of control mice (five of five) on an average with 31.7 lesions per kidney sectioned (Fig. 5E). In drastic contrast, melanomas derived from FZD3 knockdown cells failed to produce detectable kidney metastases in corresponding animals examined (Fig. 5E and SI Appendix, Fig. S4A). In addition to kidneys, we also detected metastases in lungs of the control group of mice transplanted with shSC M727 cells (Fig. 5F and SI Appendix, Fig. S4B). Lungs from control mice contained multiple metastatic lesions throughout the epithelia of the lung structures in 80% of the cases (four of five), with ∼87 lesions per lung section on average. At the same time mice transplanted with shFZD3 M727 cells had lungs that were virtually free of metastases (Fig. 5H and SI Appendix, Fig. S4B).
We next investigated whether metastatic cells could still be present in the organs of mice transplanted with shFZD3 melanoma cells despite the absence of microscopically detectable metastatic lesions. To answer this important question, we utilized an established technique of human cell detection based on the human-specific Alu sequences and a real-time PCR (29). Using human Alu-specific primers and the DNA isolated from the lungs and kidneys of the mice transplanted with either shSC or shFZD3 M727 cells, we performed multiple qRT-PCR reactions. Human melanoma cells in each mouse organ were quantified based on the standard curve detection method generated by spiking a wide range of known human cell quantities into the separate PCR reactions, as previously described (29). Our results reveal that a small fraction of metastatic cells (<103 in kidneys and <7 × 103 in lungs) can be found in the visceral organs of mice transplanted with shFZD3_M727 melanomas, compared with a significantly larger fraction of metastatic cells (1.4 × 106 in kidneys and 106 in lungs) found in the organs of mice transplanted with control, shSC_M727 melanomas (Fig. 5 F and I). These findings indicate that, while FZD3 down-regulation inhibits metastatic disease, it does not completely abolish the ability of the rare tumor cells to migrate and seed into visceral locations. Nonetheless, shFZD3-transduced cell populations that are capable of distant seeding completely failed to proliferate and establish any microscopically detectable metastatic lesions in the target organs (Fig. 5 D and G).
Taken together, the above results demonstrate that FZD3 suppression leads to the significant reduction of M727 melanoma invasiveness and inhibits proliferative capacity of seeded metastatic cells in vivo.
FZD3 Expression Is Clinically Associated with Melanoma Progression and Reduced Patient Survival.
Given the critical importance of FZD3 in melanoma growth and metastasis, we evaluated clinical significance of FZD3 expression levels in melanoma patients by using gene-expression datasets for the cohorts of patients whose disease stage was individually linked to their transcriptome profile. Analysis of independent groups of patients (GSE8401, n = 83 and GSE46517, n = 104) (30) revealed that FZD3 expression is significantly elevated in metastatic lesions compared with primary tumors (P = 0.00895 and P = 0.000239, respectively) (SI Appendix, Fig. S5 A and B). We next investigated groups of melanoma patients whose transcriptome profiles were linked to their survival outcomes (GSE19234, n = 44 and GSE65904, n = 130) (31, 32). When these patients were stratified by either high or low expression levels for FZD3, significant differences (P = 0.01541 and P = 0.016, respectively) between Kaplan–Meier survival curves of the two patient groups were observed in a 40-wk study period, with a hazard ratio of high FZD3 expression group vs. low FZD3 expression group equals to 3.3 and 1.78, respectively (SI Appendix, Fig. S5 C and D).
In addition, transcriptomics and somatic copy number analysis of the TCGA database (n = 471) (https://cancergenome.nih.gov/) revealed that in skin-cutaneous melanoma and uveal melanoma, both cancer tissues related to melanocytes, showed up-regulation, amplification, or copy number gain in 28.0% of the cases. To confirm that up-regulation of FZD3 correlates with melanoma progression on the protein level, we analyzed the human protein atlas (33) to learn that FZD3 overexpression can be detected in 91,7% of the microarrays of melanoma tissue specimens with more than 50% staining (n = 12 with single replicates) (SI Appendix, Fig. S5E).
In summary, the above results, while mostly limited to the mRNA expression levels in several patient cohorts, provide indication that an increase in FZD3 expression can be associated with melanoma progression to the advanced metastatic stages and represents an adverse survival factor for melanoma patients. Future experiments that include a large number of early-stage melanoma biopsies along with the matched samples collected from the same patients that progressed to metastatic stage 3–4 phase will allow in depth assessment of the messenger and protein–tissue expression levels of FZD3 to firmly establish its role as a marker of melanoma progression and survival.
FZD3 Control of Human Melanoma Cell Proliferation Is Independent of the Canonical WNT Signaling.
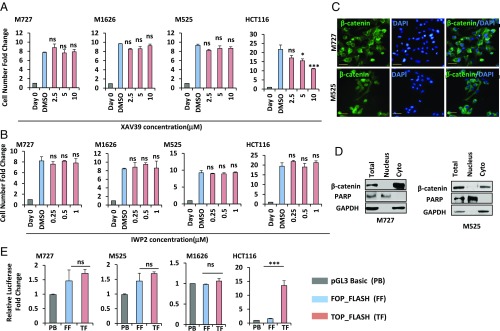
FZD receptors often signal through the canonical WNT pathway, which is stimulated by their biding to the WNT ligands, leading to the stabilization and nuclear translocation of β-catenin protein, where it acts as an important costimulatory factor initiating transcription of downstream target genes (8). Having established that multiple aggressive patient-derived melanoma cells required FZD3 activity to activate the specific transcription programs to maintain their tumorigenic properties, we wished to determine whether it was dependent on the canonical WNT signaling. In these experiments we used upstream (IWP2) and downstream (XAV939) inhibitors of the WNT pathway to measure proliferation of M727, M525, and M1626 cells, which were found to be sensitive to FZD3 down-regulation. Briefly, cells were seeded at the density of 104 cells per well in six-well plates, and their growth was assessed in the presence of XAV939 (at the concentration of 2.5, 5, and 10 μM) or IWP2 (at the concentration of 0.25, 0.5, and 1 μM). As a control for these experiments, we also included a well characterized, β-catenin–dependent, colon cancer cell line, HCT116. At the end of the incubation period, we observed that while XAV939 had significantly inhibited proliferation of HCT116 cells, it had no effect on the growth of either melanoma samples analyzed under the same conditions (Fig. 6A). Similarly to XAV939, IWP2 also failed to arrest proliferation of M727-, M525-, and M1626-derived cells (Fig. 6B), indicating that in these cases melanoma growth was not sensitive to the inhibition of the canonical WNT pathway. To understand the reason behind this phenotypic insensitivity to the canonical WNT-inhibiting molecules, we analyzed cellular distribution and localization of β-catenin in these cells during an actively growing phase. Immunohistochemical and cell fractionation approaches revealed that β-catenin was expressed in all melanoma-derived cells but was mainly localized in the cytoplasm and virtually undetectable in the nucleus of actively proliferating melanoma cells (Fig. 6 C and D). Poly(ADP-ribose) polymerase (PARP) protein was used as a control for the cytoplasmic and nuclear fractionation assays and was efficiently detected only in the nuclear fraction (Fig. 6D).
Fig. 6.
FZD3 control of human melanoma cell proliferation is independent of the canonical WNT signaling. Cell growth kinetics of indicated melanoma cells and HCT-116 colon cancer cells in the presence of potent Tankyrase inhibitor, XAV939 (A) and Porcn inhibitor, IWP2 (B) at indicated concentrations. Error bars represent SEM of duplicate experiments with two replicates each. *P < 0.05, **P < 0.005, ***P < 0.0005. (C) Immunofluorescence analysis of patient-derived melanoma cells using β-catenin–specific antibodies, following their transduction shSC or shFZD3 lentivectors. Green color indicates positive β-catenin staining. Blue color indicates nuclear staining by DAPI. (Scale bars, 100 μm.) (D) Analysis of β-catenin protein in the nuclear and cytosolic cell fractions using Western blot and indicated antibodies. (E) Analysis of β-catenin–mediated transcriptional activity using Luciferase assay following transient transfection of TOPFLASH and FOPFLASH constructs into the indicated melanoma cells. HCT-116 colon cancer cells were used as a positive control. Firefly luciferase activity was normalized to Renilla luciferase activity and pGL3 basic negative control to obtain the fold-change. Error bars represent SEM of duplicate experiments with two replicates each. *P < 0.05, **P < 0.005, ***P < 0.0005; ns, not significant.
Finally, to confirm the absence of nuclear β-catenin activity in the analyzed melanoma cells, we utilized the Super8x TOPFLASH/FOPFLASH luciferase reporter system, which contains seven T cell factor/lymphoid enhancer factor binding sites driving expression of the luciferase enzyme, whose activity was quantified using Luc colorometric assay. Super8x FOPFLASH luciferase reporter with mutated binding sites was used in the same set-up as a negative control. Introduction of these reporters into the HCT116 cells resulted in ∼14-fold induction of TOPFLASH luciferase activity normalized to FOPFLASH luciferase, reflecting an active transcriptional program regulated by nuclear β-catenin (Fig. 6E, rightmost panel). However, none of the FZD3-dependent melanoma-derived cell cultures displayed any significant induction of β-catenin–dependent transcription of Luc enzyme upon its introduction (Fig. 6E). Taken together, the above results indicate that FZD3 tumorigenic signaling is independent of the canonical WNT pathway.
Discussion
In malignant melanoma, one of the most aggressive cancers, the WNT pathway is frequently deregulated (10, 34–36). WNT signaling represents a critical determinant for disease progression and can be mediated by multiple Frizzled receptors and their corresponding ligands. Furthermore, changes in the activity of Wnt network components have been shown to drive resistance of melanoma cells to the targeted and immune therapies (37, 38). Therefore, identifying tumorigenic properties of key WNT signaling components in the pathogenesis of melanoma represents a pivotal task against this devastating disease.
In the present study, we characterized FZD3 as a critical factor underlying tumorigenic properties of aggressive human melanoma. The functional genomics analysis of FZD3 knockdown in human patient-derived cells highlighted regulation of the cell proliferation and invasiveness as the key processes positively regulated by the FZD3 receptor during malignant transformation. Using global transcriptome profiling and protein-expression analysis, we established that a tumorigenic role of FZD3 is linked to its ability to positively regulate (i) major components of the cell cycle machinery, such as cyclins and CDKs, and (ii) genes enabling aggressive metastatic properties of these cells. Up-regulation of the cyclins and CDKs is a hallmark of many melanomas with altered MAPK signaling pathway (mutated in ∼80% of melanomas). The most common genetic alteration augmenting MAPK signaling network activity in melanoma resides within the BRAF kinase, which is mutated in ∼65% of the patients (39). Mutations in this protein at the V600 amino acid position enable cell proliferation independent of the upstream regulatory signals and receptors. Significantly, in the present study we discovered that even in the presence of BRAF oncogenic mutation activity of FZD3 is still required for melanoma cell cycle progression and an induction of key proliferative proteins, such as cyclins D1, E2, and B1, as well as, CDKs 1, 2, and 4. Moreover, intradermal xenotransplantation of BRAF mutant melanoma cells with reduced FZD3 expression levels prevented ability of these cells to initiate and sustain tumor growth in vivo. This indicates that FZD3 plays a dominant role in the regulation of melanomagenesis with an altered MAPK pathway. A number of previous reports established that rewiring of the MAPK pathway represents a key node for melanoma progression (40–43). Therefore, identification of MAPK regulatory mechanisms represents a critical task in the field. In our study, enriched pathway analysis revealed that FZD3 inhibits transcriptional networks controlled by CREB5, FOXD1, and ATF3, which suppress the activity of MAPK-mediated signaling. Thus, FZD3 establishes a positive feedback mechanism that activates the MAPK signal transduction network, critical to melanoma carcinogenesis. Importantly, in our present study we also demonstrate that high FZD3 expression levels correlate with melanoma progression and reduced patient survival, making this molecule an ideal candidate for targeted therapeutic approaches in the clinic.
In addition to MAPK, protein networks controlled by E2F, MYC, AP1, and EZH2 also became deregulated in FZD3 knockdown cells and reflected pleotropic ability of FZD3 to control key transcriptional nodes vital for the tumorigenic phenotype. This was functionally confirmed in our present study, when we assayed metastatic properties of aggressive patient-derived melanoma cells. We identified a specific gene signature, consisting of the proteins previously associated with cell invasion and migration, such as TWIST1, EDNRB, and members of the MCM family (44, 45), to be significantly down-regulated following FZD3 knockdown. Thus, as a result of FZD3 inhibition, target melanoma cells had greatly reduced invasive potential and failed to form metastases in vivo when intradermally transplanted into the skin of experimental animals. In depth pathological and molecular analysis of mouse organs from the group of mice transplanted with shFZD3 cells revealed the presence of rare melanoma cells in them, which nevertheless failed to establish any proliferative clones, leading to the formation of microscopically detectable metastatic lesions. This indicates that while FZD3 down-regulation inhibits metastatic disease, it does not completely prevent initial seeding of the rare metastatic cells in the case of the M727 PDX model. Further in vivo experiments that include a large panel of patient-derived melanoma cells capable of spontaneous metastases in mice, combined with the live luciferase-based imaging approaches for extended period of time, will reveal a more specific requirement for FZD3 signaling during the multistage process of metastasis. Interestingly enough, expression of the hyperactive FZD3 mutant, Fz37A, insensitive to the inhibitory C-terminal phosphorylation, results in the up-regulation of the of the neural crest markers, such as TWIST and Slug (46). EDNRB, TWIST1, and SLUG have been previously defined as early markers that promote migration and invasion of neural crest cells, using dorsolateral pathway (47, 48). These cells eventually give rise to melanocytic precursors, which invade developing embryo skin layers and differentiate into pigment-synthesizing cells, melanocytes. Studies of the mouse brain, neural crest, and perepherial nervous system demonstrated that FZD3 functions through the noncanonical WNT pathway to play a critical role in the development of those normal tissues (13, 16, 49). Taken previously described observations and the results presented in this paper on the critical involvement of FZD3 in melanoma pathogenesis together, we further confirm the paradigm that tumors recapitulate, to a certain degree, growth and developmental programs of their original tissues by selectively augmenting activity of those genes that convey proliferation and invasion advantages.
Previous studies of the WNT signaling network demonstrated an important role of the canonical, nuclear β-catenin–driven, arm of this pathway in melanoma development, as well as its resistance to the multiple types of therapeutic approaches (24, 37, 38). However, a number of other studies using clinical and pathological analysis of human tumors demonstrated that the lack of a nuclear β-catenin was indicative of an adverse prognosis for melanoma patients, while its transcriptional activity was associated with a less-aggressive phenotype (22, 23, 50). Besides species-related variations during disease development, these seemingly contradictory results can also be explained by the differences in the genetic make-up of melanoma subtypes. Depending on the context and tumor microenvironment, β-catenin is able to induce expression of not only the positive regulators of cell proliferation but also augment differentiation and cell death pathways in melanoma as well. This points to the fact that contingent on the disease subtype substantial proportion of human melanomas can utilize receptors of the WNT signaling network that bypass activation and nuclear localization of β-catenin to positively regulate tumor development. Thus, identification of molecular mechanisms that enable β-catenin–independent tumorigenic initiation and progression of melanoma represents an important task in the field. Significantly, in our present study we identified FZD3 as a critical mediator of oncogenic signaling in melanoma subtypes that evolved in the absence of the nuclear β-catenin activity. Determining a global gene-expression pattern regulated by FZD3 uncovered its unique property to activate transcription of proliferative and invasive genes, while at the same time avoiding up-regulation of the genes that can induce differentiation or cell death. It is also notable that FZD6, the most closely evolutionary related receptor to FZD3 in the Frizzled receptor family, has been characterized as a key mediator of noncanonical WNT signaling, playing a central role in the pathogenesis of human sarcomas and breast carcinomas (19, 51) through the regulation of cell cycle components, such as CDC25 (52).
Requirement for FZD3 function to sustain malignant properties of melanoma, combined with the fact that an up-regulation of this receptor is associated with the disease progression and a reduced patient survival, uncovers FZD3 as a critical therapeutic target. Further development of FZD3 blocking agents, including inhibitory peptides and antibodies, will provide a powerful approach that can be combined with available kinase inhibitors, as well as immune-modulating agents to significantly enhance efficacy of antimelanoma treatments and increase patient survival.
Materials and Methods
Cells and Growing Conditions.
Human melanoma samples were obtained with the consent of patients at the Stanford Hospital, per protocols approved by the Institutional Review Board of the Stanford University School of Medicine, and as previously described (53, 54). Samples were deidentified before use in the study. M727 cells were grown in DMEM) (Corning) supplemented with 10% FBS (Omega Scientific) and penicillin/streptomycin at 37°, 5% CO2. M525 cells were cultured in RPMI medium 1640 (Corning) supplemented with 5% FBS. M1626 cells were maintained in RPMI medium 1640 supplemented with 10% FBS. All animal procedures were performed in strict accordance with the University of California Institutional Animal Care and Use Committee (IACUC).
FZD3 Knockdown and Full-Length FZD3 cDNA Rescue.
Permanent FZD3 down-regulation was achieved via shRNA mediated mechanism (multiple independent targets were selected) using lentiviral transduction of target cells. The target sequences of FZD3—shFZD3-2: GCAGAGAATATCACATTCCATCT and shFZD3-3: CGCTCCTATTTGTATGGAATACT—were cloned into the lentiviral vector, plKO1 (Sigma-Aldrich). Lentiviral infectious particles were generated by introducing respective plasmid DNAs into 293T cells. The full-length ORF of FZD3 (NM_017412.3) was synthesized and cloned into lentiviral pLM-CMV-H4 plasmid via NheI-BamHI cloning sites.
RNA-Seq Analysis.
RNA sequencing was performed by the University of California, Irvine Genomics High-Throughput Facility with an Illumina Hi-Seq 2500 system (Illumina) to obtain the RNA-Seq profiles. The RNA-Seq profiles were mapped to the reference Genome Reference Consortium GRCh38 using Bowtie alignment to more than 28,000 transcripts for each specimen. Statistical testing for differential expression and multiple hypothesis correction was based on read counts and performed using EdgeR in the Bioconductor toolbox, as previously described (27). We utilized controls, shSC, and FZD3 knockdowns, shFZD3, of all available specimens for the clustering analysis. Both, rows (genes) and columns (specimens) were clustered using Pearson correlation distance and average linkage of log-transformed, normalized fragments per kilobase of transcript per million mapped reads (FPKM) RNA-Seq values. PCA separated control and knockdown specimens into two clusters with about 78% data representation in the first two principal components (PC1 = 69% and PC2 = 9%). Pathway analysis was performed by Ingenuity Pathway Analysis software (Qiagen Bioinformatics) and mapped with KEGG pathway analysis (https://www.genome.jp/).
Detection of Human Metastatic Cells in the Mouse Organs.
Mouse kidney and lung sections were cut from the respective OCT blocks at 10-μm thickness. After fixation and blocking the sections were incubated with anti-human KU80 antibody (1:100) overnight at 4 °C. Following incubation with Alexa Fluor 488 goat anti rabbit IgG (H+L), 1:400, (Life Technologies) at room temperature in the dark for 1 h. Following immunostaining, immunofluorescent images were taken using a Nikon TI-E fluorescent microscope. Genomic DNA was extracted from the respective mouse organs using PureLink Genomic DNA Mini Kit (Life Technologies). Real-time PCR was performed using human Alu primers: hALU F: 5′ - CAC CTG TAA TCC CAG CAC TTT - 3′ hALU R: 5′ - CCC AGG CTG GAG TGC AGT - 3′ and a PowerUp SYBR Green Master Mix (ThermoFisher Scientific). Normalized mean ΔCT values were used for human cell quantification according to the standard curve obtained with known human cell numbers and as previously described (29).
Supplementary Material
Acknowledgments
We thank Jing Chen for laboratory assistance and management; Melanie Oakes, Jenny Wu, and the University of California, Irvine Genomics core for excellent support in RNA-Seq experiments; and Drs. Marian Waterman, Anand Ganesan, and Peter Donovan for sharing their reagents, their valuable help and discussions, as well as, the critical reading of the manuscript. A.D.B. was supported by NIH Grant R00 CA154960, American Cancer Society/Institutional Research Grant University of California, Irvine seed Grant 98-279-10, University of California, Irvine Stem Cell Research Center (SCRC) seed Grant GU12319, and University of California, Irvine Start-up Funds (to A.D.B.). F.V.F. is grateful for the support by CA154887 from the NIH National Cancer Institute, GM115293 NIH Bridges to the Doctorate, National Science Foundation, Graduate Research Fellowship Program, CRN-17-427258 by the University of California, Office of the President, Cancer Research Coordinating Committee, and the Science Alliance on Precision Medicine and Cancer Prevention by the German Federal Foreign Office, implemented by the Goethe-Institute, Washington, DC, and supported by the Federation of German Industries (BDI), Berlin, Germany.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813802116/-/DCSupplemental.
References
- 1.Liu J, Fukunaga-Kalabis M, Li L, Herlyn M. Developmental pathways activated in melanocytes and melanoma. Arch Biochem Biophys. 2014;563:13–21. doi: 10.1016/j.abb.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seberg HE, Van Otterloo E, Cornell RA. Beyond MITF: Multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma. Pigment Cell Melanoma Res. 2017;30:454–466. doi: 10.1111/pcmr.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer. 2016;16:345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- 4.White RM, Zon LI. Melanocytes in development, regeneration, and cancer. Cell Stem Cell. 2008;3:242–252. doi: 10.1016/j.stem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Dupin E, Le Douarin NM. Development of melanocyte precursors from the vertebrate neural crest. Oncogene. 2003;22:3016–3023. doi: 10.1038/sj.onc.1206460. [DOI] [PubMed] [Google Scholar]
- 6.Bronner ME, LeDouarin NM. Development and evolution of the neural crest: An overview. Dev Biol. 2012;366:2–9. doi: 10.1016/j.ydbio.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijksterhuis JP, et al. Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. J Biol Chem. 2015;290:6789–6798. doi: 10.1074/jbc.M114.612648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 9.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 10.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reya T, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 12.Zhang B, Tran U, Wessely O. Expression of Wnt signaling components during Xenopus pronephros development. PLoS One. 2011;6:e26533. doi: 10.1371/journal.pone.0026533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deardorff MA, Tan C, Saint-Jeannet J-P, Klein PS. A role for frizzled 3 in neural crest development. Development. 2001;128:3655–3663. doi: 10.1242/dev.128.19.3655. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Saint-Jeannet JP, Klein PS. Wnt-frizzled signaling in neural crest formation. Trends Neurosci. 2003;26:40–45. doi: 10.1016/s0166-2236(02)00011-5. [DOI] [PubMed] [Google Scholar]
- 15.Hua ZL, Smallwood PM, Nathans J. Frizzled3 controls axonal development in distinct populations of cranial and spinal motor neurons. eLife. 2013;2:e01482. doi: 10.7554/eLife.01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hua ZL, Jeon S, Caterina MJ, Nathans J. Frizzled3 is required for the development of multiple axon tracts in the mouse central nervous system. Proc Natl Acad Sci USA. 2014;111:E3005–E3014. doi: 10.1073/pnas.1406399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang C-H, Tsai R-K, Tsai M-H, Lin Y-H, Hirobe T. The roles of frizzled-3 and Wnt3a on melanocyte development: In vitro studies on neural crest cells and melanocyte precursor cell lines. J Dermatol Sci. 2014;75:100–108. doi: 10.1016/j.jdermsci.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Siemers NO, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One. 2017;12:e0179726. doi: 10.1371/journal.pone.0179726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corda G, Sala A. Non-canonical WNT/PCP signalling in cancer: Fzd6 takes centre stage. Oncogenesis. 2017;6:e364. doi: 10.1038/oncsis.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee EHL, et al. Disruption of the non-canonical WNT pathway in lung squamous cell carcinoma. Clin Med Oncol. 2008;2008:169–179. [PMC free article] [PubMed] [Google Scholar]
- 21.Many AM, Brown AMC. Both canonical and non-canonical Wnt signaling independently promote stem cell growth in mammospheres. PLoS One. 2014;9:e101800. doi: 10.1371/journal.pone.0101800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien AJ, et al. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci USA. 2009;106:1193–1198. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arozarena I, et al. In melanoma, beta-catenin is a suppressor of invasion. Oncogene. 2011;30:4531–4543. doi: 10.1038/onc.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damsky WE, et al. β-Catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–754. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grossmann AH, et al. The small GTPase ARF6 stimulates β-catenin transcriptional activity during WNT5A-mediated melanoma invasion and metastasis. Sci Signal. 2013;6:ra14. doi: 10.1126/scisignal.2003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown K, et al. WNT/β-catenin signaling regulates mitochondrial activity to alter the oncogenic potential of melanoma in a PTEN-dependent manner. Oncogene. 2017;36:3119–3136. doi: 10.1038/onc.2016.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zecena H, et al. Systems biology analysis of mitogen activated protein kinase inhibitor resistance in malignant melanoma. BMC Syst Biol. 2018;12:33. doi: 10.1186/s12918-018-0554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovacs D, et al. The role of Wnt/β-catenin signaling pathway in melanoma epithelial-to-mesenchymal-like switching: Evidences from patients-derived cell lines. Oncotarget. 2016;7:43295–43314. doi: 10.18632/oncotarget.9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preston Campbell J, et al. TRIzol and Alu qPCR-based quantification of metastatic seeding within the skeleton. Sci Rep. 2015;5:12635. doi: 10.1038/srep12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu L, et al. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res. 2008;6:760–769. doi: 10.1158/1541-7786.MCR-07-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cirenajwis H, et al. Molecular stratification of metastatic melanoma using gene expression profiling: Prediction of survival outcome and benefit from molecular targeted therapy. Oncotarget. 2015;6:12297–12309. doi: 10.18632/oncotarget.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogunovic D, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci USA. 2009;106:20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 34.Guan J, Gupta R, Filipp FV. Cancer systems biology of TCGA SKCM: Efficient detection of genomic drivers in melanoma. Sci Rep. 2015;5:7857. doi: 10.1038/srep07857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mauerer A, et al. Identification of new genes associated with melanoma. Exp Dermatol. 2011;20:502–507. doi: 10.1111/j.1600-0625.2011.01254.x. [DOI] [PubMed] [Google Scholar]
- 36.Paluncic J, et al. Roads to melanoma: Key pathways and emerging players in melanoma progression and oncogenic signaling. Biochim Biophys Acta. 2016;1863:770–784. doi: 10.1016/j.bbamcr.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 37.Anastas JN, et al. WNT5A enhances resistance of melanoma cells to targeted BRAF inhibitors. J Clin Invest. 2014;124:2877–2890. doi: 10.1172/JCI70156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 39.Shain AH, et al. The genetic evolution of melanoma from precursor lesions. N Engl J Med. 2015;373:1926–1936. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
- 40.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fecher LA, Amaravadi RK, Flaherty KT. The MAPK pathway in melanoma. Curr Opin Oncol. 2008;20:183–189. doi: 10.1097/CCO.0b013e3282f5271c. [DOI] [PubMed] [Google Scholar]
- 42.Zhang G, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest. 2016;126:1834–1856. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez-Bergami P, et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11:447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bittner M, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 45.Winnepenninckx V, et al. Melanoma Group of the European Organization for Research and Treatment of Cancer Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 46.Yanfeng WA, Tan C, Fagan RJ, Klein PS. Phosphorylation of frizzled-3. J Biol Chem. 2006;281:11603–11609. doi: 10.1074/jbc.M600713200. [DOI] [PubMed] [Google Scholar]
- 47.Harris ML, Hall R, Erickson CA. Directing pathfinding along the dorsolateral path—The role of EDNRB2 and EphB2 in overcoming inhibition. Development. 2008;135:4113–4122. doi: 10.1242/dev.023119. [DOI] [PubMed] [Google Scholar]
- 48.Pla P, et al. Ednrb2 orients cell migration towards the dorsolateral neural crest pathway and promotes melanocyte differentiation. Pigment Cell Res. 2005;18:181–187. doi: 10.1111/j.1600-0749.2005.00230.x. [DOI] [PubMed] [Google Scholar]
- 49.Yanfeng W, Saint-Jeannet JP, Klein PS. Wnt-frizzled signaling in the induction and differentiation of the neural crest. Bioessays. 2003;25:317–325. doi: 10.1002/bies.10255. [DOI] [PubMed] [Google Scholar]
- 50.Kageshita T, et al. Loss of beta-catenin expression associated with disease progression in malignant melanoma. Br J Dermatol. 2001;145:210–216. doi: 10.1046/j.1365-2133.2001.04336.x. [DOI] [PubMed] [Google Scholar]
- 51.Kilander MB, Dahlström J, Schulte G. Assessment of frizzled 6 membrane mobility by FRAP supports G protein coupling and reveals WNT-Frizzled selectivity. Cell Signal. 2014;26:1943–1949. doi: 10.1016/j.cellsig.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 52.Vijayakumar S, et al. High-frequency canonical Wnt activation in multiple sarcoma subtypes drives proliferation through a TCF/β-catenin target gene, CDC25A. Cancer Cell. 2011;19:601–612. doi: 10.1016/j.ccr.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boiko AD, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ngo M, et al. Antibody therapy targeting CD47 and CD271 effectively suppresses melanoma metastasis in patient-derived xenografts. Cell Rep. 2016;16:1701–1716. doi: 10.1016/j.celrep.2016.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.