Significance
Oncogenic mutations in the BRAF gene are found in ∼50% of melanomas and drive melanoma growth. Thus, BRAF kinase inhibitors (BRAFi), such as vemurafenib and dabrafenib, have been developed and used for the treatment of BRAF-mutant metastatic melanoma in clinic. However, the clinical benefits of BRAFi are temporary and short-lived due to the emergence of drug resistance. Although several mechanisms of acquired BRAFi resistance have been identified, the basis for acquired resistance remains unknown in over 40% of melanomas. Identifying a new mechanism of acquired resistance to BRAFi may provide new opportunities to effectively treat BRAF-mutant melanoma. In this study, we have identified Block of Proliferation 1 as a new factor the loss of which results in resistance to BRAFi.
Keywords: BOP1, drug resistance, melanoma, BRAF inhibitor, RNAi
Abstract
Acquired resistance to BRAF kinase inhibitors (BRAFi) is the primary cause for their limited clinical benefit. Although several mechanisms of acquired BRAFi resistance have been identified, the basis for acquired resistance remains unknown in over 40% of melanomas. We performed a large-scale short-hairpin RNA screen, targeting 363 epigenetic regulators and identified Block of Proliferation 1 (BOP1) as a factor the loss of which results in resistance to BRAFi both in cell culture and in mice. BOP1 knockdown promoted down-regulation of the MAPK phosphatases DUSP4 and DUSP6 via a transcription-based mechanism, leading to increased MAPK signaling and BRAFi resistance. Finally, analysis of matched patient-derived BRAFi or BRAFi+MEKi pre- and progressed melanoma samples revealed reduced BOP1 protein expression in progressed samples. Collectively, our results demonstrate that loss of BOP1 and the resulting activation of the MAPK pathway is a clinically relevant mechanism for acquired resistance to BRAFi in melanoma.
Melanoma is an aggressive cancer that frequently metastasizes to various distal organs (1, 2). Although treatment of melanoma at early stages is generally effective, even with several improvements in current therapeutic approaches the median survival of patients with metastatic melanoma is only 4.5–12.5 mo (1, 3). Genomic sequencing of melanoma has identified oncogenic mutations in the BRAF gene in over 50% of tumors (4, 5). Acquiring oncogenic mutations in the BRAF gene causes constitutive activation of the BRAF → MEK → ERK pathway and is necessary for melanoma growth and progression (4, 6). These findings have led to the development and approval of several BRAF and MEK kinase inhibitors by the Food and Drug Administration for treating unresectable metastatic melanoma (7, 8). However, although melanoma patients initially respond robustly to BRAF kinase targeted therapy, they show acquired resistance within a matter of a few months, resulting in disease progression. Due to the high prevalence of this problem, intensive efforts have focused on identifying the causes of resistance to BRAF and MEK kinase inhibitors, and several mechanisms have been identified (9, 10). These mechanisms can be broadly categorized as either dependent or independent of the MAPK pathway (11, 12).
Block of proliferation 1 (BOP1) contains WD40 repeats and has been shown to be involved in 28S and 5.8S ribosomal RNA (rRNA) processing and 60S ribosome biogenesis (13). BOP1 is also part of the PES1-BOP1-WDR12 (PeBoW) complex, and inactivation of subunits from this complex inhibits rRNA processing and ribosome biogenesis (13, 14). Here, using a large-scale short-hairpin RNA (shRNA) screen, we have identified that loss of BOP1 causes resistance to BRAF kinase inhibitor (BRAFi). We show that loss of BOP1 results in reduced expression of Dual specificity phosphatase 4 (DUSP4) and Dual specificity phosphatase 6 (DUSP6), which results in activation of the MAP kinase pathway, resulting in resistance to BRAFi. Furthermore, analysis of matched patient-derived BRAFi or BRAFi+MEKi pre- and progressed melanoma samples revealed reduced BOP1 protein expression in progressed samples.
Results
A Large-Scale Epigenome-Wide Human shRNA Screen Identifies Candidates That Confer Resistance to BRAF Inhibitors.
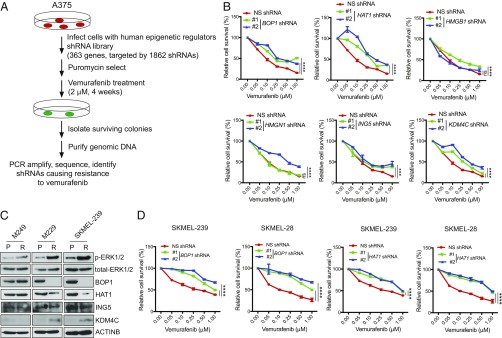
Epigenetic alterations are shown to play an important role in the regulation of cancer cell growth and their response to targeted therapies (15–17). Therefore, to determine the role of epigenetic regulators in conferring resistance to BRAFi, we performed a large-scale, unbiased, epigenome-wide shRNA screen by targeting 363 known and predicted epigenetic regulators with 1862 shRNAs (SI Appendix, Table S1). For this screen, we infected the BRAF-mutant melanoma line A375 with the epigenetic regulator shRNA library at a multiplicity of infection (MOI) of 0.2 and selected with puromycin to enrich for shRNA-expressing cells. After selection, shRNA-containing cells were treated with 2 μM of vemurafenib for 4 wk. Surviving colonies were then harvested, and genomic DNA was isolated and sequenced for shRNA identification (Fig. 1A). From this analysis, we identified shRNAs corresponding to six different epigenetic regulators in vemurafenib-resistant A375 colonies (SI Appendix, Table S2).
Fig. 1.
Large-scale shRNA screen to identify epigenetic regulators of vemurafenib resistance in BRAF-mutant melanoma. (A) Schematic for large-scale epigenetic regulator shRNA screen. (B) Relative cell survival (%) as measured by MTT assay of A375 cells expressing shRNAs targeting the indicated genes or NS control shRNAs that were treated with the indicated concentrations of vemurafenib for 3 d. (C) Immunoblotting for the indicated proteins in three pairs of BRAFi-sensitive (P) or resistant (R) BRAF-mutant melanoma cell lines. (D) Relative cell survival (%) as measured by MTT assay of SKMEL-239 or SKMEL-28 cells expressing shRNAs targeting the indicated genes or NS control shRNAs that were treated with indicated concentrations of vemurafenib for 3 d. Data are presented as the mean ± SEM. ns, not significant. ***P < 0.001 and ****P < 0.0001.
Next, we individually knocked down expression of all six genes identified from our primary screen in A375 cells (SI Appendix, Fig. S1 A and B) and measured sensitivity to vemurafenib using the MTT assay. Knockdown of four (BOP1, HAT1, ING5, and KDM4C) of six candidates conferred resistance to vemurafenib, as confirmed by two sequence-independent shRNAs (Fig. 1B). Based on these results, we analyzed expression of all four validated candidates in three pairs of BRAF-sensitive parental and BRAFi-resistant melanoma cell lines (SI Appendix, Fig. S1 C–E). We found that two of three BRAFi-resistant lines showed significant reduction in BOP1 and HAT1 protein levels (Fig. 1C). Interestingly, down-regulation of BOP1 and HAT1 was observed only in vemurafenib-resistant lines that also showed substantially increased MAP kinase signaling, as indicated by higher p-ERK1/2 levels in BRAFi-resistant parental cells compared with those in BRAFi-sensitive cells (Fig. 1C).
We then analyzed the role of BOP1 and HAT1 in mediating vemurafenib resistance in additional BRAF-mutant melanoma cells. For these experiments, we knocked down BOP1 and HAT1 expression in BRAF-mutant SKMEL-28 and SKMEL-239 cells (SI Appendix, Fig. S1 F and G) and measured vemurafenib sensitivity. Similar to A375 cells, SKMEL-28 and SKMEL-239 cells also developed vemurafenib resistance (Fig. 1D).
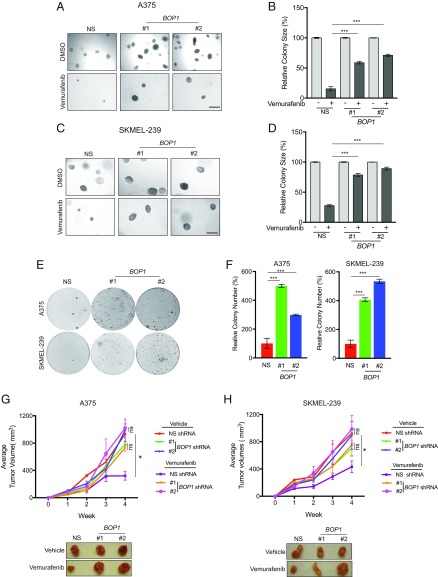
To further confirm the role of BOP1 and HAT1 in vemurafenib resistance, we performed soft-agar assays to measure anchorage-independent growth. Consistent with MTT assay results, vemurafenib-treated melanoma cells expressing BOP1 and HAT1 shRNAs showed significantly larger colonies compared with cells containing nonspecific (NS) shRNAs (Fig. 2 A–D and SI Appendix, Fig. S2 A–D). Next, to better emulate the long-term treatment scenario in the clinic, we performed clonogenic assays in shRNA knockdown cells in the presence of vemurafenib. Consistent with our previous data, we found that BOP1 or HAT1 knockdown in melanoma cells also conferred vemurafenib resistance in clonogenic assays (Fig. 2 E and F and SI Appendix, Fig. S2 E and F). However, we noted that melanoma cells expressing BOP1 shRNA formed significantly more colonies than cells expressing HAT1 shRNA. Because all phenotypes associated with vemurafenib resistance were more potent in BOP1 knockdown cells than in HAT1 knockdown cells, we focused on BOP1 for subsequent detailed studies.
Fig. 2.
Loss of BOP1 confers resistance to BRAF kinase inhibitor. (A) A375 cells expressing NS or BOP1 shRNAs were treated with vemurafenib (1 μM) or DMSO control, and anchorage-independent growth was measured by soft-agar assay. Representative soft-agar colony images are shown. (Scale bar, 500 μM.) (B) Relative colony size (%) for the data in A. (C) SKMEL-239 cells expressing NS or BOP1 shRNAs were treated with vemurafenib (1 μM) or DMSO control, and anchorage-independent growth was measured by soft-agar assay. Representative soft-agar colony images are shown. (Scale bar, 500 μM.) (D) Relative colony size (%) for the data in C. (E) A375 and SKMEL-239 cells expressing NS or BOP1 shRNAs were treated with vemurafenib (2 μM) or DMSO control and analyzed by clonogenic assay. Representative clonogenic plates for both cell lines are shown. (F) Relative colony number (%) for the data in E. (G and H) A total of 1 × 107 A375 (G) or SKMEL-239 (H) cells expressing NS or BOP1 shRNAs were injected s.c. into the flanks of athymic nude mice (n = 5), and animals were treated with vemurafenib (30 mg/kg) or vehicle control via oral gavage three times per week, starting 3 d after melanoma cell injection. (Top) Average tumor volumes (n = 5) from NS or BOP1 shRNA-expressing melanoma cells are shown. (Bottom) Representative A375 and SKMEL-239 tumors expressing NS or BOP1 shRNAs and treated with vemurafenib or vehicle control. Data are presented as the mean ± SEM. ns, not significant. *P < 0.05 and ***P < 0.001.
To further elucidate the role of BOP1 in BRAFi resistance, we next determined whether BOP1 knockdown can also confer resistance to dabrafenib, a reversible ATP-competitive kinase inhibitor that inhibits BRAFV600E (18). Notably, we found that similar to vemurafenib, BOP1 knockdown in melanoma cells promoted dabrafenib resistance (SI Appendix, Fig. S3). Based on our in vitro data, we then tested the effect of BOP1 knockdown on development of vemurafenib resistance in mice. To this end, we injected athymic nude mice s.c. with BRAF-mutant A375 or SKMEL-239 melanoma cells expressing either BOP1 or NS shRNAs. After injection, mice were treated with vemurafenib, and tumor progression was monitored. We found that BOP1 shRNA-expressing melanoma cells displayed significantly greater resistance to vemurafenib compared with NS shRNA-expressing cells (Fig. 2 G and H). Collectively, these data demonstrated that BOP1 loss conferred acquired resistance to BRAFi, both in cell culture and in mice.
BOP1-Mediated BRAF Inhibitor Resistance Does Not Rely upon Its Ability to Modulate rRNA or Ribosome Biogenesis.
Previous studies have shown that BOP1 plays an important role in the formation of mature 28S and 5.8S rRNAs and in biogenesis of the 60S ribosomal subunit (13). BOP1 is also part of the PES1-BOP1-WDR12 (PeBoW) complex, and inactivation of subunits from this complex inhibits rRNA processing and ribosome biogenesis (13, 14). Therefore, we tested whether knockdown of other components of the PeBoW complex can promote vemurafenib resistance, similar to BOP1 knockdown. Using shRNA, we knocked down the expression of WDR12 and PES1 in A375 cells; however, neither knockdown conferred significant BRAFi resistance (SI Appendix, Fig. S4).
Ribosome biogenesis defects cause a group of diseases collectively referred to as ribosomopathies, the characteristic features of which are well documented (19). Therefore, we assessed BRAFi-resistant melanoma cells and BRAF-mutant BOP1 knockdown melanoma cells for ribosomopathy-like phenotypes. Specifically, we measured p53 protein expression, a major marker of ribosomopathies (19, 20). No change in p53 expression was detected in either vemurafenib-resistant cells or BRAF-mutant melanoma cells containing BOP1 shRNAs, compared with parental or NS shRNA-expressing melanoma cells, respectively (SI Appendix, Fig. S5 A and B). To further rule out a role for rRNA biogenesis defects in BRAFi resistance or in mediating the effects of BOP1 knockdown, we measured rRNA biogenesis using tritiated uridine-based metabolic labeling in vemurafenib-resistant and BRAF-mutant melanoma cells with BOP1 knockdown. We did not, however, detect any reproducible changes in rRNA biogenesis in either cell line (SI Appendix, Fig. S5C). Finally, we measured expression of a panel of genes known to be altered in ribosomopathies in BRAFi-resistant vs. BRAFi-sensitive cells and found that a large majority remained unchanged (SI Appendix, Fig. S5 D and E). Similarly, levels of most genes were unaltered in A375 cells after shRNA-mediated BOP1 knockdown (SI Appendix, Fig. S5F). Expression of two genes, RPL11 and RSP24, was decreased both in BRAFi-resistant melanoma cells and in A375 cells expressing BOP1 shRNAs. However, shRNA-mediated gene knockdowns of RPL11 or RSP24 were unable to confer resistance to vemurafenib (SI Appendix, Fig. S6). Collectively, these results indicated that rRNA processing and ribosome biogenesis defects did not play a role in BRAFi resistance, and loss of BOP1 likely conferred BRAFi resistance via a mechanism that was independent of its ability to regulate rRNA and ribosome biogenesis.
Loss of BOP1 Causes Reduced DUSP4 and DUSP6 Expression, Which Results in Increased MAP Kinase Signaling.
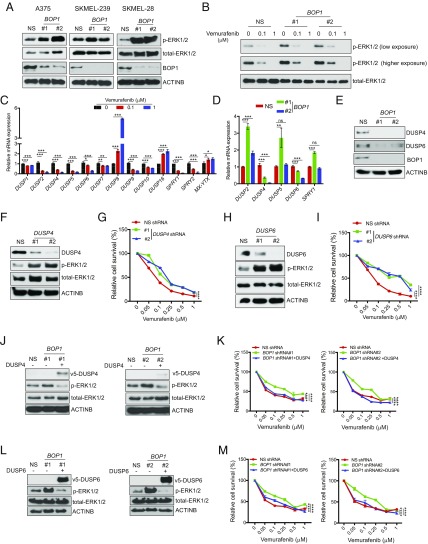
Previous studies have indicated that the majority of BRAFi-acquired resistance results from activation of the MAPK pathway by various mechanisms (11, 12, 21, 22). Therefore, we hypothesized that BOP1 knockdown might modulate this pathway. Interestingly, we found that BRAF-mutant melanoma cells expressing BOP1 shRNAs showed substantially higher p-ERK1/2 levels compared with NS shRNA (Fig. 3A). Furthermore, higher p-ERK1/2 levels in vemurafenib-treated melanoma cells expressing BOP1 shRNAs compared with those with NS shRNA indicated that vemurafenib was less effective in blocking MAP kinase signaling (Fig. 3B and SI Appendix, Fig. S7A). These results indicated that BOP1 loss enhanced MAPK pathway activation, in turn conferring BRAFi resistance.
Fig. 3.
BOP1 loss inhibits DUSP4 and DUSP6 expression, resulting in MAPK pathway activation. (A) Levels of indicated proteins measured by immunoblotting in melanoma lines expressing NS or BOP1 shRNAs. (B) Levels of p-ERK1/2 and total-ERK1/2 measured by immunoblotting in A375 cells expressing NS or BOP1 shRNAs and treated with vemurafenib (0.1 or 1.0 μM) or DMSO for 3 h. (C) Levels of indicated phosphatases measured by RT-qPCR in A375 cells treated with vemurafenib (0.1 or 1.0 μM) or DMSO for 24 h. Relative mRNA expression in vemurafenib-treated relative to DMSO-treated cells. (D) Relative mRNA expression of indicated phosphatases in BOP1 shRNA-expressing A375 cells relative to NS shRNA-expressing A375 cells. (E) Levels of indicated proteins measured by immunoblotting in A375 cells expressing NS or BOP1 shRNAs. (F) Levels of indicated proteins measured by immunoblotting in A375 cells expressing NS or DUSP4 shRNAs. (G) Relative cell survival (%) measured by MTT assay in A375 cells expressing NS or DUSP4 shRNAs, treated with indicated concentrations of vemurafenib for 3 d. (H) Levels of indicated proteins measured by immunoblotting in A375 cells expressing NS or DUSP6 shRNAs. (I) Relative cell survival (%) measured by MTT assay in A375 cells expressing NS or DUSP6 shRNAs, treated with indicated concentrations of vemurafenib for 3 d. (J) Levels of indicated proteins measured by immunoblotting in A375 cells expressing NS or BOP1 shRNAs alone or with DUSP4-ORF. (K) Relative cell survival (%) measured by MTT assay in A375 cells expressing NS or BOP1 shRNAs alone or with DUSP4-ORF, treated with indicated concentrations of vemurafenib for 3 d. (L) Levels of indicated proteins measured by immunoblotting in A375 cells expressing NS or BOP1 shRNAs alone or with DUSP6-ORF. (M) Relative cell survival (%) measured by MTT assay in A375 cells expressing NS or BOP1 shRNAs alone or with DUSP6-ORF, treated with indicated concentrations of vemurafenib for 3 d. Data are presented as the mean ± SEM. ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Based on this finding, we next investigated the mechanism by which BOP1 knockdown-mediated MAPK up-regulation. Numerous protein phosphatases can regulate the MAPK pathway by dephosphorylating either MEK1/2 or ERK1/2 (23, 24). Therefore, we assessed the role of these phosphatases in regulating BOP1 knockdown-mediated MAPK pathway activation and development of BRAFi resistance. We first measured expression of 13 MAPK phosphatases after vemurafenib treatment in BRAF-mutant melanoma lines (A375 and SKMEL-28). We found that expression of many phosphatases, including DUSP2, DUSP4, DUSP5, DUSP6, and DUSP10, as well as SPRY1 and SPRY2, were down-regulated after vemurafenib treatment (Fig. 3C and SI Appendix, Fig. S7B), whereas expression of PP2A, PP2C, and several tyrosine phosphatases (DUSP3, STEP, HePTP, and PTP-SL) remained unchanged (SI Appendix, Fig. S7 C and D).
We then measured expression of BRAF pathway-regulated DUSPs and SPRY phosphatases in vemurafenib-sensitive and -resistant melanoma cell line pairs, as well as in melanoma lines expressing BOP1 or NS shRNAs. We found decreased DUSP4 and DUSP6 expression in BRAFi-resistant cells compared with parental cell lines (SI Appendix, Fig. S7 E and F). Furthermore, expression of both DUSP4 and DUSP6 was also down-regulated in melanoma cells expressing BOP1 shRNAs compared with NS shRNA-expressing cells (Fig. 3 D and E and SI Appendix, Fig. S7 G and H).
Analysis of the promoter sequences of DUSP4 and DUSP6 genes revealed CpG islands in both loci (SI Appendix, Fig. S8A), which suggested that down-regulation of these genes in BOP1 knockdown cells occurred via a transcription-based mechanism. To test this possibility, we treated BOP1 knockdown cells with the DNA methyltransferase inhibitor 5Aza-2deoxy-cytidine (5Aza2dC), and the histone deacetylase inhibitor Tricostatin A (TSA), which restored expression of both DUSP4 and DUSP6 (SI Appendix, Fig. S8 B and C). Consistently, 5Aza2dC and TSA treatment restored DUSP4 and DUSP6 protein levels in BOP1 knockdown cells (SI Appendix, Fig. S8 D and E). Our results showed that BOP1 loss promoted DNA methylation-based epigenetic silencing of DUSP4 and DUSP6 in melanoma cells.
We then investigated the role of DUSP4 and DUSP6 down-regulation in BRAFi resistance. Consistent with previous results, we found that DUSP4 and DUSP6 knockdown in melanoma cell lines led to increased ERK1/2 phosphorylation (Fig. 3 F and H) and vemurafenib resistance (Fig. 3 G and I). Furthermore, ectopic expression of DUSP4 or DUSP6 in cells expressing BOP1 shRNAs restored sensitivity to vemurafenib (Fig. 3 J–M).
BOP1 Protein Is Down-Regulated in Patient-Derived Melanoma Samples That Are Progressed After BRAF Inhibitor Treatment.
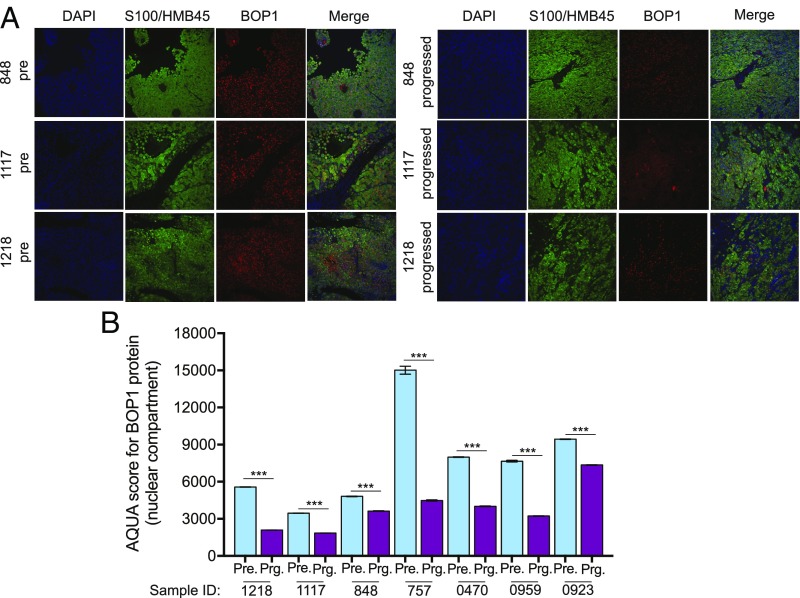
Next, we determined if the mechanism identified and validated in cell culture and mice is also clinically relevant. Toward this end, we decided to validate our findings in the clinical setting and confirm the significance of BOP1 as a mediator of BRAFi resistance. To do so, we measured BOP1 protein expression in 11 pairs of matched pre- and post-BRAFi or BRAFi+MEKi-treated melanoma samples using automated quantitative analysis (AQUA)-based immunofluorescence analysis (25) (SI Appendix, Table S3). AQUA analysis revealed that 7 of 11 cases that progressed on BRAFi or BRAFi+MEKi had lower BOP1 protein expression compared with pretreatment samples (Fig. 4). These results reveal the potentially important role of BOP1 in a clinical setting. However, we recognize the need for analyzing a large cohort of paired BRAFi-resistant samples to determine the exact percentage of cases in which BOP1 loss correlates with BRAFi resistance.
Fig. 4.
BOP1 is down-regulated in patient samples that progress on BRAF pathway-targeting therapies. (A) Representative AQUA of immunofluorescence and merge images of the indicated matched pre- and progressed melanoma samples treated with BRAF kinase inhibitors and stained for DAPI, S100/HMB45, and BOP1. (B) Average AQUA scores for pre- and progressed melanoma samples treated with BRAF kinase inhibitors. Data are presented as the mean ± SEM. ***P < 0.001.
Discussion
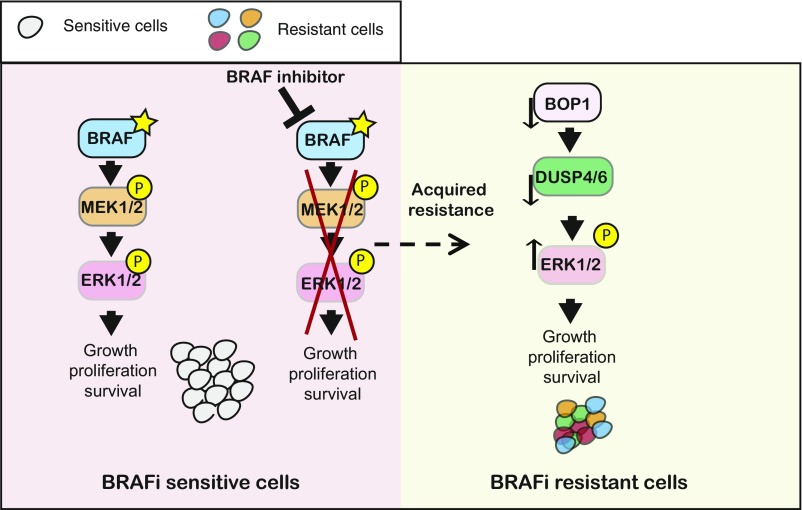
Acquired resistance to targeted therapeutic agents, such as BRAFi, is the major hurdle in the success of targeted therapies and limits the benefit of these agents. One of the approaches to overcome this limitation is to understand the mechanism that causes acquired resistance. These studies can provide important insight into the mechanisms of acquired resistance that can then be used to revise the therapeutic regimen or develop combination therapies that may forestall emergence of drug resistance. In this study, we performed a large-scale epigenome-wide human shRNA screen and identified that the loss of a protein BOP1, which was previously implicated in the regulation of rRNA biogenesis and ribosome biogenesis, conferred acquired resistance to BRAFi. Our results are summarized in Fig. 5 and described below.
Fig. 5.
Proposed model showing the role of BOP1 in vemurafenib resistance. The model indicates that loss of BOP1 results in down-regulation of DUSP4 and DUSP6 that in turn results in activation of the MAPK kinase pathway and BRAFi resistance.
BOP1 in Acquired Resistance to BRAF Kinase Inhibitors.
Due to the dependence of BRAF-mutant melanoma cells on oncogenic BRAF activity, clinical translation of BRAF and MEK kinase inhibitors for treating metastatic melanoma patients was considered a significant advance in the field of precision medicine. However, upon treatment BRAF-mutant melanoma rapidly evolved to acquire resistance to BRAF and MEK kinase inhibitors that rendered the use of these inhibitors ineffective for treating metastatic melanoma patients (9–12). Several genetic and epigenetic mechanisms that modulate BRAF inhibitor responses have been identified, which indicate the complex underlying mechanisms that cause acquired drug resistance (17, 26–28).
We have identified BOP1 using an unbiased epigenome-wide large-scale shRNA screen. Since epigenetic factors play an important role in almost all aspects of cancer biology and therapy (29, 30), our screen targeted genes that encode for known or predicted epigenetic regulators in human cells and assessed their role in conferring resistance to BRAFi. We showed that BOP1 loss was able to confer resistance to BRAFi both in cell culture and in mice. Additionally, because many paired melanoma patient samples that progress on BRAFi treatment showed BOP1 down-regulation, our results also have direct clinical implications. However, a larger cohort of matched patient melanoma samples should be analyzed to determine the percentage of melanoma in which BOP1 down-regulation contributes to BRAFi resistance. Furthermore, although we did not follow up HAT1 in detail due to HAT loss-associated phenotypes being less robust compared with BOP1 in regard to acquired resistance to BRAFi. However, future studies using mouse models and BRAFi-treated patient samples may fully reveal the importance of HAT1 in vivo and in clinical settings in conferring resistance to BRAFi.
Role of DUSP4 and DUSP6 in Conferring Resistance to BRAFi in Melanoma Downstream of BOP1.
As we mentioned above, the alterations that result in acquired BRAFi resistance can be broadly categorized to function either in a MAPK-dependent or in a MAPK-independent manner (11, 12). For example, a large number of genetic events have been identified in a study that either at the level of BRAF itself or downstream of BRAF restore the activity of the MAPK pathway (11, 12). This study analyzed BRAF inhibitor resistance mechanisms using 132 samples in a multicenter meta-analysis and was able to identify resistance mechanisms in 58% of the cases, including NRAS or KRAS mutations (20%), BRAF splice variants (16%), BRAF(V600E/K) amplifications (13%), MEK1/2 mutations (7%), and nonmitogen-activated protein kinase pathway alterations (11%) (11).
DUSP4 and DUSP6 are shown to negatively regulate the MAPK pathway (31, 32). Previous studies have identified the role of the dual-specificity phosphatases, such as DUSP6, in causing resistance to BRAFi (33). We found that BOP1 regulates DUSP4 and DUSP6 expression by a transcription-based mechanism. BOP1 knockdown cells show significantly lower expression of DUSP4 and DUSP6 and consequentially an increased MAPK pathway. Thus, our studies provide a nongenetic mechanism of DUSP4 and DUSP6 regulation and show that loss of BOP1 and the resulting activation of the MAPK pathway result in acquired resistance to BRAFi in melanoma that occurs independently of the canonical functions of BOP1.
Materials and Methods
shRNA Interference Screen.
A375 cells were transduced with eight lentiviral shRNA pools containing 1,862 shRNAs targeting 363 known or predicted epigenetic regulatory genes and including NS control shRNA. These shRNAs were administered at an MOI of 0.2 to prevent superinfection and to ensure that each cell received no more than one shRNA. Transduced A375 cells were selected with puromycin (0.5 μg/mL) for 7 d to enrich A375 shRNA-expressing cells, and after puromycin selection, cells were grown in vemurafenib (2 μM). Media was changed every 3 d, and cells were grown for 4 wk. Surviving colonies were collected, and genomic DNA was isolated. Integrated shRNAs were PCR-amplified using primers specific to the shRNA vector (pLKO.1) and cloned using the pGEM-T Easy Kit (Promega). Cloned shRNAs were amplified by colony PCR, and the products were sequenced with SP6 primers to identify candidate shRNAs. All primer sequences are listed in SI Appendix, Table S4, and the epigenetic regulator library was obtained from the University of Massachusetts shRNA facility.
Cell Culture and Plasmids.
SKMEL-28 and A375 cells were purchased from the American Type Culture Collection and grown as recommended in Dulbecco’s Modified Eagle Medium containing 10% FBS at 37 °C in 5% CO2. The M249 parent and BRAFi-resistant lines, the SKMEL-239 parent and BRAFi-resistant lines, and the M229 parent and BRAFi-resistant lines were gifts from Roger Lo, University of California, Los Angeles, and Neil Rosen, Memorial Sloan Kettering Cancer Center, New York. V5-tagged DUSP4 and DUSP6 ORF lentivirus constructs cloned in the pLX304-Blast-V5 plasmid were purchased from Dharmacon and are listed in SI Appendix, Table S4.
shRNA and Lentivirus Preparation.
pLKO.1 lentiviral vector-based shRNAs targeting specific candidate genes and NS shRNA controls were obtained from OpenBiosystems (Dharmacon); shRNA information is provided in SI Appendix, Table S4. Lentivirus particles were prepared by transfecting 293T cells with either gene-specific shRNA or NS shRNA plasmids along with the lentiviral packaging plasmids, as described in detail at https://portals.broadinstitute.org/gpp/public/resources/protocols. All lentiviral transfections were performed using Effectene (Qiagen). Stable cell lines were generated by infecting melanoma cells with lentivirus particles, followed by selection with appropriate concentrations of puromycin (0.5–1.5 μg/mL) to enrich for infected cells. For the pLX304-Blast-V5-based lentivirus, infected melanoma cells were selected with 2 μg/mL Blasticidin (Thermo Fisher Scientific).
Antibodies and Immunoblot Analysis.
Whole-cell protein extracts were prepared using Pierce IP lysis buffer (Thermo Fisher Scientific) containing protease inhibitor mixture (Roche) and phosphatase inhibitor mixture (Sigma-Aldrich). Protein concentrations were estimated using the Bradford assay (Bio-Rad). Proteins were separated by 10 or 12% SDS/PAGE and transferred onto polyvinylidene fluoride (PVDF) membrane by wet transfer. PVDF membranes were blocked with 5% nonfat dry milk or 5% BSA as recommended for specific antibodies, washed, and probed with primary antibodies. Membranes were washed again and then incubated with HRP-conjugated secondary antibodies (GE Healthcare). Immunoblots were developed using Supersignal West Pico or Femto Substrates (Pierce) as needed. All primary and secondary antibodies used in these studies are listed in SI Appendix, Table S4.
RNA Preparation, cDNA Preparation, and Quantitative PCR Analysis.
Total RNA was extracted with TRIzol Reagent (Invitrogen) and purified using the RNeasy Mini Kit (Qiagen); cDNA was generated using the M-MuLV First Strand cDNA Synthesis Kit (New England Biolabs) according to the manufacturer’s instructions. Quantitative RT-PCR was performed with gene-specific primers, using the Power SYBR-Green Master Mix (Applied Biosystems) according to the manufacturer’s instructions. Actin was used as an internal control. For measurement of 28S rRNA by RT-qPCR, primers specific to the 28S rRNA were utilized, and U1 nuclear RNA was used as control. Primer sequences are provided in SI Appendix, Table S4.
5Aza2dC and TSA Treatment and DUSP4 and DUSP6 Analysis.
A375 and SKMEL-239 cells were treated with 5Aza2dC at 5 μM for 72 h and TSA at 1 μM for 12 h or as control dimethyl sulfoxide (DMSO) was used. Treated cells were used to isolate RNA and also for preparing protein lysates. These were then used to analyzed DUSP4 and DUSP6 mRNA and protein levels by RT-qPCR and immunoblotting, respectively. Primer sequences used for DUSP4 and DUSP6 mRNA expression analysis are listed in SI Appendix, Table S4.
MTT Assays.
For MTT assays, 5 × 103 melanoma cells were plated in triplicate in a volume of 100 µL in 96-well plates. After 48 h, BRAF inhibitors (vemurafenib or dabrafenib) were administered at various concentrations, as indicated in the figures and figure legends. Cell viability was evaluated after 3 d of treatment. To this end, 20 µL of 5 mg/mL MTT solution dissolved in 1× PBS was added to each well of the 96-well plate and incubated for 1 h at 37 °C. The MTT solution was then gently removed, and 100 µL of DMSO was added to each well. After mixing well by pipetting, absorbance was measured at 590 and 630 nm using the Biotek Synergy MX Multi Format Microplate Reader. The average measurement at 630 nm was subtracted from the average at 590 nm, and relative growth rate was plotted with respect to vehicle control-treated cells.
Clonogenic Assays.
The clonogenic ability of cells stably expressing control or gene-specific shRNAs was measured in untreated and in vemurafenib- or dabrafenib-treated conditions. For these assays, 1 × 106 cells were seeded in 100-mm cell culture dishes and grown for 24 h. For drug treatment experiments, cells were then treated with DMSO, vemurafenib (2 µM), or dabrafenib (100 nM), and cell culture media was changed every 3 d, adding fresh drug each time. After 3–4 wk of treatment, surviving colonies were fixed in a solution containing 50% methanol and 10% acetic acid and then stained with 0.05% Coomassie blue (Sigma-Aldrich). The relative number of colonies was calculated by normalizing to the average colony number under control conditions and then plotting the average obtained from the triplicates under experimental conditions, as indicated in the figures and figure legends.
Soft-Agar Assay.
Soft-agar assays were performed by seeding 5 × 103 to 2 × 104 melanoma cells stably expressing the indicated shRNA or cDNA constructs onto 0.4% low-melting-point agarose (Sigma-Aldrich), layered on top of 0.8% agarose. For drug treatment experiments, cells were then treated with DMSO, vemurafenib (1 µM), or dabrafenib (50 nM), and cell culture media was changed every 3 d, adding fresh drug each time. After 3–4 wk of incubation, colonies were stained with a 0.005% crystal violet solution and imaged using a microscope. Colony size was measured using ImageJ software (https://imagej.nih.gov/ij/) and plotted.
Analysis of rRNA Biogenesis by Tritiated Uridine Labeling.
Analysis of rRNA biogenesis by tritiated uridine (H3-U) labeling was performed as described previously (34). Briefly, 2 × 106 cells per well were seeded in six-well plates in triplicate. After 24 h, [5,6-3H] uridine was added to the medium at a final concentration of 3 μCi/mL, and samples were incubated for 2 h. Total RNA was then prepared using TRIzol Reagent (Invitrogen), as described in Materials and Methods, RNA Preparation, cDNA Preparation, and Quantitative PCR Analysis. Incorporated [5,6-3H] uridine was measured by liquid scintillation counter, and equal amounts of radiolabeled RNA were separated by 1.5% agarose-formaldehyde gel electrophoresis. RNA was then transferred overnight from agarose gels to Amersham Hybond-N+ membranes via the capillary transfer method. After the transfer, the nylon membranes were sprayed with autoradiography enhancer (EN3HANCE; Perkin-Elmer) and then completely covered with a small volume of carbon tetrachloride. Finally, the membranes were air-dried for 5 min and exposed to a radiography film in a light-proof cassette at −80 °C for 72 h. Films were developed, and the ratios for various rRNA species were measured using ImageJ software (NIH).
Mouse Experiments.
Athymic nude (NCr nu/nu) mice (6 wk of age) were injected s.c. with cell lines expressing different shRNAs or control NS shRNAs. After 1 wk, mice with tumors were treated with either vehicle control or vemurafenib (30 mg/kg) via oral gavage three times a week. Tumor volume was measured every week, and the size was calculated using the formula: length × width2 × 0.5 and plotted. All animal protocols were approved by the Institutional Animal Care and Use Committee at Yale University. For all mouse experiments, vemurafenib was dissolved in DMSO and diluted to 25× in 2% Klucel-LF, and animals were dosed orally at 30 mg/kg on every alternative day.
Patient Sample Acquisition.
Melanoma samples were obtained through biopsies, and surgical resections were performed during standard clinical care of melanoma patients. Excess samples not required for surgical pathology assessment were stored in the Vanderbilt University melanoma tumor repository as FFPE. All patients provided consent through Institutional Review Board-approved protocol before tissue acquisition (Vanderbilt IRB# 030220), and samples were de-identified for the analysis.
Quantitative Immunofluorescence Analysis.
Whole-tissue sections of paired pre- and progressed patient samples that were treated with targeted BRAF inhibitors, as described in SI Appendix, Table S3, were deparaffinized at 60 °C for 30 min, incubated in xylene (soaking twice for 20 min), and rehydrated with ethanol (twice in 100% ethanol for 1 min and then in 70% ethanol for 1 min). Antigen retrieval was performed by boiling samples for 20 min at 97 °C in citrate buffer, pH 6.0 (PT module, Lab Vision; Thermo Scientific). Slides were blocked by treating with 30% hydrogen peroxide in methanol and then incubated with a blocking solution containing 0.3% BSA in Tris-buffered saline and 0.05% Tween solution (TBST) for 30 min at room temperature. Slides were then incubated overnight with a mixture of BOP1 rabbit antibodies, as well as S100 and HMB45 mouse antibodies (SI Appendix, Table S4). The next day, these were washed and treated with a mixture of Alexa 546-conjugated goat anti-mouse secondary antibody (Invitrogen), diluted 1:100 in rabbit Envision reagent (K4003, Dako), and incubated for 60 min at room temperature. For target detection, slides were treated with cyanine 5, directly conjugated to tyramide (FP1117; Perkin-Elmer) at a 1:50 dilution, for 10 min. ProLong gold mounting medium (Invitrogen), containing 4′,6-diamidino-2-phenylindole (DAPI), was used to stain nuclei. Control slides were processed for reproducibility alongside each experimental slide-staining run. Quantitative measurements of BOP1 and immunofluorescence analysis were performed using the AQUA method (25). A tumor mask was created by binarising the BOP1 signal and S100 and HMB45 signals. Quantitative immunofluorescence scores were calculated by dividing the target pixel intensity by the area of the S100 and HMB45 compartments. All patient samples were scored using AQUA software, and in each sample 40–70 different spots were analyzed; the average scores of target in the tumor mask-normalized are shown in the figures.
Statistical Analysis.
All experiments were conducted in at least three biological replicates. Results for individual experiments are expressed as the mean ± SEM. For measurement of tumor progression in mice and MTT assays, statistical analyses were performed by analyzing the area under curve using GraphPad Prism software, version 7.0, for Macintosh (GraphPad Software; https://www.graphpad.com). For the remaining experiments, P values were calculated using the two-tailed unpaired Student’s t test in GraphPad Prism software, version 7.0, for Macintosh.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01CA195077 (to N.W.), R01CA200919 (to N.W.), 1R01CA218008 (to N.W. and M.R.G.), K23CA204726 (to D.B.J.), and R03CA221926 (to R.G.). N.W. is also supported by a Research Scholar Grant from the American Cancer Society (128347-RSG-15-212-01-TBG).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821889116/-/DCSupplemental.
References
- 1.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 2.Zbytek B, et al. Current concepts of metastasis in melanoma. Expert Rev Dermatol. 2008;3:569–585. doi: 10.1586/17469872.3.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schadendorf D, et al. Melanoma. Nat Rev Dis Primers. 2015;1:15003. doi: 10.1038/nrdp.2015.3. [DOI] [PubMed] [Google Scholar]
- 4.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 7.Chapman PB, et al. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaherty KT, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong DJ, Ribas A. Targeted therapy for melanoma. Cancer Treat Res. 2016;167:251–262. doi: 10.1007/978-3-319-22539-5_10. [DOI] [PubMed] [Google Scholar]
- 11.Johnson DB, et al. Acquired BRAF inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer. 2015;51:2792–2799. doi: 10.1016/j.ejca.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizos H, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: Spectrum and clinical impact. Clin Cancer Res. 2014;20:1965–1977. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 13.Strezoska Z, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol Cell Biol. 2000;20:5516–5528. doi: 10.1128/mcb.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hölzel M, et al. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol. 2005;170:367–378. doi: 10.1083/jcb.200501141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baylin SB. Resistance, epigenetics and the cancer ecosystem. Nat Med. 2011;17:288–289. doi: 10.1038/nm0311-288. [DOI] [PubMed] [Google Scholar]
- 16.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, et al. An acquired vulnerability of drug-resistant melanoma with therapeutic potential. Cell. 2018;173:1413–1425.e14. doi: 10.1016/j.cell.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Hauschild A, et al. Dabrafenib in BRAF-mutated metastatic melanoma: A multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 19.Mills EW, Green R. Ribosomopathies: There’s strength in numbers. Science. 2017;358:eaan2755. doi: 10.1126/science.aan2755. [DOI] [PubMed] [Google Scholar]
- 20.Narla A, Ebert BL. Ribosomopathies: Human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long GV, et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat Commun. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 23.Kidger AM, Keyse SM. The regulation of oncogenic Ras/ERK signalling by dual-specificity mitogen activated protein kinase phosphatases (MKPs) Semin Cell Dev Biol. 2016;50:125–132. doi: 10.1016/j.semcdb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondoh K, Nishida E. Regulation of MAP kinases by MAP kinase phosphatases. Biochim Biophys Acta. 2007;1773:1227–1237. doi: 10.1016/j.bbamcr.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Camp RL, Dolled-Filhart M, King BL, Rimm DL. Quantitative analysis of breast cancer tissue microarrays shows that both high and normal levels of HER2 expression are associated with poor outcome. Cancer Res. 2003;63:1445–1448. [PubMed] [Google Scholar]
- 26.Sanjana NE, et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robert C, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 28.Oddo D, et al. Molecular landscape of acquired resistance to targeted therapy combinations in BRAF-mutant colorectal cancer. Cancer Res. 2016;76:4504–4515. doi: 10.1158/0008-5472.CAN-16-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8:a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao S, et al. A genetic interaction analysis identifies cancer drivers that modify EGFR dependency. Genes Dev. 2017;31:184–196. doi: 10.1101/gad.291948.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balko JM, et al. Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer. Cancer Res. 2013;73:6346–6358. doi: 10.1158/0008-5472.CAN-13-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joseph EW, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci USA. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen CH, et al. Loss of cohesin complex components STAG2 or STAG3 confers resistance to BRAF inhibition in melanoma. Nat Med. 2016;22:1056–1061. doi: 10.1038/nm.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pestov DG, Lapik YR, Lau LF. Assays for ribosomal RNA processing and ribosome assembly. Curr Protoc Cell Biol. 2008;Chapter 22:22.11.1–22.11.16. doi: 10.1002/0471143030.cb2211s39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.