Significance
The molecular mechanisms underlying cerebellar development have not yet been fully understood. Here, we analyzed and revealed a critical function of receptor for activated C kinase 1 (Rack1), a WD40-repeat domain containing scaffolding protein, in the regulation of cerebellar patterning and fissure formation. We found that during embryonic and early postnatal development, Rack1 oppositely regulates Wnt/β-catenin and Sonic hedgehog (Shh) signaling pathways in distinct developmental stages. Interestingly, we also found that Rack1-mediated stabilization of histone deacetylase 1 (HDAC1)/HDAC2 is essential for the activation of Shh signaling in granule neuron progenitors during cerebellar development. Together, we have demonstrated the crucial role of Rack1 for cerebellar morphogenesis by homeostatic regulation of two distinct morphogenic pathways.
Keywords: Rack1, neural stem cells, granule neuron progenitors, HDAC1/HDAC2, cerebellar morphogenesis
Abstract
The development of the cerebellum depends on intricate processes of neurogenesis, migration, and differentiation of neural stem cells (NSCs) and progenitor cells. Defective cerebellar development often results in motor dysfunctions and psychiatric disorders. Understanding the molecular mechanisms that underlie the complex development of the cerebellum will facilitate the development of novel treatment options. Here, we report that the receptor for activated C kinase (Rack1), a multifaceted signaling adaptor protein, regulates mammalian cerebellar development in a cell type-specific manner. Selective deletion of Rack1 in mouse NSCs or granule neuron progenitors (GNPs), but not Bergmann glial cells (BGs), causes severe defects in cerebellar morphogenesis, including impaired folia and fissure formation. NSCs and GNPs lacking Rack1 exhibit enhanced Wnt/β-catenin signaling but reduced Sonic hedgehog (Shh) signaling. Simultaneous deletion of β-catenin in NSCs, but not GNPs, significantly rescues the Rack1 mutant phenotype. Interestingly, Rack1 controls the activation of Shh signaling by regulating the ubiquitylation and stability of histone deacetylase 1 (HDAC1)/HDAC2. Suppression of HDAC1/HDAC2 activity in the developing cerebellum phenocopies the Rack1 mutant. Together, these results reveal a previously unknown role of Rack1 in controlling mammalian cerebellar development by opposite regulation of Wnt/β-catenin and Shh signaling pathways.
Cerebellum morphogenesis is critically important in normal brain development. The developmental deficits of the cerebellum can result in motor and higher cognitive dysfunctions, including impaired balance control, language processing, sensory/motor learning, and spatial memory (1–3). Cerebellar patterning critically depends on neurogenesis, an intricate process that involves the proliferation, migration, and differentiation of neural stem cells (NSCs) and progenitor cells (4, 5). The appearance of granule neuron progenitors (GNPs) over the surface of the cerebellum was identified as a key feature of cerebellar development (6). The coordinated interaction between NSCs, GNPs, Purkinje cells (PCs), and Bergmann glia cells (BGs) refines the developing cerebellum into the typical pattern of 10 folia and a three-layered cerebellar cortex, which includes the external molecular layer (ML), the middle Purkinje cell layer (PCL), and the innermost internal granular layer (IGL) (4, 7, 8). The cellular organization and signaling assembly of the cerebellar cortex constitute an ideal model for studying neuronal properties and cortical circuitry formation.
The proliferation, migration, and differentiation of GNPs is temporally and spatially regulated during cerebellum development. PCs control the proliferation of GNPs by releasing diffusible factors, such as insulin-like and epidermal growth factors (8–11) and Sonic hedgehog (Shh), a predominant player in cerebellar patterning (12–15). Constitutive activation of Shh signaling in GNPs may contribute to the formation of medulloblastoma, the most common type of pediatric malignant primary brain tumor (16, 17). In addition, GNP differentiation and cerebellar vermis formation are regulated by Wnt/β-catenin signaling in the rhombic lip (RL), in the ventricular zone, and in early migrating GNPs (18–20). Several other molecules, such as FGF8 (21, 22), contactin (23), Reelin (24), Gbx2 (25), Zic1/2 (26), En1/2 (27), and neurofibromin 1 (28), have also been implicated in cerebellar development. However, the particular intracellular signaling pathways that coordinate and integrate those morphogenic signals are not well understood. Furthermore, genetic pathways that determine the formation and patterning of cerebellar fissures remain unresolved.
The receptor for activated C kinase 1 (Rack1) is a multifaceted scaffolding protein with seven conserved WD40-repeat (WDR) domains, which was originally identified as an anchoring protein for the conventional protein kinase C (PKC) (29, 30). Mice lacking Rack1 are embryonic lethal at the gastrulation stage, suggesting it is crucial for mammalian development (31). In the central nervous system (CNS), Rack1 is involved in the regulation of neurite outgrowth and dendritic transport, long-term potentiation initiation, intracellular Ca2+ release, synaptic transmission, and neurodegenerative processes, suggesting a critical role of Rack1 for normal brain functions (32). Rack1 is abundantly coexpressed with PKC-βII, an isozyme of PKC, in select brain regions, including the cerebellum (33). Nevertheless, the exact function of Rack1 in the cerebellum remains elusive. Intriguingly, previous studies in gastric cancer cells show that Rack1 represses Wnt/β-catenin transcriptional activity and promotes β-catenin degradation by stabilizing the β-catenin destruction complex (34). In contrast, Rack1 activates Shh signaling by activating the Smoothened receptor in non-small cell lung cancer cells (35). Thus, we hypothesized that Rack1 might be critical for cerebellar development by simultaneously targeting both Wnt/β-catenin and Shh signaling pathways.
In this study, we demonstrated that Wnt/β-catenin and Shh signaling pathways were oppositely regulated by Rack1 during distinct stages of cerebellar development in a cell type-specific manner. Ablation of Rack1 expression in either NSCs or GNPs disrupts cerebellar morphogenesis. Simultaneous removal of Rack1 and β-catenin in NSCs significantly rescues cerebellar developmental deficits, consistent with the notion that Rack1 inhibits Wnt/β-catenin signaling. Moreover, we found Rack1-mediated stabilization of histone deacetylase 1 (HDAC1)/HDAC2 is essential for the activation of Shh signaling. Suppression of HDAC1/HDAC2 activity in the neonatal cerebellum severely disrupts cerebellar folia formation and GNP proliferation, phenocopying NSC- or GNP-specific Rack1 knockouts (KOs). Taken together, our findings reveal a crucial role of Rack1-mediated opposite regulation of Wnt/β-catenin and Shh signaling pathways during cerebellar development.
Results
Ablation of Rack1 in Multipotent NSCs Disrupts Cerebellar Development.
Consistent with high cerebellar proliferation in early postnatal development (5, 12), we found relatively high Rack1 expression levels in the external granular layer (EGL) at the early postnatal stage, which gradually decreased thereafter (details are provided in SI Appendix, Fig. S1). Because constitutive Rack1-null mice are embryonic lethal, the function of Rack1 in neural development has remained elusive (31). To circumvent this caveat, we generated conditional KO mice with cell-specific Rack1 deletion in NSCs to investigate the function of Rack1 in multipotent NSCs during early cerebellar development. To this end, we used a well-characterized human GFAP-Cre transgenic mouse line (hGFAP-Cre), which is specifically expressed in radial glial cells from embryonic day 12.5 (E12.5) onward, to generate NSC-specific Rack1 conditional mutant mice (hGFAP-Cre;Rack1F/F) (details are provided in SI Appendix, Fig. S2 A–C). We assessed the patterns of NSC-derived GNPs and BGs at embryonic stages, and observed significantly reduced EGL width with a decreased number of Pax6+ GNPs and disorganized radial glial cells in mutants from E16.5 to E18.5 (SI Appendix, Fig. S3 A–C), These results suggest that Rack1 depletion affects GNP proliferation and migration during embryonic stages in the cerebellum.
Next, we asked whether Rack1 depletion results in a distinctive phenotype at postnatal stages. Remarkably, compared with Rack1F/F controls, hGFAP-Cre;Rack1F/F mutants displayed typical ataxic movement disorders with reduced brain size, reduced body weight, and severely diminished cerebellar volume (SI Appendix, Fig. S2 D–F). Almost all hGFAP-Cre;Rack1F/F mutant mice died within 3 wk after birth (n = 17 of mutants and n = 29 of controls; SI Appendix, Fig. S2G). The area of the developing cerebellum in control and mutant mice appears indistinguishable at postnatal day 0 (P0) (0.88 ± 0.05 mm2 in control mice and 0.91 ± 0.09 mm2 in mutant mice; P > 0.05, n = 4; Fig. 1A). However, from P3, hGFAP-Cre;Rack1F/F mutant mice displayed significantly smaller and disorganized cerebellar histoarchitecture, which was hypoplastic and with a dramatically decreased sagittal area of the cerebellar vermis compared with controls (Fig. 1B). In the medial vermis of control mice, distinct lobules formed along the anteroposterior axis to generate the typical postnatal foliation pattern, which was completely abolished in the mutant cerebellum (Fig. 1A and SI Appendix, Fig. S2E). The mutant mice produced very few and incomplete lobules throughout the rostral, central, and caudal parts of the medial vermis, compared with control littermates at indicated postnatal stages (Fig. 1 A and C). These results indicate that Rack1 is required for postnatal cerebellar compartmentation.
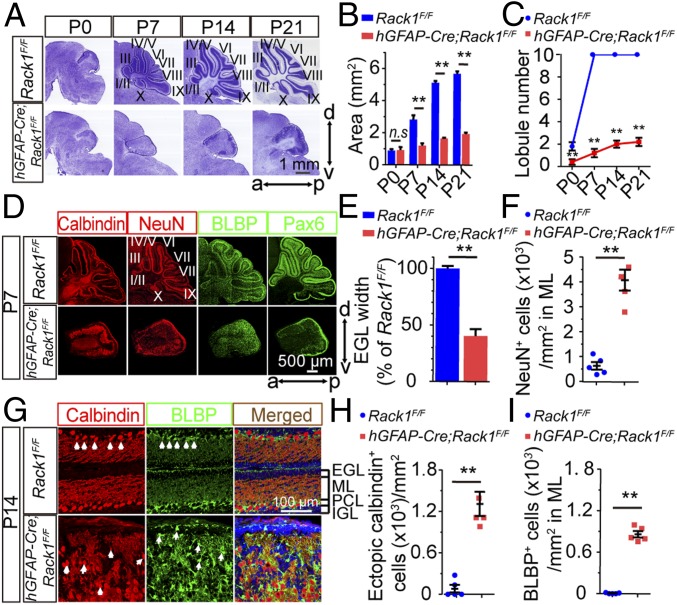
Fig. 1.
hGFAP-Cre;Rack1F/F mutant mice display impaired cerebellar morphogenesis. (A) Nissl staining of sagittal histological sections of the vermis shows considerable cerebellar foliation defects in hGFAP-Cre;Rack1F/F mutant mice at the indicated postnatal developmental stages. a, anterior; d, dorsal; p, posterior; v, ventral. (Scale bar: 1 mm.) (B) Area of sagittal sections of the cerebellar vermis in Rack1F/F control and hGFAP-Cre;Rack1F/F mutant mice (mean ± SEM; P = 0.67, **P = 0.0035, **P = 0.0022, and **P = 0.0011, respectively; n = 4). n.s., not significant. (C) Number of observed lobules in control and mutant mice at the indicated postnatal developmental stages (mean ± SEM; **P < 0.0001, n = 5). (D) Immunofluorescent staining with anticalbindin, anti-NeuN, anti-BLBP, and anti-Pax6 antibodies reveals dysmorphic PCs, granule neurons, BGs, and GNPs, respectively, in hGFAP-Cre;Rack1F/F mutant cerebellum at P7. (Scale bar: 500 μm.) (E) EGL thickness in control and mutant mice (mean ± SEM; **P = 0.0086, n = 4). (F) Number of NeuN+ granule neurons in the ML per unit area (mean ± SEM; **P < 0.0001, n = 4). (G) Immunofluorescent staining of cerebellar sections with anticalbindin and anti-BLBP antibodies. Arrowheads indicate PCs and BGs in control and mutant cerebella, respectively. (Scale bar: 100 μm.) (H) Number of calbindin+ ectopic PCs outside the PCL in control and mutant cerebella (mean ± SEM; **P < 0.0001, n = 4). (I) Number of BLBP+ ectopic BGs outside the PCL in control and mutant cerebella (mean ± SEM; **P < 0.0001, n = 4).
The above morphological defects were further confirmed by immunofluorescent staining of cerebellar sections at P7 and P14 with antibodies to calbindin, NeuN, brain lipid-binding protein (BLBP), and Pax6, which are markers for PCs, postmitotic granule neurons, BGs, and GNPs, respectively (28, 36) (Fig. 1 D–F). At P7, in the Rack1F/F control cerebellum, BGs were localized in the PC layer near the pial surface (37), NeuN+ granule cells were present in the separated fissures and restricted to granule neurons within the IGL (38), and Pax6+ GNPs were highly abundant in the apical surface of EGL (Fig. 1D). Pax6 immunostaining demonstrated that the EGL width in hGFAP-Cre;Rack1F/F mutants was significantly smaller than that of the controls (40.4 ± 2.7% of controls; P = 0.0086, n = 4; Fig. 1E), indicating impaired proliferation or survival of GNPs in mutants. In addition, although calbindin+ PCs and BLBP+ BGs were detectable in hGFAP-Cre;Rack1F/F mutants, the cells displayed aberrant localization and morphology (Fig. 1D). Furthermore, the total number of NeuN+ granule cells was significantly decreased in mutant mice, but a larger number were restricted in the disordered ML compared with controls (4,055 ± 417 cells per square millimeter in mutants vs. 628 ± 153 cells per square millimeter in controls; P < 0.0001, n = 4; Fig. 1F), suggesting potential differentiation and migration defects (38). At P14, the mutant mice showed severe dyslamination of the PCL and a disrupted ML throughout the cerebellar cortex (Fig. 1G). Immunolabeling with calbindin found disorganized and abnormal dendritic arborization of PCs, whereas BLBP staining revealed severely disorganized glial scaffolds of BGs in hGFAP-Cre;Rack1F/F mutants (Figs. 1G, arrowheads and 2A and SI Appendix, Fig. S9A). In addition, the mutant mice showed increased numbers of ectopic PCs and BGs in the disordered ML, which further illustrates disorganization in the developing cerebellar cortex (Fig. 1 H and I). Together, these results suggest that cerebellar cortical lamination and foliation require Rack1.
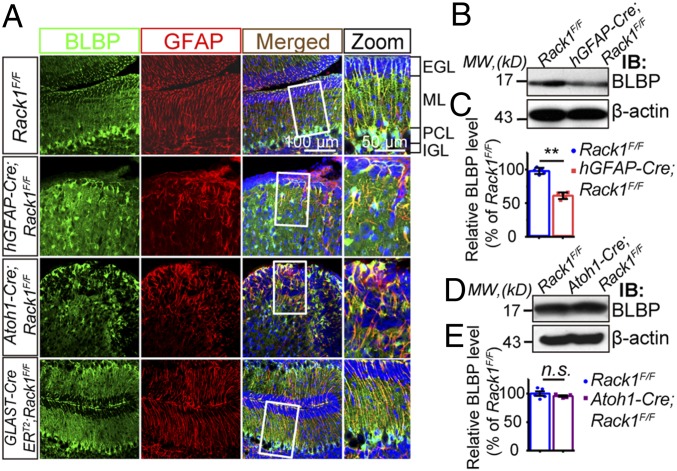
Fig. 2.
hGFAP-Cre and Atoh1-Cre mice, but not GLAST-CreERT2-driven Rack1 conditional KO mice, display abnormal BG morphology. (A) BGs colabeled for BLBP (green) and GFAP (red) were morphologically defective in P14 hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mutants, but not in GLAST-CreERT2-Rack1F/F mutant mice compared with controls. Boxed areas of MLs are shown at higher magnification to illustrate the detailed morphology of BGs (Zoom column). (Scale bars: 100 μm and 50 μm.) (B and C) Representative Western blot and quantification show significantly reduced BLBP protein expression in hGFAP-Cre;Rack1F/F mutant mice (mean ± SEM; **P = 0.0003, n = 5). IB, immunoblot; MW, molecular weight. (D and E) Representative Western blot and quantification show unperturbed BLBP protein expression in Atoh1-Cre;Rack1F/F mutant compared with control mice at P14 (mean ± SEM; P = 0.3496, n = 5). n.s., not significant.
Altered Production and Migration of GNPs in hGFAP-Cre;Rack1F/F Mutants.
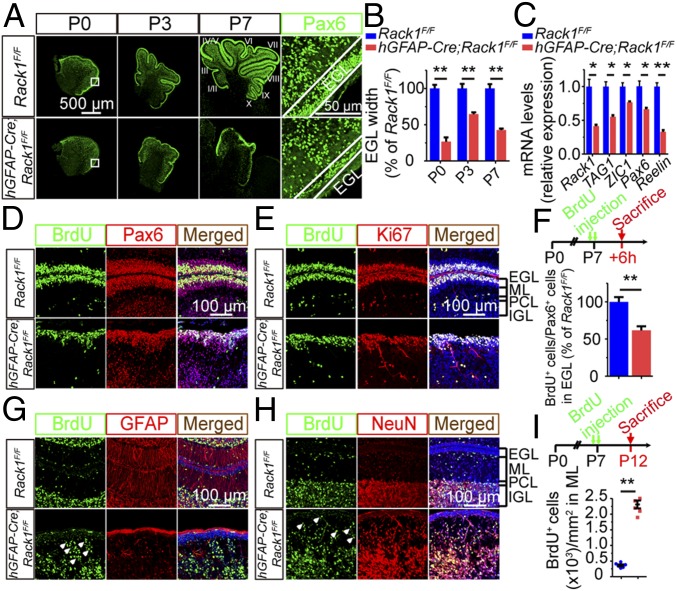
Having characterized the morphological and cellular deficits in hGFAP-Cre;Rack1F/F mice, we next asked whether impaired GNP production and migration from the EGL underlies the observed decrease in Pax6+ GNPs and NeuN+ granule cell number. We first analyzed the expression of Pax6+ neurons at different postnatal stages (Fig. 3A). We found that the widths of the Pax6+ EGL in hGFAP-Cre;Rack1F/F mutants were 26.67 ± 3.48%, 62.47 ± 3.45%, and 42.67 ± 1.20% of those in controls at P0, P3, and P7, respectively (P < 0.01, n = 5; Fig. 3B). Consistently, at P4, the transcripts of early cerebellar granule cell differentiation markers, including TAG1, ZIC1, and Reelin, were significantly decreased in the mutants (24, 39, 40) (Fig. 3C).
Fig. 3.
hGFAP-Cre;Rack1F/F mutant mice are deficient in GNP proliferation and migration. (A) Pax6 antibody was used to label GNPs in sagittal sections of the vermis at the indicated postnatal developmental stages. (Scale bars: 500 μm and 50 μm.) (B) EGL thickness in control and mutant mice (mean ± SEM; **P = 0.0076, **P = 0.0093, and **P = 0.0084, respectively; n = 3). n.s., not significant. (C) Quantitative RT-PCR analysis indicates significantly down-regulated expression of TAG1, ZIC1, Pax6, and Reelin genes in mutant mice. Rack1 was used as a positive control (mean ± SEM; *P = 0.0175, *P = 0.0137, *P = 0.0298, *P = 0.0269, and **P = 0.0044, respectively; n = 3). (D and E) Proliferating GNPs in control and hGFAP-Cre;Rack1F/F mutant cerebellum were immunolabeled with anti-BrdU, anti-Pax6, and anti-Ki67 antibodies. Deletion of Rack1 significantly decreased the number of proliferating GNPs as indicated by BrdU+/Pax6+ and BrdU+/Ki67+ cells. (F) Proliferation of GNPs was evaluated 6 h after BrdU administration in control and mutant mice at P7. Quantitative analysis of proliferating GNPs was indicated by the proportion of BrdU+ cells/total Pax6+ cells within the EGL (mean ± SEM; **P < 0.0001, n = 5). (G and H) Migrating GNPs in control and mutant cerebella were labeled with anti-BrdU. Differentiated BGs and granule cells were co-immunolabeled with anti-GFAP and anti-NeuN antibodies, respectively. Deletion of Rack1 significantly reduced GNP migration and led to ectopic accumulation of GNP descendant cells (arrowheads). (Scale bar: 100 μm.) (I) GNP migration was evaluated after 5 d following BrdU treatment at P7. Quantification of the number of ectopic BrdU+ cells per unit area of the ML is shown (mean ± SEM; **P < 0.0001, n = 6).
To assess the effects of Rack1 loss on postnatal GNP proliferation in hGFAP-Cre;Rack1F/F mutants, we labeled dividing neurons with 5-bromo-2′-deoxyuridine (BrdU) at P7, a peak stage for postnatal GNP proliferation (41). Compared with controls, the EGL of mutant mice contained significantly fewer Pax6+ and BrdU+ cells (Fig. 3 D and E), whereas the ratio of BrdU+ cells to Pax6+ cells in mutants was 60.6 ± 2.5% of that in controls (Fig. 3F). Double immunolabeling of BrdU and Ki67 further confirmed decreased GNP proliferation in hGFAP-Cre;Rack1F/F mice (Fig. 3E).
Next, we investigated the effect of Rack1 ablation on the migration of GNPs in EGL. We labeled migrating neurons with BrdU and analyzed the number of BrdU-incorporated cells in the ML (Fig. 3 G–I). At P12, a significantly increased number of BrdU-incorporated cells were retained at the disordered ML in mutants compared with controls (2,224 ± 109 cells per square millimeter in KO mice vs. 356 ± 35 cells per square millimeter in WT mice; Fig. 3 G–I, arrowheads), indicating abnormal GNP migration in mutants. We next examined the differentiation of BGs and mature granule cells by colabeling GFAP or NeuN with BrdU. In control mice, the cell bodies of BGs localized at the PCL and their radial fibers projected into the EGL. However, BGs from mutant mice were misaligned and dispersed throughout the dyslaminated cortex, displaying short and thin fibers that did not contact the pial surface at the EGL (Figs. 2A and 3G and SI Appendix, Fig. S9). Because BG radial fibers provide scaffolds for GNP migration (36, 42), the abnormal BG morphology and localization likely contribute to the impaired migration of GNPs and morphogenesis defects in hGFAP-Cre;Rack1F/F mice.
Further investigation revealed defects in NeuN+ granule cells from mutant mice. In the control cerebellum, the majority, if not all, of mature granule neurons localize in the IGL and are hardly detectable in the ML at P12. In contrast, the mutant mice had more BrdU+/NeuN+ mature granule neurons distributed within the disrupted ML and reduced NeuN+ granule neurons in the IGL (Fig. 3 H and I), suggesting impaired migration and delayed differentiation of Rack1-depleted GNPs.
Ablation of Rack1 in GNPs Is Sufficient to Disrupt Cerebellar Development.
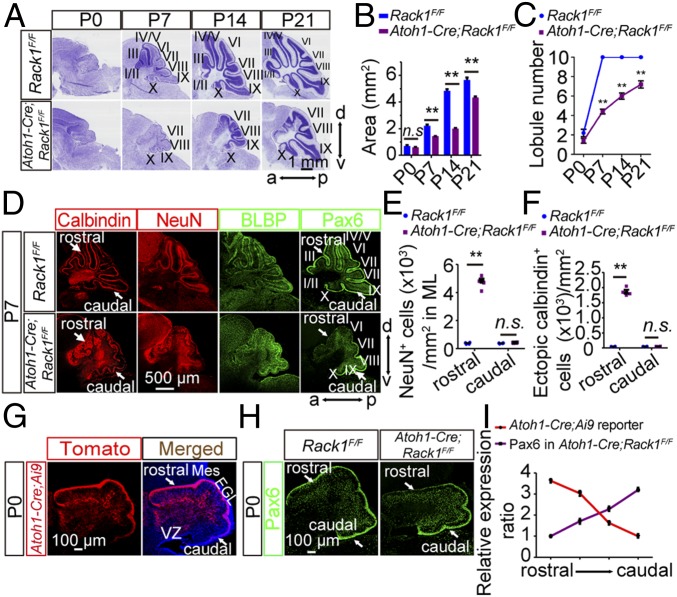
Given that Rack1 is expressed throughout the developing cerebellum (SI Appendix, Fig. S1) including GNPs (33), we next asked whether the observed hypoplastic phenotypes in the hGFAP-Cre;Rack1F/F mutant cerebellum are due to loss of Rack1 in GNPs. We generated GNP-specific Atoh1-Cre (also named Math1-Cre)–driven Rack1 conditional KO mice (details are provided in SI Appendix, Fig. S4) to investigate potential cell-autonomous effects of Rack1 KO in GNPs. Intriguingly, similar to hGFAP-Cre;Rack1F/F mutants, we found notable cerebellar foliation defects in Atoh1-Cre;Rack1F/F mutants from P7 to P21 (Fig. 4A). The area of the midsagittal sections was significantly decreased in Atoh1-Cre;Rack1F/F mutants from P7 to P21, but not at P0 (P = 0.0002, P = 0.0001, P = 0.0004, and P = 0.0846, respectively; n ≥ 4; Fig. 4B). After P7, the rostral and central regions of the cerebellum exhibited fewer lobules in Atoh1-Cre;Rack1F/F mutants, compared with control mice (Fig. 4C), and the shapes of lobules were dramatically altered (Fig. 4 A and D). Specifically, in mutant mice, lobules I to VI in the rostral and central cerebellum was nearly indistinguishable.
Fig. 4.
Conditional ablation of Rack1 in GNPs disrupts cerebellar morphogenesis. (A) Nissl staining of sagittal histological sections of the vermis shows cerebellar foliation defects in Atoh1-Cre;Rack1F/F mutant mice at indicated postnatal developmental stages. a, anterior; d, dorsal; p, posterior; v, ventral. (Scale bar: 1 mm.) (B) Area of cerebellar sagittal sections in control and Atoh1-Cre;Rack1F/F mice at the indicated postnatal developmental stages (mean ± SEM; P = 0.0846, **P = 0.0002, **P = 0.0001, and **P = 0.0004, respectively; n ≥ 4). n.s., not significant. (C) Number of observed cerebellar lobules in control and mutant mice at the indicated developmental stage (mean ± SEM; **P < 0.0001, n = 6). (D) Immunofluorescent staining with anticalbindin, anti-NeuN, anti-BLBP, and anti-Pax6 antibodies shows dysmorphic PCs, granule neurons, BGs, and GNPs, respectively, particularly in the rostral region of Atoh1-Cre;Rack1F/F cerebellum at P7. (Scale bar: 500 μm.) (E) Number of ectopic NeuN+ granule neurons per unit area in the rostral and caudal regions of the control and mutant cerebella (mean ± SEM; **P < 0.0001, n = 4). (F) Number of ectopic calbindin+ Purkinje neurons in the rostral and caudal regions of the control and mutant cerebella (mean ± SEM; **P < 0.0001, n = 4). (G) Expression of Atoh1-Cre recombinase in the cerebellum was indicated by Tomato fluorescence (red) in the P0 Atoh1-Cre;Ai9 reporter mouse. Arrows indicate high Atoh1-Cre activity in the rostral, but not caudal, region of the EGL at P0. Mes, mesencephalon; VZ, ventricular zone. (Scale bar: 100 μm.) (H) Rostral, but not caudal, region of the cerebellar EGL shows significantly reduced Pax6+ GNPs in Atoh1-Cre;Rack1F/F mutant mice. (Scale bar: 100 μm.) (I) Semiquantitative analysis of Atoh1-Cre activity and Pax6+ immunofluorescent signals in Atoh1-Cre;Rack1F/F mutant cerebellum from the rostral to caudal EGL within the cerebellum. Relative expression ratio indicates the relative intensity of fluorescence from the rostral to caudal region in the cerebellum (n = 3).
At the cellular level, we observed severe morphological alterations and defective differentiation and migration of GNPs, PCs, and BGs in Atoh1-Cre;Rack1F/F mutants at P7. Notably, the abnormalities were clear in the rostral regions but less prominent in caudal regions (Fig. 4D, arrowheads). Atoh1-Cre;Rack1F/F mutants had a significantly increased number of NeuN+ granule cells and ectopic calbindin+ PCs in the disrupted ML in rostral, but not caudal, regions compared with controls (P < 0.001, n = 4; Fig. 4 E and F), suggesting severe differentiation and migration defects of GNPs and PCs limited to rostral regions of the cerebellum. Of note, Atoh1-Cre recombinase activity is predominantly high in GNPs at rostral lobules during early embryonic stages and gradually expands to caudal lobules at late embryonic stages. At P0, our Atoh1-Cre;Ai9 reporter assay demonstrated higher Cre recombinase activity in rostral parts compared with caudal regions. Accordingly, the number of Pax6+ GNPs in the apical surface of the EGL in rostral parts was significantly decreased compared with caudal regions in Atoh1-Cre;Rack1F/F mutants (Fig. 4 G–I). Moreover, Atoh1-Cre;Rack1F/F mutants spent a significantly decreased amount of time on an accelerating rotarod compared with controls (P = 0.0275, n ≥ 6; SI Appendix, Fig. S4G), suggesting defects in motor learning and locomotor activity in GNP-specific Rack1 mutant mice.
Next, we assessed the proliferation and migration of GNPs in Atoh1-Cre;Rack1F/F mutants by 5-ethynyl-20-deoxyuridine (EdU) or BrdU labeling (details are provided in SI Appendix, Fig. S5). We found a significantly decreased number of EdU+ cells in the EGL (SI Appendix, Fig. S5 A and B), but an increased number of BrdU+ cells retained in the ML in mutants (P < 0.0001, n = 5; SI Appendix, Fig. S5 C and D, arrowheads), suggesting impaired proliferation and migration of GNPs. Moreover, our BrdU/EdU double-labeling assay further demonstrated impaired self-renewal of GNPs in both hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mutant mice (SI Appendix, Fig. S6 A–H), indicating the crucial role of Rack1 in the proliferation and migration of GNPs. Taken together, these data illustrate similar deficiencies in gross and cellular morphology and localization in the cerebellum between hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mice, indicating that Rack1 may function cell-autonomously in GNPs to regulate the production, migration, and differentiation of granule neurons during cerebellar development.
Rack1 in BGs Is Dispensable for Cerebellum Morphogenesis.
Although our experiments have identified Rack1 as a crucial regulator of NSC and GNP development, little is known about its role in BG development. We observed misaligned and morphologically abnormal BGs in both hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mutant cerebella, suggesting that the absence of Rack1 results in abnormal BGs that may contribute to cerebellar developmental deficits (Figs. 1 G and I, 2A, and 3G, and SI Appendix, Fig. S5C). Of note, only hGFAP-Cre, but not Atoh1-Cre, recombinase activity could be detected in BGs (SI Appendix, Figs. S2B and S4B). Thus, we reasoned that Rack1 may regulate BGs in three ways: first, cell-autonomously and directly in BGs; second, via its actions in NSCs, which generate BGs; or, third, through a non–cell-autonomous manner in GNPs.
To determine whether Rack1 in BGs is essential for cerebellar development, we generated tamoxifen-inducible, glial cell-specific Rack1 mutant mice using a GLAST-CreERT2 knock-in driver line (42) (details are provided in SI Appendix, Figs. S7–S9). First, we performed histoarchitecture assays at P4 and P14. Surprisingly, GLAST-CreERT2;Rack1F/F mutant cerebella appeared to have normal BG morphology and alignment when examined at P14 (Fig. 2A and SI Appendix, Figs. S8C and S9A), indicating Rack1 in BGs is dispensable for their differentiation. We next examined BLBP expression, a specific marker for BGs, in both hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mutant cerebella. Intriguingly, BLBP protein expression was significantly decreased in NSC-specific Rack1 KO mice (36.33 ± 3.07% reduction; P = 0.0003, n = 3), but not in GNP-specific KO mice (P = 0.3496, n = 3), compared with Rack1F/F controls at P14 (Fig. 2 B–E), suggesting that hGFAP-Cre;Rack1F/F mice have a reduced cell number or altered transcription of BGs in the cerebellum.
Collectively, these results suggest that cell-autonomous Rack1 depletion in either NSCs or GNPs directly contributes to the observed cerebellar developmental defects. The abnormal BGs in Atoh1-Cre;Rack1F/F mutant mice are likely due to impaired GNP production, migration, and function, whereas the abnormal BGs observed in hGFAP-Cre;Rack1F/F mutant mice may also be due to impaired proliferation and differentiation of BGs caused by hGFAP-Cre–mediated gene ablation at early embryonic stages.
Constitutive Activation of Wnt/β-Catenin but Impaired Shh Signaling Pathways in Rack1 KO GNPs.
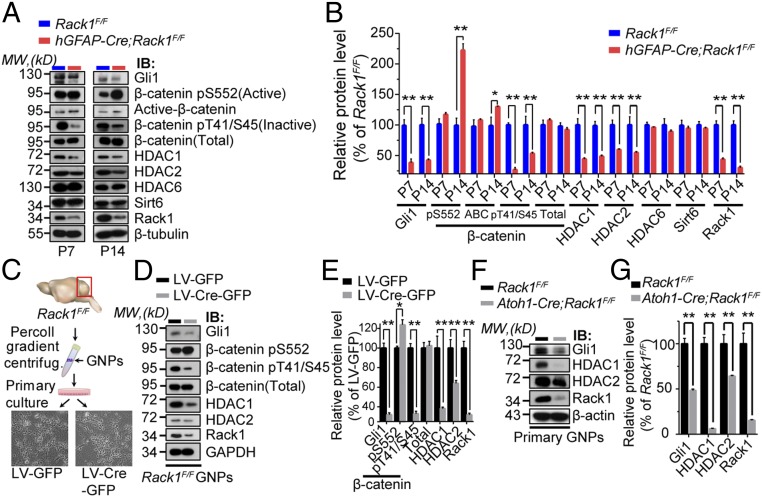
Having determined a central role of Rack1 in cerebellar development, we next explored the underlying Rack1-mediated signaling transduction networks in GNPs. Rack1 positively regulates Shh signaling but negatively regulates Wnt/β-catenin signaling pathways in cancer cells (34, 35). Moreover, HDAC1/HDAC2 critically actives Shh/Gli1 signaling and the differentiation and maturation of neural precursors (43, 44). Of note, the above signaling pathways are critical for the normal development of the cerebellum (5, 14, 20, 43); overactivation of Wnt/β-catenin signaling is observed in roughly 20% of human medulloblastomas (45). Therefore, we tested whether Rack1 depletion affects these pathways in vivo in the cerebellum and in cultured GNPs in vitro. The hGFAP-Cre;Rack1F/F mutant mice showed significantly reduced Gli1, Rack1, and HDAC1/HDAC2 protein levels but unaltered HDAC6 and Sirt6 protein levels at P7 and P14 (Fig. 5 A and B). These mice also had significantly increased active β-catenin (pS552) but decreased inactive β-catenin (pT41/S45) (Fig. 5 A and B). These results suggest that Shh signaling is impaired, whereas Wnt/β-catenin signaling is overactive, in the Rack1 mutant cerebellum.
Fig. 5.
Conditional ablation of Rack1 causes simultaneously overactivated Wnt/β-catenin signaling but impaired Shh signaling pathways. (A) Representative Western blots examining downstream Wnt/β-catenin and Shh signaling protein expression in control and hGFAP-Cre;Rack1F/F mutant cerebella at P7 and P14, respectively. IB, immunoblot; MW, molecular weight. (B) Quantitative analysis indicates elevated levels of the active form but decreased levels of the inactive form of β-catenin and reduced Gli1 and HDAC1/HDAC2 expression in hGFAP-Cre;Rack1F/F mutants. HDAC6 and Sirt6 expression was not significantly different between controls and mutants. Significantly reduced expression of Rack1 was detected in mutant (mean ± SEM; **P < 0.001, *P < 0.01, n = 3). (C) Schematic showing the primary culture of GNPs from Rack1F/F mice and infection with LV-Cre-GFP or LV-GFP control LV. (D and E) Quantitative Western blot analysis indicates the significantly increased active form (pS552), but decreased inactive form (pT41/S45), of β-catenin in primary Rack1F/F GNPs after LV-Cre-GFP infection compared with LV-GFP–infected control cells (mean ± SEM; **P = 0.0226 and **P = 0.0001, respectively; n = 3). The total level of β-catenin was unaltered (P = 0.6028, n = 3). The expression of Gli1 and HDAC1/HDAC2 was also significantly reduced after LV-Cre-GFP infection (mean ± SEM; **P = 0.0001, **P = 0.0009, and **P = 0.0025, respectively; n = 3). Rack1 was used as a positive control (**P = 0.0007, n = 3). (F and G) Representative Western blots and quantification showing reduced expression of downstream Shh signaling molecules in the Atoh1-Cre;Rack1F/F mutant cerebellum at P7 (mean ± SEM; **P < 0.0001,**P = 0.0039, and **P = 0.0002, respectively; n = 3).
The above findings suggest that Rack1 deletion oppositely regulates Shh activation and Wnt/β-catenin signaling in vivo. To test whether this is a cell-autonomous effect, we isolated and cultured primary GNPs from P7 Rack1F/F cerebellum and knocked out Rack1 using a Cre recombinase-expressing lentivirus (LV-Cre-GFP) (Fig. 5C). The expression of Rack1 was significantly reduced in LV-Cre-GFP–infected GNPs compared with LV-GFP–infected control cells (Fig. 5 D and E). Although total β-catenin expression levels were unaltered, active β-catenin was significantly up-regulated and inactive β-catenin was significantly reduced in LV-Cre-GFP–infected GNPs. In contrast, Gli1 and HDAC1/HDAC2 expression was significantly decreased in LV-Cre-GFP–infected GNPs (P < 0.01, Student’s t test; Fig. 5 D and E) and primary GNPs cultured from Atoh1-Cre;Rack1F/F mutants (P < 0.01, Student’s t test; Fig. 5 F and G). Together, these results demonstrate that Rack1 is involved in the opposite regulation of Wnt/β-catenin and Shh signaling pathways in GNPs in a cell-autonomous manner to control their proliferation and migration during cerebellar development.
We previously showed increased expression of calbindin and Shh in hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mutant mice (SI Appendix, Fig. S9 B–G). Therefore, we asked whether the observed impaired Shh signaling activation is a result of defective GNPs that cannot adequately respond to Shh stimulation in mutant mice. Treatment with the Shh signaling agonist Smoothened Agonist (SAG) significantly enhanced the interaction between Rack1 and Smoothened receptor in primary cultured GNPs (SI Appendix, Fig. S10 A–D). Moreover, LV-Cre-GFP recombinase-mediated deletion of Rack1 in primary GNPs blunted Gli1 and HDAC1/HDAC2 up-regulation in response to SAG treatment (SI Appendix, Fig. S10 E and F). These data demonstrate that impaired Shh signaling activation in hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mutant mice is due to Rack1-KO–induced GNP dysfunction.
Rack1/β-Catenin Double Mutants in NSCs, but Not GNPs, Show Genetic Rescue Effects.
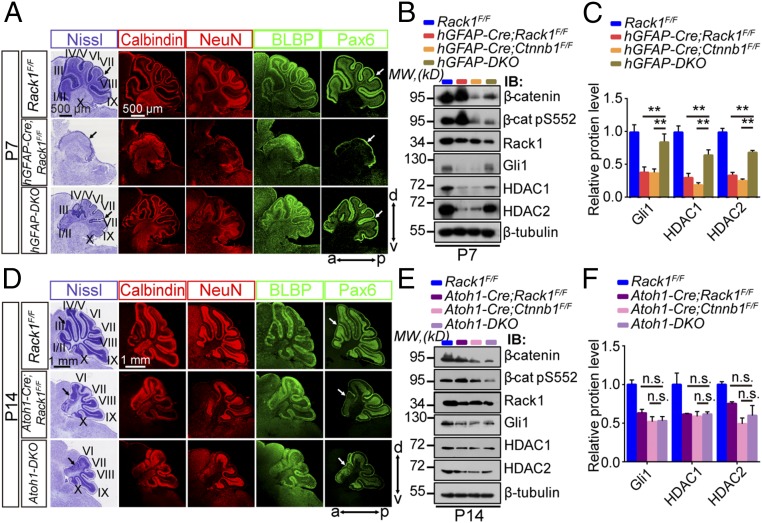
To determine whether the overactivation of the Wnt/β-catenin signaling pathway detected in hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mutants is essential for impaired cerebellar development, we simultaneously deleted Rack1 and Ctnnb1 (coding β-catenin) in NSCs by generating hGFAP-Cre;Rack1F/F;Ctnnb1F/F (hGFAP-DKO) double-KO mutant mice and in GNPs by generating Atoh1-Cre;Rack1F/F;Ctnnb1F/F (Atoh1-DKO) double-KO mutant mice, respectively (Fig. 6A). Western blot analysis confirmed significantly decreased Rack1 levels in hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F single mutants as well as in hGFAP-DKO and Atoh1-DKO mice (Fig. 6 B, C, E, and F). Similarly, significantly decreased expression of β-catenin was also confirmed in hGFAP-Cre– or Atoh1-Cre–mediated Ctnnb1F/F single-KO mice and in hGFAP-DKO or Atoh1-DKO mice (Fig. 6 B, C, E, and F). Importantly, the cerebellar foliation and lamination defects observed in hGFAP-Cre;Rack1F/F mutants were significantly recovered at P7 in hGFAP-DKO mice (Fig. 6A), suggesting double deletion of Rack1 and Ctnnb1 in NSCs could effectively rescue cerebellar morphological abnormalities in hGFAP-Cre;Rack1F/F mutants. In stark contrast, in Atoh1-DKO mutant cerebellum, the impaired foliation and lamination in rostral parts were indistinguishable from those in Atoh1-Cre;Rack1F/F mutants at P14 (Fig. 6D), indicating that double deletion of Rack1 and Ctnnb1 in GNPs is insufficient to rescue the cerebellar developmental deficits in Atoh1-Cre;Rack1F/F mutants.
Fig. 6.
Simultaneous deletion of β-catenin in NSCs, but not GNPs, significantly rescues the Rack1 mutant cerebellar phenotype. (A and D) Nissl and immunofluorescent staining of cerebellar sections for the indicated genotypes with anticalbindin, anti-NeuN, anti-BLBP, and anti-Pax6 antibodies at P7 and P14, respectively. Arrows point to cerebellar lobules. (Scale bars: 500 μm and 1 mm.) (B) Representative Western blots examining the expression of β-catenin, Rack1, and Shh signaling downstream molecules in the control, hGFAP-Cre;Rack1F/F, hGFAP-Cre;Ctnnb1F/F, and hGFAP-DKO mutant cerebellum, respectively, at P7. IB, immunoblot; MW, molecular weight. (C) Quantitative analysis indicates significantly decreased expression of Gli1 and HDAC1/HDAC2 in both hGFAP-Cre;Rack1F/F and hGFAP-Cre;Ctnnb1F/F single-mutant mice compared with Rack1F/F controls, which could be significantly rescued in hGFAP-DKO mice (mean ± SEM; **P < 0.001, n = 4). (E) Representative Western blots examining the expression of β-catenin, Rack1, and Shh signaling downstream molecules in control, Atoh1-Cre;Rack1F/F, Atoh1-Cre;Ctnnb1F/F, and Atoh1-DKO double-mutant cerebellum, respectively, at P14. (F) Quantitative analysis indicates significantly decreased expression of Gli1 and HDAC1/HDAC2 in Atoh1-Cre;Rack1F/F, Atoh1-Cre;Ctnnb1F/F, and Atoh1-DKO muta.9nt mice compared with Rack1F/F controls. The expression of Gli1 and HDAC1/HDAC2 in Atoh1-DKO double-mutant mice is indistinguishable compared with Atoh1-Cre;Rack1F/F or Atoh1-Cre;Ctnnb1F/F single-mutant mice (mean ± SEM; P > 0.05, n = 5). n.s., not significant.
We next asked if the expression of Shh signaling molecules Gli1 and HDAC1/HDAC2 is altered in either hGFAP-Cre– or Atoh1-Cre–mediated Ctnnb1 or Rack1F/F;Ctnnb1F/F KO mice. Decreased expression of Gli1 and HDAC1/HDAC2 in both Rack1F/F and Ctnnb1F/F single mutants was significantly rescued in hGFAP-DKO mice, but not in Atoh1-DKO mice (Fig. 6 C and F). In addition, using cultured primary GNPs isolated from Ctnnb1F/F single-floxed mice or Rack1F/F;Ctnnb1F/F double-floxed mice, we found that Ctnnb1 single KO in GNPs did not affect Gli1 and HDAC1 expression (SI Appendix, Fig. S11). Moreover, DKO of Rack1 and Ctnnb1 in GNPs did not rescue the decreased expression of Gli1 and HDAC1/HDAC2 (SI Appendix, Fig. S12), which was consistent with the results observed in Atoh1-DKO mutants in vivo (Fig. 6 E and F).
Although hGFAP-Cre;Rack1F/F mutant mice died around P21, hGFAP-DKO mutant mice were fertile and survived past 6 mo. Similarly, using the footprint test and rotarod assay, we found that motor behavioral deficits observed in hGFAP-Cre;Rack1F/F mutant mice were significantly rescued in hGFAP-DKO mice (SI Appendix, Fig. S13). Together, these results suggest that double deletion of Rack1 and Ctnnb1 in NSCs, but not in GNPs, sufficiently rescues disrupted cerebellar development and locomotor function in hGFAP-Cre;Rack1F/F mice.
Rack1-Mediated Stabilization of HDAC1/HDAC2 Is Essential for GNP Proliferation and Migration by Activating Shh Signaling.
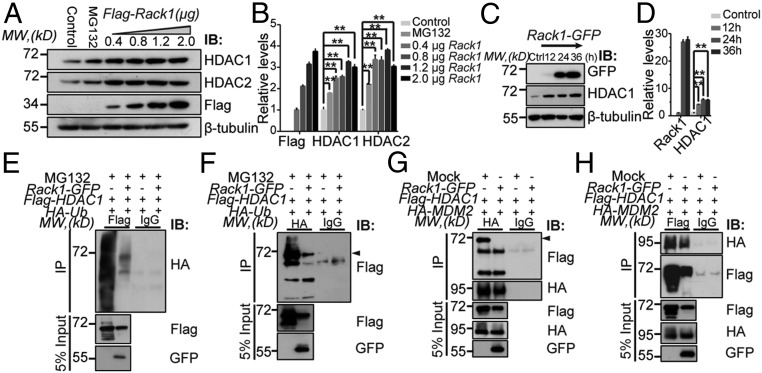
As double deletion of Rack1 and Ctnnb1 genes in GNPs cannot rescue impaired cerebellar development, we next examined whether the Shh signaling network is instead involved in regulating GNPs. Our previous results identified decreased Shh signaling, including lower Gli1 and HDAC1/HDAC2 expression, in Rack1-deleted GNPs. Given that HDAC1 up-regulates Gli1 transcription and Gli1 deacetylation enhances Shh signaling (43, 44), we asked if Rack1 deletion down-regulates HDAC1/HDAC2, and whether this response critically underlies the impaired Shh signaling activation and developmental defects in GNPs. Overexpression of Rack1 in HEK293 cells up-regulated HDAC1/HDAC2 expression at a posttranslational, but not transcriptional, level in a dose- and time-dependent manner (Fig. 7 A–D and SI Appendix, Fig. S14). HDAC1 was heavily ubiquitinated in the presence of hemagglutinin-tagged ubiquitin (HA-Ub), but ubiquitinated HDAC1 was notably reduced after cotransfection with Rack1 (Fig. 7E). The Rack1-driven reduction of ubiquitinated HDAC1 was verified using reverse coimmunoprecipitation (co-IP) (Fig. 7F), indicating that Rack1 controls the ubiquitination and stability of HDAC1/HDAC2. In addition, the interaction between HDAC1 and its E3 ligase MDM2 was strongly inhibited in the presence of Rack1 (Fig. 7 G and H). Together, these findings reveal that Rack1 regulates the stability of HDAC1/HDAC2 by suppressing the ubiquitylation and MDM2-mediated proteasomal degradation pathway.
Fig. 7.
Rack1 stabilizes HDAC1/HDAC2 protein levels by inhibiting their polyubiquitination. (A and B) Representative Western blots and quantification show a dose-dependent increase in HDAC1/HDAC2 expression in HEK293 cells transfected with exogenous Flag-tagged Rack1. MG132-treated cells were detected as a positive control (**P < 0.001, n = 3). IB, immunoblot; MW, molecular weight. (C and D) Representative Western blots and quantification show a time-dependent increase in HDAC1 expression in HEK293 cells transfected with exogenous Flag-tagged Rack1 (**P < 0.001, n = 3). (E and F) Expression vectors encoding Flag-tagged HDAC1 (Flag-HDAC1) and hemagglutinin-tagged ubiquitin (HA-Ub) were transfected either with or without GFP-tagged Rack1 (Rack1-GFP) into HEK293 cells in the presence of 20 μg/mL MG132 to inhibit proteasome-dependent degradation. Protein extracts were immunoprecipitated with either anti-Flag or anti-HA and immunoblotted for HA or Flag, respectively. IgG was used as a negative control. The black arrowhead indicates Flag-HDAC1 (n = 4). IP, immunoprecipitate. (G and H) HEK293 cells were cotransfected with Flag-HDAC1 and HA-MDM2 with or without Rack1-GFP, and protein extracts were immunoprecipitated with anti-HA or anti-Flag and immunoblotted for Flag or HA, respectively. IgG was used as a negative control. The black arrowhead indicates Flag-HDAC1 (n = 4).
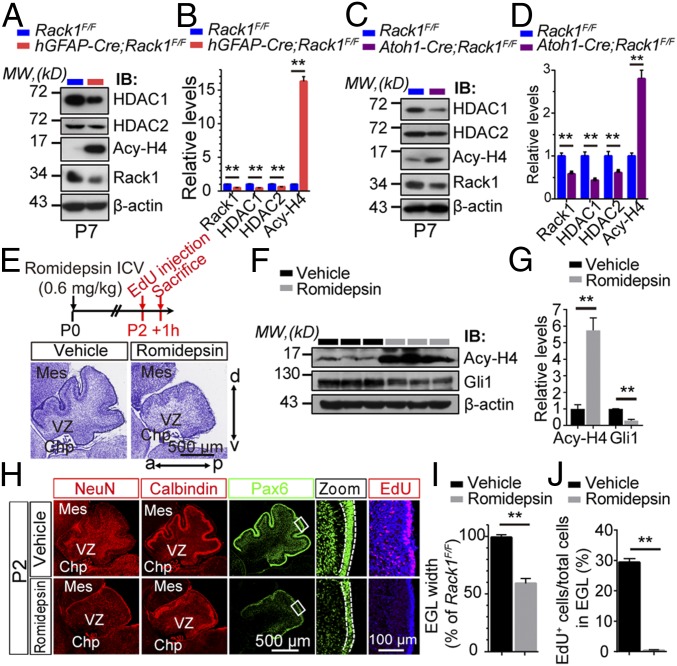
The expression of HDAC1/HDAC2 was significantly reduced in both hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mutant mice (P < 0.01, n = 3; Fig. 8 A–D). In contrast, the depletion of Rack1 led to significantly increased levels of acetylated histone H4 (Acy-H4) (P = 0.0001, n = 3; Fig. 8 A–D), suggesting that Rack1 is involved in the regulation of histone H4 deacetylation by stabilizing HDAC1/HDAC2 in GNPs. Interestingly, intracerebroventricular injection of romidepsin, a specific pharmacological inhibitor for HDAC1/HDAC2, dramatically suppresses cerebellar fissure formation (Fig. 8E), suggesting a critical role of HDAC1/HDAC2 for cerebellum development.
Fig. 8.
Pharmacological inhibition of HDAC1/HDAC2 activity in the early postnatal cerebellum phenocopies Rack1 mutants. (A and B) Representative Western blots and quantification examining the expression of HDAC1/HDAC2, Acy-H4, and Rack1 in control and hGFAP-Cre;Rack1F/F mutant cerebellum at P7. The expression of HDAC1/HDAC2 and Rack1 was significantly decreased, whereas Acy-H4 was significantly increased in hGFAP-Cre;Rack1F/F mutants (mean ± SEM; **P = 0.0006, **P = 0.0004, **P = 0.0012, and **P < 0.0001, respectively; n = 3). IB, immunoblot; MW, molecular weight. (C and D) Representative Western blots and quantification examining the expression of HDAC1/HDAC2, Acy-H4, and Rack1 in control and Atoh1-Cre;Rack1F/F mutant cerebellum at P7. Atoh1-Cre;Rack1F/F mutants show a significant decrease in HDAC1/HDAC2 and Rack1 expression, while Acy-H4 was dramatically increased (mean ± SEM; **P = 0.0008, **P = 0.0006, **P = 0.0042, and **P < 0.0001, respectively; n = 3). (E) Schematic showing the experimental design and Nissl staining of cerebellar sections at P2 following treatment with the HDAC1/HDAC2 pharmacological inhibitor romidepsin (0.6 mg/kg) at P0. (Scale bar: 500 μm.) a, anterior; Chp, choroid plexus; d, dorsal; ICV, intracerebroventricular; Mes, mesencephalon; p, posterior; v, ventral; VZ, ventricular zone. (F and G) Representative Western blots and quantification examining Acy-H4 and Gli1 expression in control and romidespin-treated groups (mean ± SEM; **P < 0.0001, n = 3 for each examined protein). (H) Representative images of immunofluorescent staining with anti-NeuN, anticalbindin, anti-Pax6, and anti-EdU antibodies showing impaired cerebellar morphogenesis at P2 and deficient cellular proliferation following romidepsin treatment. (Scale bar: 500 μm.) (I and J) Quantitative analysis shows significantly decreased Pax6+ EGL width and fewer EdU-incorporated GNPs within the EGL after treatment with romidepsin at P0 (mean ± SEM; **P < 0.0001, n = 5).
In addition, romidepsin-mediated HDAC1/HDAC2 inhibition reduced Gli1 and up-regulated Acy-H4 expression in vivo, suggesting lower Shh signaling levels (Fig. 8 F and G). Using EdU labeling to monitor proliferation and migration of GNPs after exposure to romidepsin, we found significantly reduced EGL width in treated mice (60.00 ± 1.84% of controls; P < 0.001, n = 3). The percentage of EdU+ cells in the EGL dramatically decreased to hardly detectable levels (29.88 ± 1.36% in controls vs. 0.61 ± 0.13% in romidepsin; P < 0.001, n = 3; Fig. 8 H–J). This result recapitulates our previous observations in hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mutants (Fig. 3 D–F and SI Appendix, Fig. S5 A and B), suggesting that HDAC1/HDAC2 activation is required for the development of GNPs in vivo and, more broadly, cerebellar development (44). Together, these results suggest that Rack1–mediated stabilization of HDAC1/HDAC2 is essential for the proliferation and migration of GNPs by activating Shh signaling.
Discussion
This study reveals a pivotal role of Rack1-mediated down-regulation of Wnt/β-catenin signaling in NSCs and up-regulation of Shh signaling in GNPs for controlling normal cerebellar development (Fig. 9). In the absence of Rack1, simultaneous deletion of β-catenin in NSCs significantly rescued cerebellum developmental deficits. Significantly decreased HDAC1/HDAC2 expression in Rack1-deleted GNPs prevented Shh signaling activation, resulting in abnormal GNP migration and differentiation that contributes to grossly dysmorphic cerebella.
Fig. 9.
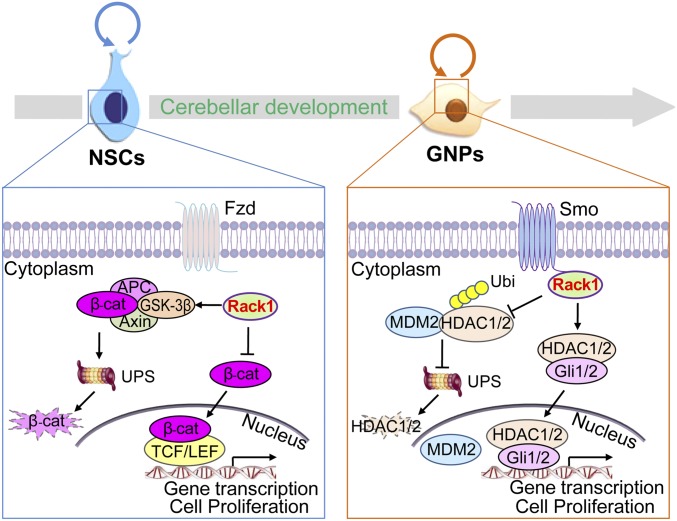
Working model for Rack1 in cerebellar development. Rack1 inhibits Wnt/β-catenin signaling by promoting the APC-, axin-, and GSK-3β–mediated proteasomal degradation of β-catenin. (Left) Lowered β-catenin transcriptional activity within the nucleus impairs NSC self-renewal at early cerebellar development. (Right) In contrast, Rack1 activates Shh/Gli signaling transduction by inhibiting MDM2-mediated proteasomal degradation of HDAC1/HDAC2, which, in turn, promotes the stabilization and transcriptional activity of Gli1/2 in GNPs during postnatal cerebellar development. Fzd, Frizzled; Smo, Smoothened; Ubi, ubiquitin; UPS, ubiquitin/proteasome system.
Rack1 Determines the Size and Foliation of the Cerebellum.
The cerebellar cortex contains distinct cell types that are positioned in a typical layered pattern. The outmost EGL contains dividing GNPs, which proliferate and migrate inward to form the terminally differentiated granule cells in the innermost IGL. Migrating GNPs pass through the middle PCL, which contains the cell bodies of PCs and BGs. As GNPs differentiate and migrate into the IGL, the EGL is eventually depleted and the ML is expanded from cells populating the area and growth of PC dendrites (3). This intricate pattern of neurogenesis and migration is tightly controlled by cell-autonomous and indirect signals in the developing cerebellum, which are not yet fully understood. In this study, we used various Cre recombinase-expressing lines to assess the function of Rack1 in cerebellar development. Our results demonstrate that Rack1 deletion in either NSCs or GNPs leads to a profound disruption of the cerebellar histoarchitecture. Furthermore, the effect of Rack1 at the cellular level is cell type-specific. In particular, deletion of Rack1 in embryonic NSCs causes severe cerebellar hypoplasia, decreased proliferation and migration of GNPs, dyslamination of PCs, and the generation of BGs with abnormal morphologies. Of note, hGFAP-Cre recombinase–expressing embryonic radial glial cells give rise to GNPs and BGs, but not PCs (28, 46) (SI Appendix, Fig. S2B). Thus, the morphological and lamination defects of PCs found in hGFAP-Cre;Rack1F/F mutants point to non–cell-autonomous regulation of Rack1 on PC development. Our analysis using GNP-specific Rack1 KO mice, Atoh1-Cre;Rack1F/F, further confirmed this notion (Fig. 1 and SI Appendix, Fig. S9). Intriguingly, Rack1 deletion in BGs results in structurally normal cerebellum, whereas disrupted BGs were observed in both hGFAP-Cre;Rack1F/F and Atoh1-Cre;Rack1F/F mutant mice, suggesting that Rack1 could directly regulate the generation of BGs by modulating NSC proliferation and differentiation or indirectly via its functions in GNPs, but not through cell-autonomous effects in BGs. Notably, Rack1 is a scaffolding protein localized within the cytoplasm and nuclei (47), and how this intracellular protein can non–cell-autonomously affect BG differentiation from its location in GNPs and granule cells requires further investigation.
Unlike development of the cerebral cortex, the size and foliation of the cerebellum occur much later in postnatal development (4), during which RL-derived GNPs proliferate and migrate tangentially above the surface of the cerebellum and sequentially give rise to the EGL and IGL (6, 48, 49), as well as determining cerebellar patterning. Previous results show that GNPs acquire their fate from the onset of their migration in the RL (1, 6, 48). Here, we show that Rack1 critically regulates GNP proliferation and migration during both embryonic and postnatal stages, which, in turn, determines the distinct foliation pattern of the mouse cerebellum.
Opposite Regulation of Wnt/β-Catenin and Shh Signaling Pathways by Rack1 During Cerebellum Development.
In cancer cells, Rack1 activates Shh signaling but suppresses Wnt/β-catenin signaling by binding and stabilizing the β-catenin destruction complex (34, 35), which includes adenomatous polyposis coli (APC). APC helps maintain the appropriate Wnt signaling activity that is necessary for radial progenitor function and cerebral corticogenesis, and double deletion of APC and β-catenin rescues cortical defects in Apc mutant mice (50). Here, we further elucidate the interplay between Rack1 and Wnt/β-catenin signaling in the control of cerebellar morphogenesis. Specifically, Rack1 deletion in NSCs or GNPs results in increased β-catenin activation. Intriguingly, the cerebellum developmental defects in NSC- or GNP-specific Rack1 KOs were similar to those in mice with constitutive activation of β-catenin or loss of APC (20, 51, 52). Surprisingly, the codeletion of β-catenin only rescued cerebellar defects in hGFAP promoter-specific, but not Atoh1 promoter-specific, Rack1 conditional KOs (Fig. 7), suggesting that increased Wnt/β-catenin signaling in the absence of Rack1 in NSCs, but not GNPs, is crucial for the disrupted cerebellar morphogenesis.
Previously, several lines of evidence have linked Wnt signaling inhibition to impaired NSC proliferation and severe brain malformations (53, 54). These seemingly contradictory results are likely due to the divergent roles that Wnt signaling plays in the CNS, and that Wnt likely requires precise time- and cell type-specific regulation by multiple molecules to maintain physiological activity levels that ensure normal development. Together with previous reports, our data suggest the following. First, Wnt/β-catenin signaling is critical for early NSC/NPC proliferation, but the repression of this signaling pathway may be an essential switch that allows the transition from proliferation to differentiation to generate the GNP pool. Second, the time-specific regulation of Wnt signaling is extremely subtle; thus, the role of Wnt/β-catenin can be divergent in the same cell population depending on different embryonic developmental phases and in postnatal stages. Third, Rack1-mediated Wnt/β-catenin signaling homeostasis seems to be essential and critical for cerebellar development during the embryonic and postnatal stages.
Interestingly, in contrast to the overactivation of Wnt/β-catenin signaling, both Gli1 and Gli2 expression was significantly decreased, indicating the impaired Shh signaling pathway in Rack1-deleted mice (Fig. 5). Shh is the major mitogen derived from PCs that promotes GNP proliferation by inducing the expression of Gli1 and downstream genes during cerebellar development (1, 12). Our results demonstrate that the impaired response to Shh signaling in Rack1 mutant mice may be caused by the abnormal activation of Smoothened receptor and destabilized HDAC1/HDAC2 expression in Rack1 mutant GNPs (43, 44) (Fig. 9). Previous studies have shown opposing activities of Shh and Wnt signaling pathways during somite and inner ear development (55, 56). Wnt signal promotes Atoh1 expression (57–59), which is an essential gene for both inner ear hair cell and cerebellar granule neuron formation. In contrast, Shh signaling in the inner ear represses or delays Atoh1 transcription, further supporting the opposite roles that Wnt and Shh play in hair cell development. Intriguingly, consistent with our findings in Rack1 mutant cerebellum, constitutively active Wnt/β-catenin induces inappropriate proliferation of Atoh1+ granule neurons, thereby preventing regular migration of granule neurons and normal cortical layering of the developing cerebellum (52). In contrast, Shh signaling critically influences the initial phases of territorial determination and regulates cerebellar progenitor cell proliferation and maturation during early embryonic development (60). Therefore, it is possible that early activation of Wnt/β-catenin is crucial for the initial development of the EGL by promoting NSC proliferation, while later on, as the Shh signaling pathway is gradually activated (12), down-regulation of the Wnt/β-catenin signaling pathway becomes important for the maintenance, proliferation, and migration of granule neurons.
Furthermore, the cross-talk between Wnt/β-catenin and Shh signaling pathways has been demonstrated in both embryonic organ morphogenesis and cancer development (61–63), indicating Shh may serve as an inhibitor of the Wnt/β-catenin signaling pathway. Together with those of others, our results support a model in which Wnt/β-catenin signaling controls NSC lineage commitment, Shh signaling stimulates GNP development, and both pathways are precisely modulated and integrated by Rack1 during cerebellar development. Finally, Shh-induced histone deacetylase activation is required for proliferation of GNPs and cerebellar formation (43, 44, 64). Here, we found that Rack1-mediated HDAC1/HDAC2 stabilization in GNPs is critical for Shh signaling activation and cerebellar development. This demonstration shows Rack1 activation of the Shh signaling pathway by inhibiting the proteasomal degradation of HDAC1/HDAC2.
Rack1 in Medulloblastoma Formation and Neural Development.
Cerebellar granule neurons account for the majority of neurons in the human brain (65). Dysfunctional Wnt/β-catenin and Shh signaling in cerebellar granule neurons are also associated with medulloblastoma, the most common malignant type of pediatric brain tumor (1, 16, 17). Defining the extracellular cues and intracellular signaling pathways that coordinate cerebellar neurogenesis, especially the proliferation, migration, and differentiation of GNPs, has been critical in unraveling the mechanisms underlying medulloblastoma formation and growth (66). In this study, we have shown severe cerebellar hypoplasia in Rack1 mutant mice as a result of abnormal GNP development. Moreover, HDACs have emerged as promising targets in preclinical antimedulloblastoma drug development (64, 67). The Rack1-mediated regulation of HDAC1/HDAC2 in GNPs described in this study reveals a potential signaling pathway in medulloblastoma pathogenesis, which requires further exploration.
As a seven-WDR domain–containing scaffolding protein, Rack1 has been previously shown to regulate neurite outgrowth, dendritic transport, and neural degeneration, suggesting a broader role in neural development and brain disorders in addition to regulating cerebellar morphogenesis (32). Recent human genetic studies have begun to recognize the importance of WDR genes in brain disorders, notably in intellectual disabilities associated with microcephaly and abnormal neural development (68). Thus, future investigations into the role of Rack1 in controlling brain development, emotion, and cognitive behaviors are warranted.
Materials and Methods
A more detailed description of material and methods is provided in SI Appendix, Materials and Methods.
Animals.
The hGFAP-Cre, Atoh1-Cre, GLAST-CreERT2, Ai9, Rack1F/F, and Ctnnb1F/F lines were generated as previously described (46, 69–72). Homozygous Rack1F/F mice were crossed with mice expressing a transgene encoding Cre recombinase driven by each particular promoter. Conditional KO mice were generated by the second generation, and Rack1F/F littermates served as controls. GLAST-CreERT2 recombinase was activated by i.p. injection of 50 mg/kg of 4-hydroxy tamoxifen (H7904; Sigma–Aldrich) once per day for four times from E17.5 to P0 (details are provided in SI Appendix). Mice genotypes were determined by PCR assay using murine tail DNA (details are provided in SI Appendix). All experiments with animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Beijing Institute of Basic Medical Sciences.
Immunofluorescent Staining.
The immunofluorescent staining of frozen cerebellar sections was performed using standard techniques as previously described (73). Briefly, frozen sections (30 μm) were washed for 10 min with 0.5% Triton X-100/PBS (PBS-T) for three times and then blocked with 3% BSA in PBS-T for 1 h. The sections were then incubated overnight at 4 °C with primary antibodies (details are provided in SI Appendix). The sections were washed for 10 min with 0.5% PBS-T for three times and subsequently treated with Alexa Fluor 594- or Alexa Fluor 488-conjugated fluorescent secondary antibody (1:500; Thermo Fisher Scientific). Nuclear staining was visualized with a mounting medium with DAPI (H-1200; VECTASHIELD). All images were processed and analyzed using FV10-ASW or Image Pro Plus software.
Western Blot and Co-IP.
The experiments were performed as previously described (74). Briefly, cerebellum tissues or cells were lysed by radioimmunoprecipitation assay (89901; Thermo) supplemented with 1× protease inhibitor mixture (A32965; Thermo) and phosphatase inhibitor mixture or immunoprecipitation lysis buffer [50 mM Hepes (pH 7.9), 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EDTA, 1 mM NaF] supplemented with 1× protease inhibitor mixture (A32965). Protein concentration was measured using the BCA Protein Assay Kit (23227; Thermo). To identify the protein levels of the bound proteins, samples (20–50 μg) were electrophoresed on SDS/PAGE and transferred to PVDF membranes (10600023; General Electric). The membranes were then blocked with 5% skim milk in 0.1% Tris-buffered saline/Tween 20 for 1 h and incubated overnight at 4 °C with primary antibodies (details are provided in SI Appendix). In each co-IP experiment, IgG was used as a negative control. Horseradish peroxidase and ECL (RPN2232; General Electric) were used to image protein bands on film. The film signal was electronically scanned and analyzed by Image Pro Plus.
Statistical Analysis.
The cerebellar immunofluorescence images were taken from 2 × 2–4 × 4 slices for confocal scanning or imaged with 20× or 40× objective multiple-layer scans. Immunofluorescence images were statistically analyzed using Image Pro Plus for fluorescence intensity, the number of positive cells per unit area, and the thickness of positive cell layers. Western blots, cerebellum area, and behavioral data were statistically analyzed using NDP.view and GraphPad Prism 6.0. The data between the two independent groups were shown as mean ± SEM for at least three independent experiments. The experimental data were analyzed using the Student’s t test, χ2 test, or one-way ANOVA. P < 0.05 (*) or P < 0.01 (**) was considered a statistically significant difference.
Supplementary Material
Acknowledgments
We thank Dr. Ying Shen at Zhejiang University for valuable input. This study was supported by the National Natural Science Foundation of China (Grants 31522029, 31770929, and 31371149), Program 973 from the State Key Development Program for Basic Research of China (Grant 2014CB542203), and the Beijing Municipal Science and Technology Commission (Grants Z181100001518001 and Z161100000216154).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813244116/-/DCSupplemental.
References
- 1.Hatten ME, Roussel MF. Development and cancer of the cerebellum. Trends Neurosci. 2011;34:134–142. doi: 10.1016/j.tins.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strick PL, Dum RP, Fiez JA. Cerebellum and nonmotor function. Annu Rev Neurosci. 2009;32:413–434. doi: 10.1146/annurev.neuro.31.060407.125606. [DOI] [PubMed] [Google Scholar]
- 3.Middleton FA, Strick PL. The cerebellum: An overview. Trends Neurosci. 1998;21:367–369. doi: 10.1016/s0166-2236(98)01330-7. [DOI] [PubMed] [Google Scholar]
- 4.Butts T, Green MJ, Wingate RJ. Development of the cerebellum: Simple steps to make a ‘little brain’. Development. 2014;141:4031–4041. doi: 10.1242/dev.106559. [DOI] [PubMed] [Google Scholar]
- 5.Sillitoe RV, Joyner AL. Morphology, molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Annu Rev Cell Dev Biol. 2007;23:549–577. doi: 10.1146/annurev.cellbio.23.090506.123237. [DOI] [PubMed] [Google Scholar]
- 6.Alder J, Cho NK, Hatten ME. Embryonic precursor cells from the rhombic lip are specified to a cerebellar granule neuron identity. Neuron. 1996;17:389–399. doi: 10.1016/s0896-6273(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nat Rev Neurosci. 2001;2:484–491. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- 8.Gao WO, Heintz N, Hatten ME. Cerebellar granule cell neurogenesis is regulated by cell-cell interactions in vitro. Neuron. 1991;6:705–715. doi: 10.1016/0896-6273(91)90168-y. [DOI] [PubMed] [Google Scholar]
- 9.Hatten ME. Neuronal regulation of astroglial morphology and proliferation in vitro. J Cell Biol. 1985;100:384–396. doi: 10.1083/jcb.100.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smeyne RJ, et al. Local control of granule cell generation by cerebellar Purkinje cells. Mol Cell Neurosci. 1995;6:230–251, and erratum (2006) 32:215. doi: 10.1006/mcne.1995.1019. [DOI] [PubMed] [Google Scholar]
- 11.Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998;21:375–382. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- 12.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 13.Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- 14.Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 15.Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development. 2004;131:5581–5590. doi: 10.1242/dev.01438. [DOI] [PubMed] [Google Scholar]
- 16.Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annu Rev Pathol. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- 17.Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol Med. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selvadurai HJ, Mason JO. Wnt/β-catenin signalling is active in a highly dynamic pattern during development of the mouse cerebellum. PLoS One. 2011;6:e23012. doi: 10.1371/journal.pone.0023012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schüller U, Rowitch DH. Beta-catenin function is required for cerebellar morphogenesis. Brain Res. 2007;1140:161–169. doi: 10.1016/j.brainres.2006.05.105. [DOI] [PubMed] [Google Scholar]
- 20.Lorenz A, et al. Severe alterations of cerebellar cortical development after constitutive activation of Wnt signaling in granule neuron precursors. Mol Cell Biol. 2011;31:3326–3338. doi: 10.1128/MCB.05718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basson MA, et al. Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development. 2008;135:889–898. doi: 10.1242/dev.011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J, Liu Z, Ornitz DM. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development. 2000;127:1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- 23.Berglund EO, et al. Ataxia and abnormal cerebellar microorganization in mice with ablated contactin gene expression. Neuron. 1999;24:739–750. doi: 10.1016/s0896-6273(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 24.Rice DS, Curran T. Mutant mice with scrambled brains: Understanding the signaling pathways that control cell positioning in the CNS. Genes Dev. 1999;13:2758–2773. doi: 10.1101/gad.13.21.2758. [DOI] [PubMed] [Google Scholar]
- 25.Li JY, Lao Z, Joyner AL. Changing requirements for Gbx2 in development of the cerebellum and maintenance of the mid/hindbrain organizer. Neuron. 2002;36:31–43. doi: 10.1016/s0896-6273(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 26.Aruga J, Inoue T, Hoshino J, Mikoshiba K. Zic2 controls cerebellar development in cooperation with Zic1. J Neurosci. 2002;22:218–225. doi: 10.1523/JNEUROSCI.22-01-00218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Y, et al. The engrailed homeobox genes determine the different foliation patterns in the vermis and hemispheres of the mammalian cerebellum. Development. 2010;137:519–529. doi: 10.1242/dev.027045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Ortiz E, et al. NF1 regulation of RAS/ERK signaling is required for appropriate granule neuron progenitor expansion and migration in cerebellar development. Genes Dev. 2014;28:2407–2420. doi: 10.1101/gad.246603.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams DR, Ron D, Kiely PA. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun Signal. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JJ, Xie D. RACK1, a versatile hub in cancer. Oncogene. 2015;34:1890–1898. doi: 10.1038/onc.2014.127. [DOI] [PubMed] [Google Scholar]
- 31.Volta V, et al. RACK1 depletion in a mouse model causes lethality, pigmentation deficits and reduction in protein synthesis efficiency. Cell Mol Life Sci. 2013;70:1439–1450. doi: 10.1007/s00018-012-1215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sklan EH, Podoly E, Soreq H. RACK1 has the nerve to act: Structure meets function in the nervous system. Prog Neurobiol. 2006;78:117–134. doi: 10.1016/j.pneurobio.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Ashique AM, et al. Localization of the scaffolding protein RACK1 in the developing and adult mouse brain. Brain Res. 2006;1069:31–38. doi: 10.1016/j.brainres.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 34.Deng YZ, et al. RACK1 suppresses gastric tumorigenesis by stabilizing the beta-catenin destruction complex. Gastroenterology. 2012;142:812–823.e15. doi: 10.1053/j.gastro.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 35.Shi S, et al. RACK1 promotes non-small-cell lung cancer tumorigenicity through activating sonic hedgehog signaling pathway. J Biol Chem. 2012;287:7845–7858. doi: 10.1074/jbc.M111.315416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sathyamurthy A, et al. ERBB3-mediated regulation of Bergmann glia proliferation in cerebellar lamination. Development. 2015;142:522–532. doi: 10.1242/dev.115931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada K, Watanabe M. Cytodifferentiation of Bergmann glia and its relationship with Purkinje cells. Anat Sci Int. 2002;77:94–108. doi: 10.1046/j.0022-7722.2002.00021.x. [DOI] [PubMed] [Google Scholar]
- 38.Weyer A, Schilling K. Developmental and cell type-specific expression of the neuronal marker NeuN in the murine cerebellum. J Neurosci Res. 2003;73:400–409. doi: 10.1002/jnr.10655. [DOI] [PubMed] [Google Scholar]
- 39.Yue Q, et al. PTEN deletion in Bergmann glia leads to premature differentiation and affects laminar organization. Development. 2005;132:3281–3291. doi: 10.1242/dev.01891. [DOI] [PubMed] [Google Scholar]
- 40.Aruga J, et al. Mouse Zic1 is involved in cerebellar development. J Neurosci. 1998;18:284–293. doi: 10.1523/JNEUROSCI.18-01-00284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leffler SR, et al. A mathematical model of granule cell generation during mouse cerebellum development. Bull Math Biol. 2016;78:859–878. doi: 10.1007/s11538-016-0163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mori T, et al. Inducible gene deletion in astroglia and radial glia–A valuable tool for functional and lineage analysis. Glia. 2006;54:21–34. doi: 10.1002/glia.20350. [DOI] [PubMed] [Google Scholar]
- 43.Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci USA. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canettieri G, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12:132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 45.Gilbertson RJ. Medulloblastoma: Signalling a change in treatment. Lancet Oncol. 2004;5:209–218. doi: 10.1016/S1470-2045(04)01424-X. [DOI] [PubMed] [Google Scholar]
- 46.Zhuo L, et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 47.Baumann M, et al. The PKC targeting protein RACK1 interacts with the Epstein-Barr virus activator protein BZLF1. Eur J Biochem. 2000;267:3891–3901. doi: 10.1046/j.1432-1327.2000.01430.x. [DOI] [PubMed] [Google Scholar]
- 48.Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 49.Wingate RJ, Hatten ME. The role of the rhombic lip in avian cerebellum development. Development. 1999;126:4395–4404. doi: 10.1242/dev.126.20.4395. [DOI] [PubMed] [Google Scholar]
- 50.Nakagawa N, et al. APC sets the Wnt tone necessary for cerebral cortical progenitor development. Genes Dev. 2017;31:1679–1692. doi: 10.1101/gad.302679.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shintani T, Takeuchi Y, Fujikawa A, Noda M. Directional neuronal migration is impaired in mice lacking adenomatous polyposis coli 2. J Neurosci. 2012;32:6468–6484. doi: 10.1523/JNEUROSCI.0590-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pöschl J, Grammel D, Dorostkar MM, Kretzschmar HA, Schüller U. Constitutive activation of β-catenin in neural progenitors results in disrupted proliferation and migration of neurons within the central nervous system. Dev Biol. 2013;374:319–332. doi: 10.1016/j.ydbio.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 53.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 54.McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- 55.Brent AE, Tabin CJ. Developmental regulation of somite derivatives: Muscle, cartilage and tendon. Curr Opin Genet Dev. 2002;12:548–557. doi: 10.1016/s0959-437x(02)00339-8. [DOI] [PubMed] [Google Scholar]
- 56.Riccomagno MM, Takada S, Epstein DJ. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005;19:1612–1623. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi F, Cheng YF, Wang XL, Edge AS. Beta-catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3′ enhancer. J Biol Chem. 2010;285:392–400. doi: 10.1074/jbc.M109.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bok J, Zenczak C, Hwang CH, Wu DK. Auditory ganglion source of Sonic hedgehog regulates timing of cell cycle exit and differentiation of mammalian cochlear hair cells. Proc Natl Acad Sci USA. 2013;110:13869–13874. doi: 10.1073/pnas.1222341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tateya T, et al. Hedgehog signaling regulates prosensory cell properties during the basal-to-apical wave of hair cell differentiation in the mammalian cochlea. Development. 2013;140:3848–3857. doi: 10.1242/dev.095398. [DOI] [PubMed] [Google Scholar]
- 60.De Luca A, et al. Sonic hedgehog patterning during cerebellar development. Cell Mol Life Sci. 2016;73:291–303. doi: 10.1007/s00018-015-2065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iwatsuki K, et al. Wnt signaling interacts with Shh to regulate taste papilla development. Proc Natl Acad Sci USA. 2007;104:2253–2258. doi: 10.1073/pnas.0607399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van den Brink GR, et al. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet. 2004;36:277–282. doi: 10.1038/ng1304. [DOI] [PubMed] [Google Scholar]
- 63.Ahn Y, Sanderson BW, Klein OD, Krumlauf R. Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development. 2010;137:3221–3231. doi: 10.1242/dev.054668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SJ, et al. Sonic hedgehog-induced histone deacetylase activation is required for cerebellar granule precursor hyperplasia in medulloblastoma. PLoS One. 2013;8:e71455. doi: 10.1371/journal.pone.0071455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herculano-Houzel S, Lent R. Isotropic fractionator: A simple, rapid method for the quantification of total cell and neuron numbers in the brain. J Neurosci. 2005;25:2518–2521. doi: 10.1523/JNEUROSCI.4526-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roussel MF, Hatten ME. Cerebellum development and medulloblastoma. Curr Top Dev Biol. 2011;94:235–282. doi: 10.1016/B978-0-12-380916-2.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sonnemann J, et al. Histone deacetylase inhibitors induce cell death and enhance the susceptibility to ionizing radiation, etoposide, and TRAIL in medulloblastoma cells. Int J Oncol. 2006;28:755–766. [PubMed] [Google Scholar]
- 68.Kannan M, et al. Sanger Mouse Genetics Project WD40-repeat 47, a microtubule-associated protein, is essential for brain development and autophagy. Proc Natl Acad Sci USA. 2017;114:E9308–E9317. doi: 10.1073/pnas.1713625114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agarwal A, et al. Transient opening of the mitochondrial permeability transition pore induces microdomain calcium transients in astrocyte processes. Neuron. 2017;93:587–605.e7. doi: 10.1016/j.neuron.2016.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Y, et al. RACK1 promotes autophagy by enhancing the Atg14L-Beclin 1-Vps34-Vps15 complex formation upon phosphorylation by AMPK. Cell Rep. 2015;13:1407–1417. doi: 10.1016/j.celrep.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 71.Matei V, et al. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu H, et al. Slit2 as a β-catenin/Ctnnb1-dependent retrograde signal for presynaptic differentiation. eLife. 2015;4:e07266. doi: 10.7554/eLife.07266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu H, et al. β-catenin gain of function in muscles impairs neuromuscular junction formation. Development. 2012;139:2392–2404. doi: 10.1242/dev.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.