Significance
Actin filaments comprise a major part of the cytoskeleton of eukaryotic cells and serve as tracks for myosin motor proteins. The filaments assemble from actin monomers with a bound ATP. After polymerization, actin rapidly hydrolyzes the bound ATP and slowly dissociates the γ-phosphate. ADP-actin filaments then disassemble to recycle the subunits. Understanding how actin filaments assemble, disassemble and interact with numerous regulatory proteins depends on knowing the structure of the filament. We determined high-quality structures of actin filaments with bound AMPPNP (a slowly hydrolyzed ATP analog), ADP and phosphate, or ADP by cryo-electron microscopy. These structures show how conformational changes during actin assembly promote ATP hydrolysis and faster growth at one end of the filament than the other.
Keywords: actin, ATPase, cryo-electron microscopy, polymerization
Abstract
We used cryo-electron microscopy (cryo-EM) to reconstruct actin filaments with bound AMPPNP (β,γ-imidoadenosine 5′-triphosphate, an ATP analog, resolution 3.1 Å), ADP-Pi (ADP with inorganic phosphate, resolution 3.1 Å), or ADP (resolution 3.6 Å). Subunits in the three filaments have similar backbone conformations, so assembly rather than ATP hydrolysis or phosphate dissociation is responsible for their flattened conformation in filaments. Polymerization increases the rate of ATP hydrolysis by changing the positions of the side chains of Q137 and H161 in the active site. Flattening during assembly also promotes interactions along both the long-pitch and short-pitch helices. In particular, conformational changes in subdomain 3 open up multiple favorable interactions with the DNase-I binding loop in subdomain 2 of the adjacent subunit. Subunits at the barbed end of the filament are likely to be in this favorable conformation, while monomers are not. This difference explains why filaments grow faster at the barbed end than the pointed end. When phosphate dissociates from ADP-Pi-actin through a backdoor channel, the conformation of the C terminus changes so it distorts the DNase binding loop, which allows cofilin binding, and a network of interactions among S14, H73, G74, N111, R177, and G158 rearranges to open the phosphate release site.
Actin, one of the most abundant proteins in eukaryotes, contributes to both cellular structure and motility. Filaments of actin form the cytoskeleton and interact with myosin motor proteins for cytokinesis, muscle contraction, and transporting particles inside cells. Decades of biochemical analysis produced a detailed explanation of actin assembly, including rate and equilibrium constants for most of the reactions (1, 2). Actin binds ATP, which is hydrolyzed when the protein incorporates into filaments (3). Subsequent slow dissociation of the γ-phosphate (4) favors depolymerization and changes the affinity of the filament for proteins such as cofilin (2).
Despite more than 100 crystal structures of actin monomers (SI Appendix, Fig. S1A) (1), understanding the structure of actin filaments progressed slowly (SI Appendix, Table S1). Holmes et al. (5) based the first atomic model of the actin filament on X-ray fiber diffraction data extending to 8.4 Å and their crystal structure of the actin molecule. Oda et al. (6) improved the model using X-ray fiber diffraction data to 3.3 Å resolution and discovered that subunits in filaments are flattened compared with monomers. Fujii et al. (7) followed with the first cryogenic electron microscopy (cryo-EM) structure of the Mg2+-ADP-actin filament, achieving a resolution of 6.6 Å before the advent of direct electron detectors. Since then, improvements in cryo-EM methods (8–11) extended the resolution of filament reconstructions to 3.3 Å. The preprint of this paper (12) and a paper from Merino et al. (13) offered the first views of the structures of AMP-PNP and ADP-Pi-actin filaments.
Here, we report the highest resolution, cryo-EM structures of actin filaments with bound ATP analog β,γ-imidoadenosine 5′-triphosphate (AMPPNP) (3.1 Å) and ADP with inorganic phosphate (ADP-Pi) (3.1 Å) as well as a 3.6-Å resolution structure of the ADP filament. These structures of the three well-characterized nucleotide states of actin monomers and filaments (14–16) provide insights about unanswered questions regarding rapid ATP hydrolysis (17, 18), faster elongation at “barbed ends” than “pointed ends” (19), and changes associated with dissociation of the γ-phosphate.
Results
Cryo-EM Reconstructions of AMPPNP-Actin, ADP-Pi-Actin, and ADP-Actin Filaments.
We obtained near atomic resolution reconstructions of actin filaments with Mg2+ and three different bound nucleotides, AMPPNP, ADP-Pi, or ADP (Fig. 1). To assure that these samples were homogeneous, we purified Ca-ATP-actin monomers from chicken skeletal muscle (which has the same sequence as the more frequently used rabbit skeletal muscle actin) and converted these monomers to the desired nucleotide states before polymerization as described in Materials and Methods. Since actin filaments hydrolyze ATP rapidly (18) and irreversibly (4), we used the slowly hydrolyzed ATP analog AMPPNP with a nitrogen atom replacing the oxygen atom bridging β-phosphate and γ-phosphate. AMPPNP-actin polymerizes with the same kinetics as ATP-actin (16), and ATP and AMPPNP have identical conformations in actin monomers (20).
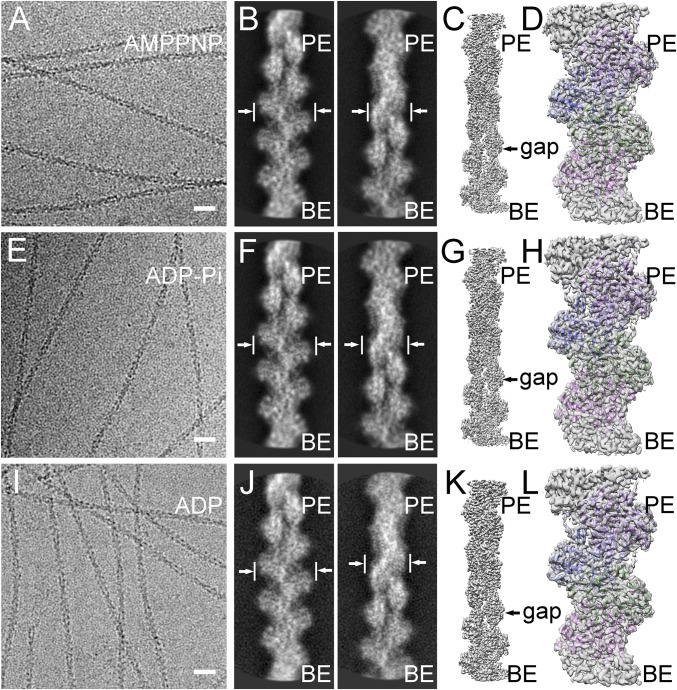
Fig. 1.
Helical reconstructions of actin filaments from low-dose electron cryo-micrographs. (A–D) AMPPNP-actin filaments reconstructed at 3.1 Å resolution. (E–H) ADP-Pi-actin filaments reconstructed at 3.1 Å resolution. (I–L) ADP-actin filaments reconstructed at 3.6 Å resolution. The maps are contoured at 0.020 V. The pointed ends (PE; top) and barbed end (BE; bottom) are labeled. (A, E, and I) Representative images. (Scale bas: 20 nm.) (B, F, and J) Pairs of contrast-inverted 2D class averages showing the wide (9 nm) and narrow (6 nm) projections of filaments. (C, G, and K) Three-dimensional reconstructions. (D, H, and L) Models fit into the density maps with the chain in each subunit colored differently.
We employed cryo-EM to collect high-quality images of actin filaments with the three different bound nucleotides (Fig. 1 A, E, and I) using a direct-electron detector after an energy filter. After drift correction and dose weighting, regions from the middles of filaments were boxed and windowed into segments of ∼12 subunits. The box was moved successively along the filament with one new subunit per particle. We used 310,000 particles of AMP-PNP-actin, 410,000 of ADP-Pi-actin, and 120,000 of ADP-actin filaments for reference-free 2D classifications (Fig. 1 B, F, and J) and 3D reconstructions (Fig. 1 C, G, and K) by iterative helical real-space reconstruction (21). The reconstructions were refined to global resolutions of 3.1 Å for AMPPNP-actin filaments, 3.1 Å for ADP-Pi-actin filaments, and 3.6 Å for ADP-actin filaments (SI Appendix, Fig. S2 A, C, and E and Table S2) estimated by Fourier shell correlation with 0.143 criterion (FSC0.143). We confirmed global resolutions with layer-line images calculated from back projected images (SI Appendix, Fig. S2 B, D, and F) and by calculating local resolution estimations (SI Appendix, Fig. S3 A–C).
The cryo-EM maps (Fig. 2 A and B) clearly resolved the bound nucleotides and side chains of most residues (SI Appendix, Fig. S4), allowing us to build atomic models (Figs. 1 D, H, and L and 2). The ADP-Pi-actin and AMPPNP-actin maps had backbone bulges for many carbonyl oxygens. The backbone densities were weakest at G48 in the DNase I-binding loop (D-loop), but the side chain densities of the adjacent residues were strong enough to build with confidence models of the whole D-loop for AMP-PNP-actin and ADP-Pi-actin filaments and all of the D-loop of the ADP-actin filament except residues G46, M47, G48, and Q49. Only residues N-terminal residues 1–3 were missing from the maps, likely due to their flexibility.
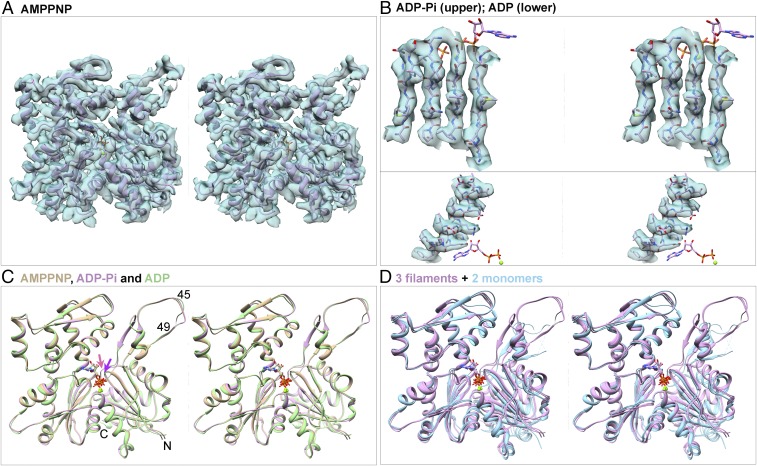
Fig. 2.
Stereoviews of models of actin filament subunits and actin monomers and of EM densities of actin filament subunits contoured at the same levels as Fig. 1. (A) Map and ribbon diagram of one subunit from the AMPPNP-actin filament. (B) Maps and stick figure models show densities for the side chains of β-strands from the ADP-Pi-actin filament (Upper) and an α-helix from the ADP-actin filament (Lower). The stick models of the nucleotides are for orientation. (C) Superimposed ribbon diagrams of subunits from the AMPPNP-actin (tan), ADP-Pi-actin (plum), and ADP-actin (green) filaments show that their backbones are nearly identical, except P1 loop (purple arrow) and sensor loop (salmon arrow). (D) Three ribbon diagrams from C are superimposed on ribbon diagrams of actin monomers (light blue) with bound Mg-ATP (PDB ID code 1NM1) or Mg-ADP (PDB ID code 3A5L) to show differences between filament subunits and monomers. Structures in C and D are aligned using subdomain 3.
Conformations of Actin Subunits in Filaments with Three Different Bound Nucleotides.
The backbone conformations of polymerized actin subunits with bound AMPPNP and ADP-Pi are identical in our models (Fig. 2C) and those of Merino et al. (13), while the ADP-actin subunits differ only slightly, so assembly is responsible for the major conformational changes during actin polymerization (6–9, 22). The root mean square deviations (rmsds) between the α-carbon atoms in our three models are <0.5 Å with the exception of the C-terminal residues T351-F375 in subdomain 1 in ADP-actin, which differ by 0.5–2 Å. The following sections explain the small but functionally important differences after phosphate release. The conformations of polymerized actin subunits all differ from the corresponding nucleotide states of crystallized actin monomers (Fig. 2D).
Our three filament structures have helical parameters (SI Appendix, Fig. S1 B and C and Table S2) similar to each other and other filament structures (SI Appendix, Fig. S5). Most previous actin filament structures with subnanometer resolutions had bound ADP and a variety of ligands including gelsolin (6), tropomyosin (9), tropomyosin and myosin-II (10), phalloidin and myosin-I (11), or coronin and BeFx (23) (SI Appendix, Table S1). Our ADP-actin model fits reasonably well (rmsd: 1.56 Å) into Fujii’s lower resolution map of Mg-ADP-actin (EMDB accession no. 5168), but the nucleotide binding cleft is closed more tightly in our structure (SI Appendix, Fig. S5 C and H). The conformation of the subunits in our model of ADP-actin filaments is similar to human ADP-α-actin decorated with tropomyosin (rmsd: 0.62 Å; SI Appendix, Fig. S5 D and I), but differs from ADP-actin filaments with phalloidin and myosin-I (11) or Jasplakinolide (13), both of which are closely similar to our models of AMPPNP-actin and ADP-Pi-actin filaments.
Flattening of Actin Subunits During Polymerization.
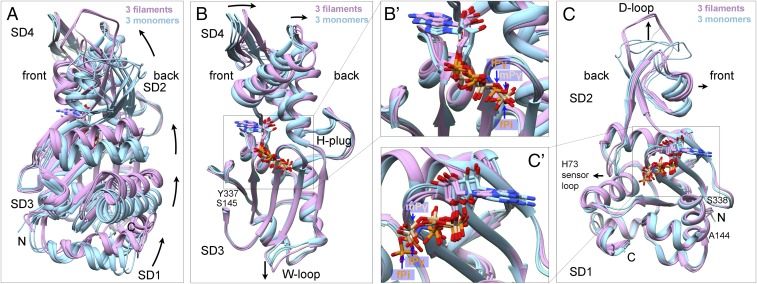
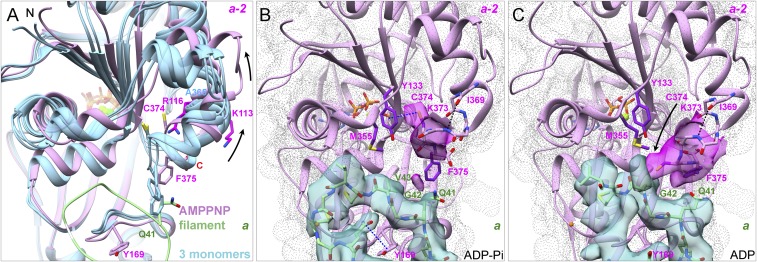
Actin subunits are flattened to the same extent in our filaments with bound Mg-AMPPNP, Mg-ADP-Pi, or Mg-ADP (Figs. 2D and 3 and SI Appendix, Figs. S5 F and G and S6A). The interdomain dihedral angle of subunits in our Mg-AMPPNP-actin filament model are rotated 13.1° relative to the crystal structure of the Ca-ATP-actin monomer bound to DNase-I (PDB ID code 2A42) (Fig. 3A) and by 18.7° relative to the TMR-labeled Ca-ADP-actin monomer (PDB ID code 1J6Z). DynDom analysis (24) confirmed that rotation occurs around a hinge helix (residues 137–145) and a hinge loop (residues 335–337) (6, 7). The catalytic residue Q137 stands on the hinge helix. The side chain of K336 in the center of the hinge loop interacts with the adenosine base as in monomers (25). We confirmed the discovery of Fujii et al. (7) that the subdomains in both halves of actin also flatten during polymerization (Fig. 3 B and C).
Fig. 3.
Ribbon diagrams showing interdomain rotation and intersubdomain (SD) bending upon filament formation. Light blue shows three actin monomer crystal structures in the closed conformation: rabbit skeletal muscle Ca-ATP-actin complexed with DNase I (PDB ID code 2A42), Dictyostelium Mg-ADP-actin complexed with human gelsolin segment 1 (PDB ID code 3A5L), and budding yeast Mg-ATP-actin complexed with human gelsolin segment 1 (PDB ID code 1YAG). Plum shows our three EM structures of actin filaments. Nucleotides are shown as stick figures with phosphorus atoms orange in filaments and tan in monomers. Arrows mark differences between monomers and filaments. (A) Dihedral angle-like interdomain rotation. The six molecules are aligned using subdomains 3 and 4 (residues 145–337). (B) Bending of subdomain 4 (181–269) relative to subdomain 3 (residues 145–180 and 270–337). The molecules are aligned using subdomain 3. Inset B′ shows the phosphates in monomers and filaments. (C) Bending of subdomain 2 relative to subdomain 1. The molecules are aligned using subdomain 1 (residues 5–32, 70–144, and 338–370). Inset C′ shows the phosphates in monomers and filaments.
Contacts Between Subunits in Filaments.
Each subunit in the middle of a filament buries 3,540 Å2 of surface area in contacts four neighbors (SI Appendix, Fig. S1B). “Interstrand” interactions along the short-pitch helix (subunits a+1 to a to a−1) (Fig. 4 and SI Appendix, Fig. S1B) each bury 490 Å2 of surface area. “Intrastrand” interactions along the long-pitch helix bury 1,180 Å2 of surface area between subdomains 2 and 4 of each subunit with subdomain 3 of the neighbor toward the pointed end. The stronger contacts along the long-pitch helix likely explain why longitudinal dimers are favored over short-pitch dimers for the first step in nucleation (26).
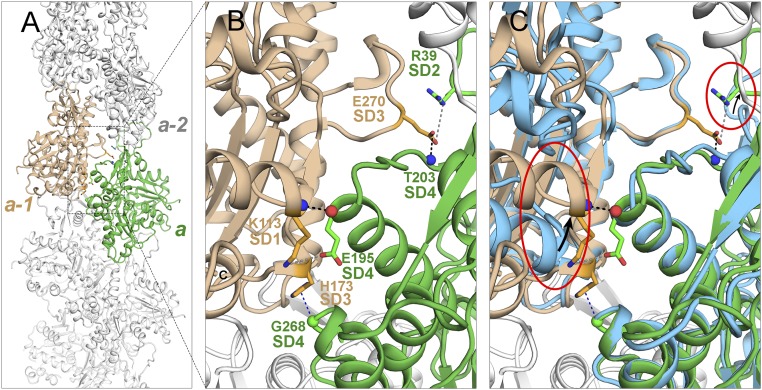
Fig. 4.
Ribbon diagrams showing lateral contacts between subunits along the short-pitch helix. (A) Overview of filament with two subunits highlighted in green and tan. (B) Detail of contacts between subunits a (green) and a-1 (tan) with labels on stick figures of the interacting residues. (C) Same as B with two overlaid actin monomers (light blue) aligned with subdomains 3 and 4. This superimposition shows that subunit flattening allows interactions between the backbone and side chain of E195 in subdomain 4 of subunit a and K113 in subdomain 1 of subunit a-1 and between side chain of E270 in subdomain 3 of subunit a-1 with side chain of R39 in subdomain 2 of subunit a. The red ovals highlight differences.
Conformational Changes on the Surface of Subunits Associated with Polymerization.
Local conformational changes during polymerization create complementary surfaces for interactions between the subunits along both the short-pitch (Fig. 4) and long-pitch helices (Figs. 5 and 6). These interfaces are similar in filaments with each bound nucleotide, so we describe them together in Figs. 4–6. The contacts in our three models are similar to those in other high-resolution structures (10, 11, 13). Our maps allow for accurate positioning of several side chains that make hydrophilic interactions between subunits.
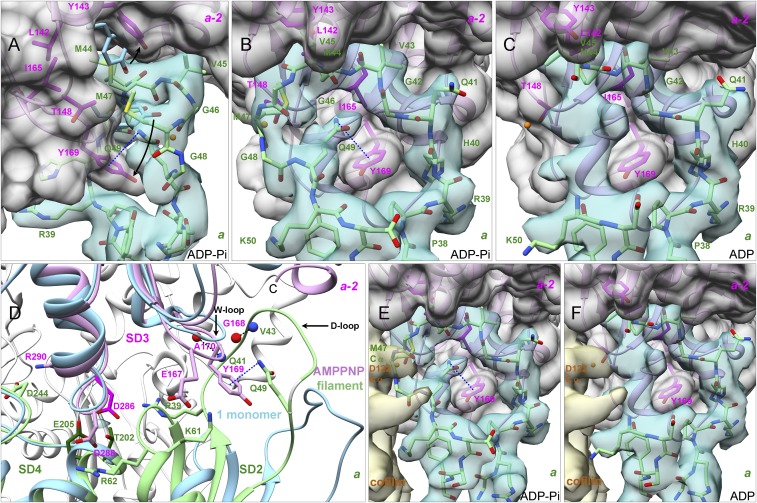
Fig. 5.
Interactions along the long-pitch helix of the actin filament between subunit a (green models) and subdomain 3 of subunit a-2 (plum models). The models are rendered as ribbon diagrams showing backbones or as stick figures showing backbones and side chains. Experimental densities are semitransparent surfaces in turquoise for subunit a. The gray surfaces for subunit a-2 were calculated from models. (A and B) Orthogonal views from the ADP-Pi-actin filament of the D-loop (residues 40–50) of subunit a wrapped snugly around the W-loop (residues 165–172) of subdomain 3 of subunit a-2 with a π–cation interaction (dashed line) between Q49 and Y169. Arrows show how subunit flattening separates the side chains of Y143 and Y169 of subunit a-2 from their position in monomers (blue stick figures; Mg-ADP-actin with human gelsolin segment 1, PDB ID code 3A5L) to open a cavity for the side chain of M44 of subunit a. (C) D-loop of subunit a from the ADP-actin filament docked on subdomain 3 of subunit a-2. Models of residues 46–49 are not included due to the ambiguous map. (D) Polar contacts along the long-pitch helix of the AMPPNP-actin filament formed by side chains: R39 in subdomain 2 with D286 in subdomain 3, K61 in subdomain 2 with E167 in subdomain 3, R62 in subdomain 2 with D288 in subdomain 3, and D244 in subdomain 4 with R290 in subdomain 3. The turquoise density among the side chains of T202, E205, and D286 is the proposed binding site for the “polymerization cation.” The balls are backbone N (blue) and O (red) atoms. The blue ribbon diagram shows a Ca-ATP-actin monomer complexed with DNase I, PDB ID code 2A42 aligned using the α-carbons of subdomains 3 and 4, residues 145–337. (E and F) Placement of cofilin from a cofilin-decorated ADP-actin filament (EMDB accession no. 6844; PDB ID code 5YU8) on our structures. (E) The α-carbon atom of cofilin D122 (orange ball in B and C) clashes with the α-carbon of M47 in our ADP-Pi-actin filament. (F) No clash in our ADP-actin filament.
Fig. 6.
Interactions of the C terminus of subunit a-2 with the D-loop of subunit a. (A) Ribbon diagrams comparing the C-terminal regions of our AMPPNP-actin filament structure (plum) with crystal structures (blue) of three actin monomers (Ca-ATP-actin with DNase I, PDB ID code 2A42; Mg-ADP-actin with human gelsolin segment 1, PDB ID code 3A5L; and Mg-ATP-actin with human gelsolin segment 1, PDB ID code 1YAG). F375 clashes with the D-loop of subunit a (green) in two monomers but not in the filament (arrows). (B and C) Experimental densities are semitransparent surfaces in turquoise for subunit a and in magenta for subunit a-2. The gray dot surface for subunit a-2 was calculated from models. Models are rendered as ribbon diagrams and stick figures for the backbone of residues 369–375 and the side chains of C374 and F375. (B) ADP-Pi-actin filament with an intramolecular SH–π interaction between the side chain of C374 and the ring of Y133 of subunit a-2. The side chain of F375 is buried in a hydrophobic pocket formed by P109, L110, I136, V139, A170, P172, and I175 of its subunit and V43 of subunit a. (C) After phosphate release in the ADP-actin filament, the side chain of C374 of subunit a-2 moves ∼8 Å close to the side chain of M355 of its own subunit and forms hydrogen bonds with the backbone of G42 or V43 of subunit a.
Interactions along the short-pitch helix.
Interstrand contacts are largely (∼87%) between subdomain 4 at the pointed end of one subunit and subdomain 3 at the barbed end of its neighbor (Fig. 4). Subunit flattening is required for three polar contacts between the subunits: (i and ii) a hydrogen bond between the backbone N of K113 of subunit a-1 and backbone O of E195 of subunit a and charge–charge interaction of their side chains; and (iii) electrostatic interaction of the side chain of E270 in the “H-plug” of subdomain 3 of subunit a-1 and side chain of R39 in subdomain 2 of subunit a. Subunit flattening is not required for two other contacts (Fig. 4C): a hydrogen bond between the side chain of E270 with the backbone of T203 in the upper part of the nucleotide-binding cleft of subunit a and a contact between the backbone of G268 in the H-plug of subunit a with the side chain of H173 in subdomain 3 of subunit a-1.
Interactions along the long-pitch helix.
Longitudinal contacts involve interactions of subdomains 2 and 4 of subunit a with subdomain 3 at the barbed end of subunit a-2 (Figs. 5 and 6). Subdomain 2 is small but makes three pairs of charge–charge interactions, a π–cation interaction, two pairs of backbone hydrogen bonds, and extensive hydrophobic contacts with its neighbor.
Subdomains 2 and 4 make multiple polar contacts with the barbed end subdomain 3 of subunit a-2 (Fig. 5D). The side chains of T202 and E205 in subdomain 4 of subunit a and D286 in subdomain 3 of subunit a-2 are proposed to bind the “polymerization cation” that stabilizes the filament (27). Our map of the Mg-AMPPNP-actin filament has weak density in the position proposed for the polymerization site cation (Fig. 5D); the densities of the surrounding acidic side chains are also weak, as usual in EM maps (28). Another divalent cation is proposed to bind subdomain 2 and stiffen the filament (27), but none of our maps has density at the proposed position. The stiffness-related cation ion might be located between the side chain of D56 in subdomain 2 and the backbone of V30 in subdomain 1, as seen in the crystal structure of TMR-labeled actin monomers (29) (PDB ID code 1J6Z).
Interaction of the D-loop (residues 40–50) of subunit a with subdomain 3 of subunit a-2 forms a major contact along the long-pitch helix (Fig. 5 A–C). The D-loop is flexible in monomers, so the electron densities are incomplete or weak in most crystal structures unless bound to associated proteins. The maps of the AMPPNP-actin and ADP-Pi-actin filaments have strong densities for the backbones and side chains of all 11 D-loop residues except for G48, both similar to a cryo-EM structure of ADP-actin with bound phalloidin and myosin-I (11). Most of the D-loop is similar in our map of ADP-actin filaments, but the densities for residues G46-M47-G48-Q49 are in a different position and are weak, perhaps due to flexibility. This difference allows cofilin to bind the ADP-actin filaments (30) without steric interference from the D-loop (Fig. 5 E and F). Interpreting that part of our map is difficult, so we did not include these four residues in our model of ADP-actin filaments. Our maps do not have densities for alternate paths of the D-loop as reported by Merino et al. (13) for filaments of ADP-Be-Fx-actin, ADP-Pi-actin with jasplakinolide, ADP-actin with jasplakinolide, and AMPPNP.
Interactions of the D-loop with the adjacent subunit depend on displacement of the W-loop (residues 165–172) in subdomain 3 with Y169 at its tip from its position in monomers toward the barbed end of the subunit (Figs. 3B and 5A). The α-carbon of Y169 moves about 2.2 Å during polymerization, similar to the 3.7 Å model of the ADP-actin-tropomyosin filament (9) (PDB ID code 3J8A), but different from the 2016 model of the same ADP-actin-tropomyosin filament (10) (PDB ID code 5JLF). The maps of Merino et al. are similar to ours in this region, but their models differ, likely due to limited map resolutions. These structures do not explain how flattening of the subunits in the filament drives this conformational change in the W-loop, which facilitates two crucial interactions of the D-loop of subunit a with subunit a-2.
First, displacement of the W-loop allows the D-loop of subunit a to wrap around the side chain of Y169 (F169 in yeast) (Fig. 5 B and C), which forms an N-H⋅⋅⋅π bond with the side chain of D-loop residue Q49 (Fig. 5B). Hydrophobic contacts with P38 and I64 of subunit a immobilize the W-loop (Fig. 5C). Molecular dynamics simulations (31) implicated Y169 in filament formation, which was confirmed by biochemical experiments (32).
Second, movement of the W-loop opens a hydrophobic pocket in subunit a-2 surrounded by L142, Y143, T148, and I165 for insertion of the side chain of M44 from the D-loop of subunit a. This contact buries 151 Å2 of surface area. The distance between the Y143 CZ atom (next to the hydroxyl group) and the α-carbon of Y169 increases from 6.6 Å in monomers to 10.9 Å in filaments, avoiding a clash of the M44 side chain with the aromatic ring of Y143 and backbone of Y169. The side chain of M47 makes hydrophobic contacts with its tip close to Cβ of M44 and the side chain of T148 in our AMPPNP-actin and ADP-Pi-actin maps (Fig. 5A).
Densities for C-terminal residues 366–375 are absent in some actin monomer crystals (i.e., DNase I-Ca-ATP-actin, PDB ID code 2A42) and vary in other crystals (PDB ID codes 3A5L and 1YAG) (Fig. 6A). The side chain of F375 in monomers would overlap with the side chain of Q41 in the D-loop of adjacent subunit a in filaments and interfere with assembly (Fig. 6A).
The C-terminal residues form α-helices in our three filament structures. Our maps include the backbone and the side chains of residues I369, V370, H371, R372 (to the γ-carbon), K373 (to the δ-carbon), and C374. The backbone density for F375 is strong in all three maps, while the side chain density is strong in our ADP map but not the AMPPNP and ADP-Pi maps. The side chain of C374 forms an SH⋅⋅⋅π interaction (33) with Y133 in AMPPNP and ADP-Pi filaments but rearranges to interact with the D-loop of the adjacent subunit in ADP-actin filaments (Fig. 6 B and C) as explained in the last section of Results.
Alteration of the Active Site by Polymerization.
Conformational changes in actin subunits during polymerization (Fig. 3) do not disrupt any interactions between the protein and base or ribose of ATP known from crystal structures of ATP–actin and ATP–Arp2/3 complex (1, 25, 34, 35), but subunit flattening repositions the side chains of Q137 and H161 relative to the γ-phosphate of ATP and promotes hydrolysis (Figs. 7 and 8) also noted by Merino et al. (13). First, rotation of the outer domain with respect to the inner domain repositions Q137 on the hinge helix of subdomain 1, bringing the side-chain OE1 atom of Q137 ∼1 Å closer to the γ-phosphorous atom (Fig. 8). Second, rotation of the H161 side chain brings the NE2 atom ∼2.4 Å closer to the γ-phosphorous atom in our filaments (Fig. 8 A, C, and D) than most monomers (9.1 Å). An exception is the Dictyostelium Li-ATP-actin monomer bound to gelsolin subdomain 1 (35) (PDB ID code 1NMD), where the side chain of H161 is in an intermediate position, 7.8 Å from the γ-phosphorous atom (Fig. 8B). Third, subunit flattening moves the backbone of S14 and G15 ∼1.8 Å toward the front side of the subunit (Fig. 8D) and the sensor loop (residues 71–77) 1.1 Å toward the back side of actin (Fig. 3C).
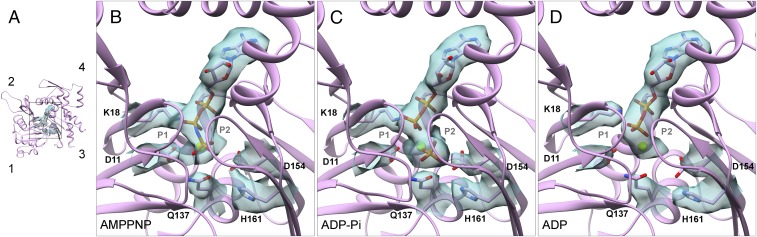
Fig. 7.
Changes in the active site during the ATPase cycle of polymerized actin. (A and B) AMPPNP-actin. (C) ADP-Pi-actin. (D) ADP-actin. (A) A small ribbon diagram looking down into the active site for orientation. (B–D) Ribbon diagrams with stick figures and map densities zoned within 2.12 Å of the nucleotide and the important side chains D11, K18, Q137, D154, and H161.
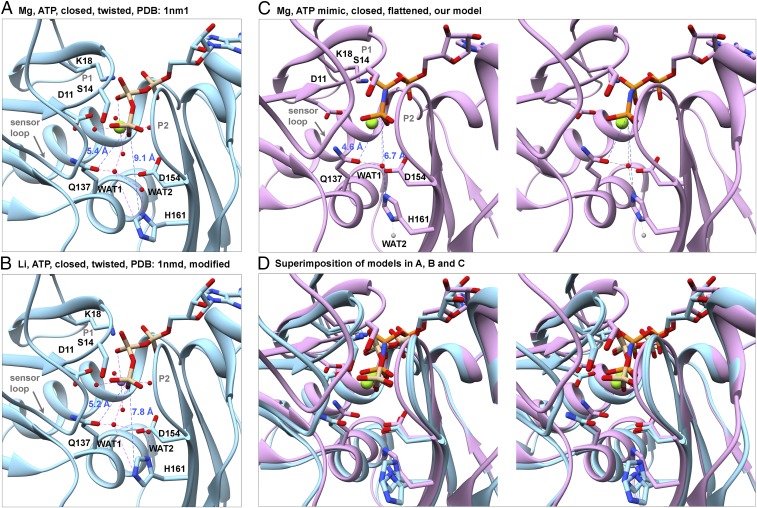
Fig. 8.
Rearrangement of the catalytic center stimulates ATP hydrolysis by polymerized actin. Ribbon diagrams of models with stick figures of the nucleotides and selected side chains compare distances between the γ-phosphate (Pγ) and the OE1 atom of Q137, NE2 atom of H161, and water molecules in actin monomers and filaments. (A and B) Crystal structure at 1.8 Å resolution of the Dictyostelium Mg-ATP-actin monomer complexed with human gelsolin segment 1 (PDB ID code 1NM1). (B) Based on an analysis of the water network, we flipped the imidazole ring of H161, which fits in the electron density equally well as its conformation in the original PDB file. This change brings the side chain ND1 atom closer to WAT2 (distance: 2.5 Å), indicating that they are hydrogen bonded. (C) Stereoview of our model of polymerized Mg-AMPPNP-actin. Compared with monomers, Pγ is 0.8 Å closer to the OE1 of Q137 and 2.4 Å closer to the NE2 atom of H161, which is rotated by ∼120° in all three filaments relative to monomers (Fig. 7). Based on the direction of protruding density from the imidazole ring of H161 in Fig. 7B, we propose that WAT1 from the X-ray structure (red ball) remains close to OE1 of Q137 and that WAT2 moves away, either downward (gray ball) or upward (not shown). (D) Stereo pair superimposing the models in A–C aligned on subdomain 3 to show their differences.
Changes in Filaments Associated with ATP Hydrolysis and Phosphate Release.
Our reconstructions of AMPPNP-actin and ADP-Pi-actin filaments and those of Merino et al. (13) are remarkably similar, including the positions of backbone and the side chains in the active site. A density protruding from the NE2 atom of H161 in our AMPPNP map may be a water molecule (Fig. 7B). The density for the γ-phosphate is similar to the α-phosphates and β-phosphates in the ADP-Pi-actin filament (Fig. 7C), but weaker in the AMPPNP-actin filament, so some subunits may have hydrolyzed AMPPNP.
The overall structures of the subunits change little upon ATP hydrolysis and phosphate release (Fig. 2C), but small changes in subdomains 1 and 2 explain why ADP-actin filaments have different properties than ATP-actin and ADP-Pi-actin filaments. After hydrolysis, the γ-phosphate moves ∼1.5 Å to a position 4.4 Å from the β-phosphate and Pβ-Mg-Pγ bond angle increases ∼30° (from 47° to 77°), while the distances between Mg and the two phosphates remain the same (∼3.3 Å) (Figs. 7C and 9C). Weak densities between residues N12 and G74 in the exit tunnel of the AMPPNP-actin and ADP-Pi-actin filaments may be partially occupied by phosphate (Fig. 9 A–D).
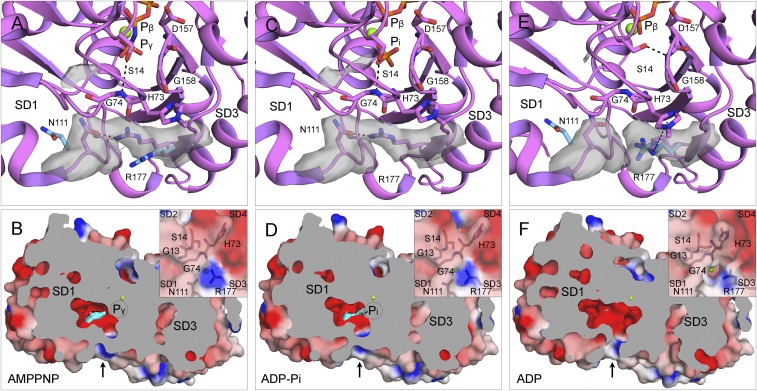
Fig. 9.
Mechanism of phosphate (Pi) release. (A, C, and E) Ribbon diagrams and (B, D, and F) cross-section views with surface representations of single subunits from filaments of AMPPNP-actin (A and B), ADP-Pi-actin (C and D), and ADP-actin (E and F). Ribbon diagrams include stick figures for nucleotides and selected residues, green spheres for Mg, and gray electron potential densities of N111 and R177 and extra density in the Pi cavity. The electrostatic potential surfaces in the cross-section view are rendered with PyMOL using a solvent radius of 1.0 Å with the extra density in the Pi cavity in cyan. (A and C) The S14 side chain forms an H-bond with the backbone of G74, and the side chains of N111 and R177 form an H-bond (∼2.5 Å) that closes the backdoor for Pi release (arrow in B and D). (A and E) The light blue side chains of N111 and R177 are separated by ∼10.0 Å in actin monomers [Ca-ATP-actin, PDB ID code 2A42 (A); Mg-ADP-actin, PDB ID code 3A5L (E)]. (E and F) After phosphate release, the pore of the tunnel lined by the backbone C=O of H73, side chain C=O of N111, and the guanidino group of R177 is open, because the side chain of R177 is turned close enough (<6.0 Å) to the side chain of largely unprotonated methylated H73 to form an N+-H⋅⋅⋅π interaction.
ADP-actin filaments lack density for the γ-phosphate (Fig. 7D) and the extra density in the exit tunnel (Fig. 9F). Phosphate dissociation results in multiple, similar changes in filaments and monomers (20, 29). First, the Mg loses its Mg-O bond to the γ-phosphate but remains ∼3.8 Å from the side chain OE1 atom of Q137 (Fig. 7D), likely coordinated by a water molecule found in crystal structures of actin monomers (35).
Second, the side chain of S14 in subdomain 1 rotates toward the β-phosphate (Fig. 9E) resulting in the loss of its H-bond with G74 in the sensor loop of subdomain 2 (Fig. 9A). The repositioned S14 side chain forms a new H-bond with the backbone of G158 in P2 loop of subdomain 3.
Third, the loss of the S14 connection between subdomains 1 and 2 is coupled to rotation of the side chain of methylated H73 in the sensor loop (Fig. 9E).
Our maps revealed details of the mechanism that releases phosphate from the active site through a tunnel with a back door (Fig. 9). The backbone C=O of methylated H73, side chain C=O of N111, and the guanidino group of R177 form the pore at the exit from the tunnel. Strong densities in our AMPPNP-actin and ADP-Pi-actin maps show the side chain of R177 with its the guanidino group H-bonded with the side chain of N111 (minimum distance: ∼2.5 Å). This bridge closes the backdoor. In our ADP map, the side chain guanidino group of R177 is close enough (<6.0 Å) to the side chain of methylated H73 for an N-H⋅⋅⋅π interaction. Methylation of H73 in most actins reduces the positive charge and favors this interaction.
Phosphate release changes the C-terminal residues beyond those driven by assembly. In AMPPNP-actin and ADP-Pi-actin filaments, the C-terminal carboxyl group of F375 is close enough to the side chains of K113 and R116 for favorable electrostatic interactions, but its side chain appears to be divided between two rotamer positions (Fig. 6B). In the ADP filament (Fig. 6C), the F375 side chain is rigidly buried in a large, intramolecular hydrophobic pocket formed by subdomain 1 residues P109, L110, I136, V139, and subdomain 3 residues A170, P172, I175, as well as V43 from the D-loop of subunit a+2. Furthermore, rearrangement of the backbone of C374 in the ADP filament moves its side chain SH ∼8 Å toward the D-loop where it contacts the backbone oxygen atoms of G42 and V43. This interaction appears to distort the backbone of D-loop residues G46, M47, and G48 (Fig. 6C).
Discussion
When Do Conformational Changes Take Place in Actin Subunits During Polymerization, ATP Hydrolysis, and Phosphate Release?
The high-resolution structures in our preprint (12) and those of Merino et al. (13) established that the major conformational changes take place when ATP-actin is incorporated into a filament rather than being associated with ATP hydrolysis or phosphate release. However, more subtle conformational changes in subdomain 1 and the D-loop after phosphate release impact subunit dissociation, filament flexibility, and cofilin binding.
Strong backbone and side chain densities in our maps show that the D-loops occupy the same position, tightly opposed to subdomain 3 of the adjacent subunit, in AMPPNP-actin, ADP-Pi-actin, and ADP-actin filaments and differ only in the conformation of residues G46-M47-G48-N49 in ADP-actin filaments. These findings differ from reports of conformational changes in subdomain 2 associated with phosphate release in early reconstructions actin filaments (36) and spectroscopic assays (37). Merino et al. (13) concluded that the entire D-loop dissociates from the neighboring subunit in an “open conformation” under four conditions: fully open in ADP-BeFx-actin filaments, ADP-Pi-actin filaments with jasplakinolide, and ADP-actin filaments with jasplakinolide, and partially open in their AMPPNP-actin filament. Their maps have densities for D-loops similar to our three maps, but our maps do not have densities for alternative D-loop conformations.
Why Does the Barbed End Elongate Faster than the Pointed End?
Flattening actin subunits during incorporation into a filament is associated with conformational changes that facilitate interactions within the filament. First, displacement of the W-loop (Figs. 3B and 5A) of subunit a-2 creates a knob around which the D-loop of subunit a binds (Fig. 5 B and C). Second, repositioning the W-loop of subunit a-2 opens a hydrophobic cavity for insertion of the side chain of highly conserved M44 from the D-loop of subunit a (Fig. 5 B and C). The side chain of M47 also makes hydrophobic contacts with subunit a-2. The importance of these interactions is confirmed by the effects of the enzyme MICAL, which oxidizes both M44 and M47, causing rapid filament disassembly (38) due to the incompatibility of the sulfoxide groups with their hydrophobic binding sites. Third, rearranging the C terminus during polymerization eliminates steric interference between the F375 side chain of subunit a-2 and the D-loop of the neighboring subunit a (Fig. 6A). Fourth, flattening enables two interactions between K113 and E195 and other polar residues along the short-pitch helix (Fig. 4).
The conformational changes associated with polymerization offer an explanation for the different rates of subunit association at the two ends of filaments (14). Incoming subunits at the barbed end of a filament bind to the side of the terminal subunit (n) and the barbed end of the penultimate subunit (n-1) (SI Appendix, Fig. S7B), so the rate of the reaction depends on the conformations of these two subunits. We suggest that interactions with the surrounding subunits flatten the conformations of these two terminal subunits creating a favorable, filament-like binding site for the incoming subunit. For example, flattened subunits at the barbed end of a filament would have a preformed cavity for M44 on the flexible D-loop of the incoming subunit. Similarly, flattening of the terminal subunits can explain why profilin has a lower affinity for the barbed end of filaments (especially AMPPNP-actin filaments) than actin monomers (16).
Adding a subunit to the pointed end of a filament is unfavorable (14) for three reasons. Most importantly, incoming monomer are not flattened so their barbed ends lack the features required for favorable interactions with the penultimate subunit at the pointed end of a filament. Second, the D-loops of the two subunits exposed at the pointed end of a filament have no lateral or longitudinal interactions (SI Appendix, Fig. S7C), so they likely remain as flexible as in monomers and these subunits may not be flattened. Third, a reconstruction of the pointed end suggested other differences (39), but the resolution was not good enough to define the details.
ADP-actin may dissociate faster from the barbed end than ATP-actin or ADP-Pi-actin, because its D-loop is not as fully engaged with its neighbor. Factors that are not apparent in our structures of the middle of filaments must contribute to why ATP-actin and ADP-Pi-actin monomers have larger association rate constants than ADP-actin at both ends (14–16).
How Does Polymerization Increase the Rate of ATP Hydrolysis?
Polymerization stimulates ATP hydrolysis (3) by 42,000-fold from 0.000007 s−1 by monomers (17) to 0.3 s−1 in filaments (18). Water is important for ATP hydrolysis by actin (40). High-resolution crystal structures of actin monomers revealed two water molecules that may contribute to hydrolysis (Fig. 8 A and B). Water 1 is hydrogen-bonded to the OE1 atom of Q137 (distance: ∼3.0 Å) and positioned by Q137, the imidazole ring of H161 and the γ-phosphate. Water 2 is located between ND1 atom of H161 and Water 1. H161 is the best candidate to activate the attacking water by extracting a proton from Water 1 directly or via the bridging Water 2 (35). Water molecules are underrepresented in electron potential maps compared with electron density maps from X-ray crystallography (28), but our map of AMPPNP-actin filaments has a water-like density protruding Pγ from the side chain of H161 (Fig. 7B). Previous work showed that the side chain of Q137 is closer to the nucleotide in filaments of ADP-actin than in monomers (6–9), but the mechanism of hydrolysis was uncertain without structures of ATP- actin and ADP-Pi-actin. Conformational changes in our filament structures and those of Merino et al. (13) bring Q137 and H161 closer to the γ-phosphate where they will position water for hydrolysis. Quantum mechanical simulations (41, 42) should provide more mechanistic details about the hydrolysis mechanism.
How Does γ-Phosphate Dissociate from an ADP-Pi-Actin Subunit?
Polymerized actin dissociates the γ-phosphate very slowly after hydrolysis (4). Our maps of AMPPNP and ADP-Pi-actin filaments show that the side chain of R177 blocks the opening of the phosphate release channel to the exterior. The map of the ADP-actin filament offers a plausible mechanism for how the pore is opened. The hydrolyzed γ-phosphate initially occupies a position close to the β-phosphate with the side chain of S14 H-bonded to the backbone of G74. When the γ-phosphate occasionally moves into the adjacent negatively charged cavity, the side chain of S14 breaks its H-bond with G74, flips into the space left by the γ-phosphate, and forms a new H-bond with G158 as observed in ADP-actin monomers (29). This reorganization allows the sensor loop to move and reposition methyl-H73 to form an N+-H⋅⋅⋅π interaction with the side chain of R177, which opens the backdoor for the γ-phosphate to escape. The open backdoor of ADP-actin filaments allows phosphate to rebind to the ADP-actin filament (4).
How Does Phosphate Release Change the Affinity of Filaments for Cofilin?
The conformations of the subunits in AMPPNP-actin, ADP-Pi-actin, and ADP-actin filaments are remarkably similar, but our maps have two clues about why phosphate dissociation changes the affinity for cofilin (43). First, the conformation of the D-loop residues G46-M47-G48-N49 changes, which allows more space for cofilin to bind the filament (Fig. 5F). Second, dissociation of the γ-phosphate results in the loss of hydrogen bonds and a divalent cation bond that might account for ADP-actin filaments being more flexible than ATP or ADP-Pi filaments (44). Flexibility is important, because cofilin is unlikely to bind to the standard conformation of polymerized actin, given the tighter helical twist (163°) of cofilin-decorated filaments than undecorated filaments (167°) (30, 45, 46). Since the association rate constant for cofilin binding filaments is <1% the expected value (47), Blanchoin proposed that <1% of the subunits in ADP-actin filaments are in the high energy (163°) conformation. Reconstructions of undecorated filaments from electron micrographs established that the subunits have a range of twist angles and that cofilin binding stabilizes a minor, high-energy state (48). Cofilin binding to subunits with highly twisted conformations is an example of the conformational selection theory for protein interactions (49). The energy barrier between the equilibrium state and the 163° state will be higher and thus less populated for stiff ATP-actin and ADP-Pi-actin filaments than flexible, ADP-actin filaments. After cofilin binds, the D-loop is disordered (30).
Materials and Methods
Actin Purification and Polymerization.
Muscle acetone powder was made using flash-frozen chicken muscle from a local Trader Joe’s grocery store (50). Actin was purified using one cycle of polymerization and depolymerization followed by gel filtration through Sephacryl S-300 and stored in Ca-G-buffer (2 mM Tris⋅HCl, pH 8.0; 0.2 mM ATP; 0.1 mM CaCl2; 1 mM NaN3; 0.5 mM DTT). Ca-ATP-actin was converted to Mg-ATP actin by incubation in G-Buffer with 50 µM MgCl2 and 0.2 mM EGTA (14). We made AMPPNP-actin (16) by first removing free ATP with neutralized AG1-X4 resin (Bio-Rad) and then incubating actin in 1 mM AMPPNP pH 7.0 for 3 min at 4 °C in Mg-G-buffer. We made Mg-ADP-actin by incubating Mg-ATP actin monomers with 1 mM glucose and 5 units/mL hexokinase at room temperature for 10 min followed by adding 0.01 volumes of 100 mM ADP (pH 7.0) alone or with 0.04 volumes of 500 mM potassium phosphate (pH 7.0) (14–16). We purchased ATP (A2383), AMPPNP (A2647), ADP (A2754), and hexokinase (H6380) from Sigma-Aldrich, AG 1-X4 resin (1431345) from Bio-Rad, and glucose (167454) from Thermo Fisher Scientific.
Sample Vitrification and Image Acquisition.
We used holy carbon C-flat 2/1 400-mesh Cu grids for ADP-actin (Protochips) and Quantifoil 2/1 300-mesh Au grids for AMPPNP-actin and ADP-Pi-actin (Structure Probe Inc.). The C-flat grids were glow-discharged for 6 s in a Solarus plasma cleaner (Gatan) in Ar (75%)/O2 (25%) (pressure: 70 mTorr) at 25 W and the Quantifoil grids for 30 s in a Bal-Tec SCD 005 sputter coater (Leica Biosystem Inc.) in air (pressure: 0.05 mBar) at 25 mA. To vitrify the samples, 3 µL of actin filament solution polymerized from 4 µM (C-flat grids) or 9 µM (Quantifoil grids) actin monomers was applied onto the carbon side of the grid in Mark III or IV Vitrobot (FEI company) at 20 °C and >95% humidity. After incubating the grid for 15 s, extra solution was blotted off using standard Vitrobot paper (grade 595; Ted Pella) for 4.5 s at offset 1 or 2.5 s at blot force −15. Grid prescreening was performed on an F20 microscope operated at 200 kV and equipped with a K2 Summit camera (FEI company). The three datasets were collected on a Titan Krios microscope equipped with an XFEG at 300 kV, a nanoprobe, and a Gatan image filter (slit width: 20 eV). Image stacks were recorded on a K2 Summit camera in superresolution mode, controlled by SerialEM (51). The dose rate was set to ∼8.0 counts/pixel per s. For ADP-actin, image stacks were recorded at a defocus value between −1 and −2.5 µm (one image per hole). For AMPPNP-actin and ADP-Pi-actin, image stacks were recorded using beam image shift (four images per hole) (52) at a defocus value between −1.5 and −2.5 µm. For all three datasets, each image was fractionated into 32 frames (0.25 s per frame) and the physical pixel size was 1.045 Å.
Image Processing.
Dose-fractioned image stacks were dose-averaged, magnification-corrected, motion-corrected, and summed with MotionCor2 (53) using 9 × 9 patches. The frames in the first second of image recording (with large drifts) and those in the last second (with high radiation damage) were discarded. CTF parameters were estimated with Gctf (54) using the unweighted sums. Filaments were manually boxed out with sxhelixboxer.py in SPARX (55). The filaments coordinates were exported from SPARX and imported into RELION3 (56) for further analysis. Filaments were windowed into square segments using a box size of 328 × 328 pixels, and an interparticle distance along the long axis of 26 pixels. First, we worked on a subset (∼20,000 particles) of the whole dataset for each sample. Following 2D classifications, we reconstructed 3D maps using the known helical parameters (rise: 27.3 Å; twist: −166.5°) and a simulated model, in which each actin subunit is depicted as a ball. The maps from small datasets were filtered to 10 Å before being used as reference models for reconstructions using the whole datasets. After postprocessing, the resolution of all three maps was better than 3.8 Å at this point. Later, we performed 2D and 3D classifications to remove bad particles (∼10%). The well-resolved classes from each 3D classification were similar. Particles in good classes of 2D and 3D classifications were pooled together, and their local CTFs were used in final reconstructions. A soft-edged 3D mask with a radius of 45% of the box size was created for postprocessing. The B-factors for map sharpening were first determined by RELION3 itself. The B-factors were −114.8 Å2 for AMPPNP-actin, −99.7 Å2 for ADP-Pi-actin, and −113.9 Å2 for ADP-actin. The Fourier shell correlation 0.143 criterion (FSC0.143) was used for resolution estimation. Layer-line images were calculated from map projections with SPARX (project and periodogram commands) (55). Local resolutions were calculated with ResMap (57). All of the image processing was carried out on Yale High Performance Computing servers.
Model Building and Refinement.
Atomic models were built with Coot (58). Most residue side chains were built unambiguously. When there was an ambiguity, we referred the local conformations of the corresponding residue in the 1.5-Å resolution crystal structure of rabbit actin (PDB ID code 1J6Z) (29). The primary sequence of chicken actin is the same as rabbit actin. Refinements were carried out for several rounds in reciprocal space with REFMAC (59) and then in real space with PHENIX (60). The models from REFMAC had slightly better fitting statistics, while the models from PHENIX had better geometry as analyzed with Coot. At last, we chose the structures from PHENIX as the final models.
Structure Analysis and Presentation.
Interdomain rotation angles were calculated with the DynDom web server (24). The rise (translation) and twist (rotation) for the helices were calculated with Chimera (match showMatrix command) using two interstrand adjacent subunits in models, which are pixel-size independent. rmsds were also calculated with Chimera (rmsd command) (61). Electrostatic potentials were calculated with DelPhi 5 (62) and mapped onto the molecule surface with PyMOL. Figures of structures were generated with MolScript/Raster3D (63) (SI Appendix, Fig. S1A), PyMOL (64) (Figs. 4 and 9 and SI Appendix, Figs. S6 and S7), and Chimera (61) (all other figures).
Supplementary Material
Acknowledgments
We thank the two reviewers for helpful suggestions; Dr. Shenping Wu, Dr. Marc Llaguno, and Dr. Xinran Liu for managing the microscopes; Yale High Performance Computing support team for guidance and use of the computing infrastructure; Moon Chatterjee and Shoshana Zhang for commenting on the manuscript; and members of our laboratory for helpful discussions. Research in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under Grants R01GM026132 and R01GM026338. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The electron density maps have been deposited in Electron Microscopy Data Bank (EMDB) (accession numbers: 7936, 7937, and 7938). The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org/pdb (PDB ID codes: 6DJM, 6DJN, and 6DJO).
See Commentary on page 3959.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807028115/-/DCSupplemental.
References
- 1.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard TD. Actin and actin-binding proteins. Cold Spring Harb Perspect Biol. 2016;8:a018226. doi: 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Straub FB, Feuer G. Adenosinetriphosphate. The functional group of actin. Biochim Biophys Acta. 1950;4:455–470. [PubMed] [Google Scholar]
- 4.Carlier MF, Pantaloni D. Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry. 1986;25:7789–7792. doi: 10.1021/bi00372a001. [DOI] [PubMed] [Google Scholar]
- 5.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 6.Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 7.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 8.Galkin VE, Orlova A, Vos MR, Schröder GF, Egelman EH. Near-atomic resolution for one state of F-actin. Structure. 2015;23:173–182. doi: 10.1016/j.str.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von der Ecken J, et al. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature. 2016;534:724–728. doi: 10.1038/nature18295. [DOI] [PubMed] [Google Scholar]
- 11.Mentes A, et al. High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc Natl Acad Sci USA. 2018;115:1292–1297. doi: 10.1073/pnas.1718316115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou SZ, Pollard TD. 2018. Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. bioRxiv:10.1101/309534. Preprint, posted April 27, 2018.
- 13.Merino F, et al. Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat Struct Mol Biol. 2018;25:528–537. doi: 10.1038/s41594-018-0074-0. [DOI] [PubMed] [Google Scholar]
- 14.Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiwara I, Vavylonis D, Pollard TD. Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc Natl Acad Sci USA. 2007;104:8827–8832. doi: 10.1073/pnas.0702510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courtemanche N, Pollard TD. Interaction of profilin with the barbed end of actin filaments. Biochemistry. 2013;52:6456–6466. doi: 10.1021/bi400682n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J Biol Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- 18.Blanchoin L, Pollard TD. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 2002;41:597–602. doi: 10.1021/bi011214b. [DOI] [PubMed] [Google Scholar]
- 19.Woodrum DT, Rich SA, Pollard TD. Evidence for biased bidirectional polymerization of actin filaments using heavy meromyosin prepared by an improved method. J Cell Biol. 1975;67:231–237. doi: 10.1083/jcb.67.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graceffa P, Dominguez R. Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J Biol Chem. 2003;278:34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 21.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 22.Murakami K, et al. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell. 2010;143:275–287. doi: 10.1016/j.cell.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 23.Ge P, Durer ZA, Kudryashov D, Zhou ZH, Reisler E. Cryo-EM reveals different coronin binding modes for ADP- and ADP-BeFx actin filaments. Nat Struct Mol Biol. 2014;21:1075–1081. doi: 10.1038/nsmb.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model. 2002;21:181–183. doi: 10.1016/s1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 26.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang H, Bradley MJ, Elam WA, De La Cruz EM. Regulation of actin by ion-linked equilibria. Biophys J. 2013;105:2621–2628. doi: 10.1016/j.bpj.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Moore PB. On the interpretation of electron microscopic maps of biological macromolecules. Protein Sci. 2017;26:122–129. doi: 10.1002/pro.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka K, et al. Structural basis for cofilin binding and actin filament disassembly. Nat Commun. 2018;9:1860. doi: 10.1038/s41467-018-04290-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng X, Diraviyam K, Sept D. Nucleotide effects on the structure and dynamics of actin. Biophys J. 2007;93:1277–1283. doi: 10.1529/biophysj.107.109215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudryashov DS, Grintsevich EE, Rubenstein PA, Reisler E. A nucleotide state-sensing region on actin. J Biol Chem. 2010;285:25591–25601. doi: 10.1074/jbc.M110.123869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forbes CR, et al. Insights into thiol-aromatic interactions: A stereoelectronic basis for S-H/π interactions. J Am Chem Soc. 2017;139:1842–1855. doi: 10.1021/jacs.6b08415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vorobiev S, et al. The structure of nonvertebrate actin: Implications for the ATP hydrolytic mechanism. Proc Natl Acad Sci USA. 2003;100:5760–5765. doi: 10.1073/pnas.0832273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belmont LD, Orlova A, Drubin DG, Egelman EH. A change in actin conformation associated with filament instability after Pi release. Proc Natl Acad Sci USA. 1999;96:29–34. doi: 10.1073/pnas.96.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moraczewska J, Wawro B, Seguro K, Strzelecka-Golaszewska H. Divalent cation-, nucleotide-, and polymerization-dependent changes in the conformation of subdomain 2 of actin. Biophys J. 1999;77:373–385. doi: 10.1016/S0006-3495(99)76896-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grintsevich EE, et al. Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat Commun. 2017;8:2183. doi: 10.1038/s41467-017-02357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narita A, Oda T, Maéda Y. Structural basis for the slow dynamics of the actin filament pointed end. EMBO J. 2011;30:1230–1237. doi: 10.1038/emboj.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saunders MG, Voth GA. Water molecules in the nucleotide binding cleft of actin: Effects on subunit conformation and implications for ATP hydrolysis. J Mol Biol. 2011;413:279–291. doi: 10.1016/j.jmb.2011.07.068. [DOI] [PubMed] [Google Scholar]
- 41.McCullagh M, Saunders MG, Voth GA. Unraveling the mystery of ATP hydrolysis in actin filaments. J Am Chem Soc. 2014;136:13053–13058. doi: 10.1021/ja507169f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun R, Sode O, Dama JF, Voth GA. Simulating protein mediated hydrolysis of ATP and other nucleoside triphosphates by combining QM/MM molecular dynamics with advances in metadynamics. J Chem Theory Comput. 2017;13:2332–2341. doi: 10.1021/acs.jctc.7b00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. J Mol Biol. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 44.Isambert H, et al. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J Biol Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 45.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galkin VE, et al. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci USA. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- 48.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boehr DD, Wright PE. Biochemistry. How do proteins interact? Science. 2008;320:1429–1430. doi: 10.1126/science.1158818. [DOI] [PubMed] [Google Scholar]
- 50.MacLean-Fletcher S, Pollard TD. Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem Biophys Res Commun. 1980;96:18–27. doi: 10.1016/0006-291x(80)91175-4. [DOI] [PubMed] [Google Scholar]
- 51.Mastronarde D. SerialEM: A program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microsc Microanal. 2003;9(Suppl 2):1182–1183. [Google Scholar]
- 52.Cheng A, et al. High resolution single particle cryo-electron microscopy using beam-image shift. J Struct Biol. 2018;204:270–275. doi: 10.1016/j.jsb.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 56.He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Rocchia W, et al. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: Applications to the molecular systems and geometric objects. J Comput Chem. 2002;23:128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 63.Merritt EA, Bacon DJ. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 64.DeLano WL. PyMOL: An open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography. 2002;40:82–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.