Significance
Digital assays have enormous untapped potential for diagnostics, environmental surveillance, and biosafety monitoring, but are currently confined to laboratory settings due to the instrumentation necessary to generate, control, and measure millions of droplets. We instead use a mobile phone-based imaging technique that is 100× faster than conventional microfluidic droplet detection, does not require expensive optics, is invariant to flow rate, and can simultaneously measure multiple fluorescent dyes in droplets. By using this time domain modulation with cloud computing, we overcome the low frame rate of digital imaging, and achieve throughputs as high as 1 million droplets per second. We integrate on-chip delay lines and a microbead processing unit, resulting in a robust device, suitable for low-cost implementation, with ultrasensitive measurement capabilities.
Keywords: digital, ELISA, portable, multiplex, cell phone
Abstract
Digital droplet assays—in which biological samples are compartmentalized into millions of femtoliter-volume droplets and interrogated individually—have generated enormous enthusiasm for their ability to detect biomarkers with single-molecule sensitivity. These assays have untapped potential for point-of-care diagnostics but are currently mainly confined to laboratory settings, due to the instrumentation necessary to serially generate, control, and measure tens of millions of droplets/compartments. To address this challenge, we developed an optofluidic platform that miniaturizes digital assays into a mobile format by parallelizing their operation. This technology is based on three key innovations: (i) the integration and parallel operation of a hundred droplet generators onto a single chip that operates 100× faster than a single droplet generator, (ii) the fluorescence detection of droplets at 100× faster than conventional in-flow detection using time domain-encoded mobile phone imaging, and (iii) the integration of on-chip delay lines and sample processing to allow serum-to-answer device operation. To demonstrate the power of this approach, we performed a duplex digital ELISA. We characterized the performance of this assay by first using spiked recombinant proteins in a complex media (FBS) and measured a limit of detection, 0.004 pg/mL (300 aM), a 1,000× improvement over standard ELISA and matching that of the existing laboratory-based gold standard digital ELISA system. We additionally measured endogenous GM-CSF and IL6 in human serum from n = 14 human subjects using our mobile duplex assay, and showed excellent agreement with the gold standard system ().
Digital droplet-based assays achieve 1,000× improved sensitivity over conventional assays by performing millions of assays in parallel within femtoliter volume droplets. This parallelization converts the traditionally analog problem of quantifying biomarkers into a digital one, where each droplet contains either one copy or zero copies of the target molecule. Digital assays have demonstrated enormous utility as a platform for the ultrasensitive detection of nucleic acids (1–4) and proteins (5–11), as well as the analysis of single cells (12–15) and single exosomes (16). Digital enzyme-linked immunosorbent assays (dELISA) (9, 17–19) and digital polymerase chain reaction (20) have found broad utility and have been successful in achieving attogram per milliliter sensitivity and high levels of multiplexing for a broad range of targets (20). In a particularly exciting demonstration, digital assays were recently used to measure both protein and mRNA simultaneously from single cells (21). The improvement in sensitivity of digital assays over conventional assays has allowed measurement of previously undetectable concentrations of clinical biomarkers, opening new opportunities for improved diagnostics and prognostics for applications such as traumatic brain injury, HIV, and early cancer detection (2, 22–25).
Due to digital assays’ high sensitivity, their capability for absolute quantification without calibration, and the robustness of digital detections to reaction conditions, they are particularly well suited for point-of-care diagnostics. However, the instrumentation currently required to generate, process, and detect the many independent reaction vessels for ultrasensitive digital assays has proven cumbersome to implement. The gold standard commercial implementation of dELISA is Quanterix’s Simoa (17, 26), which uses a microfabricated array of 200,000 wells that are each 40 fL. The Simoa HD-1 Analyzer provides a fully automated sample-to-answer readout, capable of being loaded with up to four 96-well ELISA plates. The machine has automated the entire digital ELISA, minimizing the time required to process multiple samples through their workflow, resulting in a throughput of 66 samples per hour (17). Furthermore, the Simoa HD-1 can perform a multiplexed 10-plex assay on each sample. While the Simoa system has demonstrated the value of ultrasensitive protein detection in a laboratory setting, it requires bulky optics and bulky fluid handling, resulting in a technology not suitable for portable use and that has an instrumentation cost of more than $100,000. Point-of-care systems have been developed that typically use smaller numbers (10,000) of nanoliter wells, much larger than the femtoliter wells used in the ultrasensitive systems (27), and, as a result, do not achieve the same sensitivity, dynamic range, or capability for multiplexing (28–30).
Compared with static arrays, continuous flow microfluidic droplet systems allow much greater numbers of partitions to be analyzed (1 million), allowing increased multiplexing, sensitivity, and the capability for downstream sorting of the droplets (7, 10, 14, 15, 31). However, droplet microfluidic systems are currently hindered by both (i) the throughput ( droplets per s) at which droplets can be serially generated in microfluidic systems and be monodisperse (32, 33) and (ii) the throughput ( droplets per s) at which the fluorescence of droplets can be detected by flowing them one by one through a micrometer-scale laser spot (14, 15, 31) (Fig. 1A and SI Appendix, Table S1). An emerging approach to overcome these limitations has been to incorporate many replica generators or detectors that can operate on the same chip in parallel to increase throughput (34–40). However, it has not yet been possible to fully implement ultrasensitive digital assays into a mobile format, due to the required instrumentation to generate the highly controlled flows required for conventional droplet microfluidics (41), the difficulty of parallelizing the optics necessary for multicolor fluorescence detection, and the challenge of integrating sample preparation.
Fig. 1.
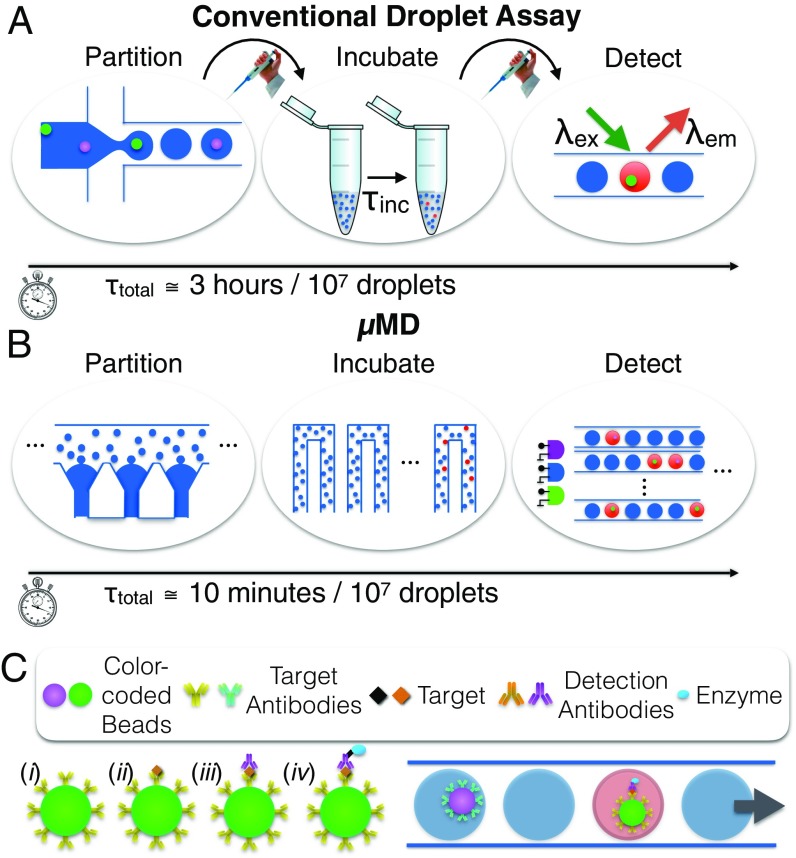
Miniaturization and parallelization of droplet dELISA. (A) A schematic of the conventional workflow for dELISA, which requires multiple hands-on steps and is rate-limited by the serial partitioning of the sample into droplets and the serial detection of the fluorescence of each individual droplet. (B) The MD parallelizes droplet generation, incubation, and detection to miniaturize dELISA fully onto a mobile platform and increase its throughput by 100×. (C) Antibody-functionalized, color-coded beads are used in a duplex dELISA assay, wherein individual beads are encapsulated into droplets and read out if they have captured a single target protein.
To address this challenge, we have developed an optofluidic platform, the microdroplet Megascale Detector (MD), that miniaturizes digital droplet assays into a mobile device, while matching the limit of detection of the current laboratory-scale gold standard technology. To achieve this limit of detection in a robust, mobile device, the MD is built on three key innovations (Fig. 1B). (i) Rather than generate droplets one at a time, we instead incorporate a parallelized microfluidic droplet generator that operates 100× faster than a single-droplet generator. Moreover, by making use of the recently published Millipede geometry (36), the monodispersity of the generated droplets are invariant to flow rate, which allows use of inexpensive peristaltic pumps that can be incorporated into a mobile device. (ii) To rapidly read out the fluorescence of the droplets ( drops per s), we use a mobile phone-based imaging technique that is 100× faster than conventional detection, wherein droplets are detected one by one (15, 31). Our approach does not require expensive optics and is invariant to flow rate, making it well suited for a mobile implementation (35). The key innovation of this approach is that it overcomes the low frame rate of digital imaging, and can achieve multicolor fluorescence detection, by modulating multiple, differently colored LED/laser diode excitation sources with unique nonperiodic signals. The video feed can be decoded to accurately measure each droplet’s fluorescent signals at throughputs far exceeding that of the frame rate of the camera, as high as 1 million droplets per second. This work builds on a previously published proof-of-concept device that demonstrated the measurement of only a fluorescent dye in passing droplets (35, 42), and is here extended to measure three fluorescence channels in each droplet to implement dELISA. (iii) We integrate a microbead processing unit, droplet generators, on-chip delay lines for droplet incubation, and droplet fluorescence detection, resulting in a robust device, suitable for a low-cost implementation, that allows raw serum to be input and molecular data to be output.
To demonstrate the power of this approach, we implement multiplexed dELISA using microbeads color-coded with different fluorescent dyes, where the color code corresponds to the protein targeted by its antibody (Fig. 1C). We performed a duplex cytokine assay (GM-CSF and IL6) in serum using UV and green fluorescent beads, where droplets containing a bead with a complete immunocomplex fluoresce red. We accurately measured IL6 and GM-CSF simultaneously in complex media (bovine serum) over four orders of magnitude with a limit of detection as low as 0.004 pg/mL (300 aM)—a thousand-fold improvement over standard ELISA and matching that of the current gold standard digital platform (5, 6). Our chip is designed for minimal user interaction (Movie S1); has a total droplet processing time of 10 min for 10 million droplets, where the workflow encompasses droplet generation, droplet incubation, and fluorescence droplet detection for each sample; and has a prototype instrumentation cost of $500 and a disposable cost of $5.
Results and Discussion
μMD Design.
The complete workflow of dELISA is incorporated onto our chip (Fig. 2A) and consists of (i) a microbead processor where microbeads capture their target proteins from serum, are tagged with enzyme labeled immunocomplexes for downstream amplification within droplets, and are iteratively washed between each labeling step; (ii) a droplet generator, where the microbeads are mixed with the enzyme’s substrate and encapsulated into water-in-oil droplets; (iii) a 3D microfluidic channel that takes 3.2 min for the droplets to pass, allowing time for the enzymatic amplification of the fluorescence signal; and (iv) a mobile phone-based detector, where the droplets’ fluorescence are rapidly detected using time domain-encoded optofluidics.
Fig. 2.
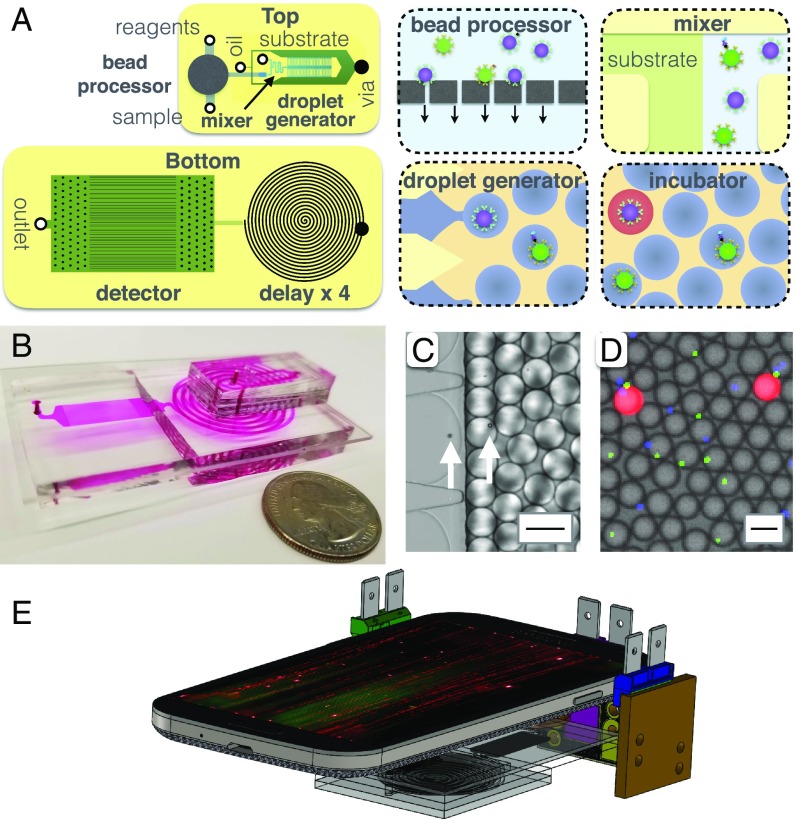
Integrated MD workflow. (A) A schematic of the MD chip, showing both a top view and a bottom view. Each cartoon shows a schematic of the modules that are incorporated onto the MD. (B) A photograph of the disposable MD chip, with the channels filled with dye to make them visible. (C) A micrograph showing the droplet generator encapsulate microbeads into d = 40 m droplets. The arrows highlight the microbeads. (Scale bar = 50 m.) (D) A fluorescence micrograph of the droplets after the delay line. (Scale bar = 50 m.) (E) A schematic of the MD platform, consisting of a mobile phone, three light sources, and the disposable MD chip.
The microbead processor unit consists of a semipermeable membrane to immobilize the beads. Multiple reagents and washing buffers are sequentially delivered to the immobilized beads, after which the beads are released for downstream analysis (Fig. 2B). One of the populations of color-coded microbeads (d = 5.4 m, / = 470/490 nm, CFH-5052-2; Spherotech) is functionalized with antibody for GM-CSF (MAB2172; R&D). The other population of beads (d = 4.5 m, / = 370/410 nm, CFP-4041-2; Spherotech) is functionalized with antibody for IL6 (MAB206; R&D). The beads are first incubated with the sample for 60 min, and then immobilized on the membrane. Subsequently, the beads are washed with 1 mL of T20 Buffer at 10 mL/h, incubated with 0.1 mL of 0.7 nM detection antibody (BAF206, BAM215; R&D) in T20 buffer for 30 min, washed in 1 mL of T20 Buffer at 10 mL/h, and subsequently released from the membrane by reversing the flow at 6 mL/h. The semipermeable membrane is an A = 300 mm2 track etched polycarbonate membrane with d = 3 m pores (Fig. 2A). The membrane is incorporated into the microfluidic chip using laser-cut Mylar membrane microfluidics (43, 44) (SI Appendix, Figs. S1 and S2). For testing, a syringe pump (Harvard Apparatus) or a low-cost peristaltic (; Intllab) were used.
Downstream of the microbead processor, the released microbeads are mixed with the ELISA substrate (QuantaRed Enhanced Chemifluorescent HRP Substrate; Thermo) (Fig. 2 C and D) and encapsulated into d = 40 m droplets suspended in QX200 Droplet Generation Oil (1864006; Biorad) (Fig. 2D). A channel length of 14 mm with a staggered herringbone design is used to ensure proper mixing of the beads and the substrate, while minimizing background signal that comes from enzymes generating fluorescence signal before they are encapsulated into droplets (45, 46) (SI Appendix, Fig. S3). To generate monodisperse droplets that are robust to flow rate, we used the Millipede geometry described by Amstad et al. (36). In brief, the Millipede uses step emulsification, where the droplet diameter depends only on the channel geometry and not the flow rates of the dispersed or continuous phase over a large range of flow rates. Our device consists of 100 droplet generators to achieve a throughput of 100,000 droplets per s. The droplet generator layer has a height h = 10 m, and the continuous phase layer has a height h = 120 m (SI Appendix, Fig. S2). Each droplet encapsulates one or zero beads (Fig. 2D) by setting the concentration of beads such that there are 10× more droplets than beads, resulting in a 0.5% probability of a droplet containing two beads based on Poisson statistics.
Downstream of the droplet generator, the droplets pass through a delay line (Fig. 2A), which we have designed to hold droplets for a precise minutes-scale duration in continuous flow, without the need for active valves. To achieve a precise minute-scale delay, a channel is required that has both a large cross-sectional area, to reduce velocity, and a long length. To achieve a large cross-sectional area, we mold the polydimethylsiloxane (PDMS) microfluidics using a laser-cut acrylic mold rather than conventional SU-8 to achieve channels with width w = 1.8 mm and height h = 1.5 mm. To achieve a large channel length, without leading to an overly large device footprint, we stack n = 4 spiral channels vertically by plasma-bonding multiple PDMS pieces with punched hole vias (SI Appendix, Fig. S1). Using a flow rate of = 67 mL/h, it takes droplets 3.2 min to traverse the entire channel, allowing the enzymes time to generate a measurable fluorescence signal (Fig. 2D).
Time Domain-Encoded Optofluidic Fluorescence Detection.
To achieve high-throughput, multicolor, fluorescence droplet detection on a mobile platform, we modulate the excitation light in time with a pseudorandom sequence that allows individual droplets to be resolved that would otherwise overlap due to the limited frame rate of digital cameras. Using conventional excitation that is constant in time, a droplet moving across a camera’s field of view is imaged as a streak (, where v is the droplet velocity and is the exposure time of the camera). This streak length L sets the minimum distance between droplets, and thus severely limits throughput. We overcome this limitation by modulating the excitation light source with a pseudorandom sequence at a rate 10 faster than the exposure time of the camera, modulating the streak so that it can be resolved among neighboring droplets as close as three droplet diameters via correlation detection, and do so in 120 parallel channels in the camera’s field of view. In our previous work in this area (35, 38, 42), we only interrogated a single fluorescent dye in each droplet, which is not sufficient to read out the multiplexed dELISA assays carried out in this paper. We had previously presented a proof-of-concept demonstrating that two distinct dyes could be detected (42). Here, we expand this approach by using three light sources, each of which emits a wavelength tuned to excite a different dye and that is modulated in time with a unique maximum length sequence (MLS) that can be decoded independently to read out each fluorescence channel. A band-pass filter is placed on the camera to diminish the effects of scattered excitation light (#87-241; Edmund Optics). We implemented a three-color system using two diode lasers (blue, green) and one LED (UV). This MD platform is invariant to flow rate, has a maximum throughput of 160 mL/h ( droplets per s), and a dynamic range of 1: to 1:40 fluorescent:nonfluorescent droplets.
To decode the videos taken by our cell phone camera, we perform a correlation detection for the three expected modulation patterns m, corresponding to each of the three light sources. By doing so, we generate the correlation vectors , where the indices k are the video frames, n are the n = 1:120 channels in the device, R, G, B corresponds to the color channels of the digital camera, and r, g, b corresponds to the three unique excitation sources (Fig. 3A). We chose to pattern the droplets using MLS with m = 63 bits, where each bit corresponds to 10 pixels (px) in the digital image. Thus, 63 bits would correspond to 630 px, or 1/3 of a 1,920-px-wide video frame. To create a set of MLS with minimal autocorrelation and cross-correlation from each other, we followed the process in MacWilliams and Sloane (47) to create a pseudorandom vector with elements, that we folded into a 63×65 matrix, and chose the first three rows to select the three MLS patterns.
Fig. 3.
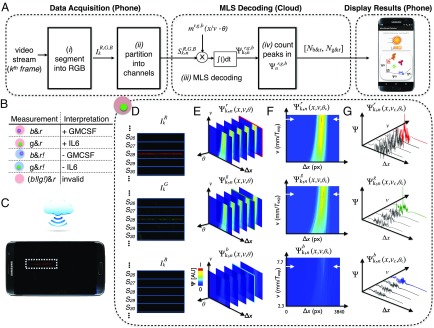
Software workflow for phase and velocity invariant optofluidic fluorescence droplet detection. (A) The algorithm for detecting droplets. (B) Truth table for interpreting the readout of the MD’s three-color (r, red ELISA signal; g, green beads; b, blue beads) fluorescence measurement. (C) Schematic showing the MD platform collecting data, which are sent to the cloud to be processed, and then returned to the mobile phone to report the results of the assay to the user. (D–F) A sample workflow for a droplet that contains a green bead and is positive for its target. (D) The video’s image frames are segmented into 1D vectors. (E) A 3D correlation results in a data matrix where the phase is first identified. (F) From this 2D “slice” of the data matrix, the velocity of the droplet is found. (G) The position is recorded for each peak in the correlation space.
The goal of the fluorescence detector is to inspect each droplet and determine (i) whether the droplet contains a microbead, and, if so, determine its color (UV or green), which indicates the protein target the droplet is measuring (GM-CSF and IL6, respectively) (Fig. 3B), and (ii) whether the droplet fluoresces red, which indicates whether the droplet has detected one molecule of its target. The workflow to extract this information from each droplet is as follows: (i) The th frame of the video is separated into its red, green, and blue components based on the camera’s red, green, and blue sensors (Fig. 3D). (ii) A line average is taken along the direction of each of the n = 120 microchannels . (iii) To simplify the hardware of the system, rather than control the droplet velocity v or phase , relative to the MLS excitation, of the passing droplets, we instead use cloud computing to computationally detect droplets with unknown phase and velocity (Fig. 3C). We generate a 3D matrix by correlating each of the modulated signals with expected emission patterns that scans the range of velocities and phase at which the LED strobes , corresponding to the three excitation sources (Fig. 3E). (iv) By selecting the optimal phase and velocity of every droplet, we can identify peaks in the correlation space (Fig. 3 F and G). These detected signals are tabulated as , where corresponds to droplets that contain a UV bead and fluoresce red and thus contain a molecule of GM-CSF, and corresponds to droplets that contain a green bead and fluoresce red and thus contain a molecule of IL6. The data are collected using our custom Android app, sent into the cloud, processed using MATLAB in a remote server, and then sent back to the smartphone and reported to the user in an easy-to-interpret format. For each target molecule, the active enzymes per bead (5) (AEB) is calculated by quantifying the number of droplets that contained a bead and that fluoresced red, normalized to the total number of beads. The values that we report are calculated by subtracting the AEB measured when we ran a blank sample from the measured AEB of the real sample, which does not contain the target protein, and corrected for the precalculated loss factor, obtained in the measurements of spiked proteins into PBS, multiplied by the molecular weight of the target protein.
Droplet Generation and Integrated Incubation Line.
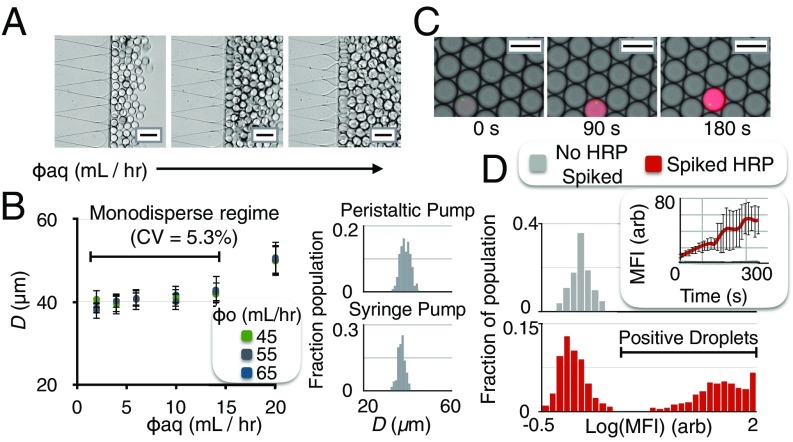
Droplet uniformity is critical for digital assays, because variance in droplet diameter leads to variance in fluorescence after the delay line, confounding the ability to discriminate positive and negative droplets. To evaluate the droplet generator’s capability to generate monodispersed droplets in a mobile setting, we scanned the continuous phase over flow rates = 45 mL/h to 65 mL/h, and we scanned the dispersed phase over flow rates = 2 mL/h to 14 mL/h (Fig. 4A). We generated droplets with a diameter d = 40 m and a coefficient of variation CV 6% with both syringe pumps and inexpensive peristaltic pumps (Fig. 4B). The greater the ratio of the aqueous flow rate to the continuous flow rate, the larger the volume fraction of droplets was, allowing increased throughput.
Fig. 4.
Flow rate-invariant droplet generation and detection allow inexpensive, compact implementation of dELISA. (A) By using the Millipede geometry, droplet size is invariant to dispersed phase flow rate. (B) For a range of continuous flow rates (45 mL/h to 65 mL/h) and dispersed flow rates (2 mL/h to 14 mL/h), the generated droplets remained monodispersed with syringe pumps (CV = 5.3) and with inexpensive peristaltic pumps (CV = 6.0). (C) To evaluate the enzymatic amplification of captured protein in the droplets, we inspected the droplets after the delay line with fluorescence microscopy. (D) After a 3.2-min delay, the distribution of droplets positive and negative for enzyme were measured. (Scale bar for A and C = 50 m.)
To evaluate and optimize the delay line and the enzymatic amplification of captured protein in the droplets, we inspected the droplets after the delay line, with fluorescence microscopy (Leica DM4200) (Fig. 4C). We calculated the distribution of fluorescence intensities of droplets with and without an enzyme to identify the delay time that minimized their overlap. For a 3.2-min delay, the ratio of the average droplet with an enzyme had a 30× greater mean fluorescence intensity than a droplet without an enzyme (Fig. 4D).
Ultrasensitive, Duplex Protein Detection in Complex Media.
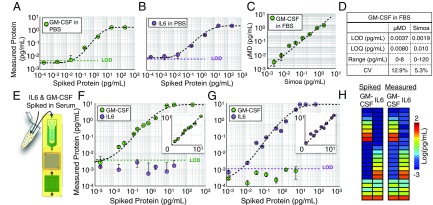
To evaluate our integrated MD’s capability for sensitively detecting proteins, we first measured IL6 and GM-CSF in PBS, complex media (FBS), and human serum, and compared the results with those from a commercial dELISA device (Quanterix Simoa). Nonhuman serum provides a good model to simulate human serum (5, 48, 49), because it has the convenient property that it does not include any human IL6 or GM-CSF, allowing titration experiments to be performed down to our device’s limit of detection (LOD). In these initial experiments, we performed bead processing off-chip, such that the droplet generator, incubator, and detector could be evaluated. In PBS, we first performed separate single-plex measurements on GM-CSF (Fig. 5A) and IL6 (Fig. 5B) by measuring serial dilutions from pg/mL to . We achieved an LOD = 0.0045 pg/mL (320 aM) and LOD = 0.0070 pg/mL (350 aM) for GM-CSF and IL6, respectively. We next performed the same titration measurement for GM-CSF in 1:4 FBS solution. In this experiment, we split the sample between our MD platform and Simoa (GM-CSF 2.0 kit) to perform a head-to-head comparison. We found excellent agreement between the output of our chip and that of Simoa ( = 0.95) (Fig. 5C). The LOD, limit of quantification (LOQ), dynamic range, and average CV were tabulated for MD and Simoa (Fig. 5D) and showed similar performance.
Fig. 5.
Benchmarking and characterization of ultrasensitive, duplex protein detection in complex media. (A) Single-plex detection of GM-CSF spiked into PBS. The limit of detection LOD = 0.0045 pg/mL (320 aM). (B) Single-plex detection of IL6 spiked into PBS. LOD = 0.0070 pg/mL. (C) The same samples of FBS spiked with varying concentrations of GM-CSF were measured using the MD and Quanterix’s Simoa. Good agreement was found between the two measurements, = 0.95. (D) The LOD, LOQ, dynamic range, and CV are reported for the MD’s and Simoa’s measurement of GM-CSF in FBS. (E) The duplex assay is tested by measuring various concentrations of GM-CSF and IL6 spiked into FBS. (F) Varying concentrations of GM-CSF into FBS resulted in insignificant cross-talk with the measurement of IL6 and did not significantly change the LOD for GM-CSF. (G) Conversely, varying concentrations of IL6 into FBS resulted in insignificant cross-talk with the measurement of GM-CSF and did not significantly change the LOD for IL6. Insets for F and G show these measurements on a linear scale. (H) Twenty-two various concentrations of GM-CSF and IL6 were spiked into FBS and measured. Good agreement was found between the spiked and measured results, for both GM-CSF () and IL6 ().
Next, we evaluated the MD chip’s capability to simultaneously measure two protein levels in a duplex measurement of GM-CSF and IL6 in complex media. To this end, we first spiked various quantities of GM-CSF into FBS, keeping IL6 concentrations at 0 pg/mL. In these samples, we measure both GM-CSF and IL6 using our duplex microbead assay and evaluate cross-talk and compare with our single-plex results (Fig. 5 F and G). We also performed the same experiment but, instead, spiked various levels of IL6 and kept GM-CSF concentrations at 0 pg/mL. In both cases, the LOD, for GM-CSF or IL6, did not change significantly from the single-plex measurement (p 0.88 for GM-CSF, p 0.90 for IL6). To further verify our capability to simultaneously measure both GM-CSF and IL6, we evaluated our chip’s accuracy in measuring n = 22 separate titrations of various quantities of GM-CSF and IL6 spiked into FBS (Fig. 5H). We found excellent agreement between the expected spiked concentrations and the measured concentrations () for GM-CSF and () for IL6.
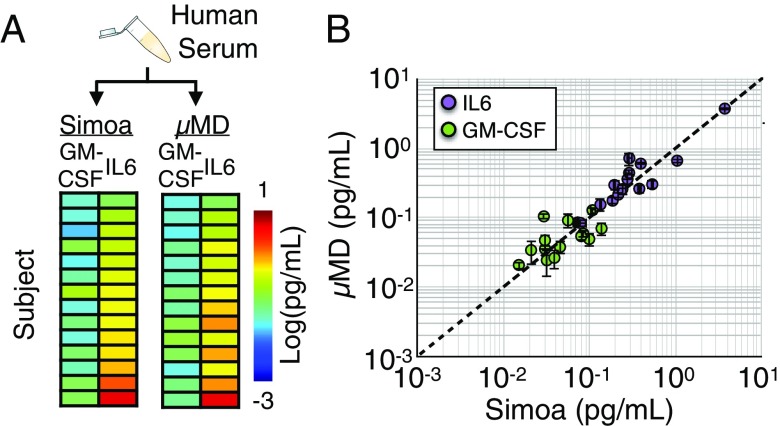
We next validated that the MD can measure endogenous protein in human serum. We collected serum from n = 14 healthy subjects, and, for each subject, measured an aliquot using our mobile MD platform’s IL6 and GM-CSF duplex assay, and we measured an aliquot using Quanterix’s commercial assay (Fig. 6A), allowing us to compare our results to the commercial gold standard. We saw excellent agreement between measurements on our mobile platform and that performed on Quanterix’s Simoa () (Fig. 6B), demonstrating that our microfluidic device can perform on human serum.
Fig. 6.
The measurement of endogenous protein in human serum. (A) Human serum was collected from n = 14 healthy controls, and an aliquot was measured using our MD’s duplex IL6, GM-CSF assay and was measured on Quanterix’s commercial assay. (B) Good agreement between Simoa and the MD was found for measurements of both IL6 and GM-CSF ().
Conclusion
Our MD platform, with its integrated and miniaturized implementation, its high sensitivity, and its high droplet throughput, allows digital assays containing millions of droplets to be performed on a mobile platform. By integrating and miniaturizing digital assays, the MD can translate the benefits of dELISA assays to a mobile diagnostic platform. While, in this paper, we performed a duplex assay, we can further leverage improvement in droplet throughput and multicolor detection to increase multiplexing to hundreds of markers. Multiplexing can be increased by the following approaches. (i) M assays can be run in parallel by leveraging our chip’s high droplet throughput and dividing the sample to be mixed with different reagents in either individual channels or sets of channels of the n = 120 detection channels. This approach comes at the expense of device throughput and a reduction in sensitivity that comes from splitting the sample volume for each additional assay. (ii) Microbeads with varying concentrations of multiple dyes can be used to barcode the microbeads for M assays in a single pot, as has been done by groups such as Luminex (50). Moreover, these two approaches can be combined on the same chip to achieve M 100 multiplexed assays. Similarly, multiple samples can be processed by running them either serially through the MD or in parallel by dividing up the n = 120 detection channels. In either approach, the samples would be incubated with beads, labeled, and washed in separate wells to avoid cross-contamination. In serial operation, the incubation time would remain the same (currently 3 h), and the total processing time would increase linearly with the number of samples (five additional minutes per sample, given a droplet throughput of 100,000 droplets/s and a 4-min droplet incubation time). The additional processing time per sample could be reduced by parallelizing the droplet incubation stage. Our miniaturized technology matches the limit of detection to existing dELISA; however, it is important to highlight that the MD does not currently have the ability to sequentially load samples in an automated fashion that the Simoa HD-1 has demonstrated for up to 384 samples (17). By making use of recent developments of applying dELISA to microRNA (miRNA) detection (19), multiplexed detection of miRNA and protein can be measured on the same chip for multimodal characterization of complex disease states (51, 52). By automating and incorporating dELISA onto a mobile platform, the MD allows ultrasensitive, multiplexed biomarker detection to be brought directly to the point of use, where digital assays can have the greatest impact.
Materials and Methods
Device Fabrication.
The MD is composed primarily of four components, all integrated into a monolithic chip (Fig. 2A): (i) a bead processor where beads are incubated and washed in successive steps, (ii) droplet generators, (iii) a delay line for the enzymatic amplification reaction, and (iv) the fluorescence detection region. The bead filtration unit consists of a 3-m polycarbonate filter (Sterlitech) sandwiched between laser-cut layers of adhesive-coated Mylar (7602A54; McMaster Carr). The top PDMS piece contains the droplet generators, and is fabricated using multilayer soft lithography, consisting of a layer that contains the nozzles and the spine (h = 10 m), a second layer that contains only the spine delivery channels (h = 120 m), and a third layer that contains herringbone structures for enhanced mixing (h = 30 m). The bottom PDMS piece was also fabricated using soft lithography, and consists of the spiral delay line (h = 1.5 mm) and the detector channels (h = 40 m). The three pieces are assembled by bonding the droplet generators PDMS piece to the top of a glass slide and bonding the delay line PDMS piece to the bottom, using plasma bonding. The glass slide (Glass Slide 26005; Corning) is etched with a through-hole d = 200 m using a CO2 laser, to serve as a via between the top and bottom pieces of PDMS microfluidics (Fig. 2A). The bead processing unit is adhered to the top PDMS piece using adhesive-coated Mylar (7602A54; McMaster Carr). The PDMS portion of the chip was made hydrophobic by running 1% silane [Trichloro(1H,1H,2H,2H-perfluorooctyl)silane; Sigma] in Novec 7500 (Oakwood Chemical) and flushed with Novec 7500 alone. The filter portion of the MD was soaked in 1% F127 in PBS, flushed with PBS, and then soaked in T20 buffer to reduce adhesion with antibodies and labeling enzyme.
Measurement of Endogenous Protein in Human Serum.
We collected blood from n = 14 healthy subjects (age = 20 to 43, 71% male, 29% female), in which all experiments involving human subjects were approved by University of Pennsylvania Institutional Review Board protocols (Protocol 828435). Informed consent was obtained from the eligible subject directly. A written document was provided to the subject detailing the procedure involved and the rationale for the study. The risks and benefits of study participation were explained. After a consent document was signed, an 8-mL blood draw was acquired, along with information regarding gender and age. Blood was collected in yellow cap tubes which contain Acid Citrate Dextrose Solution and serum-separating gels (BD Vacutainer Venous Blood Collection Tubes: SST Serum Separation Tubes: Hemogard, BD 368013). Blood was allowed to sit for 15 min after collection, and was then centrifuged at 1,500 rcf (relative centrifugal force) for 15 min to isolate the serum. After centrifugation, serum was frozen at −80 °C. Samples were thawed immediately before use, ensuring that all samples experienced the same freeze–thaw cycling. For Simoa measurements, we followed Quanterix’s protocol for the single-plex GM-CSF 2.0 and IL-6 2.0 kits, using the standard protocol (53, 54). For the MD measurements, we followed the same procedure described for the measurements of spiked proteins in FBS.
Supplementary Material
Acknowledgments
We thank Erika Silverman and Nimay Kulkarni for helping with blood draws for human subject experiments. We thank Margalit Haber for experimental setup with the Simoa and Brian Chow for advice on our manuscript. We thank Dave Meaney, Hakho Lee, Daeyeon Lee, Chris Fang-Yen, and Brian Chow for reading our manuscript and providing valuable feedback. This work was supported by the Department of Bioengineering, University of Pennsylvania. Funding was provided by the National Institutes of Health Awards 1R33CA206907 and 5R21CA182336. Funding was also provided by The Hartwell Foundation and the Pennsylvania Consortium on Traumatic Brain Injury. D.I. was supported by American Cancer Society CEOs Against Cancer CA Division Research Scholar Grant RSG-15-227-01-CSM, and by National Science Foundation CAREER Award 1554200. V.Y. is supported by the Microsoft Graduate Fellowship.
Footnotes
Conflict of interest statement: D.I. is the founder and D.I. and V.Y. currently hold equity in Chip Diagnostics. J.R.B. is employed by Chip Diagnostics.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814110116/-/DCSupplemental.
References
- 1.Prakadan SM, Shalek AK, Weitz DA. Scaling by shrinking: Empowering single-cell ’omics’ with microfluidic devices. Nat Rev Genet. 2017;18:345–361. doi: 10.1038/nrg.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pekin D, et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab A Chip. 2011;11:2156. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- 3.Hindson CM, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rissin DM, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rissin DM, Walt DR. Digital readout of target binding with attomole detection limits via enzyme amplification in femtoliter arrays. J Am Chem Soc. 2006;128:6286–6287. doi: 10.1021/ja058425e. [DOI] [PubMed] [Google Scholar]
- 7.Shim J-u, et al. Ultrarapid generation of femtoliter microfluidic droplets for single-molecule-counting immunoassays. ACS Nano. 2013;7:5955–5964. doi: 10.1021/nn401661d. [DOI] [PubMed] [Google Scholar]
- 8.Leirs K, et al. Bioassay development for ultrasensitive detection of influenza a nucleoprotein using digital ELISA. Anal Chem. 2016;88:8450–8458. doi: 10.1021/acs.analchem.6b00502. [DOI] [PubMed] [Google Scholar]
- 9.Chang L, et al. Simple diffusion-constrained immunoassay for p24 protein with the sensitivity of nucleic acid amplification for detecting acute HIV infection. J Virol Methods. 188:153–160. doi: 10.1016/j.jviromet.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Guan Z, et al. A highly parallel microfluidic droplet method enabling single-molecule counting for digital enzyme detection. Biomicrofluidics. 8:014110. doi: 10.1063/1.4866766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Obayashi Y, et al. A single-molecule digital enzyme assay using alkaline phosphatase with a cumarin-based fluorogenic substrate. Analyst. 2015;140:5065–5073. doi: 10.1039/c5an00714c. [DOI] [PubMed] [Google Scholar]
- 12.Shembekar N, Hu H, Eustace D, Merten CA. Single-cell droplet microfluidic screening for antibodies specifically binding to target cells. Cell Rep. 2018;22:2206–2215. doi: 10.1016/j.celrep.2018.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, van Zee M, Goda K, Di Carlo D. Size-based sorting of hydrogel droplets using inertial microfluidics. Lab A Chip. 2018;18:2575–2582. doi: 10.1039/c8lc00568k. [DOI] [PubMed] [Google Scholar]
- 14.Dennis JE, Adam S, Adam RA. Ultrahigh-throughput mammalian single-cell reverse-transcriptase polymerase chain reaction in microfluidic drops. Anal Chem. 2013;85:8016–8021. doi: 10.1021/ac402057q. [DOI] [PubMed] [Google Scholar]
- 15.Dennis JE, Adam S, Adam RA. Identification and genetic analysis of cancer cells with PCR-activated cell sorting. Nucleic Acids Res. 2014;42:e128. doi: 10.1093/nar/gku606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C, et al. Single-exosome-Counting immunoassays for cancer diagnostics. Nano Lett. 2018;18:4226–4232. doi: 10.1021/acs.nanolett.8b01184. [DOI] [PubMed] [Google Scholar]
- 17.Wilson DH, et al. The Simoa HD-1 analyzer. J Lab Autom. 2016;21:533–547. doi: 10.1177/2211068215589580. [DOI] [PubMed] [Google Scholar]
- 18.Korley FK, et al. Performance evaluation of a multiplex assay for simultaneous detection of four clinically relevant TBI biomarkers. J Neurotrauma. 2018;36:182–187. doi: 10.1089/neu.2017.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rissin DM, et al. Polymerase-free measurement of microRNA-122 with single base specificity using single molecule arrays: Detection of drug-induced liver injury. PLoS One. 2017;12:e0179669. doi: 10.1371/journal.pone.0179669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDermott GP, et al. Multiplexed target detection using DNA-binding dye chemistry in droplet digital PCR. Anal Chem. 2013;85:11619–11627. doi: 10.1021/ac403061n. [DOI] [PubMed] [Google Scholar]
- 21.Albayrak C, et al. Digital quantification of proteins and mRNA in single mammalian cells. Mol Cel. 2016;61:914–924. doi: 10.1016/j.molcel.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 22.Caroline Pereira Bittencourt Passaes et al. Ultrasensitive HIV-1 p24 assay detects single infected cells and differences in reservoir induction by Latency reversal Agents. J Virol. 2017;91:e02296-16. doi: 10.1128/JVI.02296-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivera A, et al. Peripheral total tau in military personnel who sustain traumatic brain injuries during deployment. JAMA Neurol. 2015;72:1109–1116. doi: 10.1001/jamaneurol.2015.1383. [DOI] [PubMed] [Google Scholar]
- 24.Schubert SM, et al. Ultra-sensitive protein detection via single molecule arrays towards early stage cancer monitoring. Scic Rep. 2015;5:11034. doi: 10.1038/srep11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Descours B, et al. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature. 2017;543:564–567. doi: 10.1038/nature21710. [DOI] [PubMed] [Google Scholar]
- 26.McGuigan W, et al. The optics inside an automated single molecule array analyzer. Proc SPIE. 2014;8935:89350X. [Google Scholar]
- 27.Du W, Liang L, KP Nichols, Ismagilov RF. Slipchip. Lab A Chip. 2009;9:2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen F, Du W, Kreutz JE, Fok A, Ismagilov RF. Digital PCR on a slipchip. Lab A Chip. 2010;10:2666–2672. doi: 10.1039/c004521g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatch AC, et al. 1-million droplet array with wide-field fluorescence imaging for digital PCR. Lab A Chip. 2011;11:3838–3845. doi: 10.1039/c1lc20561g. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Q, et al. Digital PCR on an integrated self-priming compartmentalization chip. Lab A Chip. 2014;14:1176–1185. doi: 10.1039/c3lc51327k. [DOI] [PubMed] [Google Scholar]
- 31.Baret J-C, et al. Fluorescence-activated droplet sorting (FADS): Efficient microfluidic cell sorting based on enzymatic activity. Lab A Chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 32.Utada AS, Fernandez-Nieves A, Stone HA, Weitz DA. Dripping to jetting transitions in coflowing liquid streams. Phys Rev Lett. 2007;99:094502. doi: 10.1103/PhysRevLett.99.094502. [DOI] [PubMed] [Google Scholar]
- 33.Yadavali S, Jeong H-H, Lee D, Issadore D. Silicon and glass very large scale microfluidic droplet integration for terascale generation of polymer microparticles. Nat Commun. 2018;9:1222. doi: 10.1038/s41467-018-03515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim M, et al. Optofluidic ultrahigh-throughput detection of fluorescent drops. Lab A Chip. 2015;15:1417–1423. doi: 10.1039/c4lc01465k. [DOI] [PubMed] [Google Scholar]
- 35.Yelleswarapu VR, et al. Ultra-high throughput detection (1 million droplets per second) of fluorescent droplets using a cell phone camera and time domain encoded optofluidics. Lab Chip. 2017;17:1083–1094. doi: 10.1039/c6lc01489e. [DOI] [PubMed] [Google Scholar]
- 36.Amstad E, et al. Robust scalable high throughput production of monodisperse drops. Lab Chip. 2016;16:4163–4172. doi: 10.1039/c6lc01075j. [DOI] [PubMed] [Google Scholar]
- 37.Jeong H-H, Yelleswarapu VR, Yadavali S, Issadore D, Lee D. Kilo-scale droplet generation in three-dimensional monolithic elastomer device (3d med) Lab A Chip. 2015;15:4387–4392. doi: 10.1039/c5lc01025j. [DOI] [PubMed] [Google Scholar]
- 38.Muluneh M, Kim B, Buchsbaum G, Issadore D. Miniaturized, multiplexed readout of droplet-based microfluidic assays using time-domain modulation. Lab A Chip. 2014;14:4638–4646. doi: 10.1039/c4lc00819g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martini J, et al. Time encoded multicolor fluorescence detection in a microfluidic flow cytometer. Lab A Chip. 2012;12:5057–5062. doi: 10.1039/c2lc40515f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muluneh M, Issadore D. Hybrid soft-lithography/laser machined microchips for the parallel generation of droplets. Lab A Chip. 2013;13:4750–4754. doi: 10.1039/c3lc50979f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward T, Faivre M, Abkarian M, Stone HA. Microfluidic flow focusing: Drop size and scaling in pressure versus flow-rate-driven pumping. Electrophoresis. 2005;26:3716–3724. doi: 10.1002/elps.200500173. [DOI] [PubMed] [Google Scholar]
- 42.Yelleswarapu V, Issadore D. 2017 IEEE Healthcare Innovations and Point of Care Technologies (HI-POCT) Inst Electr Electron Eng; New York: 2017. Multicolor detection of fluorescent droplets on a cell phone using time domain encoded optofluidics; pp. 245–248. [Google Scholar]
- 43.Weigl BH, Bardell R, Schulte T, Battrell F, Hayenga J. Design and rapid prototyping of thin-film laminate-based microfluidic devices. Biomed Microdevices. 2001;3:267–274. [Google Scholar]
- 44.Shaffer SM, et al. Multiplexed detection of viral infections using rapid in situ RNA analysis on a chip. Lab A Chip. 2015;15:3170–3182. doi: 10.1039/c5lc00459d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee C-Y, Chang C-L, Wang Y-N, Fu L-M. Microfluidic mixing: A review. Int J Mol Sci. 2011;12:3263–3287. doi: 10.3390/ijms12053263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abraham DS, et al. Chaotic mixer for microchannels. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 47.MacWilliams FJ, Sloane NJA. Pseudo-random sequences and arrays. Proc IEEE. 1976;64:1715–1729. [Google Scholar]
- 48.Gaster RS, et al. Matrix-insensitive protein assays push the limits of biosensors in medicine. Nat Med. 2009;15:1327–1332. doi: 10.1038/nm.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu D, Milutinovic MD, Walt DR. Single molecule array (Simoa) assay with optimal antibody pairs for cytokine detection in human serum samples. Analyst. 2015;140:6277–6282. doi: 10.1039/c5an01238d. [DOI] [PubMed] [Google Scholar]
- 50.Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the luminex labmap system. J Microbiol Methods. 2003;53:245–252. doi: 10.1016/s0167-7012(03)00028-9. [DOI] [PubMed] [Google Scholar]
- 51.Ko J, et al. Combining machine learning and nanofluidic technology to diagnose pancreatic cancer using exosomes. ACS Nano. 2017;11:11182–11193. doi: 10.1021/acsnano.7b05503. [DOI] [PubMed] [Google Scholar]
- 52.Ko J, et al. Machine learning to detect signatures of disease in liquid biopsies–A user’s guide. Lab A Chip. 2018;18:395–405. doi: 10.1039/c7lc00955k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quanterix . IL-6 Advantage Kit 101622 Datasheet. Quanterix; Lexington, MA: 2018. [Google Scholar]
- 54.Quanterix . Human GM-CSF 2.0 102329 Datasheet. Quanterix; Lexington, MA: 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.