Significance
Triggered release of calcium from intracellular stores is crucial for diverse cellular processes, including excitation–contraction coupling in muscle. Trimeric intracellular cation (TRIC) channels are essential components of endoplasmic reticulum/ sarcoplasmic reticulum (ER/SR) membranes, but their physiological roles have been unclear. Studies reported here elucidate a counter-ion mechanism by which TRIC channels facilitate calcium release. Calcium depletion during release through inositol 1,4,5-trisphosphate or ryanodine receptors depolarizes the ER/SR membrane potential, which retards continued calcium release. We show that Ca2+ binding to TRIC channels stabilizes a nonconductive conformation; however, Ca2+-free TRIC, as is generated during calcium release, frees a gating residue on a voltage-sensing helix to move out of blockage in response to membrane depolarization. Resulting K+ counter-ion currents can restore polarization for continued calcium release.
Keywords: counter-ion mechanism, Ca2+ modulation, lipid modulation, X-ray crystallography, electrophysiology
Abstract
Trimeric intracellular cation (TRIC) channels are thought to provide counter-ion currents that facilitate the active release of Ca2+ from intracellular stores. TRIC activity is controlled by voltage and Ca2+ modulation, but underlying mechanisms have remained unknown. Here we describe high-resolution crystal structures of vertebrate TRIC-A and TRIC-B channels, both in Ca2+-bound and Ca2+-free states, and we analyze conductance properties in structure-inspired mutagenesis experiments. The TRIC channels are symmetric trimers, wherein we find a pore in each protomer that is gated by a highly conserved lysine residue. In the resting state, Ca2+ binding at the luminal surface of TRIC-A, on its threefold axis, stabilizes lysine blockage of the pores. During active Ca2+ release, luminal Ca2+ depletion removes inhibition to permit the lysine-bearing and voltage-sensing helix to move in response to consequent membrane hyperpolarization. Diacylglycerol is found at interprotomer interfaces, suggesting a role in metabolic control.
Ca2+ released from the sarcoplasmic reticulum (SR) and endoplasmic reticulum (ER) participates in a wide variety of physiological cellular functions, including muscle contraction, neurotransmitter release, and cell growth and death (1, 2). Ion channels in SR/ER membranes, notably ryanodine receptors (RyRs) and inositol 1,4,5-trisphosphate receptors (IP3Rs), mediate intracellular Ca2+ release and play key roles in maintaining Ca2+ homeostasis. SR membranes are also rich in SR K+ channels (3, 4), which are thought to provide counter-ion currents that facilitate continued Ca2+ release from intracellular stores (5–7). A corresponding gene, Tric-a, was cloned, and its recombinant product was characterized as the trimeric intracellular cation (TRIC) channel (8–10). Mammalian TRICs have two subtypes: TRIC-A couples with RyRs to mediate Ca2+ release in excitable cells, especially within striated muscle cells and brain (8), whereas TRIC-B synchronizes with IP3Rs to mediate Ca2+ release in various tissue cells (9, 11). Tric-a–knockout mice developed hypertension due to enhanced myogenic tone in resistance arteries (12). In contrast, Tric-b–knockout mice exhibited neonatal lethality due to respiratory failure (13, 14). Furthermore, knockout mice lacking both TRIC channel subtypes exhibited embryonic heart failure. SNPs in Tric-a are associated with increased hypertension risk and decreased efficiency of antihypertensive drugs (15), whereas mutations in TRIC-B have been linked to bone and pulmonary diseases (14, 16–21).
Besides mammals, TRIC-family proteins occur in other domains of life, and structural studies have been reported on TRIC homologs from bacteria, archaea, and Caenorhabditis elegans. These structures revealed that TRIC represents a class of novel cation channel, with a unique architecture formed by two inverted structurally similar triple-helix bundles (THBs) (22–26). Mammalian TRIC channels are selective for cations, with slightly higher permeability for K+ over Na+, and their activities are mediated by the elaborate interplay of voltage sensing, Ca2+ modulation, and lipid regulation (3, 11, 24, 27–30); however, the underlying mechanisms remain unknown.
In this study, our bioinformatics analyses on TRIC sequences revealed that eukaryotic TRICs bifurcate into the TRIC-A and TRIC-B subtype at the stage of the vertebrate. We then determined crystal structures of two vertebrate TRICs: Gallus gallus (chicken) TRIC-A (GgTRIC-A) and Xenopus laevis (frog) TRIC-B (XlTRIC-B), in both Ca2+-bound and Ca2+-free states. Ca2+ binds to a conserved site on the trimeric threefold axis at the luminal side and is connected to inhibit channel activity. Structural comparison of the Ca2+-bound with Ca2+-free proteins, together with electrophysiology results, show that the channel is blocked by high Ca2+ (as the resting state in SR) and opened in lowered Ca2+ (as during RyR activity). Further structural analyses and related conductance measurements from mutants confirm the gating role of a highly conserved lysine residue. The disulfide-trapping experiments together with the electrophysiological measurement reveal the role of a putative voltage-sensitive helix TM4 in controlling channel gating. Diacylglycerol (DAG) binds at membrane-exposed subunit interfaces, and its identity suggests a role in metabolic control. Altogether, the sensing and movement of voltage-sensitive helix TM4, in concert with Ca2+ modulation, regulates TRIC channel activity to facilitate intracellular Ca2+ release. This study provides a structural and functional framework for understanding the counter-ion mechanism in TRIC channels.
Results
Sequence Analysis of TRIC Proteins.
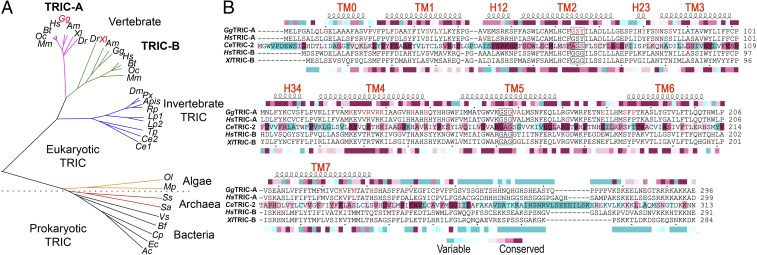
To better understand how TRIC proteins are represented in the animal kingdom, which includes various vertebrates and invertebrates, we clustered 425 nonredundant sequences into a superfamily of animal TRICs at the Position-Specific Iterated BLAST level E ≤ 10−3, then separated them into distinct clusters of vertebrate and invertebrate TRICs at an initial threshold of E ≤ 10−60, and finally into the vertebrate TRIC-A and TRIC-B subfamilies at a typical initial threshold of E ≤ 10−90, as detailed in SI Appendix, Table S1.
The vertebrate sequence includes proteins from mammals, amphibia, birds, and fish, each species typically containing two paralogous TRIC proteins, namely TRIC-A and TRIC-B, which have distinctive cellular distributions and functional characteristics. Among the vertebrate family, the orthologous TRIC-A proteins share ∼50 to 70% pairwise sequence identity, whereas the orthologous TRIC-B proteins are somewhat more divergent, sharing ∼40% sequence identity. The invertebrate family comprises a remote set of TRIC proteins including those from worms and various insects. These invertebrate TRIC proteins share merely ∼30% sequence identity with either mammalian TRIC-A or TRIC-B. However, the invertebrate TRIC proteins have not evolved into distinctive subsets as in the vertebrate family, although some organisms do contain two distinctive TRIC sequences, for example Ce1 and Ce2 from the worm C. elegans, with 54% sequence identity to each other, and with similar relatedness to human TRIC-A (31% and 31% identity) and human TRIC-B (32% and 30% identity), respectively.
Representative sequences from both vertebrate and invertebrate TRICs, together with sequences from green algae, archaea, and bacteria, were used to calculate the phylogenetic tree shown in Fig. 1A. The structure-based alignment of GgTRIC-A, XlTRIC-B, CeTRIC-2, HsTRIC-A, and HsTRIC-B sequences is shown in Fig. 1B, from which GgTRIC-A shares 76% and 40% sequence identity with human HsTRIC-A and HsTRIC-B whereas XlTRIC-B shares 46% and 54% sequence identity with human HsTRIC-A and HsTRIC-B, respectively.
Fig. 1.
Structure-based sequence alignment of TRIC Family. (A) Family tree. The presentation was computed by the program COBALT (42) from representative sequences from subfamilies. (B) Structure-based sequence alignments of eukaryotic TRICs from GgTRIC-A, XlTRIC-B, CeTRIC-2, HsTRIC-A, and HsTRIC-B. The structures of GgTRIC-A, CeTRIC-2, and XlTRIC-B have been used to restrict sequence gaps to interhelical segments. Superior coils define extents of the helical segments, boxes are drawn for the highly conserved glycine-containing motifs, red letters mark residues in the TRICs that involved in the ion conduction pathway, and the colored inferior bar encodes ConSurf (43) sequence variability for the vertebrate TRIC-A of 117 nonredundant proteins (Top), the invertebrate TRICs of 133 nonredundant proteins (Middle), and vertebrate TRIC-B of 126 nonredundant proteins (Bottom).
Biochemistry and Structure Determination.
Eight pairs of vertebrate TRIC-A and TRIC-B were selected for gene synthesis with codon optimization for expression in the yeast Schizosaccharomyces pombe. Both chicken GgTRIC-A and Xenopus XlTRIC-B were stable candidates for structural and functional characterization. As fused into planar lipid bilayers, the purified GgTRIC-A and XlTRIC-B proteins conduct K+ currents with characteristics similar to those for mammalian TRIC-A and TRIC-B, respectively (SI Appendix, Fig. S1).
When solubilized in octyl glucose-neopentyl glycol amphiphiles, GgTRIC-A and XlTRIC-B each crystallized in multiple lattices from various conditions (SI Appendix, Table S2). The GgTRIC-A structure was solved by selenomethionyl single-wavelength anomalous diffraction phasing and used to solve other structures as isomorphs or by molecular replacement. Structures were determined for each protein both in the presence and absence of Ca2+ ions, in differing conditions, and in differing lattices at resolutions down to 1.8 Å for GgTRIC-A and 3.1 Å for XlTRIC-B (SI Appendix, Table S3); structures were also determined for GgTRIC-A mutant proteins K129A and K129Q. We refined f′′ scattering factors against data measured at low X-ray energy (7k eV, λ = 1.77 Å) to identify bound ions, such as for Ca2+.
Structures of TRIC-A and TRIC-B.
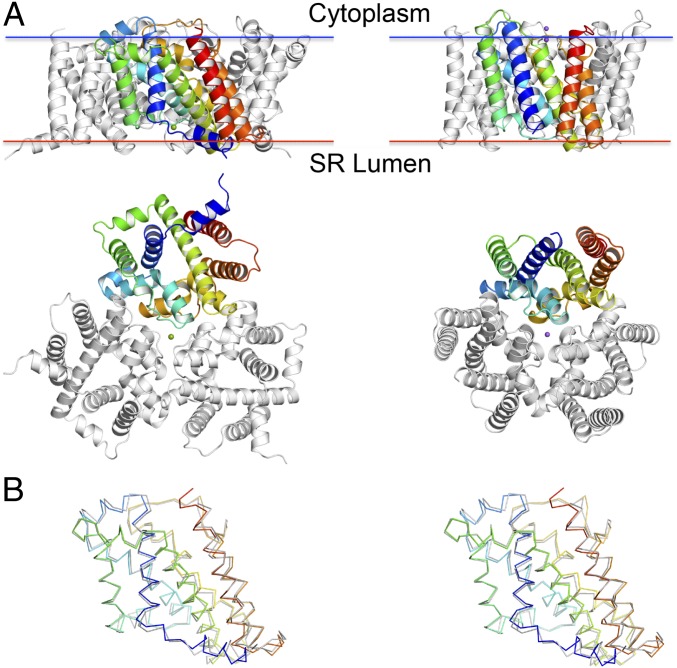
Both GgTRIC-A and XlTRIC-B are symmetrical trimers of 7-TM subunits (SI Appendix, Fig. S2A), and each protomer is composed of inverted quasi-repeats of THBs plus an additional TM7, forming an apparent ion-conduction pathway across the membrane. Despite low sequence identity (∼19.5%), TM1–3 resembles TM4–6, each adopting a THB fold, as in other TRICs (SI Appendix, Fig. S2B), while TM7 separately stands on the lateral side of the trimer, making contact with the pre-TM helix, TM0. The electrostatic potential surface generally is negative on the luminal side and largely positive on the cytoplasmic side (SI Appendix, Fig. S2 C and D). Compared with prokaryotic TRICs, most of the TMs in vertebrate TRICs have conserved proline residues located at the boundary of transmembrane segments. Their transmembrane helices are more inclined within the membrane and adopt a more expanded conformation (Fig. 2A). The XlTRIC-B shares 48.0% sequence identity with that of GgTRIC-A. The overall structure of XlTRIC-B is similar to that of GgTRIC-A, with an rmsd of 0.92 Å for 221 superimposed Cα of the protomer and 1.14 Å for 665 superimposed Cα of the trimer (Fig. 2B). They both share similar overall topological organization pattern, trimeric assembly architecture, and surface electrostatic characteristics with prokaryotic TRICs and invertebrate TRICs from C. elegans (SI Appendix, Fig. S3 A–F).
Fig. 2.
Structures of TRIC channels. (A) Comparison of vertebrate GgTRIC-A (Left) with prokaryotic and SaTRIC (Right). Ribbon drawings are viewed from the membrane (Top), and from the luminal or extracellular side (Bottom). Bound Ca2+ ion in GgTRIC-A is shown as a green sphere, and two bound Na+ ions in SaTRIC are shown as purple spheres. One protomer is colored spectrally from dark blue at its N terminus to red at its C terminus. Membrane boundaries were calculated by OPM server. (B) Superimposition of the protomer structure of GgTRIC-A and XlTRIC-B. Stereo view of the superimposed Cα backbones, oriented as in A. The coloring for GgTRIC-A protomer is as in A, and XlTRIC-B protomer is in gray.
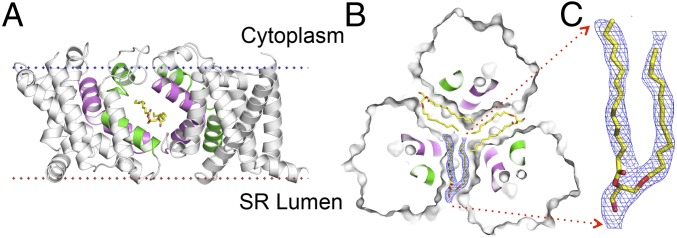
Lipid Binding in the Lateral Fenestration.
Similar to the prokaryotic TRICs (23, 25, 26) and CeTRICs (24), lateral fenestrations are found at the protomer interfaces for both GgTRIC-A and XlTRIC-B. These interfacial openings are largely hydrophobic, formed mainly by the conserved glycine-mediated kinked TM2 and TM5 from neighboring protomers (Fig. 3A). Electron density features of the lipid in GgTRIC-A (Fig. 3B) are distinct from those of the endogenous phosphatidylinositol 4,5-bisphosphate (PIP2) found in CeTRIC. Considering the shape of these densities, we speculated that the lipid molecule could be DAG, a hydrolyzed product of PIP2 (Fig. 3C). Further evidence came from the observation of DAG enrichment in the protein dissolved from crystals by lipidomics analysis via mass spectrometry (SI Appendix, Fig. S4 A and B). The DAG molecules interact extensively with the hydrophobic residues of the kinked TM2 and TM5 helices, with head groups’ crevice entrances and acyl chains embedded toward the center of the channel.
Fig. 3.
Llipid binding in the lateral fenestration. (A) Ribbon drawing of the GgTRIC-A, with one protomer removed. The TM2 and TM5 helices from two protomers are colored purple and green, respectively. The lipid molecules within the lateral fenestrations are shown as stick, with carbon, nitrogen, and oxygen atoms colored yellow, blue, and red, respectively. (B) Cross-section view of GgTRIC-A. Three DAG molecules were superimposed into the lateral fenestrations, with one lipid covered by the 2Fo-Fc electron density map, contoured at 2.0σ, and colored as in A. (C) Close-up view of the lipid molecule, as in B.
Both PIP2 and DAG are second messengers important in regulating channels, notably TRPC (31), CNG (32), Kir (33), and K2P (34). The discovery of PIP2 and/or DAG binding to the TRIC channel uncovered a link between cellular lipid signaling and Ca2+ homeostasis. In PIP2-bound CeTRIC, the head group of PIP2 protrudes into the pore, and the inositol ring is hydrogen-bonded to highly conserved residues (K129/R133 on TM4), thus blocking the pore. DAG lacks the inositol ring of PIP2, which leaves the pore unblocked in GgTRIC-A (SI Appendix, Fig. S4 C and D). Superimposition of PIP2-bound CeTRIC-2 onto the DAG-bound GgTRIC-A reveals that the TM5b segment serves as a conformational switch for binding different lipids. The indole rings of W173 residues in DAG-bound GgTRIC-A tightly associate at the threefold axis on the cytoplasmic side of the channel, whereas they swing away in PIP2-bound CeTRIC-2 (SI Appendix, Fig. S4 E and F). Altogether, our data suggested DAG plays a key role in the conformational control of pore-forming helices of TM2 and TM5, thus cooperatively coupling to channel gating.
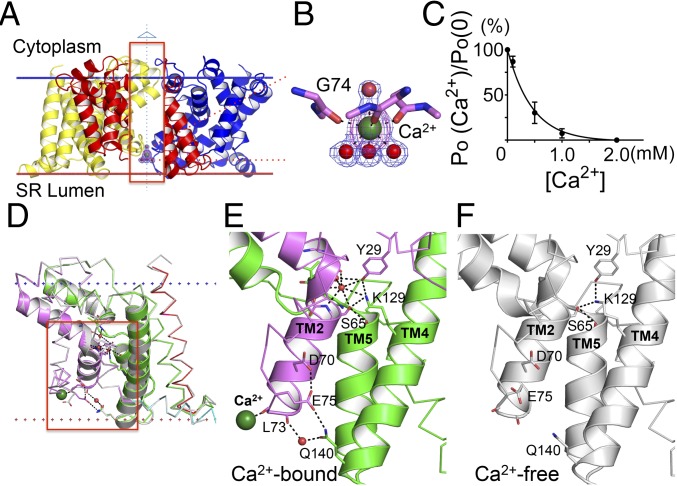
Ca2+ Binding and Channel Modulation.
Several groups have reported that Ca2+ modulates the activity of cardiac SR K+ channel (3, 27–29). The activities of TRIC-B have been shown to be regulated by Ca2+ on both the cytoplasmic and luminal sides (30); however, the underlying molecular mechanism remains unclear. Here, we show that a well-ordered Ca2+ ion, with associated water molecules, is bound at a luminal site in GgTRIC-A. The site is formed by backbone carbonyl oxygen atoms from highly conserved glycine 74 residue on the threefold axis of the trimeric channel (Fig. 4 A and B). The Ca2+ site was located easily in a Bijvoet-different map (SI Appendix, Fig. S5B), and its identification was verified by f′′ refinement in the 2.0-Å resolution structure. The fitted f′′ (Ca2+) 1.82e is close to the theoretical value 1.60e. Similarly, Ca2+ is found to bind to the same site in XlTRIC-B, which is formed by highly conserved alanine residues in the TRIC-B family.
Fig. 4.
Calcium binding and modulation in TRIC channels. (A) Ribbon diagram of Ca2+ binding in the structure of GgTRIC-A, as viewed from membrane. To have a better view for the Ca2+ binding, a section of the front protomer (inside the red box) is removed. The bound Ca2+ ion (dark green) and water molecules (red) are shown as spheres. 2Fo-Fc electron density contours are shown for both water molecules and Ca2+ ion, contoured at 1.5σ and colored in blue. The anomalous density is also shown for Ca2+ ion, colored in purple and contoured at 5.0σ. (B) A close-up view of the Ca2+ binding to the backbone carbonyl oxygen atom of G74. Water molecules and Ca2+ ion are shown, as in A. G74 O to Ca2+, distance = 2.4 Å; Ca2+ to water molecules, distance = 2.5 Å (lower) or 2.6 Å (upper). (C) GgTRIC-A channel modulation by luminal Ca2+. Relative channel open probabilities (PO) were determined from planar lipid bilayer recordings at +50 mV voltages (200 mM KCl in both trans and cis solutions) in the presence of varied Ca2+ concentrations in the cis side (n = 3). (D) Superimposition of GgTRIC-A, the Ca2+-bound structure vs. the Ca2+-free structure. The residues involved in hydrogen bonding interactions are shown as sticks. The hydrogen bonds (distance <3.2 Å) are indicated as a black dashed line. Water molecules involved in interaction are shown as red spheres. Ca2+ ion is shown as a sphere colored dark green. (E and F) Close-up views of hydrogen bonding interactions in Ca2+-bound GgTRIC-A (E) and Ca2+-free GgTRIC-A (F).
In keeping with the observation of Ca2+ binding on the luminal side of the channel, we found that luminal Ca2+ inhibits the channel activity in single-channel experiments. When 2 mM Ca2+ was applied at the luminal side of GgTRIC-A (orientation established as the asymmetrically charged TRIC protein is fused to the lipid bilayer held in an applied cis-bilayer potential), voltage-activated channels became inhibited, as evidenced by elimination of current in single-channel recordings. Gratifyingly, the inhibition was reversed when 5 mM EGTA was added to the bath solution to chelate the luminal Ca2+ (SI Appendix, Fig. S6A). Similar results were also found for XlTRIC-B, suggesting a conserved mechanism for TRIC channel modulation by Ca2+ (SI Appendix, Fig. S6B). Conductance measurements at varied luminal Ca2+ concentrations showed that inhibition occurs with Ki ∼ 0.4 mM (Fig. 4C).
The comparison of Ca2+-bound with Ca2+-free TRICs revealed substantial pore stabilization by Ca2+ (Fig. 4 D–F) despite having nearly identical overall structures (rmsd values of 0.51 Å/226Cα and 0.24 Å/224Cα for GgTRIC-A and XlTRIC-A, respectively; SI Appendix, Fig. S7 A and B). A shift of 0.49 Å in the coordinating carbonyl oxygen atom of G74 upon Ca2+ binding (SI Appendix, Fig. S7C) leads to a rotameric shift in the side chain of E75 in Ca2+-bound GgTRIC-A such that E75 then hydrogen-bonds to D70 and to Q140, stabilizing interactions between TM2 and TM4. This contact is strengthened by a water-mediated hydrogen bond between Q140 and the backbone carbonyl oxygen of L73.
Evidence that these Ca2+-mediated interactions are stabilizing is seen in comparing the atomic-mobility (B-factor) profiles from the two structures (SI Appendix, Fig. S7D). Whereas the mobility profile for the Ca2+-free TRIC-A is relatively uniform, that for the Ca2+-bound structure has its B-factors appreciably reduced for the TM2–TM3 and TM4–TM5 helix pairs associated with the Ca2+-binding site. This stabilization extends through the lengths of these helices, and consequently two backbone-coordinated water molecules bind near S65 at a kink in TM2. The resulting network of added interactions strengthens pore blockage by the K129 plug (Fig. 4E). The number of stabilizing contacts is decreased from 22 in the Ca2+-bound state to 3 in the Ca2+-free state. Only the hydrogen bonds connecting K129 with Y29 and S65 are retained in Ca2+-free GgTRIC-A (Fig. 4F). As discussed below, channel opening is voltage-dependent; thus, not surprisingly, all crystal structures are in a closed, resting state. The K129 latch is expected to loosen with Ca2+ depletion on the luminal side, which we see leads to increased PO for the TRIC channel (Fig. 4C).
Ion-Conducting Pore.
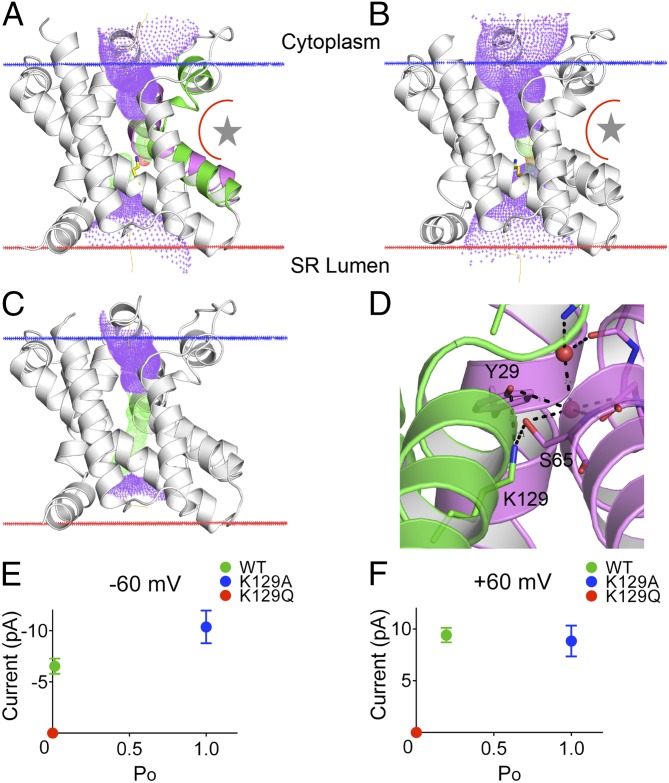
The pores in both GgTRIC-A and XlTRIC-B have similar hourglass-shaped tunnels across the membrane, with a narrow constriction in each near the middle of the membrane, as calculated by the HOLE program (Fig. 5 A and B). The pore is lined with highly conserved residues and is largely hydrophobic and electropositive (SI Appendix, Fig. S8 A and B). The basic character of the pore is a puzzle; however, conformational changes may occur during gating. Sequence analyses revealed well-conserved glycine-containing signature motifs in both TM2 and TM5 helices among all TRICs. It is highly conserved as “FGSY” in TM2 and “GWXKGXGVA” in TM5 for TRIC-As, whereas it is “FGGG” in TM2 and “GWARGXGGG” in TM5 for TRIC-Bs. These signature motifs generate structural kinks in the middle of transmembrane segments, thus disrupting the regular hydrogen-bonding patterns in TM2 and TM5, and consequently exposing backbone amines for coordinating water molecules. The site of a disease-causing mutation in human TRIC-B (VL insertion after G152) is located at the glycine-mediated kink in TM5, which could alter properties of the channel pore to cause autosomal recessive osteogenesis imperfect (21).
Fig. 5.
The ion conduction pore of TRIC channels. (A and B) The pore lining surface as computed by the program HOLE is drawn into a ribbon model of the protomer structure of GgTRIC-A (A) and XlTRIC-A (B), and the kinked TM2 (in purple) and TM5 (in green) helices are shown. The lateral fenestration interface and lipid-binding site is indicated, shown as a star. We used a simple van der Waals surface for the protein and the program default probe radius of 1.15 Å. The lining surface is shown in blue dots except at narrow restrictions (pore radius < 2.0 Å), which are in green dots. A yellow line through the channel marks the calculated centerline of the pore. (C) The pore lining surface of GgTRIC-A K129A mutant is drawn into a ribbon model, oriented and details as in A. (D) Close-up view of the conserved K129 and its hydrogen-bonded interaction with Y29 and S65. Water molecules observed within the ion-conduction pathway are shown as red spheres. (E and F) Analysis of single-channel properties for GgTRIC-A, wild type, K129A, and K129Q mutants (n = 3).
In prokaryotic TRICs, a network of interhelical hydrogen bonds (residues D99–R139–D140–Y155 in SaTRIC) locks the channel pore in a closed state (26). By comparison, this interaction network is not observed in either GgTRIC-A or XlTRIC-B. Instead, a highly conserved lysine residue (K129 in GgTRIC-A) at the neck of the hourglass-shaped pore interacts with Y29 (TM1) and S65 (TM2) and forms a hydrogen-bonded plug to block the pore. The K129A mutation generates an apparently open pore, with a uniform diameter of ∼4 Å across the membrane (Fig. 5 C and D). In keeping with these structural observations, we found that the K129A mutant resulted in a fully open pore at all voltages whereas wild-type TRIC-A channels are a voltage-dependent, with higher open probability and higher current amplitude at positive potential than at negative potential (SI Appendix, Fig. S8 C and D). This rectification characteristic of the wild-type channel is eliminated for K129A. These observations not only point to the gating role for the highly conserved K129 but also indicate its role in sensing voltage across membrane (Fig. 5 E and F). Furthermore, the channel pore of mutant K129Q remained largely closed, showing occasional openings only at high voltage (90 mV), which is consistent with stabilizing contacts of Q129 with Y66 and K155 in the structure of K129Q (SI Appendix, Fig. S8 E and F).
Voltage-Sensing and Gating Mechanism.
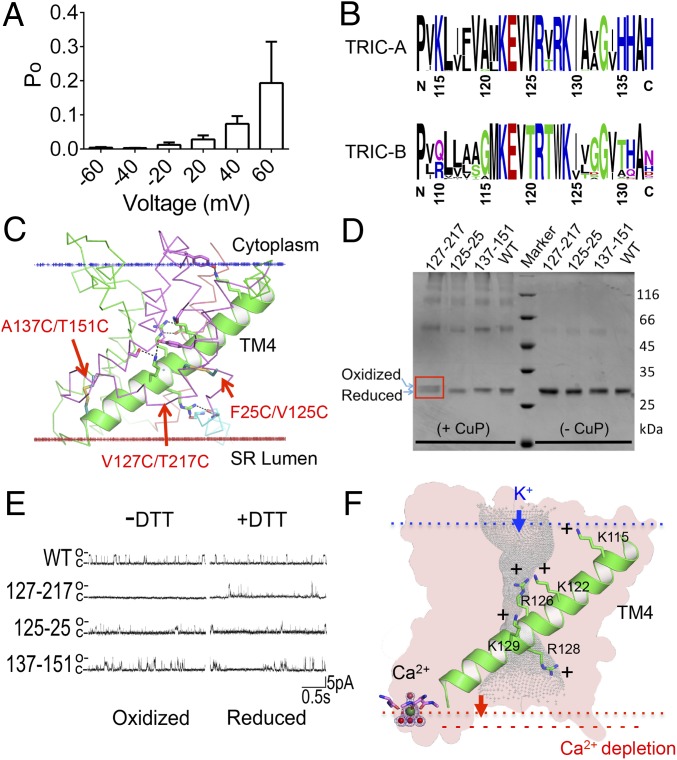
Similar to mammalian TRICs, GgTRIC-A can sense the voltage potential across the membrane and is regulated in a voltage-dependent manner (Fig. 6A). Voltage-regulated ion channels, such as Nav (35), Kv (36), and TPC1 (37), usually have a separated voltage-sensing domain (VSD), with some positively charged lysine or arginine residues on their voltage-sensing S4 helix. There is no such VSD in either TRIC-As or TRIC-Bs, but these channel proteins do have TM4 helices that are enriched in highly conserved basic residues (Fig. 6B). This composition is as in VSDs, but the TRIC TM4 segments are otherwise structurally distinct (SI Appendix, Fig. S9), being long, straight, and inclined (Fig. 6C). The five conserved basic residues on TM4 in GgTRIC-A are K115, K122, R126, R128, and K129, four of which line the pore to generate the unique electropositive characteristic. These observations raise the possibility that the conformation of the basic region within the pore could change, perhaps as part of the gating process.
Fig. 6.
The voltage sensor and gating mechanism of TRIC channels. (A) Voltage dependency of channel gating. The Po of the full conductance open state at a series of holding potentials is shown. TRIC-A channel activity was reduced at negative potential compared with positive potential (n = 5). (B) Conserved motifs on the voltage-sensing TM4 for both TRIC-A and TRIC-B. (C) The cartoon model for TM4 in the protomer structure of GgTRIC-A. The five highly conserved positive-charge residues and their stabilizing residues are shown as sticks. Three pairs of substituted cysteine mutations (F25C/V125C, V127C/T217C, and A137C/T151C) are also shown as sticks. (D) Analysis of substituted cysteine cross-linking experiment, promoted by CuP-mediated oxidation. The A137C/T151C pair mutant show shift on the SDS/PAGE, indicating the formation of disulfide bond. (E) Single-channel recording analysis of substituted cysteine cross-linking mutants. (F) A model for TRIC channels voltage sensing and its coupling to the gating process.
To tackle the possible gating mechanism, we have designed paired cysteine mutations based on the structural analysis, including F25C/V125C, V127C/T217C, and A137C/T151C, which aimed to introduce disulfide bridges to test the role of a putative voltage-sensitive helix TM4 in regulating channel gating. Oxidative cross-linking (38) of designed double-cysteine mutants was expected to restrain the conformation of the voltage-sensitive helix TM4 relative to other adjacent transmembrane helix (e.g., TM1, TM5, and TM6) upon disulfide formation, and thus lock the channel in a closed state (Fig. 6C). Interestingly, the V127C/T217C (TM4–TM6) pair mutants resulted in a small but nonnegligible shift in the SDS/PAGE upon CuP-promoted oxidation, indicating interhelical disulfide formation of C127/C217 within the protomer (Fig. 6D). Just as expected, the V127C/T217C double mutant does not show any channel opening, even at voltages higher than +50 mV, most likely because the C127/C217 disulfide bond locks the voltage-sensing TM4 helix in a restrained conformation, thus causing the channel to remain closed upon depolarization. Reduction of the disulfide bond with DTT leads to robust channel opening (Fig. 6E). Other pairs of mutants [F25C/V125C (TM1–TM4) and A137C/T151C (TM4–TM5)] did not show any obvious shift in the cross-linking analysis or different electrophysiological responses between oxidizing and reducing conditions. Presumably the possible disulfide bonds were unable to form, due to nonideal geometry (SI Appendix, Fig. S10). All above observations point to an essential role for the voltage-sensing TM4 helix and its coupling to the channel activation.
Discussion
Excitation–contraction coupling in muscle depends on rapid release of Ca2+ ions from SR stores through RyR channels. SR/ER Ca2+-ATPase pumps will ultimately restore the SR Ca2+ concentration level, but not on a time scale that could avert inevitable SR membrane hyperpolarization that would detrimentally slow Ca2+ release. It has long been appreciated that a counter-ion mechanism is needed to ensure continued Ca2+ release (5, 6); however, which channels predominantly carry such counter ions has been unclear (10, 39). SR K+ channels (3), now generally accepted to be TRIC channels, are highly abundant in SR membranes and these are logical candidates; alternatively, RyR channels themselves have been posited as the primary route for counter-ion flow (39). Supposing that TRIC channels are indeed the main conveyers of the counter ions, mechanistic questions of how TRIC channels might serve this function have also been open.
Recent studies have clarified our understanding of the counter-ion mechanism appreciably. One study measured SR membrane potentials using a novel FRET assay to provide compelling evidence that there is virtually no change in potential during muscle fiber activation (40), and detailed simulations of activity at the SR junctions with transverse tubules from the plasma membrane in cardiac muscle concluded that, although several channels participate, TRIC K+ channels predominate in conducting counter-ion currents (41). These results are consistent with conclusions from the permeability of isolated SR vesicles that the SR membrane cannot develop a potential greater than a few millivolts under physiological conditions (7).
What, then, is the mechanism by which TRIC channels provide the counter currents of K+ ions that efficiently balance the losses from Ca2+ release? Our studies show that TRIC channels are controlled by Ca2+ ion concentrations and membrane potentials as observed in the physiological range. We find that GgTRIC-A is substantially inhibited from carrying currents when with Ca2+ ions as for SR at rest (0.5 to 1.0 mM; Fig. 4C) but that TRIC-A is opened when the Ca2+ level is reduced to the 0.1 to 0.2 mM level as happens during Ca2+ release in active muscle.
TRIC-A channel opening is potentiated at nonphysiological membrane potentials; however, channel openings also occur at relevant SR potentials near zero (Fig. 5A). We find that TRIC channels are gated by a lysine residue (K129) on voltage-sensitive helix TM4, and that channel activity is blocked reversibly when TM4 is immobilized by a disulfide link. We also observe that the K129 latch is highly sensitive to structural details: The K129Q channel has a closed pore and does not conduct whereas K129A is fully open, structurally and functionally; similarly, Ca2+-bound TRIC-A has a network of hydrogen-bonded interactions surrounding the K129 latch whereas stabilizing interactions are greatly diminished in Ca2+-free TRIC-A. Pore blockage is not released in the Ca2+-free crystal, but we propose that destabilization of the K129 latch leaves it poised for displacement as voltage-sensitive TM4 moves in a hyperpolarized membrane (Fig. 6F). Such TM4 movement likely affects the electrostatic potential within the pore as well.
Our structural results also suggest that TRIC channels may be under overall metabolic control. We identify DAG as a stabilizing ligand that copurified with GgTRIC-A and bound to lateral fenestrations between protomers. It is well established that PIP2 is a membrane phospholipid of importance in cellular signaling pathways. Hydrolysis of PIP2 yields IP3 and DAG; IP3 in turn activates IP3 receptors on the ER and facilitates intracellular Ca2+ release and other activities, and DAG has been shown to activate TRPC channels and here to stabilize the TRIC-A channel. In a parallel observation, PIP2 was found copurified with C. elegans TRIC-2 and to be bound at protomer interfaces (24), as similarly found here for DAG with GgTRIC-A.
Materials and Methods
Bioinformatics analysis, cloning and expression, scaled-up production and purification, crystallization and data measurement, structure determination, f′′ refinement for ion identification, electrophysiology, and cysteine cross-linking procedures are described in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Bailong Xiao, Jie Geng, and Qi Yuan for helpful discussions and initiating preliminary lipid bilayer experiments; Yanan Deng and Meihua Wang for help with protein chemistry and crystallography; and Wen-ming Qin, De-qiang Rao, and Raj Surajit for help with synchrotron experiments at various beamlines, including BL19U and BL17U at the Shanghai Synchrotron Radiation Facility and 24-IDC at the Advanced Photon Source. This project is financially supported by National Key R&D Program of China Grants 2016YFA0500503 and 2015CB910102; Chinese Academy of Sciences Strategic Priority Research Program XDB08020301; and National Natural Science Foundation of China Grants 31872721, 31470728, and 31322005 (to Y.-h.C.), 31728010 (to Y.-h.C. and F.L.), and 11672226 (to W.X.). Y.-h.C. is supported in part by the National Thousand Young Talents program from the Office of Global Experts Recruitment in China. W.X. is supported in part by the Young Talent Support Plan of Xi’an Jiao-tong University and State Key Laboratory of Molecular Developmental Biology, China Grant 2018-MDB-KF-02. F.L. is supported in part by NIH Grant R01GM106037. W.A.H. is supported in part by NIH Grant GM 107462. This work was also benefited from NIH support to the Center on Membrane Protein Production and Analysis (COMPPÅ, Grant P41 GM116799).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6IYU, 6IYX, 6IZF, 6IYZ, 6IZ0, 6IZ1, 6IZ5, 6IZ3, 6IZ4, and 6IZ6).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817271116/-/DCSupplemental.
References
- 1.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: Dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 2.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 3.Miller C. Voltage-gated cation conductance channel from fragmented sarcoplasmic reticulum: Steady-state electrical properties. J Membr Biol. 1978;40:1–23. doi: 10.1007/BF01909736. [DOI] [PubMed] [Google Scholar]
- 4.Coronado R, Rosenberg RL, Miller C. Ionic selectivity, saturation, and block in a K+-selective channel from sarcoplasmic reticulum. J Gen Physiol. 1980;76:425–446. doi: 10.1085/jgp.76.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melzer W. No voltage change at skeletal muscle SR membrane during Ca2+ release-just Mermaids on acid. J Gen Physiol. 2018;150:1055–1058. doi: 10.1085/jgp.201812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fink RH, Veigel C. Calcium uptake and release modulated by counter-ion conductances in the sarcoplasmic reticulum of skeletal muscle. Acta Physiol Scand. 1996;156:387–396. doi: 10.1046/j.1365-201X.1996.212000.x. [DOI] [PubMed] [Google Scholar]
- 7.Garcia AM, Miller C. Channel-mediated monovalent cation fluxes in isolated sarcoplasmic reticulum vesicles. J Gen Physiol. 1984;83:819–839. doi: 10.1085/jgp.83.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yazawa M, et al. TRIC channels are essential for Ca2+ handling in intracellular stores. Nature. 2007;448:78–82. doi: 10.1038/nature05928. [DOI] [PubMed] [Google Scholar]
- 9.Zhou X, et al. Trimeric intracellular cation channels and sarcoplasmic/endoplasmic reticulum calcium homeostasis. Circ Res. 2014;114:706–716. doi: 10.1161/CIRCRESAHA.114.301816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeshima H, Venturi E, Sitsapesan R. New and notable ion-channels in the sarcoplasmic/endoplasmic reticulum: Do they support the process of intracellular Ca2+ release? J Physiol. 2015;593:3241–3251. doi: 10.1113/jphysiol.2014.281881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venturi E, et al. TRIC-B channels display labile gating: Evidence from the TRIC-A knockout mouse model. Pflugers Arch. 2013;465:1135–1148. doi: 10.1007/s00424-013-1251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao X, et al. Ca2+ overload and sarcoplasmic reticulum instability in tric-a null skeletal muscle. J Biol Chem. 2010;285:37370–37376. doi: 10.1074/jbc.M110.170084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamazaki D, et al. TRIC-A channels in vascular smooth muscle contribute to blood pressure maintenance. Cell Metab. 2011;14:231–241. doi: 10.1016/j.cmet.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 14.Zhao C, et al. Mice lacking the intracellular cation channel TRIC-B have compromised collagen production and impaired bone mineralization. Sci Signal. 2016;9:ra49. doi: 10.1126/scisignal.aad9055. [DOI] [PubMed] [Google Scholar]
- 15.Yamazaki D, et al. Essential role of the TRIC-B channel in Ca2+ handling of alveolar epithelial cells and in perinatal lung maturation. Development. 2009;136:2355–2361. doi: 10.1242/dev.036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaheen R, et al. Study of autosomal recessive osteogenesis imperfecta in Arabia reveals a novel locus defined by TMEM38B mutation. J Med Genet. 2012;49:630–635. doi: 10.1136/jmedgenet-2012-101142. [DOI] [PubMed] [Google Scholar]
- 17.Volodarsky M, et al. A deletion mutation in TMEM38B associated with autosomal recessive osteogenesis imperfecta. Hum Mutat. 2013;34:582–586. doi: 10.1002/humu.22274. [DOI] [PubMed] [Google Scholar]
- 18.Rubinato E, et al. A novel deletion mutation involving TMEM38B in a patient with autosomal recessive osteogenesis imperfecta. Gene. 2014;545:290–292. doi: 10.1016/j.gene.2014.05.028. [DOI] [PubMed] [Google Scholar]
- 19.Cabral WA, et al. Absence of the ER cation channel TMEM38B/TRIC-B disrupts intracellular calcium homeostasis and dysregulates collagen synthesis in recessive osteogenesis imperfecta. PLoS Genet. 2016;12:e1006156. doi: 10.1371/journal.pgen.1006156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichimura A, Takeshima H. TRIC-B mutations causing osteogenesis imperfecta. Biol Pharm Bull. 2016;39:1743–1747. doi: 10.1248/bpb.b16-00612. [DOI] [PubMed] [Google Scholar]
- 21.Lv F, et al. Two novel mutations in TMEM38B result in rare autosomal recessive osteogenesis imperfecta. J Hum Genet. 2016;61:539–545. doi: 10.1038/jhg.2016.11. [DOI] [PubMed] [Google Scholar]
- 22.Silverio AL, Saier MH., Jr Bioinformatic characterization of the trimeric intracellular cation-specific channel protein family. J Membr Biol. 2011;241:77–101. doi: 10.1007/s00232-011-9364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasuya G, et al. Crystal structures of the TRIC trimeric intracellular cation channel orthologues. Cell Res. 2016;26:1288–1301. doi: 10.1038/cr.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H, et al. Pore architecture of TRIC channels and insights into their gating mechanism. Nature. 2016;538:537–541. doi: 10.1038/nature19767. [DOI] [PubMed] [Google Scholar]
- 25.Ou X, et al. Ion- and water-binding sites inside an occluded hourglass pore of a trimeric intracellular cation (TRIC) channel. BMC Biol. 2017;15:31. doi: 10.1186/s12915-017-0372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su M, et al. Structural basis for conductance through TRIC cation channels. Nat Commun. 2017;8:15103. doi: 10.1038/ncomms15103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu QY, Strauss HC. Blockade of cardiac sarcoplasmic reticulum K+ channel by Ca2+: Two-binding-site model of blockade. Biophys J. 1991;60:198–203. doi: 10.1016/S0006-3495(91)82043-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uehara A, Yasukohchi M, Ogata S, Imanaga I. Activation by intracellular calcium of a potassium channel in cardiac sarcoplasmic reticulum. Pflugers Arch. 1991;417:651–653. doi: 10.1007/BF00372965. [DOI] [PubMed] [Google Scholar]
- 29.Uehara A, Yasukochi M, Imanaga I. Calcium modulation of single SR potassium channel currents in heart muscle. J Mol Cell Cardiol. 1994;26:195–202. doi: 10.1006/jmcc.1994.1022. [DOI] [PubMed] [Google Scholar]
- 30.Pitt SJ, et al. Charade of the SR K+-channel: Two ion-channels, TRIC-A and TRIC-B, masquerade as a single K+-channel. Biophys J. 2010;99:417–426. doi: 10.1016/j.bpj.2010.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storch U, et al. Dynamic NHERF interaction with TRPC4/5 proteins is required for channel gating by diacylglycerol. Proc Natl Acad Sci USA. 2017;114:E37–E46. doi: 10.1073/pnas.1612263114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crary JI, Dean DM, Nguitragool W, Kurshan PT, Zimmerman AL. Mechanism of inhibition of cyclic nucleotide-gated ion channels by diacylglycerol. J Gen Physiol. 2000;116:755–768. doi: 10.1085/jgp.116.6.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Logothetis DE, et al. Phosphoinositide control of membrane protein function: A frontier led by studies on ion channels. Annu Rev Physiol. 2015;77:81–104. doi: 10.1146/annurev-physiol-021113-170358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feliciangeli S, Chatelain FC, Bichet D, Lesage F. The family of K2P channels: Salient structural and functional properties. J Physiol. 2015;593:2587–2603. doi: 10.1113/jphysiol.2014.287268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 37.Guo J, et al. Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana. Nature. 2016;531:196–201. doi: 10.1038/nature16446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mancia F, Assur Z, Herman AG, Siegel R, Hendrickson WA. Ligand sensitivity in dimeric associations of the serotonin 5HT2c receptor. EMBO Rep. 2008;9:363–369. doi: 10.1038/embor.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gillespie D, Fill M. Intracellular calcium release channels mediate their own countercurrent: The ryanodine receptor case study. Biophys J. 2008;95:3706–3714. doi: 10.1529/biophysj.108.131987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez C, et al. Tracking the sarcoplasmic reticulum membrane voltage in muscle with a FRET biosensor. J Gen Physiol. 2018;150:1163–1177. doi: 10.1085/jgp.201812035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zsolnay V, Fill M, Gillespie D. Sarcoplasmic reticulum Ca2+ release uses a Cascading network of intra-SR and channel countercurrents. Biophys J. 2018;114:462–473. doi: 10.1016/j.bpj.2017.11.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papadopoulos JS, Agarwala R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 43.Landau M, et al. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.