Abstract
Summary
Over the last decade, genome-wide association studies (GWAS) have generated vast amounts of analysis results, requiring development of novel tools for data visualization. Quantile–quantile (QQ) plots and Manhattan plots are classical tools which have been utilized to visually summarize GWAS results and identify genetic variants significantly associated with traits of interest. However, static visualizations are limiting in the information that can be shown. Here, we present Assocplots, a Python package for viewing and exploring GWAS results not only using classic static Manhattan and QQ plots, but also through a dynamic extension which allows to interactively visualize the relationships between GWAS results from multiple cohorts or studies.
Availability and Implementation
The Assocplots package is open source and distributed under the MIT license via GitHub (https://github.com/khramts/assocplots) along with examples, documentation and installation instructions.
1 Introduction
Advances in genotyping, sequencing, and phenotyping techniques have resulted in large quantities of genome-wide association studies (GWAS) data. The results of GWAS are commonly summarized and displayed on a Manhattan plot and a quantile–quantile (QQ) plot and help identify single nucleotide polymorphisms (SNPs) that are significantly associated with a given phenotype. Over the years, several standalone programs such as WGAViewer (Ge et al., 2008), web applications (summarized in Zeigler et al., 2015) such as LocusZoom (Pruim et al., 2010) and packages (mostly written in R) such as qqman (Turner, 2014) have been developed for producing these types of plots. Although dynamic and interactive visualization has recently become more widely adopted in genomic tools such as R/qtlcharts (Broman, 2015) and LDlink (Machiela and Chanock, 2015), it has not yet become a routine part of GWAS data analysis. Interactive data visualization not only allows clearer representation of multidimensional data, but also encourages a viewer’s engagement from simple data browsing to providing a platform for answering specific scientific questions, in ways that static data cannot. We present a Python package for viewing GWAS results not only using classic static Manhattan and QQ plots, but also through an interactive extension which allows a user to visualize data interactively, e.g.: zoom into SNP dense regions, obtain underlying details (e.g. SNP rs number, base pair position, P value) by selecting a peak of interest, and visualizing the relationships between GWAS results from multiple cohorts or studies. For example, our tool allows exploration of GWAS results from: (i) multiple phenotypes in a single group of individuals, (ii) a phenotype measured among distinct cohorts, (iii) expression quantitative trait loci measured across different tissues or cohorts, and (iv) various experimental conditions such as before and after drug treatment. Thus, our tool makes it possible to browse multiple charts in real-time to better understand the relationships among groups.
2 Implementation and features
Assocplots is implemented as a package for the Python programming language. Its basic functionality includes plotting interactive data visualization for viewing in the browser as well as static publication quality plots using matplotlib. Interactive visualization is implemented via a Python interactive visualization library, bokeh (http://bokeh.pydata.org/), that targets modern web browsers; and data wrangling is implemented with Numpy and Pandas scientific computing Python libraries. All of these tools are open source. The use of Python for this package makes it easily accessible to bioinformaticians, as it is one of the commonly used programming languages in the field. The package is designed to be used both in Jupyter notebooks (http://jupyter.org/) and in command line. Visualizing GWAS data in a web-based notebook, ensures data analysis reproducibility and makes it conveniently sharable via online repositories such as GitHub. The Assocplots package is open source and distributed under the MIT license. Below we present the package’s features and give an example of the plots (Fig. 1) using data from the Genetic Investigation of ANthropometric Traits (GIANT) consortium (Randall et al., 2013).
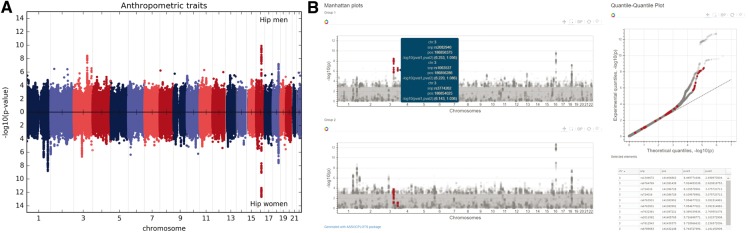
Fig. 1.
(A) Example of an inverted Manhattan plot generated using the static module and the data from the GIANT consortium, where each dot represents a single nucleotide polymorphism with the −log10(P value) on the y-axis and chromosome and base pair position on the x-axis. (B) A static view of interactive Manhattan plots for two groups (same as in A). Selecting a group of SNPs in one of the groups highlights the same SNPs in the other group, and hovering over a dot shows the information associated with the SNP: chromosome, rs number, base pair position, and a −log10(P value) for both groups. For the interactive version of this figure, see http://khramts.github.io/output.html
2.1 Static module features
2.1.1 Classic Manhattan plot
a. X-axis: chromosome and base pair (both numeric and alphabetical names, so various chromosome labeling (e.g. 1, 2, chr1, X) is acceptable).
b. Y-axis: Although −log10(P value) is the most commonly used value for the y-axis, other values such as the effect size can be specified.
c. Inverted Manhattan plot for two groups (Fig. 1A).
2.1.2 Classic QQ plot
a. Multiple groups plotting: Multiple groups can be visualized on the same QQ plot for easier comparison.
b. Genomic Inflation Factor, λgc, calculation: In GWAS, population substructure and cryptic relatedness among subjects can lead to spurious errors, and genomic control method is commonly used to correct the underlying population stratification (Devlin and Roeder, 1999).
c. Confidence interval (CI) estimation: The package allows to plot CIs for either the null distribution or the experimental data. When multiple groups are plotted, a CI can be displayed for each group.
2.1.3 Figure generation
Assocplots supports all matplotlib plotting backends and can save figures in raster format (i.e. png and jpg) and vector format (i.e. pdf and ps).
2.2 Interactive module features
2.2.1 Dynamic Manhattan and QQ plot
a. Info pop-up: Hovering over a point reveals information about the SNP, such as the SNP rs number, chromosome, base pair location, and the statistic reported on the y-axis (−log10(P value) or effect size) (Fig. 1B).
b. Group comparison: Selecting a set of SNPs in one graph automatically highlights those same SNPs in the other graph (for example, a different phenotype, population, or condition). Additionally, a table is generated below the graphs, listing all the selected SNPs and information about those SNPs including the position, and the test statistic across groups.
c. Zoom-in and -out: Plotting many points on the same graph makes it difficult to discern one point from another, as it may be in a peak or in the lower portion of the Manhattan plot which often is densely packed. To overcome this issue, the plot can be zoomed-in using a mouse scroller when the mouse pointer is placed on the Manhattan plot.
However, due to limitations described in section 3, at this moment, it is possible to visualize only a few thousand points, and thus filtering of GWAS output is required prior to plotting, which can be accomplished with the built-in data_reduce function.
2.2.2 Visualization sharing
Interactive plots can be saved as notebooks and self-contained html files that can be shared with colleagues via usual sharing platforms (GitHub and Dropbox) and opened in any web browsers on any operation system.
3 Limitations
In general, interactive visualization made through web browsers are limited by the number of objects they can smoothly display. In the current example (Fig. 1B), we have selected the top 1,000 SNPs in each of the two groups and are visualizing at most 2,000 dots if there is no overlap between those SNPs. To address this limitation, the package can be extended to a web application with dynamic data loading from a database/server. Dynamic data loading would allow a user to load SNP data in real time for a specific region of interest as the user zooms-in. By making this an open source package that is accessible via GitHub, we invite members of the scientific community to contribute and enhance the package’s capabilities.
Funding
This work has been supported by the NIH 3P50MH094267-04S1 and 1R01MH101820-02S1 grants.
Conflict of Interest: none declared.
References
- Broman K.W. (2015) R/qtlcharts: interactive graphics for quantitative trait locus mapping. Genetics, 199, 359–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Roeder K. (1999) Genomic control for association studies. Biometrics, 55, 997–1004. [DOI] [PubMed] [Google Scholar]
- Ge D. et al. (2008) WGAViewer: software for genomic annotation of whole genome association studies. Genome Res., 18, 640–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela M.J., Chanock S.J. (2015) LDlink a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics, 31, 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim R.J. et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall J.C. et al. (2013) Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet., 9, e1003500.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S.D. (2014) qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. bioRxiv, doi: 10.1101/005165. [Google Scholar]
- Zeigler G.R. et al. (2015) Zbrowse: an interactive GWAS results browser. PeerJ Comput. Sci., doi: 10.7717/peerj-cs.3. [Google Scholar]