Abstract
Background
Bulk tumor tissue samples are used for generating gene expression profiles in most research studies, making it difficult to decipher the stroma–cancer crosstalk networks. In the present study, we describe the use of microdissected transcriptome profiles for the identification of cancer–stroma crosstalk networks with prognostic value, which presents a unique opportunity for developing new treatment strategies for ovarian cancer.
Methods
Transcriptome profiles from microdissected ovarian cancer–associated fibroblasts (CAFs) and ovarian cancer cells from patients with high-grade serous ovarian cancer (n = 70) were used as input data for the computational systems biology program CCCExplorer to uncover crosstalk networks between various cell types within the tumor microenvironment. The crosstalk analysis results were subsequently used for discovery of new indications for old drugs in ovarian cancer by computational ranking of candidate agents. Survival analysis was performed on ovarian tumor–bearing Dicer/Pten double-knockout mice treated with calcitriol, a US Food and Drug Administration–approved agent that suppresses the Smad signaling cascade, or vehicle control (9–11 mice per group). All statistical tests were two-sided.
Results
Activation of TGF-β-dependent and TGF-β-independent Smad signaling was identified in a particular subtype of CAFs and was associated with poor patient survival (patients with higher levels of Smad-regulated gene expression by CAFs: median overall survival = 15 months, 95% confidence interval [CI] = 12.7 to 17.3 months; vs patients with lower levels of Smad-regulated gene expression: median overall survival = 26 months, 95% CI = 15.9 to 36.1 months, P = .02). In addition, the activated Smad signaling identified in CAFs was found to be targeted by repositioning calcitriol. Calcitriol suppressed Smad signaling in CAFs, inhibited tumor progression in mice, and prolonged the median survival duration of ovarian cancer–bearing mice from 36 to 48 weeks (P = .04).
Conclusions
Our findings suggest the feasibility of using novel multicellular systems biology modeling to identify and repurpose known drugs targeting cancer–stroma crosstalk networks, potentially leading to faster and more effective cures for cancers.
With increasing evidence demonstrating the importance of stromal involvement in ovarian cancer pathogenesis (1), identification of stroma-derived factors associated with aggressive phenotypes and chemoresistance presents a unique opportunity for development of new treatment strategies. As tumor-supportive roles of cancer-associated fibroblasts (CAFs) have been increasingly recognized, researchers have begun to evaluate the potential of inhibiting tumor progression through stromal ablation. Several studies have shown that ablation of activated myofibroblasts does not suppress tumor progression (2,3), whereas other studies showed that silencing CAF-derived mediators could inhibit ovarian cancer progression (4–6). Reprogramming CAFs by targeting the CAF–cancer crosstalk networks presents a new opportunity for development of novel cancer treatment strategies (7,8).
Gene expression profiling and genome-wide screening have accelerated the discovery of differentially expressed genes in high-grade serous ovarian cancers (HGSOCs) (9–12). However, a clinically useful interpretation of a transcriptome-based signature that guides treatment of HGSOC is lacking. In addition, most previous studies used bulk tumor tissue samples with various degrees of stromal contamination, which could skew the resulting transcriptome profiles. Without comprehensive but separate transcriptome information generated from stromal cells and cancer cells, it is impossible to decipher the stroma–cancer crosstalk networks.
Here, we report the use of transcriptome profiles generated from microdissected CAFs and epithelial cancer cells from HGSOCs, and an advanced systems biology modeling program, Cell-Cell Communication Explorer (CCCExplorer), to predict activation of TGF-β-dependent and TGF-β-independent Smad crosstalk networks in CAFs. In addition, we queried our in-house and public databases to identify a US Food and Drug Administration (FDA)–approved drug, calcitriol, to target Smad signaling preferentially to suppress tumor progression and improve survival rates.
Methods
Microdissection and Microarray Analysis of Tissue Samples
RNA was extracted from microdissected frozen tissue samples collected under protocols approved by the MD Anderson Cancer Center Institutional Review Board (IRB), with written informed consent from patients. All tumor tissue samples were resected from the primary tumor site of previously untreated HGSOC patients. Transcriptome profiling was performed using the GeneChip Human Genome U133 Plus 2.0 microarrays (Affymetrix). The Supplementary Methods (available online) include the details for microdissection, microarray processing, quality control, and data analysis.
Hierarchical Clustering and Silhouette Analysis
Hierarchical clustering based on Euclidean distance and Wards linkage was performed for the clustering analysis of transcriptome profiles. Silhouette analysis was performed to determine the optimal number of clusters.
Crosstalk Prediction With CCCExplorer
The Activated Transcriptional Factor discovery module and the Activated Ligand–Receptor interaction discovery module were used in the prediction of crosstalk signaling pathways between cancer cells and CAFs. Details on database construction, computational analysis, and robustness assessment are provided in the Supplementary Methods (available online).
Repositioned Drug Identification
Drug prediction was performed based on gene set enrichment analysis (GSEA) and affinity propagation clustering of 2202 FDA-approved drugs on the Smad signaling pathway. The analysis is described in Supplementary Methods (available online).
Statistical Analysis
The SPSS 19 (IBM Corporation) and Prism 5.0 (GraphPad Software) software programs were used. In vitro experiments were repeated independently in triplicate. A two-tailed Student t test was used to determine the statistical significance of differences in sample means for data with normally distributed means. The Mann-Whitney U test was used for analysis of nonparametric data. For survival analysis, the nonparametric Kaplan-Meier method and log-rank tests were performed. P values of less than .05 were considered statistically significant. All statistical tests were two-sided.
Accession Numbers
Data files from transcriptome profiling analysis were deposited on GEO and were assigned the accession number GSE115635.
All other detailed methods, including those for the mouse experiments, are in the Supplementary Methods (available online).
Results
Prognostic Significance of CAF Gene Expression Profile in Ovarian Cancer
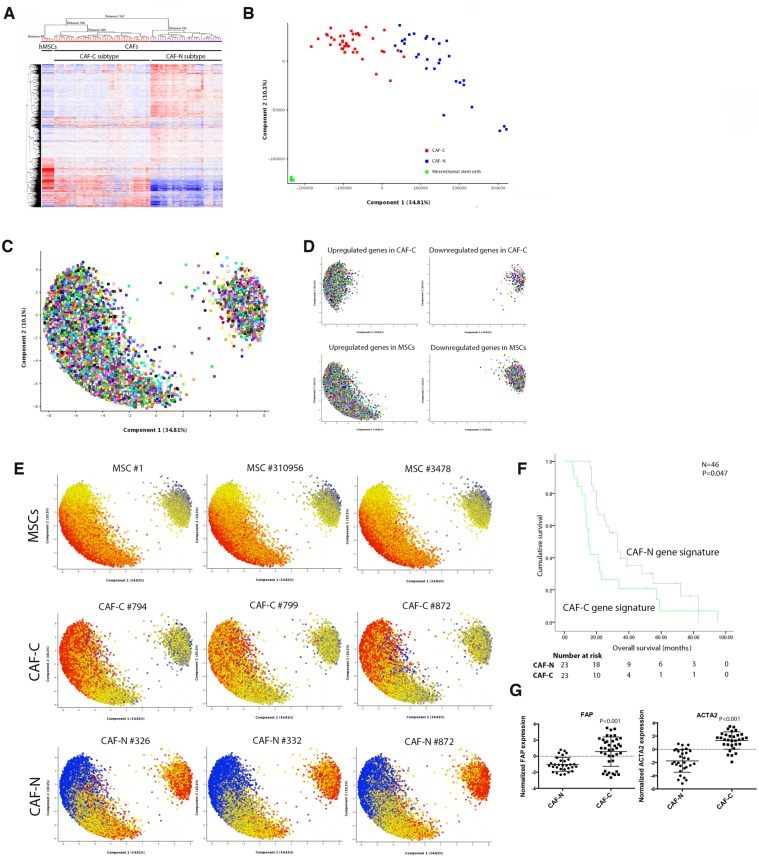
To evaluate the roles of ovarian CAFs in tumor progression and their impact on patient survival rates, transcriptome profiling was performed on laser-microdissected epithelial tumor cells and stromal CAFs from HGSOC patient samples collected under protocols approved by MD Anderson Cancer Center IRB (n = 70). The CAF-N subtype has a gene expression signature distinct from that of the MSCs, and the CAF-C subtype has a gene expression signature similar to that of the MSCs (Figure 1A). Silhouette analysis identified k = 3 as the optimal number of clusters for k = 3 through 10 for both hierarchical (Wards linkage) and k means clustering (Supplementary Table 1, available online). Therefore, transcriptome profiles were further analyzed as three distinct clusters (MSCs, CAF-C, and CAF-N). Principal component analysis (PCA) also identified MSC, CAF-C, and CAF-N as three distinct clusters (Figure 1B). Probe sets representing differentially expressed genes were visualized on the PCA loading plot (Figure 1C). Upregulated and downregulated genes in CAF-C compared with CAF-N (Supplementary Table 2, available online) largely overlapped with the upregulated and downregulated genes in MSCs, with CAF-N as a baseline (Figure 1D;Supplementary Table 3, available online). To facilitate visualization, probe sets represented in the PCA loading plots were colored according to their expression levels in a particular sample; this analysis further supports the greater similarity between CAF-C and MSCs than between CAF-N and MSCs (Figure 1E). Kaplan-Meier analysis on patients with both CAF and cancer cell gene expression profiles available demonstrated that patients with the CAF-C signature in CAFs had shorter median overall survival than patients with the CAF-N signature (16 vs 33 months, 95% confidence interval [CI] = 14.1 to 17.9 months and 95% CI = 28.6 to 37.4 months, respectively, n = 46, P = .047) (Figure 1F; additional clinical and histopathological information described in Supplementary Table 4, available online). These results suggest that CAF heterogeneity may play a role in modulating survival rates in HGSOC patients. Transcriptome analysis of the expression levels of CAF-specific markers, fibroblast activation protein (FAP) and alpha smooth muscle actin (ACTA2), found that they were expressed at statistically significantly higher levels in CAF-C than in CAF-N (both P < .001) (Figure 1G), suggesting that although CAF-C has a global expression pattern more like that of MSCs, the presence of the CAF-C subtype is due to the activation of myofibroblasts, not contamination by MSCs.
Figure 1.
The prognostic significance of the cancer-associated fibroblast (CAF) gene expression profile in ovarian cancer. A) Laser microdissection was performed on isolated epithelial tumor cells and stromal CAFs from frozen tissue samples obtained from patients with high-grade serous ovarian cancer (HGSOC; n = 70). As undifferentiated bone marrow–derived mesenchymal stem cells (MSCs) that have migrated to the tumor site have been shown to be a source of CAFs, transcriptome profiling was performed on primary bone marrow MSCs isolated from five healthy individuals. Hierarchical clustering analysis of transcriptome data generated from laser-microdissected CAFs and purified bone marrow MSCs identified two major CAF subtypes. B) Principal component analysis (PCA) on microdissected transcriptome profiles. C) With a fold change of 2 set as the cutoff, probe sets representing differentially expressed genes were visualized on the PCA loading plot. D) According to their expression levels, up- and downregulated genes in CAF-C, CAF-N, and MSCs were colored in red and blue, respectively. E) Differentially expressed probe sets represented in the PCA loading plots in three representative samples for each of the gene signature subtypes. F) The overall survival durations of patients with the CAF-C and CAF-N gene expression signatures (n = 46) were evaluated by Kaplan-Meier analysis and log-rank test. G) Transcriptome analysis of the expression levels of FAP and ACTA2 in CAFs with CAF-C and CAF-N gene expression signatures. CAF = cancer-associated fibroblast; MSC = mesenchymal stem cell.
Prediction of Stromal–Epithelial Crosstalk Signaling
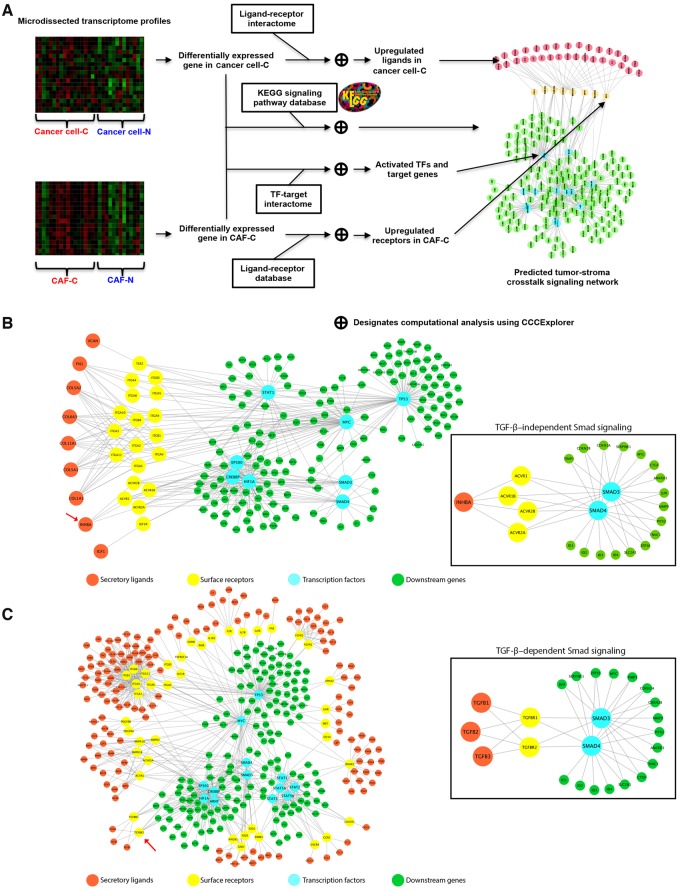
We employed a streamlined workflow for the prediction of survival-associated signaling crosstalk between ovarian cancer cells and CAFs with CCCExplorer (Figure 2A). Microdissected transcriptome profiles were analyzed, and overexpressed secretory ligands from cancer cells (Supplementary Table 5, available online) and activated transcription factors from CAFs in the CAF-C patient cohort (Supplementary Table 6, available online) were identified. Furthermore, through reconstruction of receptor–transcription factor crosstalk, differentially activated crosstalk signaling pathways in the CAF-C patient cohort were predicted (Supplementary Table 7, available online). Among them, overexpression of cancer cell–derived INHBA binding to CAF-derived ACVR2A, the receptor of INHBA (13), was associated with activated Smad signaling in CAFs in the CAF-C patient group (P < .001) (Figure 2B). As ovarian cancer patients with the CAF-C signature had lower survival rates than those with the CAF-N signature, activation of Smad signaling in CAFs may be associated with poor survival rates in patients with HGSOCs.
Figure 2.
Identification of stromal–epithelial crosstalk signaling by the CCCExplorer. A) Schematic diagram showing the workflow for the identification of differentially activated signaling pathways in ovarian cancer patients with cancer-associated fibroblast (CAF)–C and CAF-N signatures and the cellular crosstalk with the corresponding cancer cells (cancer cell–C and cancer cell–N, respectively) using transcriptome profiles generated from microdissected ovarian cancer cells and CAFs at the stromal–epithelial interface from the same patient. Such an intercellular crosstalk identification cannot be achieved by using transcriptome profiles generated from bulk tumor tissue alone. To delineate the crosstalk between CAFs and ovarian cancer cells and to identify the signaling pathways contributing to the poor survival rates in the CAF-C patient cohort, new computational functions were added to the multicellular systems biology software CCCExplorer to discover 1) activated transcription factors (TFs) based on increased expression levels of downstream target genes revealed by the transcriptome profile, 2) activated ligand–receptor interaction based on the expression levels of secretory ligands, cell surface receptors, or both, and 3) reconstruction of crosstalk signaling between cell types by linking activated ligand–receptor and TF–downstream target gene information. B) Microdissected transcriptome profiles of CAFs and epithelial tumor cell samples from CAF-C and CAF-N subtype patients were analyzed. Activated signaling pathways in CAF-C were predicted through the identification of overexpressed secretory ligands in cancer cells and activated transcription factors in CAFs in the CAF-C patient cohort. C) Similarly, activated signaling pathways in CAF-C were predicted through the identification of overexpressed receptors and activated transcription factors in CAFs in the CAF-C patient cohort. CAF = cancer-associated fibroblast; TF = transcription factor.
Activation of Smad signaling in CAFs could be due to differentially expressed receptors. To test this hypothesis, CCCExplorer was modified to allow the input of upregulated expressed receptor information in CAFs and cancer cells to predict the differential crosstalk signaling networks of CAF-C and CAF-N samples (Supplementary Tables 8 and 9, available online). Expression levels of TGFBR2 in CAFs and activation of Smad signaling were statistically significantly greater in CAF-C samples than in CAF-N samples. These data suggest that Smad signaling may be preferentially activated in CAFs in the CAF-C patient group, but not in the CAF-N patient group, through the binding of cytokine TGF-β to the overexpressed TGFBR2 (P < .001) (Figure 2C).
In sum, CCCExplorer suggested that both TGF-β-independent and -dependent Smad signaling networks were preferentially activated in CAF-C compared with CAF-N. To evaluate Smad signaling pathway activation in patients with different CAF gene expression signatures, immunostaining of phosphorylated Smad2/3 was performed on formalin-fixed, paraffin-embedded tissue sections from HGSOC patients with CAF-C and CAF-N tumors. CAFs in patients with CAF-C had higher levels of phosphorylated Smad2/3 in CAFs than in patients with CAF-N (Supplementary Figure 1, available online).
Robustness assessment of CCCExplorer using randomly generated gene lists (Supplementary Table 10, available online) or the aforementioned differentially expressed gene lists with random addition/deletion of genes (Supplementary Table 11, available online) as input data showed that randomly selected unimportant genes had a minimal effect on the prediction of truly important pathways, confirming the robustness of CCCExplorer.
INHBA-Induced Smad Signaling in Ovarian CAFs
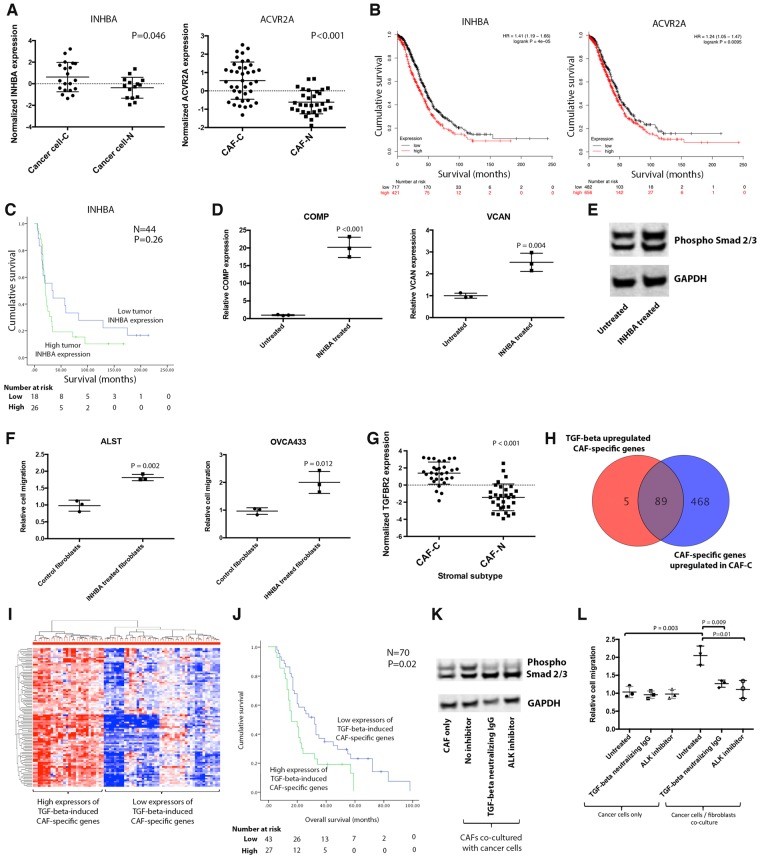
As CCCExplorer found ovarian cancer cell–derived INHBA-mediated Smad signaling activation in CAFs via its engagement of the ACVR2A receptor, we evaluated the expression levels of INHBA and ACVR2A in ovarian cancer cells and CAFs. Both INHBA expression in cancer cells and ACVR2A expression in CAFs were statistically significantly higher in patients with the CAF-C signature than in those with the CAF-N signature (Figure 3A). Analysis of 1138 transcriptome profiles of bulk HGSOC tissue samples using KMplot (14) revealed that high expression levels of INHBA and ACVR2A were each statistically significantly associated with reduced overall survival (P < .001 and P = .01, respectively) (Figure 3B). In our patient cohort, high expression of INHBA by cancer cells was associated with poor clinical outcomes. Patients with high and low levels of INHBA expression had a median overall survival (95% CI) of 21 (18.5 to 23.5) months and 33 (0 to 66.3) months, respectively (P = .26) (Figure 3C). INHBA has been shown to activate Smad signaling by binding to ACVR2A (15), and we previously demonstrated that Smad signaling activation in CAFs contributes to the aggressive phenotypes of ovarian cancer cells and is associated with poor patient survival (6). To demonstrate that INHBA activates Smad signaling in ovarian CAFs, CAFs were treated with recombinant activin A, and Smad signaling activation was determined. Quantitative real-time polymerase chain reaction (qRT-PCR) results showed that compared with control, activin A statistically significantly increased the expression levels of the Smad signaling downstream targets COMP and VCAN (P < .001 and P = .004, respectively) (Figure 3D) (6). Furthermore, immunoblot analysis showed that activin A induced phosphorylation of Smad2/3 in CAFs (Figure 3E). These results suggest that activin A activates the Smad signaling pathway in ovarian CAFs.
Figure 3.
Tumor cell–activated Smad signaling in cancer-associated fibroblasts (CAFs). A) Expression levels of INHBA and ACVR2A in microdissected tissue samples were evaluated. B) Survival analysis on INHBA and ACVR2A expression using 1138 GEO transcriptome profiles. C) In the MD Anderson patient cohort, survival analysis on tumor INHBA expression was performed. D) Expression levels of the Smad downstream genes COMP and VCAN were evaluated in CAFs treated with 10 ng/mL recombinant activin (mean ± SD of three independent experiments; two-tailed Student t test). E) Expression of phosphorylated Smad2/3 in untreated CAFs and CAFs treated with 10 ng/mL recombinant activin A was evaluated by immunoblot analysis. F) The effect of exogenous INHBA treatment of CAFs on ovarian cancer cell motility was studied using a co-culture system (mean ± SD of three independent experiments; two-tailed Student t test). G) To validate that TGF-β-dependent Smad signaling activation in CAF-C is mediated through increased TGFBR2 expression in CAFs in CAF-C tumors compared with that in CAF-N tumors, quantitative real-time polymerase chain reaction analysis of TGFBR2 was performed on microdissected CAFs (n = 62, P < .001). H) The list of TGF-β-inducible genes identified in normal fibroblasts treated with TGF-β was compared with the list of differentially expressed genes identified in CAFs with the CAF-C gene expression signature. I) Heatmap showing the unsupervised clustering results for 70 high-grade serous ovarian cancer patients according to their expression levels of TGF-β-inducible genes in CAFs. J) The prognostic significance of the expression levels of TGF-β-inducible genes by CAFs was evaluated by Kaplan-Meier analysis and log-rank tests. K) To demonstrate that the activation of TGF-β/Smad signaling in CAFs is mediated by binding of the ovarian cancer cell–secreted ligand TGF-β to TGFBR2, co-culture experiments were performed by co-culturing CAFs with cancer cells in the presence or absence of TGF-β-neutralizing antibody or TGF-β receptor 2 inhibitor. The expression level of phosphorylated Smad2/3 was evaluated by immunoblot analysis. L) To evaluate the effect of TGF-β–treated fibroblasts on ovarian cancer cell motility potential, cancer cells were cultured alone or with fibroblasts in the presence or absence of TGF-β, and in the presence or absence of TGF-β–neutralizing antibody or TGF-β receptor 2 inhibitor (mean ± SD of three independent experiments; two-tailed Student t test). CAF = cancer-associated fibroblast.
To demonstrate that activation of Smad signaling in ovarian fibroblasts induced by INHBA promotes cancer cell aggressiveness, CAFs treated with activin A were co-cultured with ALST and OVCA433 ovarian cancer cells. Cancer cells co-cultured with activin A–treated fibroblasts demonstrated a statistically significant increase in motility potential in Boyden chambers compared with those co-cultured with untreated fibroblasts (Figure 3F).
TGF-β-Induced Smad Signaling in CAFs in CAF-C Tumors
To validate that TGF-β-dependent Smad signaling activation in CAF-C is mediated through increased TGFBR2 expression, qRT-PCR analysis of TGFBR2 was performed. TGFBR2 mRNA expression levels were statistically significantly higher in CAF-C than in CAF-N (P < .001) (Figure 3G). Upon engagement of TGF-β receptor 1/2 heterodimeric complex to TGF-β, TGF-β receptor 2 undergoes phosphorylation and activates the downstream Smad signaling cascade. The results suggest that the activated TGF-β/Smad signaling in ovarian CAFs in the CAF-C tumors may be due in part to the increased TGF-β receptor 2 expression levels. Further, the list of TGF-β-inducible genes identified in normal fibroblasts treated with TGF-β was compared with the list of differentially expressed genes identified in CAFs in CAF-C and CAF-N tumors. A vast majority (89 out of 94 genes) of TGF-β-inducible genes in ovarian fibroblasts were upregulated in CAFs of the CAF-C tumors compared with CAF-N tumors (Figure 3H).
To determine the prognostic significance of the TGF-β-inducible CAF-C signature in HGSOC patients, hierarchical clustering was performed in the original cohort of 70 patients based on the expression levels of the 89 TGF-β-inducible genes. Two distinct groups were identified (Figure 3I). Kaplan-Meier analysis and log-rank tests demonstrated that patients with higher levels of TGF-β-inducible genes expressed by CAFs had poorer overall survival rates. Patients with high and low levels of TGF-β-inducible gene expression had a median overall survival (95% CI) of 16 (7.5 to 24.5) months and 32 (24.9 to 39.1) months, respectively (P = .02) (Figure 3J).
To demonstrate that the activation of TGF-β/Smad signaling in CAFs is mediated by binding of the ovarian cancer cell–secreted ligand TGF-β to TGFBR2, co-culture experiments were performed using CAFs and OVCA433 ovarian cancer cells. Co-culturing CAFs with cancer cells increased the phosphorylation of SMAD2/3 in CAFs, which was abrogated by the addition of TGF-β-neutralizing antibody or the addition of TGF-β receptor 2 inhibitor (ALK inhibitor) (Figure 3K).
To evaluate the effect of TGF-β-treated fibroblasts on ovarian cancer cell motility potential, cancer cells were cultured alone or with fibroblasts in the presence or absence of TGF-β. Cancer cell motility increased when cancer cells were co-cultured with ovarian fibroblasts in the presence of TGF-β, whereas this effect was abrogated in the presence of TGF-β-neutralizing antibody or with the addition of TGF-β receptor 2 inhibitor. Treating ovarian cancer cells with exogenous TGF-β along with TGF-β-neutralizing antibody or with TGF-β receptor 2 inhibitor had no effect on cancer motility (Figure 3L).
Activation of Smad Signaling in CAFs and its Association With Patient Survival
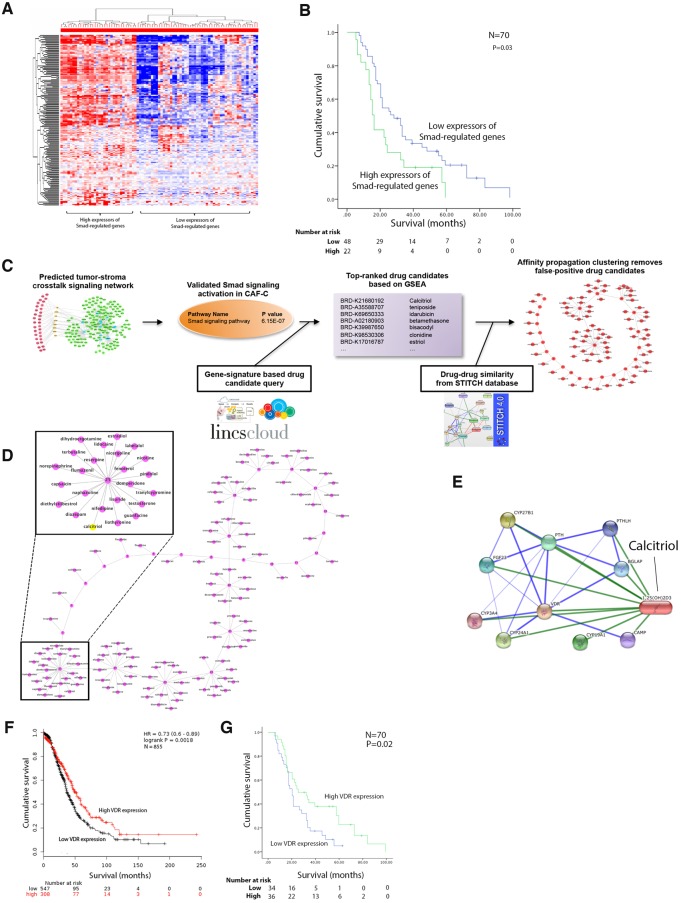
As patients with CAF-C tumors have poorer survival rates than those with CAF-N tumors, we further investigated the clinical significance of Smad signaling activation in CAFs using a list of Smad2-regulated and Smad3-regulated genes based on the information from the Ingenuity Pathway Analysis database (Supplementary Table 12, available online). Hierarchical clustering identified two distinct groups of patients on the basis of the Smad-regulated gene signature on microdissected CAF expression profiles (Figure 4A). Patients with higher levels of Smad-regulated gene expression by CAFs had a median overall survival duration (95% CI) of 15 (15.8 to 17.3) months, whereas patients with lower levels of Smad-regulated gene expression had a longer median overall survival of 26 (15.9 to 36.1) months (n = 70, P = .03) (Figure 4B).
Figure 4.
Identification of repositioned drugs that target activated Smad signaling in cancer-associated fibroblasts (CAFs). A) From the Ingenuity Pathway Analysis database, a list of Smad2-regulated and Smad3-regulated genes was generated. Hierarchical clustering was performed to stratify high-grade serous ovarian cancer (HGSOC) patients based on the expression levels of Smad-regulated genes. B) Kaplan-Meier analysis and log-rank test were performed to determine the prognostic significance of Smad-regulated gene expression by CAFs in HGSOC patients. C) The computational approach used to identify and repurpose known drugs to target activated Smad signaling networks in CAF-C-based gene set enrichment analysis and affinity propagation clustering. D) A list of US Food and Drug Administration–approved repositioned drugs that potentially target Smad signaling in CAFs was refined by filtering possible false-positive candidates through affinity propagation clustering. E) Network analysis on the binding of calcitriol to vitamin D receptor (VDR). F) The prognostic significance of VDR expression in CAFs was evaluated by Kaplan-Meier analysis and log-rank test using 855 GEO transcriptome profiles (P = .002). G) Similarly, the prognostic significance of VDR expression in CAFs was evaluated in the MD Anderson patient cohort (P = .02). CAF = cancer-associated fibroblast; VDR = vitamin D receptor.
Identification of Smad Signaling Targeting Repositioning Drugs
We used a computational approach to predict known drugs that have passed phase I safety trials to target activated Smad signaling networks in CAF-C (Figure 4C). Using the Connectivity Map database, computational modeling, and GSEA, we generated a list of FDA-approved drugs that potentially target Smad signaling. The drug list was refined by filtering possible false-positive candidates through affinity propagation clustering (Figure 4D;Supplementary Table 13, available online). Calcitriol, also known as vitamin D3, was selected for further functional studies because it is known to target both TGF-β-dependent and TGF-β-independent Smad signaling by inhibiting the binding of Smads to their target genes (16). In addition, calcitriol can bind to the vitamin D receptor (VDR) (Figure 4E) and activate downstream events that inhibit Smad signaling in CAFs (17,18). High levels of VDR expression in HGSOCs were associated with better patient survival rates via analysis of 855 transcriptome profiles deposited to GEO (P = .002) (Figure 4F) and of the microdissected transcriptome of our original cohort of 70 patients. Patients with high and low levels of VDR expression in CAFs had a median overall survival (95% CI) of 25 (10.7 to 39.3) months and 20 (16.2 to 23.8) months, respectively (P = .02) (Figure 4G).
Inhibition of Smad Signaling in CAFs
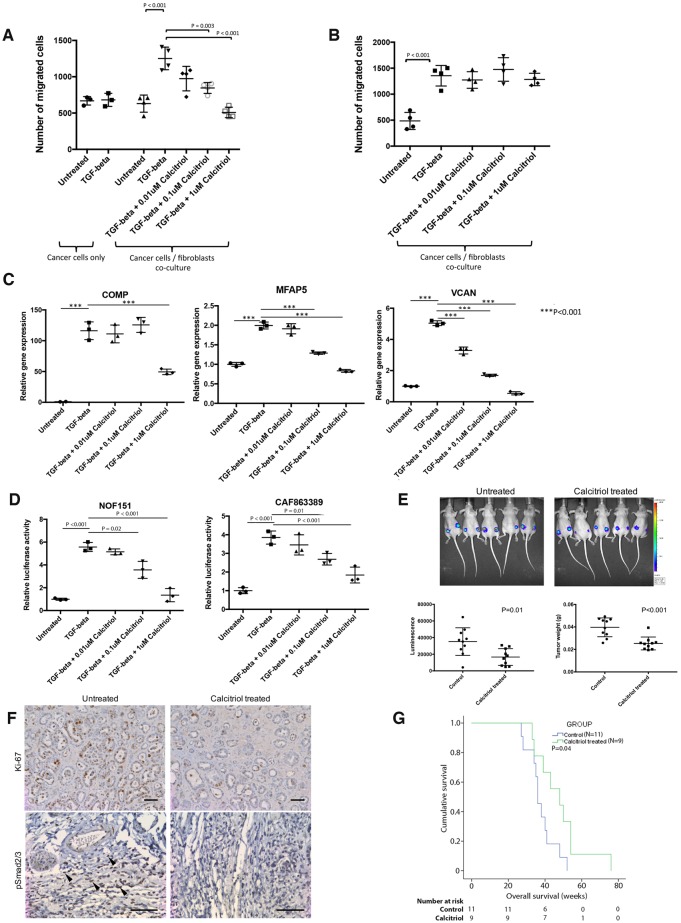
To evaluate the tumor-suppressive effects of calcitriol in vitro, calcitriol was added to the co-culture of CAFs and ALST ovarian cancer cells or to ovarian cancer cells alone in the presence of exogenous TGF-β. TGF-β induced ovarian cancer cell motility in the presence of CAFs, and the effect was abrogated by the addition of calcitriol (Figure 5A), suggesting that the inhibitory effect of calcitriol on cancer cell motility is mediated through inhibition of TGF-β/Smad signaling in CAF. To show that this effect is mediated through the VDR on CAFs, ALST ovarian cancer cells were co-cultured with CAFs, with VDR expression silenced by siRNAs (Supplementary Figure 2, available online). VDR knockdown abrogated the motility-suppressive effect of calcitriol on ovarian cancer cells (Figure 5B). To demonstrate that calcitriol inhibits TGF-β/Smad signaling in CAFs, primary CAFs treated with calcitriol were analyzed by qRT-PCR for the expression of CAF-C-derived mediators, including COMP, MFAP5, and VCAN, which have been shown to be associated with ovarian cancer progression (4,6) and poor patient survival rates (Supplementary Figure 3, available online). Although TGF-β upregulated these genes in CAFs, the stimulating effect of TGF-β was abrogated by calcitriol in a dosage-dependent manner (Figure 5C). In addition, reporter assays showed that although TGF-β treatment induced Smad promoter–driven luciferase activity, this induction was abrogated by the addition of calcitriol (Figure 5D). These data are further summarized in Supplementary Figure 4 (available online).
Figure 5.
Effects of calcitriol on ovarian cancer progression in vitro and in vivo. A) To evaluate the tumor-suppressive effects of calcitriol in vitro, calcitriol was added to the co-culture of cancer-associated fibroblasts (CAFs) and ALST ovarian cancer cells or to ovarian cancer cells alone in the presence of exogenous TGF-β (mean ± SD of four independent experiments; two-tailed Student t test). B) To show that the inhibitory effect of calcitriol was mediated through the vitamin D receptor (VDR) on CAFs, the co-culture study was repeated using CAFs with VDR expression silenced (mean ± SD of four independent experiments; two-tailed Student t test). C) Expression levels of the Smad downstream genes COMP, MFAP5, and VCAN in CAFs were evaluate in CAFs treated with various dosages of calcitriol (mean ± SD of three independent experiments; two-tailed Student t test). D) Reporter assays were performed on two different ovarian fibroblast lines, with the luciferase reporter driven by Smad promoter stably integrated into the genome to investigate the effects of calcitriol treatment on Smad signaling pathway inhibition (mean ± SD of three independent experiments; two-tailed Student t test). E) To evaluate the use of calcitriol as a novel therapeutic agent repositioned for ovarian cancer treatment, 2 × 106 OVCA432 and 2 × 106 primary ovarian CAFs were mixed, resuspended in sterile phostphate-buffered saline (PBS), and co-injected subcutaneously into athymic nude mice. Tumors were allowed to develop for five days before calcitriol treatment. On day 5, mice were randomized, and the control group received intratumoral injection of sterile PBS, whereas the treatment group received calcitriol treatment at 60 μg/kg. All mice were treated twice weekly for a total of 3 weeks. At the experimental end point, luciferase-based tumor imaging was performed. Subsequently, all mice were killed and had subcutaneous tumor nodules removed, weighed, and fixed for further histologic analysis (mean ± SD; n = 10 per group; Mann-Whitney U test). F) Immunolocalization of Ki-67 and phosphorylated Smad2/3 on tumor tissue samples harvested from control mice and mice treated with calcitriol (phosphorylated Smad2/3–positive CAFs are indicated by arrows in the untreated tumor nodule; bar = 100 μm). G) To study the effects of calcitriol treatment on the survival of transgenic mice, the Amhr2-Cre-driven Dicer/PTEN double-knockout genetically engineered mouse model was used. These transgenic mice have ovarian tumor onset at the age of three to four months and a disease progression highly similar to that of human high-grade serous ovarian cancer. The 9 mice from the treatment group were given 60 μg/kg of calcitriol whereas the 11 mice from the control group were given PBS twice per week via intraperitoneal injections until the mice became moribund. The survival duration for each mouse was recorded, and Kaplan-Meier survival analysis was performed (P = .04).
Effects of Calcitriol Treatment on In Vivo Tumor Growth
The effects of calcitriol treatment on ovarian tumor progression were evaluated in OVCA432 ovarian tumor–bearing mice. Cancer cell luciferase signals and tumor weights were statistically significantly lower in calcitriol-treated mice than in control mice (P = .01 and P < .001, respectively), suggesting that calcitriol effectively suppressed tumor growth in mice co-injected with ovarian cancer cells and CAFs (Figure 5E). Furthermore, immunohistochemical analysis of Ki-67 and phosphorylated Smad2/3 showed that mice treated with calcitriol had fewer Ki-67–positive cancer cells per unit of tumor area than did control-treated mice. Phosphorylated Smad2/3 signals in CAFs were markedly decreased in mice treated with calcitriol compared with control mice (Figure 5F).
Effects of Calcitriol Treatment on Survival of Spontaneous Ovarian Cancer–Developing Transgenic Mice
Based on the publication by Ding et al. (16) and our preliminary studies in which calcitriol dosages at 20, 40, and 60 μg/kg were tested for inhibition of Smad signaling activation, the dosage of 60 μg/kg was used for treatment using the Amhr2-Cre-driven Dicer/Pten double-knockout genetically engineered mouse model (19). The calcitriol-treated mice had a longer median survival than the control mice (48 vs 36 weeks, 95% CI = 33.4 to 62.6 weeks and 95% CI = 32.8 to 39.2 weeks, respectively, P = .04) (Figure 5G), suggesting that calcitriol can be included in the treatment regimen for HGSOC patients who develop CAF-C type tumors.
Discussion
Using crosstalk systems biology modeling, we demonstrated that advanced HGSOCs can be divided into two subtypes based on CAFs with and without the activation of TGF-β-dependent and TGF-β-independent Smad signaling pathways. We showed that activation of Smad signaling in CAFs is associated with poor survival rates in HGSOC patients. Furthermore, treating HGSOCs with the repurposed drug calcitriol suppressed HGSOC progression in both co-culture and in vivo models, suggesting that calcitriol may be used as an alternative treatment regimen to improve survival rates.
We identified two distinct subtypes of CAFs: CAF-N and CAF-C. The transcriptome signature of CAF-C resembles that of MSCs, one of the known precursor cells of CAFs (20). These findings suggest that CAF heterogeneity in the tumor microenvironment occurs within HGSOCs. Using transcriptome data generated from microdissected CAFs, we showed that patients with the CAF-C signature had worse survival rates than those with the CAF-N signature, implicating that tumor cell signatures and CAF subtype signatures have prognostic and predictive values in ovarian cancer. Additional analyses showed higher expression of TGF-β-inducible genes in CAFs with the CAF-C signature than in the CAF-N signature. Silencing genes such as VCAN and MFAP5 in CAF-C subtypes suppressed ovarian cancer progression and angiogenesis in vitro and in vivo (4,6,21), suggesting that TGF-β-regulated genes in CAFs may play a role in conferring the aggressiveness of ovarian cancer cells.
CCCExplorer is a multicellular computational systems biology platform that was first applied to uncover the stroma–tumor crosstalk in lung cancer (22). In this study, we expanded the modeling capability of the CCCExplorer by 1) adding signaling pathways activated by upregulated receptors and, more importantly, 2) adding a novel drug repositioning module to block the stroma–tumor interactions based on the uncovered novel stroma–tumor crosstalk signaling pathways. With these two additional modules, CCCExplorer predicted activation of both TGF-β-dependent and TGF-β-independent Smad pathways in CAFs in patients with the CAF-C subtype of ovarian cancer. The TGF-β-independent pathway involves the binding of INHBA, which is overexpressed in ovarian cancer cells of the CAF-C type tumors, to its receptor, ACVR2A, in CAFs. INHBA has been shown to activate Smad signaling by binding to ACVR2A (15). The TGF-β-dependent Smad signaling pathway involves the binding of TGF-β in the tumor microenvironment to the overexpressed TGFBR2 in CAFs of tumors with the CAF-C expression signature. These findings suggest that reprogramming CAFs by targeting the Smad signaling pathway in CAFs of CAF-C type tumors can suppress ovarian cancer progression and improve patient survival rates.
Our in vitro and in vivo studies demonstrated that calcitriol is an effective therapeutic agent for ovarian cancer, preferentially targeting Smad-regulated genes. Several studies show that targeting the vitamin D signaling pathway has the potential for developing anticancer therapeutics (17). Activation of the VDR axis by synthetic vitamin D analogues reprograms stromal fibroblasts to suppress pancreatitis and enhance pancreatic cancer therapy (18). In addition, synthetic vitamin D analogues can suppress kidney fibrosis by binding VDR and preventing the Smad complex from binding to the SMAD3-binding element in the nucleus (23,24). Low vitamin D3 correlates with poor survival in ovarian cancer patients (25). The identification of CAF heterogeneity in this study suggests that stratification of patients based on the CAF signature for calcitriol treatment would extend patient survival.
To our knowledge, this study is the first to use a multicellular systems biology approach combining advanced computational biology methods and rigorous experimental biology to identify novel stroma–tumor crosstalk signaling networks and known drugs targeting the networks in HGSOCs. We generated comprehensive transcriptome profiles from microdissected CAFs and ovarian cancer cells and identified signatures in CAFs with prognostic significance. We added new functionality to CCCExplorer to discover activation of TGF-β/Smad signaling pathways in a subtype of CAFs associated with poor patient survival rates. The activated Smad signaling and identified crosstalk enabled us to reposition known drugs by querying drug information databases and performing computational analysis to rank candidates for ovarian cancer.
Our study has limitations that should be considered. To further address the complexity of the tumor microenvironment, an update of the software that supports crosstalk analysis between three or more cell types is currently under development. In addition, although laser capture microdissection provided us with the spatial information of the different cell types in our study, the information is not currently being utilized. Incorporation of spatial information, like using distance between cell types as one of the input parameters, could further improve prediction accuracy.
In summary, our study opens a new vista in the design of therapeutic strategies based on transcriptional reprogramming of CAFs for the treatment of ovarian cancer. In addition, the CCCExplorer platform can be extended to uncover novel crosstalk targets and drug candidates of other cancer types.
Funding
This study was supported in part by grants R01CA133057, R01CA142832, RC4CA156551, U01188388, U54CA151668, U54CA149196, and UH2 TR000943; by The University of Texas MD Anderson Cancer Center Ovarian Cancer Specialized Program of Research Excellence (SPORE) grant P50CA083639; by The University of Texas MD Anderson Cancer Center Uterine SPORE grant P50CA098258; by MD Anderson’s Cancer Center Support Grant P30CA016672 from the National Institutes of Health; by the US Department of Health and Human Services; by W81XWH-17-1-0126, W81XWH-17-1-0146, and W81XWH-16-1-0038 from the Ovarian Cancer Research Program, Department of Defense; by the Gilder Foundation; by grants RP100094 and RP110532 from the Cancer Prevention and Research Institute of Texas (CPRIT); by CPRIT Core Facility Support Award RP160805; and by funding support from Mr. Carl L. Norton, the Anne and John J. Sie Foundation, the Mary K. Chapman Foundation, the Ovarian Cancer Research Fund, the Ting Tsung and Wei Fong Chao Center for Bioinformatics Research and Imaging for Neurosciences (BRAIN), Cancer Fighters of Houston, and the John S. Dunn Research Foundation.
Notes
Affiliations of authors: Department of Gynecologic Oncology and Reproductive Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX (TLY, CSL, SYH, KHL, SCM); Department of Biomedical Informatics, The Ohio State University, Columbus, OH (FL); Department of Systems Medicine and Bioengineering, Houston Methodist Cancer Center, Weill Cornell Medicine, Houston, TX (JS, STCW); Center for Modeling Cancer Development, Houston Methodist Cancer Center, Houston, TX (JS, STCW); Department of Biochemistry and Molecular Biology, Melvin and Bren Simon Cancer Center, Indiana University, Indianapolis, IN (JK); Department of Pathology and Immunology and Center for Drug Discovery, Baylor College of Medicine, Houston, TX (MMM); The University of Texas Graduate School of Biomedical Sciences at Houston, Houston, TX (SCM).
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
The authors declare no conflicts of interest.
Supplementary Material
References
- 1. Tlsty TD, Coussens LM.. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–50. [DOI] [PubMed] [Google Scholar]
- 2. Ozdemir BC, Pentcheva-Hoang T, Carstens JL, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25(6):719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rhim AD, Oberstein PE, Thomas DH, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25(6):735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leung CS, Yeung TL, Yip KP, et al. Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat Commun. 2014;5:5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moran-Jones K, Gloss BS, Murali R, et al. Connective tissue growth factor as a novel therapeutic target in high grade serous ovarian cancer. Oncotarget. 2015;6(42):44551–44562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yeung TL, Leung CS, Wong KK, et al. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013;73(16):5016–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yeung TL, Leung CS, Li F, et al. Targeting stromal-cancer cell crosstalk networks in ovarian cancer treatment. Biomolecules. 2016;6(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yeung TL, Leung CS, Mok SC.. CAF reprogramming inhibits ovarian cancer progression. Cell Cycle. 2014;13(24):3783–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verhaak RG, Tamayo P, Yang JY, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123(1):517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Creighton CJ, Hernandez-Herrera A, Jacobsen A, et al. Integrated analyses of microRNAs demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PLoS One. 2012;7(3):e34546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patch AM, Christie EL, Etemadmoghadam D, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521(7553):489–494. [DOI] [PubMed] [Google Scholar]
- 13. Olsen OE, Wader KF, Hella H, et al. Activin A inhibits BMP-signaling by binding ACVR2A and ACVR2B. Cell Commun Signal. 2015;13:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gyorffy B, Lanczky A, Szallasi Z.. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19(2):197–208. [DOI] [PubMed] [Google Scholar]
- 15. Peng C, Mukai ST.. Activins and their receptors in female reproduction. Biochem Cell Biol. 2000;78(3):261–279. [PubMed] [Google Scholar]
- 16. Ding N, Yu RT, Subramaniam N, et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153(3):601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deeb KK, Trump DL, Johnson CS.. Vitamin D signalling pathways in cancer: Potential for anticancer therapeutics. Nat Rev Cancer. 2007;7(9):684–700. [DOI] [PubMed] [Google Scholar]
- 18. Sherman MH, Yu RT, Engle DD, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159(1):80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim J, Coffey DM, Creighton CJ, et al. High-grade serous ovarian cancer arises from fallopian tube in a mouse model. Proc Natl Acad Sci U S A. 2012;109(10):3921–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mishra PJ, Mishra PJ, Glod JW, et al. Mesenchymal stem cells: Flip side of the coin. Cancer Res. 2009;69(4):1255–1258. [DOI] [PubMed] [Google Scholar]
- 21. Mok SC, Bonome T, Vathipadiekal V, et al. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: Microfibril-associated glycoprotein 2. Cancer Cell. 2009;16(6):521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi H, Sheng J, Gao D, et al. Transcriptome analysis of individual stromal cell populations identifies stroma-tumor crosstalk in mouse lung cancer model. Cell Rep. 2015;10(7):1187–1201. [DOI] [PubMed] [Google Scholar]
- 23. Leyssens C, Verlinden L, Verstuyf A.. The future of vitamin D analogs. Front Physiol. 2014;5:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bonventre JV. Antifibrotic vitamin D analogs. J Clin Invest. 2013;123(11):4570–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walentowicz-Sadlecka M, Grabiec M, Sadlecki P, et al. 25(OH)D3 in patients with ovarian cancer and its correlation with survival. Clin Biochem. 2012;45(18):1568–1572. [DOI] [PubMed] [Google Scholar]
- 26. Jiang C, Xuan Z, Zhao F, et al. TRED: A transcriptional regulatory element database, new entries and other development. Nucleic Acids Res. 2007;35(Database issue):D137–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanehisa M, Goto S.. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J Royal Stat Soc. 1992;85(1):87–94. [Google Scholar]
- 29. Graeber TG, Eisenberg D.. Bioinformatic identification of potential autocrine signaling loops in cancers from gene expression profiles. Nat Genet. 2001;29(3):295–300. [DOI] [PubMed] [Google Scholar]
- 30. Peri S, Navarro JD, Kristiansen TZ, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32(Database issue):D497–D501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 33. Wishart DS, Knox C, Guo AC, et al. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–D672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wishart DS, Knox C, Guo AC, et al. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–D906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. [DOI] [PubMed] [Google Scholar]
- 36. Subramanian A, Tamayo P, Mootha V, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuhn M, Szklarczyk D, Franceschini A, et al. STITCH 3: Zooming in on protein-chemical Interactions. Nucleic Acids Res. 2012;40(Database issue):D876–D880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frey BJ, Dueck D.. Clustering by passing messages between data points. Science. 2007;315(5814):972–976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.