PSIP1/p75/LEDGF promotes the expression of key cell-cycle genes by facilitating the association of RNA pol II to their promoters. PSIP1 over-expresses in TNBC patients and patient-derived cell lines, and its silencing significantly decreases the migration, invasion and tumorigenicity of TNBC cells.
Abstract
Breast cancer (BC) is a highly heterogeneous disease, both at the pathological and molecular level, and several chromatin-associated proteins play crucial roles in BC initiation and progression. Here, we demonstrate the role of PSIP1 (PC4 and SF2 interacting protein)/p75 (LEDGF) in BC progression. PSIP1/p75, previously identified as a chromatin-adaptor protein, is found to be upregulated in basal-like/triple negative breast cancer (TNBC) patient samples and cell lines. Immunohistochemistry in tissue arrays showed elevated levels of PSIP1 in metastatic invasive ductal carcinoma. Survival data analyses revealed that the levels of PSIP1 showed a negative association with TNBC patient survival. Depletion of PSIP1/p75 significantly reduced the tumorigenicity and metastatic properties of TNBC cell lines while its over-expression promoted tumorigenicity. Further, gene expression studies revealed that PSIP1 regulates the expression of genes controlling cell-cycle progression, cell migration and invasion. Finally, by interacting with RNA polymerase II, PSIP1/p75 facilitates the association of RNA pol II to the promoter of cell cycle genes and thereby regulates their transcription. Our findings demonstrate an important role of PSIP1/p75 in TNBC tumorigenicity by promoting the expression of genes that control the cell cycle and tumor metastasis.
Introduction
Breast cancer (BC) is one of the most common cancers and a leading cause of death in women worldwide. Cellular levels of various receptors such as estrogen receptor, progesterone receptor and human epidermal growth factor 2 receptor (HER2) are used as biomarkers, and along with clinical parameters like tumor size, histological grade and lymph node status, they are routinely used for BC diagnosis and treatment (1,2). This is complemented by gene signature expression profiling in BC for subtype classification and diagnosis (3). Gene expression studies in patient samples over the past decades have uncovered large sets of genes, the expression of which is found to be altered during cancer initiation, progression and metastasis (4,5). For example, expression of genes involved in key regulatory pathways, including chromatin organization, transcription, post-transcriptional RNA processing and translation, is found to be deregulated in BC patient samples (6–8).
Transcriptional cofactors/coregulators regulate transcription of genes by fine-tuning the interaction of transcriptional machinery, including RNA polymerase II (RNA pol II) with gene-specific transcription factors. Transcription cofactors modify chromatin structure in order to make the associated DNA more or less accessible to transcription. Examples of transcription cofactors include histone-modifying enzymes, chromatin remodelling proteins, mediators and general cofactors that transmit regulatory signals between gene-specific transcription factors and general transcriptional machinery (9,10). Recent studies have reported aberrant expression of transcription cofactors and chromatin regulatory proteins in BC tissue samples, and demonstrated the involvement of several candidate proteins in BC progression and metastasis (11,12). PC4 and SF2-interacting protein 1 (PSIP1) is a chromatin associated protein that is shown to act as a transcriptional coactivator as well as an RNA-binding protein (13). The PSIP1 gene encodes several alternatively spliced isoforms such as PSIP1/p75 (also known as LEDGF) and PSIP1/p52 and minor p52 variant. PSIP1/p75 shares a common 325 amino acids with PSIP1/p52 at the N-terminal and has a unique Integrase binding domain at its C-terminal. The integrase-binding domain of PSIP1/p75 plays vital role in HIV integration and viral replication. On the other hand, the N-terminal PWWP domain of PSIP1 facilitates its binding to chromatin (14). PSIP1 was initially identified as an interactor of the PC4 general coactivator. In addition, PSIP1/p75 has been reported to interact with several proteins such as the menin/MLL complex, CtIP, JPO2, PogZ, Cdc7 activator of S-phase kinase (ASK), HIV1 integrase and MeCP2, and facilitates their association to chromatin (15–20). p75 is known to act as a co-activator to regulate the expression of several stress response genes as well as the developmentally regulated Hox genes (21–23). A recent study also demonstrated direct interaction of PSIP1 with poly A + RNA, implicating its potential involvement in RNA metabolism (24). PSIP1/p52 is known to regulate transcription of Hoxa genes and also alternative splicing of several pre-mRNAs by modulating the activity of SRSF1 and other proteins involved in the pre-mRNA processing (25,26).
In this study, we analyzed the expression of PSIP1 in TCGA (The Cancer Genome Atlas) RNA-seq data from hundreds of BC patient samples (n = 633) representing various subtypes. We found PSIP1 to be expressed at elevated levels in BC samples. We observed a positive correlation between PSIP1 levels and BC of basal-like subtype or triple negative breast cancer (TNBC) with a significant impact on patient survivability. Our gain- and loss-of-function studies in TNBC cells revealed that PSIP1/p75 acts as an oncogene. It influenced the tumorigenic properties of basal-like BC cells by regulating the expression of genes that control cellular growth and proliferation, cell death and survival and cellular movement. Based on our results, we propose that in BC cells, chromatin-associated PSIP1/p75 modulates the expression of cell cycle genes by regulating the interaction of RNA pol II to the promoters of these genes.
Materials and methods
Tumor progression model cell lines
M1 are benign non-tumorigenic MCF10A cells. M2 (MCF10AT1k.cl2) are H-ras transformed MCF10A cells, isolated from xenografts and possess extremely low tumorigenic potential. M3 (MCF10CA1h) and M4 (MCF10CA1a.cl1) cells are derivatives of M2 cells, isolated from tumors that were xenografted for several generations. M3 cells are highly tumorigenic in nature and form well differentiated tumors in xenografts but have low metastatic potential. On the other hand, M4 (MCF10CA1a.cl1) cells are highly tumorigenic (usually form undifferentiated tumors) and metastatic in nature (27,28).
Cells and culture medium
M1-M4 cells were provided by Dr. Ashish Lal (NCI, NIH) and were authenticated by analyzing the RNA sequencing data of genes deleted or mutated in M1–M4 cells (see supplementary information). M1–M4 cells were cultured as described previously (27). ME16C, BT20, MDA-MB231, SUM149, Hs578, HCC1937, MCF7, T47D and SKBR3 were obtained from the ATCC and maintained as per the recommendations.
Migration and invasion assays
Migration and invasion assays using transwell migration chambers (Corning, Cat# 354578) and Matrigel invasion chambers (Corning, Cat#354483) were performed as described previously (29).
Soft agar anchorage-independent and anchorage-dependent plastic colony formation assays
Anchorage-independent colony formation (5 × 103 cells) and plastic colony formation (1 × 103 M4 and 2 × 103 HCC1937 cells) assays were performed as described previously (29).
Microarray and data analysis
Total RNA isolated from control, PSIP1 (both p75 and p52) and p75 depleted M4 cells were labeled using Illumina TotalPrep RNA amplification kit (Applied Biosystems). Microarrays were performed using a HumanHT-12 v4 Expression Bead Chip kit (Illumina) and analyzed with the R/Bioconductor packages (Lumi.limma). Data is presented in Supplementary Table 2, available at Carcinogenesis Online (The GEO accession number is GSE99699).
Nascent RNA capture assay
Nascent RNAs were isolated by Click-iT Nascent RNA capture kit (Invitrogen, Cat # C10365) following the recommended protocol. The nascent transcript levels were quantified by RT-qPCR analyses using exon–intron junction or exon primer pairs.
Chromatin immunoprecipitation
Chromatin immunoprecipitation using RNA polymerase II (Millipore, Cat # 05-623) and PSIP1 (Bethyl Laboratories, Cat # A300-848) antibody were performed as described previously (30). ChIP primers are detailed in Supplementary Table 3, available at Carcinogenesis Online.
Transcription factor analysis
Combination of sequence motifs and ENCODE ChIP-seq tracks were used to identify the transcription factors enriched in the promoters of PSIP1 downregulated genes. Common transcription factors for PSIP1 knockdown downregulated genes were identified using iRegulon and Cytoscape by searching a 10 kb span centered around transcription start site (31).
Results
PSIP1 levels are elevated in basal-like subtype of BC
PSIP1/p75 and PSIP1/p52 are chromatin-associated and/or RNA-binding proteins that have been previously implicated in cell survival, autoimmune diseases, HIV pathogenesis and cancer (32). Aberrant expression of PSIP1/p75 was found to be involved in the development of several cancers, including subcutaneous angiogenesis and lymphangiogenesis of ovarian carcinoma tumors and leukemia (15,33,34). In most cases, PSIP1 was found to regulate the expression of a specific set of genes via modulating the association of proteins to chromatin. Also, transcriptome and protein analyses in different types of cancer samples revealed significant upregulation of PSIP1/p75 in various cancer types, including prostate, colon, thyroid and BCs (35,36). However, the molecular basis remains to be understood.
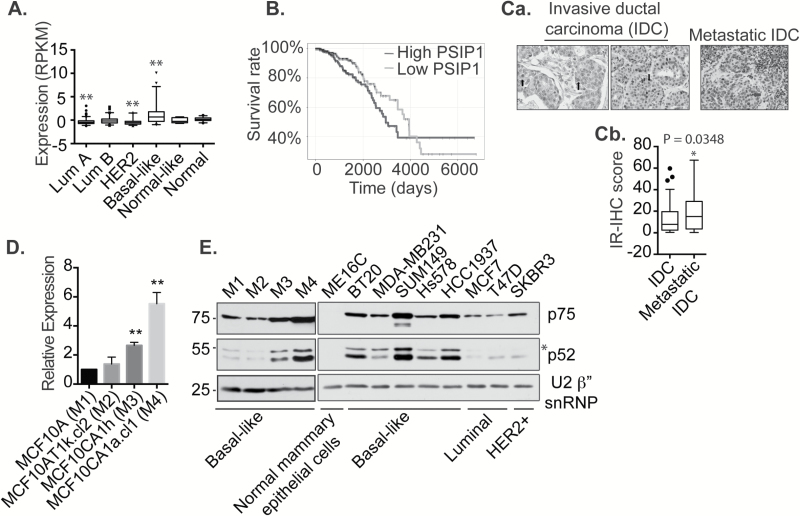
To understand the involvement of PSIP1 in BC, we analyzed the mRNA level of PSIP1 in the TCGA RNA-seq data from 633 BC patients of all subtypes. In particular, basal-like subtype BC (BSBC) or triple-negative BC patients showed higher levels of PSIP1 mRNA (Figure 1A; Supplementary Table 1, available at Carcinogenesis Online). Next, we compared the level of PSIP1 mRNA in all stages (stages I–IV) of BC. Though the expression of PSIP1 mRNA levels did not show a significant dependence on stage (Supplementary Figure 1, available at Carcinogenesis Online), the corresponding Kaplan–Meier curve demonstrated that higher expression was significantly correlated with reduced patient survival (Figure 1B). The survival graph shows a boundary effect for ~40% of the patients. Long-term survival data for TCGA patients is limited due to poor follow up information for longer periods. It is typical to see such boundary effects in survival plots. It is possible that for the 40% surviving patients, higher expression of PSIP1 is not a dominant prognostic factor or more prolonged survival data are required to see the impact of higher PSIP1 expression on these patients.
Figure 1.
PSIP1 mRNA and protein levels are elevated in basal-like subtype breast cancer. (A) Box plot shows elevated levels of PSIP1 mRNA in basal-like subtype BC patient samples. Data for the boxplot is derived from the TCGA dataset (patient number = 633; P < 0.01). (B) Kaplan–Meier curve of survivability of patients with high and low levels of PSIP1 mRNA (P < 0.05). (Ca) PSIP1 immunohistochemistry in invasive ductal carcinoma (IDC) and matched metastatic IDC in patient TMA samples. Black arrows represent the mild/weak (light brown) PSIP1 staining of scattered tumoral nuclei, red arrow denotes strong staining of scattered tumoral nuclei, blue arrow denotes the strong staining (dark brown) in mitotic nuclei, green stars represent stromal cell staining, and red star denotes the staining in lymphoid cells. Negative nuclei are stained blue-purple. (Cb) IR-IHC scores were plotted from 0 to 100% based on IHC intensity in cancerous region within the TMA. (D) RT-qPCR analysis to show PSIP1 mRNA levels in M1-M4 cell lines (*P < 0.05, **P < 0.01). (E) Western blot to detect the levels of PSIP1 (p75, p52 and p52 variant [*]) in various BC cell lines and a normal mammary epithelial cell line. U2β” snRNP is used as loading control.
Since cancer metastasis generally leads to reduced patient survival, we sought to examine whether metastatic cells demonstrated high expression of PSIP1 protein compared to the primary tumor. A tissue microarray (TMA) with 50 anonymized patient tissue samples was analyzed for expression of PSIP1 in invasive ductal carcinoma (IDC) and their matching metastatic IDC biopsies. We detected PSIP1 in both IDC and metastatic IDC but significantly elevated levels (P = 0.0348) of PSIP1 were observed in metastatic IDC samples (Figure 1Ca and b).
While we have noted the overall PSIP1 expression as typical with most translational studies in tissue, as opposed to cell line samples, we are not certain if expression level is truly measured in epithelial cells or may be repressed/enhanced by tumor microenvironment cells. In order to visualize PSIP1 expression at the protein level in patient samples, we utilized infrared (IR) spectroscopic imaging to perform automated segmentation of tissue in conjugation with IHC (37–40) (please see supplementary information for more details). Briefly, in this approach, the molecular spectral data is used as a pattern to differentiate epithelial cells from stromal cells. In order to visualize PSIP1 distribution in epithelial cells, we first performed IR imaging of the TMA and computationally labeled epithelium in each of the cores. Next, we overlaid the IHC images so that PSIP1 expression in epithelial cells could be measured with spatial specificity. This technique enabled us to visualize cores for expression of PSIP1 in epithelium while ignoring other PSIP1 positive non-cancerous cell types in the TMA such as lymphocytes (please see red asterisk in Figure 1Ca). The IHC image was digitally labelled as positive PSIP1 stain or negative PSIP1 stain (positive hematoxylin stain and no stain) by marking regions in the IHC image (Supplementary Figure 2A, available at Carcinogenesis Online) corresponding to the three classes by using color intensity in the IHC image and then applying supervised classification to perform labelling (Supplementary Figure 2B, available at Carcinogenesis Online) with 86.4% accuracy (Supplementary Figures 2C, available at Carcinogenesis Online). The labelled IR image was overlaid with the labelled IHC image to identify the regions where cancerous epithelium showed higher levels of PSIP1 (Supplementary Figure 2D, available at Carcinogenesis Online). Based on the IR-IHC score, matched patient samples collected from breast and lymph node showed a significantly greater percentage of metastatic cancer cells staining positive with PSIP1 as compared to IDC (Figure 1Cb). On a different note, patient specific PSIP1 IR-IHC score (Supplementary Figure 2E, available at Carcinogenesis Online) did not correlate with BC subtype but cannot be removed from consideration since the patient sample size (n = 50) used for IHC analysis was small compared to the sample size analyzed for RNA-seq (n = 633).
To gain mechanistic insights into the differential expression of PSIP1 in BC, we pursued our investigation using an isogenic basal-like/TNBC subtype mammary cell line-based tumor progression model. These cell lines (M1, M2, M3 and M4) were originally derived from non-tumorigenic human mammary epithelial MCF10A cells and are well characterized in several studies (27–29,41,42). The isogenic cell lines represent the complete spectrum of cancer progression ranging from non-tumorigenic (M1), hyperproliferative with low tumorigenic potential (M2) and highly tumorigenic with low metastatic potential (M3) to highly tumorigenic with metastatic potential (M4) (please see supp. Information for details). RT-qPCR analysis showed increased mRNA levels in highly tumorigenic M3 and M4 cells compared to non-tumorigenic M1 cells (Figure 1D). Next, we examined the levels of PSIP1 protein in a panel of BC cell lines of all subtypes, including basal-like/TNBC: M1-M4, BT20, MDA-MB231, SUM149, Hs578 and HCC1937, luminal: MCF7, T47D, Her2+ve: SKBR3, and normal epithelial breast cells: ME16C. We observed significantly higher levels of all the known isoforms (p75, p52 and p52 variant [*]) of PSIP1in BC cell lines compared to normal mammary ME16C cells (Figure 1E and Supplementary Figure 3, available at Carcinogenesis Online). Further, consistent with the RNA expression data, all of the PSIP1 isoforms showed elevated levels in M3 and M4 cells compared to M1 and M2 cells (Figure 1E). Among the BC cell lines, both p75 and p52 isoforms were highly up-regulated, specifically in basal-like subtype cell lines. Based on these results, we conclude that the mRNA and protein levels of PSIP1 are found to be elevated in the basal-like subtype of BC patients and cell lines.
PSIP1 contributes to the aggressive phenotype of BC cells
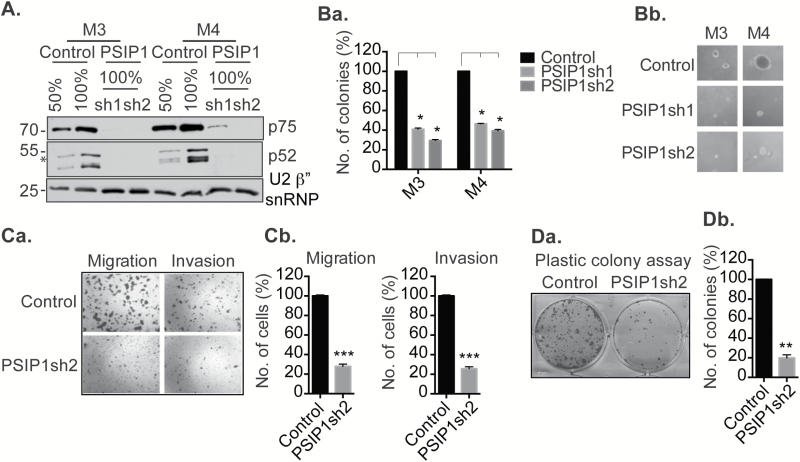
To test the involvement of PSIP1 in BC, we performed loss of function studies in M3 (MCF10CA1h; highly tumorigenic with low metastatic potential) and M4 (MCF10CA1a.cl1; highly tumorigenic and metastatic in nature) cells by stably depleting PSIP1 using shRNAs (PSIP1sh1 and PSIP1sh2) that target all the isoforms of PSIP1 (Figure 2A and Supplementary Figures 4A and B, available at Carcinogenesis Online). Depletion of PSIP1 led to significant decrease (more than 60%) in the ability of M3 and M4 cells to form discrete colonies in soft agar in the anchorage-independent colony formation assays (Figure 2Ba). PSIP1-depleted cells showed reduction in both the number of colonies as well as the size of individual colonies (Figure 2Bb). M4 cells display characteristic properties of metastatic cells, including enhanced migration and invasion in Boyden chamber assays (43). PSIP1-depleted M4 cells showed a significant reduction in their ability to migrate (up to 70%) and to invade (up to 75%) in the Boyden-chamber assays (Figures 2Ca and b). These results suggest the involvement of PSIP1 in regulating the tumorigenic and metastatic properties of BC cells.
Figure 2.
PSIP1 enhances the aggressive properties of TNBC cells. (A) Western blot showing the efficiency of shRNA mediated knockdown of PSIP1 (both p75 and p52 isoforms) in M3 and M4 cells. U2β” snRNP is used as loading control. (Ba and Bb) M3 and M4 cells depleted of PSIP1 showing reduction in colony number and size in soft agar anchorage-independent colony formation assay. Colonies are counted from three independent experiments. (Ca and Cb) M4 cells depleted of PSIP1 showing reduction in migration and invasion. Cells are stained and counted from three independent experiments. (Da and Db) M4 cells depleted of PSIP1 showing reduction in long term cell proliferation, assayed by anchorage-dependent plastic colony formation assay. Cells are stained and counted from three independent experiments. Error bars in the graphs represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Flow cytometry analyses revealed that PSIP1-depleted M3 and M4 cells showed defects in cell proliferation, including a significant increase in the G1 or G1/S population (Supplementary Figure 4C, available at Carcinogenesis Online). Next, we performed plastic colony formation assays to test the role of PSIP1 in the long term cell proliferation of BC cells. Plastic colony formation assays showed significant defects in the proliferation ability of M4 cells that were depleted of PSIP1 isoforms (Figures 2Da and b). In addition to M3 and M4 cells, depletion of PSIP1 in HCC1937, another BC cell line of basal-like subtype, displayed similar defects in cell proliferation and cell migration, further signifying the involvement of PSIP1 in controlling tumorigenic properties of TNBC cells (Supplementary Figure 5A, 5Ba and b and 5Ca and b, available at Carcinogenesis Online).
PSIP1/p75 promotes cancer cell proliferation
To test the involvement of individual isoforms of PSIP1 in cell proliferation, we generated M4 cells in which either PSIP1/p75 or PSIP1/p52 was stably depleted using isoform specific shRNAs (p75sh1 and 2 for p75 and p52sh for p52 isoforms, respectively) (Supplementary Figure 4A and 6A, available at Carcinogenesis Online). p75 specific shRNA (p75sh1 and sh2)-treated cells showed significant reduction in the levels of p75 but not p52 isoforms (Supplementary Figure 6A, available at Carcinogenesis Online). The p52 isoform specific shRNA (p52sh)-treated cells showed moderate levels of depletion of p52 isoforms (Supplementary Figure 6A, available at Carcinogenesis Online). M4 cells depleted of only PSIP1/p75 showed dramatic reduction in their ability to proliferate in the plastic colony assay (Supplementary Figure 6Ba and b, available at Carcinogenesis Online). On the other hand, p52 alone-depleted cells did not show significant change in their ability to proliferate (Supplementary Figure 6Ba and b, available at Carcinogenesis Online). This could be due to the inefficient depletion of p52 in M4 cells.
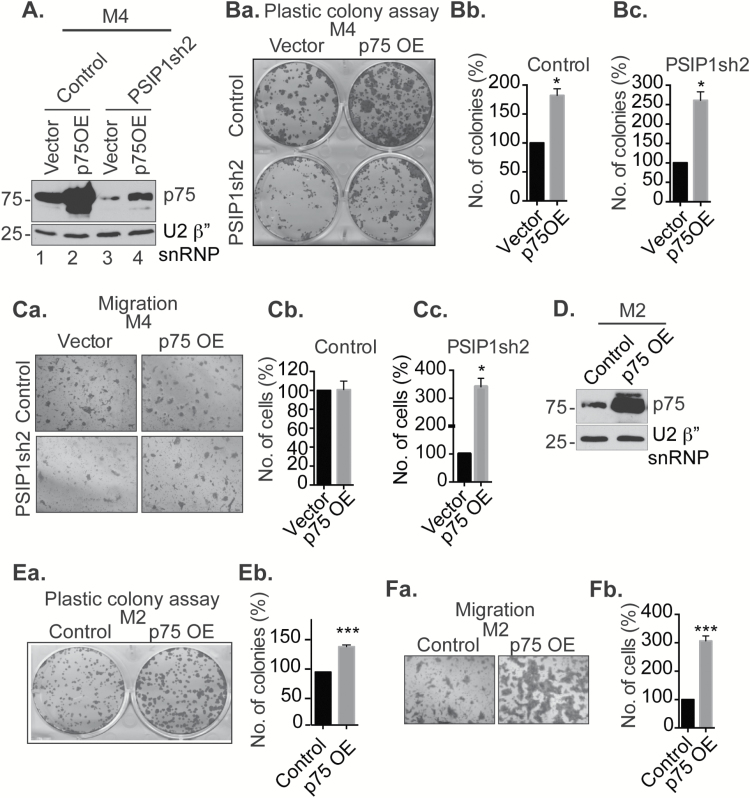
In order to gain insights into the involvement of PSIP1/p75 in cell proliferation, we overexpressed the PSIP1/p75 isoform and addressed if this could rescue the cell proliferation phenotype observed in PSIP1-depleted cells (Figure 3A and Supplementary Figure 7A, available at Carcinogenesis Online). Anchorage-dependent plastic colony formation assays revealed that empty vector expressing M4 cells that were stably depleted of PSIP1 isoforms (Figure 3A, lane 3) showed reduced cell proliferation relative to control M4 cells expressing vector plasmid (Figure 3A, lane 1 and Figure 3Ba). On the other hand, PSIP1/p75 over-expressing control M4 cells (Figure 3A, lane 2) showed enhanced cell proliferation (Figure 3Ba and b). Finally, exogenous expression of PSIP1/p75 (Figure 3A, lane 4) in M4 cells that were stably depleted of both endogenous PSIP1/p75 and p52 isoforms rescued cell proliferation defects (Figure 3Ba and c).
Figure 3.
Over-expression of PSIP1/p75 promotes BC cell proliferation, migration and invasion. (A) Immunoblot showing the over-expression of transiently expressed T7-tagged p75 isoform in control and PSIP1-depleted M4 cells. (Ba–c) Anchorage-dependent colony formation assay performed in control and PSIP1-depleted M4 cells that are transiently transfected with empty vector or T7-p75. Graphs are derived from three independent experiments. (Ca–c) Migration assay performed in control and PSIP1-depleted M4 cells that are transiently transfected with empty vector or T7-p75. Migrated cells are stained and counted from three independent experiments. (D) Western blot showing the levels of stably overexpressed PSIP1/p75 in M2 cells. U2β”-snRNP is used as loading control in A and D. (Ea–b) Plastic colony formation assay performed in M2 cells after PSIP1/p75 over-expression. (Fa–b) Migration assay performed in PSIP1/p75 stably overexpressed M2 cells. Graph is derived from three independent experiments. Error bars in the graphs represent SEM. *P < 0.05 and ***P < 0.001.
Further, p75-overexpressed M4 cells showed increased tumorigenicity in the anchorage-independent soft agar colony formation assay (Supplementary Figure 7B). Finally, overexpression of p75 in M4 cells that were depleted of endogenous PSIP1 isoforms rescued the cell migration phenotype (Figure 3Ca–c). On the other hand, p75-overexpressed control M4 cells did not show any significant change in their migratory properties (Figure 3Ca and b). We infer that the higher levels of endogenous PSIP1 in M4 cells make them insensitive to exogenous overexpression of p75. Finally, stable over-expression of PSIP1/p75 in the non-tumorigenic M2 cells led to a significant increase in the anchorage-dependent colony formation and cell migration (Figure 3D–F).
In order to test the involvement of PSIP1/p52 in cell proliferation and migration, we over-expressed PSIP1/p52 in control and PSIP1-depleted M4 cells (Supplementary Figure 7C, available at Carcinogenesis Online). Over-expression of p52 increased cell proliferation of control M4 cells and also rescued the cell proliferation defects in PSIP1-depleted cells (Supplementary Figure 7D, available at Carcinogenesis Online). Further, control M4 cells overexpressing PSIP1/p52 showed increased cell migration (Supplementary Figure 7E, available at Carcinogenesis Online). However, the migration defects observed in PSIP1-depleted M4 cells could not be rescued by PSIP1/p52 (Supplementary Figure 7E, available at Carcinogenesis Online). Altogether, our data indicates that both p75 and p52 regulate cell proliferation. However, PSIP1/p75 seems to play a dominant role in promoting tumorigenic properties in BC cells.
PSIP1/p75 utilizes its PWWP domain to interact with chromatin containing H3K36me3 modifications (23). In order to test whether the chromatin association of PSIP1/p75 is essential for its role in regulating cell proliferation, we exogenously expressed full length (FL) p75 or PWWP deletion mutant of p75 (PSIP1/p75ΔPWWP) in control and PSIP1 (both p75 and p52)-depleted M4 cells, and examined the effect on cell proliferation by plastic colony formation assay (Supplementary Figure 8A–C, available at Carcinogenesis Online). Both FL and ΔPWWP mutant p75 expressing cells showed an increased number of colonies compared to vector-transfected cells, indicating that both the constructs promote cell proliferation (Supplementary Figure 8B and C, available at Carcinogenesis Online). However, PSIP1/p75ΔPWWP-expressing cells displayed a significantly less number of colonies in comparison to cells expressing FL p75. These results imply that p75 promotes cell proliferation in part through its association with chromatin.
PSIP1 regulates the expression of genes involved in cell cycle progression and cell proliferation
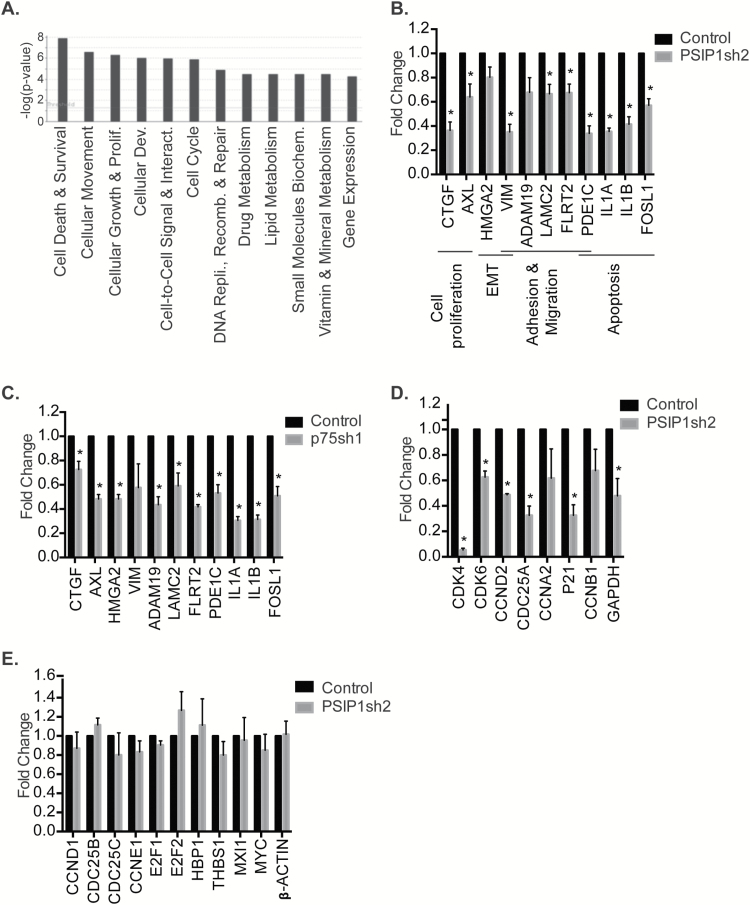
In order to gain mechanistic insights into the role of PSIP1 in cell proliferation and tumor progression, we examined the changes in gene expression profile by transcriptome microarray in control versus PSIP1 (both isoforms)-depleted M4 cells. Microarray data revealed that ~324 genes were down-regulated and 373 genes were upregulated (>2-fold) upon PSIP1 depletion (Supplementary Table 2, available at Carcinogenesis Online). Gene Ontology analysis revealed that the top affected pathways of down-regulated genes included cell death and survival, cell growth and proliferation and cell movement (Figure 4A), whereas pathways such as energy production and cellular movement were enriched in the list of overexpressed genes (Supplementary Figure 9A, available at Carcinogenesis Online). RT-qPCR experiments demonstrated that genes controlling several of the crucial pro-tumorigenic pathways such as cell proliferation, cell adhesion and migration, apoptosis and epithelial to mesenchymal transition showed altered expression in M4 cells that were depleted of both PSIP1 isoforms (Figure 4B and Supplementary Figure 9B, available at Carcinogenesis Online). Next, we performed microarray using RNA from control and PSIP1/p75 isoform alone-depleted M4 cells. Interestingly, GO analyses indicated that both p75 alone-depleted or PSIP1/p75 and p52-depleted cells affected a similar set of pathways (cell death, cell cycle and cell movement), and the downregulation of these genes was confirmed by RT-qPCR analyses (Figure 4C and Supplementary Figure 9C and D). These results indicate that the PSIP1/p75 isoform plays a crucial role in regulating the expression of genes involved in cell proliferation and tumor progression. In addition to several of the cancer-associated genes, we also confirmed reduced mRNA levels of several cell cycle regulated genes (CDK4, CDK6, CCND2 and CDC25A) in PSIP1-depleted cells (Figure 4D). For example, PSIP1-depleted M4 cells showed dramatic reduction in both mRNA and protein levels of CDK4 (Figure 4D and Supplementary Figure 10A, available at Carcinogenesis Online). In addition, PSIP1-depleted cells showed reduced levels of cyclin D2 and not cyclin D1 mRNA (Figure 4D), indicating that PSIP1 could be involved in G1 or G1/S progression. On the other hand, PSIP1-depleted cells did not show downregulation of all of G1- or G1/S-regulated genes, including several genes that are direct targets of E2F (Figure 4E and Supplementary Figure 10A, available at Carcinogenesis Online), suggesting that the changes in the expression of specific cell cycle genes observed upon PSIP1 depletion were not a consequence of the cell cycle arrest. It was previously reported that PSIP1/p75 interacted with Cdc7-Activator of S-phase kinase (Cdc7-ASK or Dbf4 kinase) and stimulated the enzymatic activity of Dbf4 (18). Cdc7-ASK/Dbf4 activity is essential for the cells to enter S-phase (44). To test whether PSIP1-depleted M4 cells show defects in Cdc7-ASK/Dbf4 activity, we examined the phosphorylation status of MCM2, a known Dbf4 substrate (18), in cell extracts of control and PSIP1-depleted cells. Phos-tag analyses revealed that both control and PSIP1-depleted cells showed comparable levels of phosphorylated as well as the total expression level of MCM2, indicating that in BC cells, depletion of PSIP1 does not seem to affect the activity of Cdc7-ASK/Dbf4 (Supplementary Figure 10Ba and b, available at Carcinogenesis Online).
Figure 4.
PSIP1 controls the expression of genes involved in cell-cycle progression, cell growth, proliferation, death and survival. (A) Gene ontology analysis of genes downregulated after PSIP1 knockdown in M4 cells. (B and C) RT-qPCR analyses to validate the microarray analyses showing downregulation of genes involved cancer progression in PSIP1- or p75-depleted M4 cells. (D and E) RT-qPCR analyses to validate the microarray analyses showing differential expression of cell cycle genes in PSIP1-depleted M4 cells. Graphs are plotted from three independent experiments and error bar represents SEM. *P < 0.05.
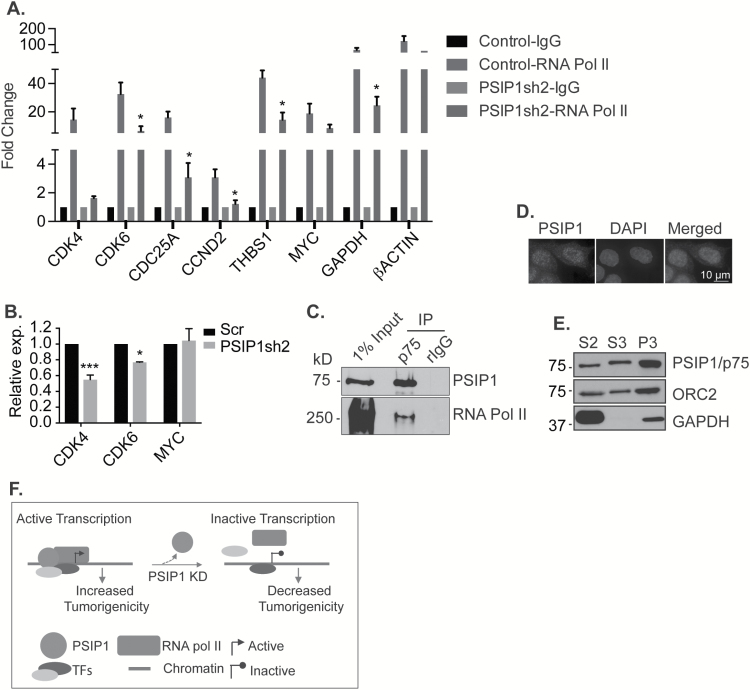
In vitro studies have shown that PSIP1/p75 acts as a transcriptional coactivator of RNA polymerase II (RNA pol II) (13). In order to test whether PSIP1/p75-depleted cells display defects in transcription of cell cycle genes, we examined the status of RNA pol II occupancy on the promoters of cell-cycle genes in the presence and absence of PSIP1. RNA pol II ChIP-qPCR displayed significant decrease in the association of RNA pol II on the promoters of several of the cell cycle genes (CDK4, CDK6, CDC25A) in PSIP1-depleted cells (Figure 5A). To quantify the level of newly synthesized RNA and therefore to determine the RNA Pol II-mediated transcription activity, we performed nascent RNA pull down assays followed by RT-qPCR in control and PSIP1-depleted cells. We observed a significant decrease in the level of nascent RNA of cell cycle genes in PSIP1-depleted cells, confirming reduced transcriptional activity (Figure 5B). Consistent with these results, we found that PSIP1 interacts with RNA pol II (Figure 5C). Finally, we examined whether PSIP1/p75 associates with the promoter region of cell cycle genes that showed reduced expression in PSIP1-depleted cells. It is known that PSIP1/p75 associates with chromatin through its interaction with H3K4me3 and H3K36me3-modified histones (21,22,45). We observed that a major fraction of PSIP1/p75 in M4 BC cells localized in the nucleus and was associated with the micrococcal nuclease resistant chromatin fraction (Figure 5D and E; P3 fraction). PSIP1/p75 ChIP-qPCR revealed that p75 interacted with the chromatin of all of the tested cell cycle genes (Supplementary Figure 10C, available at Carcinogenesis Online). However, we consistently observed enhanced association of PSIP1/p75 on the promoters of genes, the expressions of which were altered upon PSIP1/p75 depletion (CDK4 and CDK6). Altogether, our results indicate that PSIP1/p75, by interacting with the promoter region of several of the cell cycle genes, facilitates the association and/or stabilization of RNA pol II to the promoters, thereby enhancing their transcription.
Figure 5.
PSIP1 modulates the association of RNA pol II to the promoters of cell cycle genes. (A) RNA pol II ChIP-qPCR to determine the association of RNA pol II on the promoters of genes in control and PSIP1-depleted M4 cells. β-actin is used as positive control. Graph is plotted from three independent experiments, and error bars represent SEM. (B) Nascent RNA capture assay shows the level of newly synthesised RNA from representative genes in control and PSIP1-depleted M4 cells. *P < 0.05 and ***P < 0.001. (C) Endogenous co-IP reveals interaction between endogenous PSIP1/p75 and RNA pol II. (D) Immunofluorescence staining showing the localization of PSIP1 in M4 cells. DNA is counterstained with DAPI. Scale represents 10 μm. (E) Chromatin fractionation of M4 cells showing the enrichment of PSIP1 in MNase resistant chromatin fraction (P3). S2 and S3 represent cytoplasmic and soluble nuclear fractions respectively. Orc2 and GAPDH are used as control for MNase resistant chromatin and cytoplasmic fraction, respectively. (F) Proposed model depicting the involvement of PSIP1/p75 in regulating the association of RNA pol II on the promoters of cell cycle genes. It is possible that PSIP1 could regulate promoter association of RNA pol II through recruiting and/or stabilizing other transcription factors.
Discussion
Out of all the BC subtypes, basal-like subtype or TNBC has the worst prognosis, and presently lacks effective targeted therapeutic options. There is an urgent need to identify prognostic markers to better stratify TNBC patients, and to develop therapeutic targets that specifically target TNBC patients. Here, by utilizing TCGA BC patient RNA-seq datasets, we find differential subtype-specific expression of PSIP1. PSIP1, originally identified as a transcription coactivator, is shown to play a crucial role in several cellular processes, including transcriptional regulation, anti-apoptosis, DNA repair, pre-mRNA splicing and HIV viral DNA integration into the host genome (20,21,23,25,36,46–48). PSIP1/p75 levels are also found to be elevated in several cancer samples, including prostate, colon, thyroid, liver, uterus and BC (35). Since PSIP1 showed elevated expression in various cancers and is known to act as a pro-survival gene, we were interested in determining its role in BC progression and metastasis. In BC cells of basal-like subtype/TNBC, we observed that PSIP1/p75 promotes cell proliferation, migration, invasion and tumorigenicity. We further demonstrate potential involvement of PSIP1/p75 in regulating the expression of genes controlling cell cycle progression and cell growth in BC cells. In support of PSIP1’s role in tumorigenicity, we observed an association between higher expression of PSIP1 and poor progression-free survival in BC patients. Our data signifies the oncogenic nature and potentially important role of PSIP1 in BC cancer initiation and progression.
PSIP1-depleted BC cells showed altered expression of genes involved in cell cycle, cell movement, cell proliferation, survival and death. PSIP1/p75, also known as LEDGF, is known to function as a pro-survival gene, protects cells from various types of damages (including oxidative damage) and prevents apoptosis (21,22,49). These functions have been ascribed to the role of p75 in regulating the expression of genes involved in cellular stress response. For example, in prostate cancer cells, p75 is known to induce the expression of HSP27 and activate anti-apoptotic pathways (46). Silencing of p75 in these cells reduced the levels of HSP27, which resulted in defects in cell proliferation and tumorigenicity, and implicated an essential role for PSIP1/p75 in regulating the expression of stress responsive genes such as HSP27 (46). Both our microarray and RT-qPCR analyses in M4 cells revealed that PSIP1-depleted BC cells did not show significant change in the levels of HSP27 mRNA (Supplementary Figure 11A and Table 2, available at Carcinogenesis Online), suggesting that PSIP1/p75 does not seem to regulate the expression of HSP27 in BC cells.
PSIP1/p75/LEDGF is known to induce the expression of pro-angiogenic VEGF-C in glioma, non-small cell lung carcinoma and ovarian cancer cells (33,34). Finally, using mouse xenografts, the authors showed that p75-induced overexpression of VEGF-C enhanced angiogenesis and lymphangiogenesis (33). We analyzed potential changes in the levels of VEGF-C mRNA in control and PSIP1-depleted M4 cells. Unlike other studies, we did not find any difference in the levels of VEGF-C mRNA in control and PSIP1-depleted BC cells, indicating that the changes in migration and invasion observed upon PSIP1 depletion in BC cells are not due to changes in VEGF-C expression (Supplementary Figure 11A and Table 2, available at Carcinogenesis Online).
RNA pol II ChIP analyses revealed that PSIP1-depleted BC cells showed reduced association of RNA pol II to the promoters of several of the tested cell cycle genes. Both PSIP1/p75 and p52 are known to function as co-activators for RNA polymerase II-mediated transcription and therefore could influence the expression of a wide array of genes (50). Earlier, in vitro studies revealed interaction between PSIP1 and several subunits of RNA pol II, further supporting the notion that PSIP1 could influence RNA pol II activity (13,50). Our endogenous co-IP confirmed the association between PSIP1 and RNA pol II. However, it is not clear how PSIP1 controls the association of RNA pol II on certain but not all of the gene promoters. One explanation could be that PSIP1/p75 influences the binding of certain proteins, such as transcription factors (TFs) or coactivators to specific gene promoters, thereby modulating the association of RNA pol II to these promoters. For example, PSIP1 is known to interact with TFs such as TFIIF and SP1 and regulate their activity (51), and PSIP1 interacting protein PC4 interacts with GTF2A1 (or TFIIA) (50,52). In addition, PSIP1 is known to recruit H3K4 methyltransferase (MLL1) to chromatin (15). Finally, PSIP1 regulates the activity of Myc TF by controlling the association of Myc-inhibitor protein Jpo2 to the chromatin (16). Based on this, it is reasonable to assume that in BC cells PSIP1, in addition to its involvement in regulating RNA pol II association to promoters, could also modulate the association of key TFs to the regulatory elements of these genes. This prompted us to look for the common TFs that interact with the promoters of genes, the expressions of which were found to be deregulated in PSIP1-depleted M4 cells. To identify potential TF binding sites, we used the computational method iRegulon to detect enriched TFs on the regulatory elements of PSIP1-regulated genes (31). We used the existing TF ChIP-seq data from ENCODE and searched for consensus TF binding sequences within a 10 kb window centered around the transcription start site of PSIP1-regulated genes. Based on these analyses, we identified ~8 TFs, each of which potentially interacts with the promoters of 10 or more of the genes that were down-regulated in PSIP1-depleted cells (Supplementary Figure 11B, available at Carcinogenesis Online). For example, we observed that ~118 down-regulated gene promoters contain the binding sites of GTF2A1 or TFIIA. Similarly, we observed binding consensus sites for several of the key cancer-associated TFs, such as SP1 in the regulatory elements of genes that were upregulated in PSIP1-depleted cells (Supplementary Figure 11B, available at Carcinogenesis Online). Based on our results, we propose that PSIP1 directly or indirectly, via the chromatin binding of specific TFs, dictates the association of RNA pol II to the promoters of key cell cycle genes (Figure 5F). Future studies will determine the potential relationship between PSIP1/p75 and these TFs, and whether p75 regulates the chromatin association or activity of any of these TFs in BC cells.
PSIP1/p75 is a nuclear-restricted protein, and most of it is tightly associated with chromatin through the help of the PWWP domain and AT-hook motif within the protein (53). In leukemia cells, p75 aids the interaction between Menin and the MLL1 (histone methyltransferase) complex and further facilitates the association of the complex to chromatin (15,54). Specific involvement of PSIP1/p75 in modulating the association of MLL1 and polycomb proteins to the chromatin is known to influence the transcriptional activity of developmentally regulated Hox-gene clusters (23). In addition, PSIP1 is also known to interact with several other proteins such as Jpo2, CtIP and HIV integrase, and regulate the association of these proteins to chromatin (53,55). All of these studies recognized p75 as a chromatin adaptor protein that facilitated the recruitment of cellular or viral proteins to chromatin. p75 mutants lacking the chromatin-associated PWWP domain, when exogenously expressed in PSIP1-depleted BC cells, showed that it could moderately rescue the cell proliferation defects, reiterating the functional importance of the chromatin binding of PSIP1 in controlling cell proliferation.
Various adaptor proteins and nuclear-restricted long non-coding RNAs play a crucial role in transcription regulation by influencing the recruitment or stabilization of several of the chromatin-associated factors, including transcription factors, cofactors and histone-modifying enzymes to gene regulatory elements (56,57). Abnormal expression of these proteins and RNAs in cells are known to alter the expression of a large number of genes, ultimately resulting in aberrant cellular functions and causing diseases such as cancer. In this study, we have demonstrated that PSIP1 positively regulates the migration, invasion and tumorigenicity of cancer cells, and its higher expression negatively correlates with patients’ survival. Our results showing the involvement of the PSIP1/p75 adaptor protein in BC progression in the most aggressive TNBC are very promising. The specific roles of PSIP1 in TNBC, especially its involvement in regulating the expression of cell cycle genes merits further investigation, as this might have crucial implications for therapeutic interventions in BC.
Supplementary material
Supplementary material is available at Carcinogenesis online.
Funding
This work was supported by grants from National Institute of Health (1RO1GM088252 to K.V.P., 1RO1GM099669 to S.G.P.); American Cancer Society (RSG-11-174-01-RMC to K.V.P.); National Science Foundation (1723008 to K.V.P., 1243372 to S.G.P).
Supplementary Material
Acknowledgements
We would like to thank Drs. Ray P. (Carle Foundation Hospital) and members of Prasanth KV and SG laboratories for their valuable suggestions.
Conflict of Interest Statement: None declared.
Abbreviations
- BC
breast cancer
- ChIP
chromatin immunoprecipitation
- IDC
invasive ductal carcinoma
- TNBC
triple negative breast cancer
- TMA
tissue microarray
References
- 1. Schnitt S.J. (2010) Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod. Pathol., 23(Suppl 2), S60–S64. [DOI] [PubMed] [Google Scholar]
- 2. Singletary S.E., et al. (2003) Revision of breast cancer staging: the 6th edition of the TNM classification. Semin Surg Oncol., 21, 53–59. [DOI] [PubMed] [Google Scholar]
- 3. Parker J.S., et al. (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol., 27, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perou C.M., et al. (2000) Molecular portraits of human breast tumours. Nature, 406, 747–752. [DOI] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Network. (2012) Comprehensive molecular portraits of human breast tumours. Nature, 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glisovic T., et al. (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett., 582, 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerstberger S., et al. (2014) A census of human RNA-binding proteins. Nat. Rev. Genet., 15, 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dreyfuss G., et al. (2002) Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol., 3, 195–205. [DOI] [PubMed] [Google Scholar]
- 9. Krasnov A.N., et al. (2016) On the way of revealing coactivator complexes cross-talk during transcriptional activation. Cell Biosci., 6, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomas M.C., et al. (2006) The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol., 41, 105–178. [DOI] [PubMed] [Google Scholar]
- 11. Morgan M.A., et al. (2015) Chromatin signatures of cancer. Genes Dev., 29, 238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Helin K., et al. (2013) Chromatin proteins and modifications as drug targets. Nature, 502, 480–488. [DOI] [PubMed] [Google Scholar]
- 13. Ge H., et al. (1998) Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J., 17, 6723–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Llano M., et al. (2006) Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol., 360, 760–773. [DOI] [PubMed] [Google Scholar]
- 15. Yokoyama A., et al. (2008) Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell, 14, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maertens G.N., et al. (2006) Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J. Cell Sci., 119(Pt 12), 2563–2571. [DOI] [PubMed] [Google Scholar]
- 17. Bartholomeeusen K., et al. (2009) Lens epithelium-derived growth factor/p75 interacts with the transposase-derived DDE domain of PogZ. J. Biol. Chem., 284, 11467–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hughes S., et al. (2010) Transcriptional co-activator LEDGF interacts with Cdc7-activator of S-phase kinase (ASK) and stimulates its enzymatic activity. J. Biol. Chem., 285, 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cherepanov P., et al. (2003) HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem., 278, 372–381. [DOI] [PubMed] [Google Scholar]
- 20. Leoh L.S., et al. (2012) The stress oncoprotein LEDGF/p75 interacts with the methyl CpG binding protein MeCP2 and influences its transcriptional activity. Mol. Cancer Res., 10, 378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shinohara T., et al. (2002) LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res., 21, 341–358. [DOI] [PubMed] [Google Scholar]
- 22. Singh D.P., et al. (2001) LEDGF binds to heat shock and stress-related element to activate the expression of stress-related genes. Biochem. Biophys. Res. Commun., 283, 943–955. [DOI] [PubMed] [Google Scholar]
- 23. Pradeepa M.M., et al. (2014) Psip1/Ledgf p75 restrains Hox gene expression by recruiting both trithorax and polycomb group proteins. Nucleic Acids Res., 42, 9021–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Castello A., et al. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell, 149, 1393–1406. [DOI] [PubMed] [Google Scholar]
- 25. Pradeepa M.M., et al. (2012) Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet., 8, e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pradeepa M.M., et al. (2017) Psip1/p52 regulates posterior Hoxa genes through activation of lncRNA Hottip. PLoS Genet., 13, e1006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dawson P.J., et al. (1996) MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am. J. Pathol., 148, 313–319. [PMC free article] [PubMed] [Google Scholar]
- 28. Weiger M.C., et al. (2013) Real-time motion analysis reveals cell directionality as an indicator of breast cancer progression. PLoS One, 8, e58859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jadaliha M., et al. (2016) Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget, 7, 40418–40436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Giri S., et al. (2015) The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin. Elife, 4. doi:10.7554/eLife.06496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Janky R., et al. (2014) iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol., 10, e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Daugaard M., et al. (2007) Lens epithelium-derived growth factor is an Hsp70-2 regulated guardian of lysosomal stability in human cancer. Cancer Res., 67, 2559–2567. [DOI] [PubMed] [Google Scholar]
- 33. Sapoznik S., et al. (2009) Gonadotropin-regulated lymphangiogenesis in ovarian cancer is mediated by LEDGF-induced expression of VEGF-C. Cancer Res., 69, 9306–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen B., et al. (2009) Transcriptional regulation of vascular endothelial growth factor C by oxidative and thermal stress is mediated by lens epithelium-derived growth factor/p75. Neoplasia, 11, 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Basu A., et al. (2012) Expression of the stress response oncoprotein LEDGF/p75 in human cancer: a study of 21 tumor types. PLoS One, 7, e30132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Daniels T., et al. (2005) Antinuclear autoantibodies in prostate cancer: immunity to LEDGF/p75, a survival protein highly expressed in prostate tumors and cleaved during apoptosis. Prostate, 62, 14–26. [DOI] [PubMed] [Google Scholar]
- 37. Mayerich D., et al. (2015) Stain-less staining for computed histopathology. Technology (Singap. World. Sci)., 3, 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martin F.L., et al. (2010) Distinguishing cell types or populations based on the computational analysis of their infrared spectra. Nat. Protoc., 5, 1748–1760. [DOI] [PubMed] [Google Scholar]
- 39. Fernandez D.C., et al. (2005) Infrared spectroscopic imaging for histopathologic recognition. Nat. Biotechnol., 23, 469–474. [DOI] [PubMed] [Google Scholar]
- 40. Bhargava R., et al. (2006) High throughput assessment of cells and tissues: Bayesian classification of spectral metrics from infrared vibrational spectroscopic imaging data. Biochim. Biophys. Acta, 1758, 830–845. [DOI] [PubMed] [Google Scholar]
- 41. Drabsch Y., et al. (2013) Transforming growth factor-β signalling controls human breast cancer metastasis in a zebrafish xenograft model. Breast Cancer Res., 15, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elangovan S., et al. (2013) Molecular mechanism of SLC5A8 inactivation in breast cancer. Mol. Cell. Biol., 33, 3920–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weiger M.C., et al. (2013) Real-time motion analysis reveals cell directionality as an indicator of breast cancer progression. PLoS One, 8, e58859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Masai H., et al. (2002) Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J. Cell. Physiol., 190, 287–296. [DOI] [PubMed] [Google Scholar]
- 45. De Rijck J., et al. (2010) High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res., 38, 6135–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bhargavan B., et al. (2012) LEDGF gene silencing impairs the tumorigenicity of prostate cancer DU145 cells by abating the expression of Hsp27 and activation of the Akt/ERK signaling pathway. Cell Death Dis., 3, e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ciuffi A., et al. (2005) A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med., 11, 1287–1289. [DOI] [PubMed] [Google Scholar]
- 48. Daugaard M., et al. (2012) LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol., 19, 803–810. [DOI] [PubMed] [Google Scholar]
- 49. Matsui H., et al. (2001) Lens epithelium-derived growth factor: increased survival and decreased DNA breakage of human RPE cells induced by oxidative stress. Invest. Ophthalmol. Vis. Sci., 42, 2935–2941. [PubMed] [Google Scholar]
- 50. Ge H., et al. (1994) Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell, 78, 513–523. [DOI] [PubMed] [Google Scholar]
- 51. Ge H., et al. (1998) A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell, 2, 751–759. [DOI] [PubMed] [Google Scholar]
- 52. Kretzschmar M., et al. (1994) A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell, 78, 525–534. [DOI] [PubMed] [Google Scholar]
- 53. Llano M., et al. (2009) Virological and cellular roles of the transcriptional coactivator LEDGF/p75. Curr. Top. Microbiol. Immunol., 339, 125–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang J., et al. (2012) The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature, 482, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tesina P., et al. (2015) Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif. Nat. Commun., 6, 7968. [DOI] [PubMed] [Google Scholar]
- 56. Berger S.L. (2007) The complex language of chromatin regulation during transcription. Nature, 447, 407–412. [DOI] [PubMed] [Google Scholar]
- 57. Singh D.K., et al. (2013) Functional insights into the role of nuclear-retained long noncoding RNAs in gene expression control in mammalian cells. Chromosome Res., 21, 695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.