Abstract
Glycan or carbohydrate structures can be pictorially represented using symbolic nomenclatures. The symbol nomenclature for glycans (SNFG) contains 67 different monosaccharides represented using various colors and geometric shapes. A simple tool to convert International Union of Pure and Applied Chemistry (IUPAC) format text to SNFG will be useful for sketching glycans and glycopeptides. Such code can also enable the development of more sophisticated applications, where the visual representation of carbohydrate structures is necessary. To address this need, the current manuscript describes DrawGlycan-SNFG, a freely available, platform-independent, open-source tool. It allows: i. the display of glycans and glycopeptides from IUPAC-condensed text inputs and ii. the depiction of glycan and glycopeptide fragments. The online version of this program is provided with a user-friendly web interface at www.virtualglycome.org/DrawGlycan. Downloadable, stand-alone GUI (Graphical User Interface) version and the program source code are also available from this repository. DrawGlycan-SNFG will be useful for experimentalists looking for a ready to use, simple program for sketching carbohydrates and for software developers interested in incorporating SNFG into their program suite.
Keywords: carbohydrates, freeware, glycoscience, sketch glycans, systems glycobiology
Introduction
The representation of carbohydrate structures using symbolic nomenclature enables easy visualization of the underlying biology. Several groups have presented such schematics starting with the symbolic form put forth by Kornfeld (Kornfeld et al. 1978), related refinements undertaken in the “Essentials of Glycobiology” (Varki et al. 1999), expansions with color in the second edition of the text (Varki et al. 2009) and also the oxford nomenclature (UOXF) (Royle et al. 2006). In this regard, the version proposed in the “Essentials” text has been widely adopted by a number of groups including the Consortium for Functional Glycomics (CFG). While early versions of such representation were limited to vertebrate glycans, the editors of the third edition of the “Essentials” have expanded the nomenclature to also include monosaccharides from other species (Varki et al. 2015). This new nomenclature is called symbol nomenclature for glycans (SNFG) and it contains 67 monosaccharides, made of 11 shapes and 10 colors. Compared with the previous version, it includes 49 new monosaccharides mostly from microbes and also plants.
A variety of computer programs have been developed in order to render glycan drawing in symbolic form. KCaM (KEGG Carbohydrate Matcher) was one of the earliest glycan editors where users could either manually assemble glycans in a manner similar to ChemDraw or load glycan sequence file in KCF (KEGG Chemical Function) format (Aoki et al. 2004). The monosaccharides displayed here used text format and the primary purpose of this program was to search a given glycan for similarity against other structures previously deposited in the KEGG (Kyoto Encyclopedia of Genes and Genomes) or CarbBank databases. Ceroni et al. (2007) later developed a more versatile program called GlycanBuilder that supports the construction and display of glycan structures in symbolic form. This open-source code has been integrated with other programs, e.g. GlycoWorkbench to assist glycan mass spectrometry (MS) analysis (Ceroni et al. 2008). Several updates have been made to GlycanBuilder to enable its incorporation into web applications, and to allow the user to both draw glycans manually using a palette of structures, and to import and export glycan structures in XML (eXtensible Markup Language) format (Damerell et al. 2015). SimGlycanTM is another tool for drawing glycans and analysis of MS data, though it is proprietary and it is not open source (Apte and Meitei 2010). The authors of KCaM have developed a Java-based applet called DrawRINGS, where users can sketch 2D-glycan structures by dragging-and-dropping various structures from a family of options (Akune et al. 2010). Finally, 3D-SNFG is a new tool for presenting glycans in 2D and 3D form for integration into protein structures (Thieker et al. 2016).
While the above approaches have merit, we were motivated to create DrawGlycan-SNFG so that in addition to simply drawing glycans, we can also automate the sketching of glycopeptides. In addition, we were interested in developing a simple and robust program where users can rapidly display glycan and peptide bond fragmentations using simply IUPAC format input (McNaught 1997). Such functions are necessary for other code being developed in our laboratory that automate the in silico generation of glycan biosynthetic pathways using MS data (Liu et al. 2013; Liu and Neelamegham 2014), and also high-throughput glycoproteomics data analysis (Neelamegham et al. 2014). DrawGlycan-SNFG was also designed to support the SNFG nomenclature as it may be part of future IUPAC recommendations (Varki et al. 2015). The program is available with a web interface at www.virtualglycome.org/DrawGlycan. A downloadable, stand-alone Graphical User Interface (GUI) version of the application with more functionality is also provided at this site. Neither the web nor GUI version requires the MATLAB software for operation. Additionally, the platform-independent source code is also provided at this website as it can aid investigators interested in incorporating DrawGlycan-SNFG into their own projects.
Results
The input for DrawGlycan-SNFG follows from IUPAC-condensed nomenclature
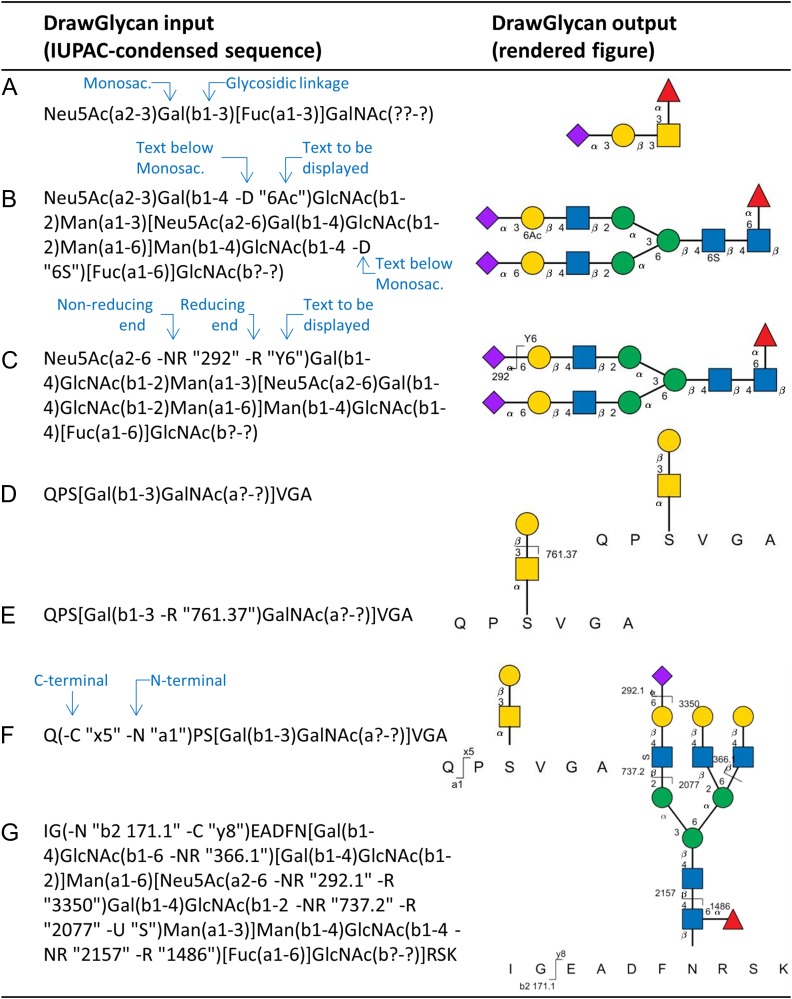
Figure 1 shows various types of inputs accepted by DrawGlycan-SNFG (left column) with the corresponding outputs (right column). All the inputs use the standard IUPAC-condensed nomenclature with linkage information presented within parenthesis and branched structures enclosed in square brackets (McNaught 1997). Bond fragmentation data are incorporated into the IUPAC string using -R and -NR options to depict fragmentations occurring at the reducing and non-reducing glycan ends, respectively. The options -N and -C adjacent to specific amino acids show corresponding N- and C-terminal peptide fragmentations. Finally, -U (“up”) and -D (“down”) options are also enabled for the monosaccharide residues in order to enable the depiction of modified monosaccharides, like the acetylated Gal and sulfated GlcNAc residues in Figure 1B. The overall DrawGlycan-SNFG input thus follows the format:
PEP[Mono1(a/bi-j) …[…glycan branch…]… Monon(a/bi-j -NR “text” -R “text” -U “text” -D “text”)….]TI(-N “text” -C “text”)DE
Fig. 1.
Glycan and glycopeptide structures generated using DrawGlycan-SNFG. (A) O-linked glycan; (B) N-linked glycan; (C) N-glycan with fragmentation and modification information; (D) simple glycopeptide. (E) Glycopeptide with glycan fragment annotation; (F) glycopeptide with peptide fragmentation annotation. (G) N-linked glycopeptide with bond breakage at multiple locations. In all cases, left column presents the IUPAC input used in DrawGlycan-SNFG to sketch the corresponding schematic. Linkage information is shown in parentheses, with reducing and non-reducing fragments displayed using -R and -NR options. Monosaccharide modifications are shown above and below monosaccharide symbols using -U and -D options within parenthesis. C- and N-terminal peptide fragments are similarly annotated using -C and -N options for corresponding peptide bond fragments. “?” is used when glycosidic bond data are unavailable. Monosaccharide symbols follow the SNFG nomenclature. This figure is available in black and white in print and in color at Glycobiology online.
Here, the amino acids that form the peptide backbone appear in capital letters (“PEPTIDE” in the example above). Monosaccharides use case-sensitive SNFG recommendations. The glycosidic bond that links the ith anomeric carbon to the jth carbon on the adjacent monosaccharide is depicted as either “ai-j” for α-glycosidic bonds or “bi-j” for β-type linkages. The question mark symbol is used for unknown bond and linkage information, for example “??-?” is used when neither the bond type nor linkage information are available.
All structures in Figure 1 are displayed with the bond linkage data, though this can be hidden using DrawGlycan-SNFG options. The examples include an O-linked glycan (panel A), N-linked glycan (panel B) and the same N-glycan with a single bond break that releases a terminal sialic acid (panel C). Here, any text can be included within inverted commas after the -R/-NR option to include either fragment type, mass or other information that the user may wish to display. The -U and -D options can be used to annotate specific monosaccharide modifications using text that appears either above or below the particular monosaccharide symbol. While the examples presented in panels A–C follow from the IUPAC-condensed recommendations, DrawGlycan-SNFG is more flexible in that it also accepts abbreviated bond descriptions (e.g. “a3” instead of “a1-3” for the α1-3 bond), and commas instead of hyphens (e.g. α1,3 instead of α1-3). Additionally, unknown bonds may be represented in short as “()” instead of “(??-?)”. Also, the closing bracket “)” at the glycan anomeric position can be omitted if desired, in order to be fully consistent with the IUPAC-condensed recommendation (McNaught 1997). The remaining inputs are case sensitive and monosaccharide inputs not recognized by the program will appear in white flat hexagons with the first letter of the unknown entity. Finally, the program can also handle the string input format used in the recently released 3D-SNFG program (Thieker et al. 2016).
An O-linked glycopeptide is shown in Figure 1D and the corresponding structures with glycosidic and peptide bond fragmentations appear in panels E and F. Here, the entire glycan is enclosed within square brackets, flanked by the peptide backbone sequence. Figure 1G shows a more complete example with multiple bond fragmentations both in the N-linked glycan and in the underlying peptide. Besides the glycopeptide examples in Figure 1 that bear a single carbohydrate chain, DrawGlycan-SNFG also supports drawing of multiple glycans on a single peptide backbone.
Overall, DrawGlycan-SNFG enables the rapid and automated sketching of glycan and glycopeptide structures along with bond fragmentation data. The code uses IUPAC-condensed format text inputs since this is widely used by the scientific community, and it closely follows the SNFG guidelines. Additionally, the software is flexible as it also accepts certain variations of the IUPAC nomenclature that are commonly used in the literature. A variety of programs are available for the interconversion between different glycan formats (Ceroni et al. 2007; Akune et al. 2010; Liu et al. 2013). These can be used along with DrawGlycan-SNFG to enable usage of other glycan descriptors.
DrawGlycan-SNFG program algorithm
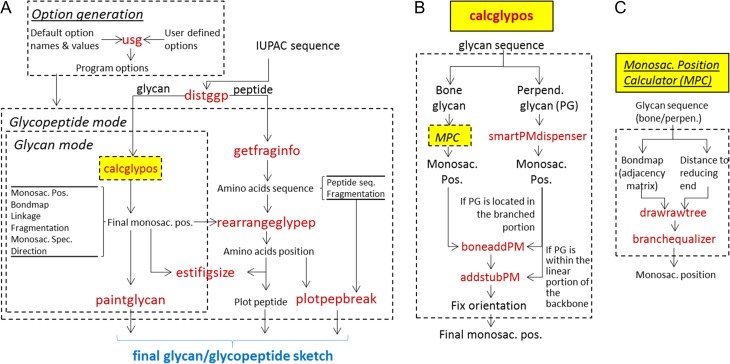
Figure 2 shows the workflow for DrawGlycan-SNFG along with selected function names. The primary program input is provided as an IUPAC string, with minor modifications made to facilitate inclusion of glycan and peptide fragmentation data as discussed in the section The input for DrawGlycan-SNFG follows from IUPAC-condensed nomenclature. The subroutine usg modifies the default program options with user preferences that define monosaccharide size, glycan orientation, font size, etc. (Figure 2A). Distggp then parses the input string to separate the portions of the sequence that correspond to the peptide from that describing the glycan(s). Calcglypos is an important program, detailed in Figure 2B. It determines the position of each of the monosaccharides in the input glycan for final rendering on the DrawGlycan-SNFG canvas. Glycan bondmap and linkage are additional outputs from this program that are used for depicting glycosidic bonds and breakage events. Getfraginfo similarly parses the peptide input to obtain backbone amino-acid sequence and fragmentation data. Rearrangeglypep uses the results emerging from calcglypos and getfraginfo to define amino-acid positions relative to the glycan. This program also adjusts the relative glycan positions if a single glycopeptide is decorated by more than one carbohydrate structure. Together, these calculations are used by estifigsize to define the final DrawGlycan-SNFG canvas size. Paintglycan then sketches the glycan including glycan fragmentation data. Plotpepbreak completes the drawing by annotating peptide fragments.
Fig. 2.
Program workflow. (A) Overall workflow for DrawGlycan-SNFG shows that the glycan and peptide portions of the initial IUPAC input are first separated by distggp. The subroutine calcglypos (explained in panel B) calculates the position of the monosaccharides in the glycan, for sketching on the canvas. Other routines calculate the positions for the individual amino acids. Paintglycan and plotpepbreak then integrate this information to render the final structure. (B) Calcglypos contains the detailed machinery for monosaccharide position calculation. This routine separates the basic glycan backbone or “bone glycan” from other glycan substructures called “perpendicular glycan” that have to be rendered perpendicular to the “bone glycan”. The position of the monosaccharides in both these structures is then independently calculated before the components are merged together to define the final monosaccharide position. (C) Monosaccharide position calculator (MPC) represents a set of routines within Calcglypos that use graph theory to draw the basic glycan tree structure and then adjust their spacing to prevent structural overlaps. In panels (A–C), function names appear in red fonts and the flow of data among the subroutines is shown using arrows. Blocks of related code are enclosed within dashed boxes for easy visualization. This figure is available in black and white in print and in color at Glycobiology online.
Figure 2B outlines the calcglypos algorithm. This routine parses the input glycan sequence to pick out substructures that are initiated by Fuc and Xyl. These structures, which are conventionally drawn perpendicular to the basic glycan backbone (“bone glycan”), are called “perpendicular glycan”. The position of the individual monosaccharides in the bone glycan is calculated using a series of subroutines that are collectively called the monosaccharide position calculator (MPC, Figure 2C). In MPC, graph theory is applied to calculate the unidirectional adjacency matrix (i.e. the bondmap), which defines the links between the different monosaccharides. The distance of each monosaccharide to the reducing end of the glycan is also enumerated. Based on this, drawrawtree determines the basic tree structure of the “bone” and “perpendicular” glycans, with branchequalizer adjusting bond angles at branching points and monosaccharide position to realize compactness and symmetry in the final figure (Supplementary Movie A illustrates this algorithm for a tetra-antennary N-linked glycan). The final monosaccharide positions are then merged using either boneaddPM or addstubPM depending on whether the perpendicular glycan is located within the branched portion of the carbohydrate (e.g. on the antenna of branched N-glycans) or within the unbranched linear portion (e.g. in the case of core fucosylation on N-glycans). Following this, the position of the individual monosaccharides is finalized after accounting for the orientation of the glycan: “up”, “down”, “right” or “left”. Note that while glycans can be oriented in any direction based on user preference (up, down, etc.), the carbohydrate structures always point “up” in the case of glycopeptides. In this case, the peptide backbone stretches out at the bottom of the DrawGlycan-SNFG canvas.
DrawGlycan-SNFG interfaces
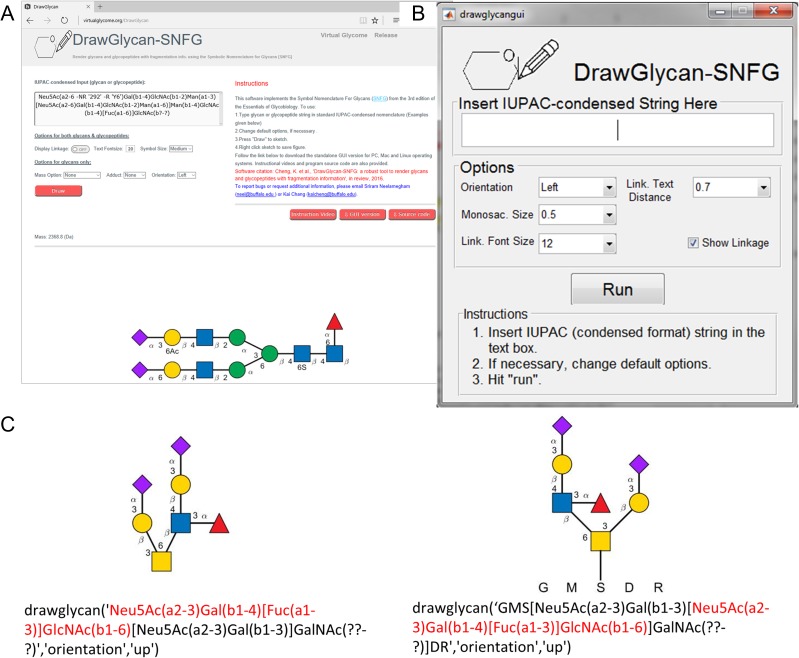
DrawGlycan-SNFG is platform independent in that it can be launched on Windows, Mac or Linux machines. It is presented with three interfaces that enable software usage from: the web, the GUI and command line operations (Figure 3). With respect to these formats, the web interface is readily accessible to any user without the need for program download or installation (Figure 3A). This is likely to satisfy the needs of a majority of users. However, many features of the final figure cannot be manually changed to suit specific user needs.
Fig. 3.
DrawGlycan-SNFG program interfaces. (A) Web interface. (B) Graphical User Interface (GUI). (C) Command line operations. In the example in panel (C), the same glycan is shown in two alternative forms with the sialofucosylated substructure (shown using red text) appearing at either the right or left side of the GalNAc residue. This is done by simply modifying the program's input string as illustrated below the drawings. All monosaccharide symbols follow the SNFG nomenclature. This figure is available in black and white in print and in color at Glycobiology online.
There is greater flexibility in the stand-alone GUI version that can be downloaded and installed (Figure 3B, Supplementary Movie B). In this case, facilities are available to change glycan orientation, monosaccharide symbol size, overall text font size and line thickness. The MATLAB software is not necessary in order to use either the web or stand-alone GUI versions.
The DrawGlycan-SNFG GUI can also be directly launched from the MATLAB command prompt by executing the “drawglycangui” command (Supplementary Movie C). In this case, virtually all aspect of the final figure can be refined including the manual addition of new annotations, and alterations of individual text font sizes, positions or contents.
The most versatile version of DrawGlycan-SNFG uses command line operations since multiple features of the program can be seamlessly integrated together using custom scripts (Figure 3C). As shown in this example, the layout of branched structures depends on the IUPAC input string. Since the same glycan can have alternative IUPAC representations, the final rendered presentation can be modified by controlling the specific text input. In the example presented here, the sialofucosylated glycan (sialyl Lewis-X, Figure 3C) appears to the right of the GalNAc residue if this sequence appears first in the input string (left half of Figure 3C), and to the left if the corresponding sLeX string appears later in the string input (right half of Figure 3C). While the default DrawGlycan-SNFG program provided in the GUI and web formats offers this flexibility, the raw source code also contains functions that can restrict the sequence of branches based on increasing carbon positions at each branching monosaccharide.
Discussion
This manuscript describes a software that renders glycans and glycopeptides including bond fragmentation data. The algorithm used is different from that employed by GlycanBuilder (Ceroni et al. 2007), a program that represents the current state-of-the-art for many in the field. In this regard, GlycanBuilder uses a bottom-up rule-based approach to progressively add one monosaccharide at a time. Here, the addition of each monosaccharide triggers a recalculation over all previously added monosaccharides. Therefore, the number of steps required for calculation is ~n(n−1)/2, where n equals the number of monosaccharides. The time necessary for rendering using GlycanBuilder thus scale as O(n2). As opposed to this, DrawGlycan-SNFG uses a top-down approach where glycans are first divided into subglycans that include the bone structure and perpendicular glycans. Each of these is individually constructed before the entire final glycan is assembled together. The number of steps required for such calculations is ~, and thus the computational time of DrawGlycan-SNFG scales as O(n). Due to these differences, DrawGlycan-SNFG is likely to scale more efficiently for larger, more complex carbohydrates.
At the current time, DrawGlycan-SNFG is designed to address a majority of the needs of the community in terms of simplicity of usage, free availability, implementation of the SNFG nomenclature, and robustness. While it can automate a majority of the drawing needs of typical experimentalist, there are some instances where manual intervention is necessary in order to improve the DrawGlycan-SNFG output. Specifically: 1. The program cannot automatically draw cyclical oligosaccharides like cyclodextrin, or polymeric glycans that have repeat units. Implementation of such structures will require some manual intervention. 2. The current version does not support the rendering of groups of similar glycans, using curly bracket notations to depict ambiguity in structure assignment. 3. While some facilities are available for the display of modified monosaccharides using the -U and -D options, manual intervention may be necessary if monosaccharides outside the SNFG collection have to be displayed.
Taken together, we anticipate that DrawGlycan-SNFG will be a useful tool for the Glycoscience community. It can either serve as a stand-alone application or be a part of a larger program suite.
Methods
The overall DrawGlycan-SNFG algorithm is presented in Figure 2. Essential elements of the program are discussed in Results. MATLAB (Mathworks, Natick, MA) code with detailed commenting and examples are also available with the downloadable, open-source software. The program uses the MATLAB programming language since it has easy-to-use functions for rendering geometric shapes, performing calculations, integration with other languages (C++, JAVA, etc.) and building GUIs, without the need for extensive coding. It can also be complied for incorporation in other web interfaces and programs in a variety of platforms. The current program has been developed using MATLAB version 2015a, and it has been tested on PC (Windows7/10, MATLAB version R2014b and higher), Mac (OS X version 10.11.5, MATLAB version R2014b and higher) and LINUX (Ubuntu 16.04.1 LTS, MATLAB version R2014b and higher) platforms.
Supplementary Material
Acknowledgements
We are grateful to Srirangaraj Setlur and Edward J. Sobczak (University at Buffalo) for setting up the VirtualGlycome server using the HUBzero platform.
Funding
National Institutes of Health grant (HL103411); Program for Excellence in Glycosciences award (HL107146).
Conflict of interest statement
None declared.
Abbreviations
IUPAC, International Union of Pure and Applied Chemistry; MS, Mass Spectrometry; SNFG, Symbol Nomenclature for Glycans.
References
- Akune Y, Hosoda M, Kaiya S, Shinmachi D, Aoki-Kinoshita KF. 2010. The RINGS resource for glycome informatics analysis and data mining on the Web. OMICS. 14(4):475–486. [DOI] [PubMed] [Google Scholar]
- Aoki KF, Yamaguchi A, Ueda N, Akutsu T, Mamitsuka H, Goto S, Kanehisa M. 2004. KCaM (KEGG Carbohydrate Matcher): a software tool for analyzing the structures of carbohydrate sugar chains. Nucleic Acids Res. 32(Web Server issue):W267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte A, Meitei NS. 2010. Bioinformatics in glycomics: Glycan characterization with mass spectrometric data using SimGlycan. Methods Mol Biol. 600:269–281. [DOI] [PubMed] [Google Scholar]
- Ceroni A, Dell A, Haslam SM. 2007. The GlycanBuilder: A fast, intuitive and flexible software tool for building and displaying glycan structures. Source Code Biol Med. 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM. 2008. GlycoWorkbench: A tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 7(4):1650–1659. [DOI] [PubMed] [Google Scholar]
- Damerell D, Ceroni A, Maass K, Ranzinger R, Dell A, Haslam SM. 2015. Annotation of glycomics MS and MS/MS spectra using the GlycoWorkbench software tool. Methods Mol Biol. 1273:3–15. [DOI] [PubMed] [Google Scholar]
- Kornfeld S, Li E, Tabas I. 1978. The synthesis of complex-type oligosaccharides. II. Characterization of the processing intermediates in the synthesis of the complex oligosaccharide units of the vesicular stomatitis virus G protein. J Biol Chem. 253(21):7771–7778. [PubMed] [Google Scholar]
- Liu G, Neelamegham S. 2014. A computational framework for the automated construction of glycosylation reaction networks. PLoS One. 9(6):e100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Puri A, Neelamegham S. 2013. Glycosylation Network Analysis Toolbox: A MATLAB-based environment for systems glycobiology. Bioinformatics. 29(3):404–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaught AD. 1997. Nomenclature of carbohydrates (recommendations 1996). Adv Carbohydr Chem Biochem. 52:43–177. [PubMed] [Google Scholar]
- Neelamegham S, Lo C, Cheng K, Li J, Qu J, Liu G. 2014. GlycoPAT: An open-source MATLAB based toolbox for glycoproteomics analysis. Glycobiology. 24(11):1166. [Google Scholar]
- Royle L, Dwek RA, Rudd PM. 2006. Determining the structure of oligosaccharides N- and O-linked to glycoproteins. Curr Protoc Protein Sci. Unit 12.6:12.6.1-12.6.45. http://onlinelibrary.wiley.com/doi/10.1002/0471140864.ps1206s43/pdf. [DOI] [PubMed] [Google Scholar]
- Thieker DF, Hadden JA, Schulten K, Woods RJ. 2016. 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology. 26(8):786–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T et al. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology. 25(12):1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Freeze HH, Hart G, Marth JD. 1999. Essentials of Glycobiology Cold Spring Harbor, NY, Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. 2009. Essentials of Glycobiology, 2nd ed.Cold Spring Harbor, NY, Cold Spring Harbor Press. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.