Beytebiere et al. show that the ability of BMAL1 to drive tissue-specific rhythmic transcription is associated with not only the activity of BMAL1-bound enhancers but also the activity of neighboring enhancers.
Keywords: circadian clock, enhancer–enhancer interactions, tissue-specific cistromes, transcription
Abstract
The mammalian circadian clock relies on the transcription factor CLOCK:BMAL1 to coordinate the rhythmic expression of thousands of genes. Consistent with the various biological functions under clock control, rhythmic gene expression is tissue-specific despite an identical clockwork mechanism in every cell. Here we show that BMAL1 DNA binding is largely tissue-specific, likely because of differences in chromatin accessibility between tissues and cobinding of tissue-specific transcription factors. Our results also indicate that BMAL1 ability to drive tissue-specific rhythmic transcription is associated with not only the activity of BMAL1-bound enhancers but also the activity of neighboring enhancers. Characterization of physical interactions between BMAL1 enhancers and other cis-regulatory regions by RNA polymerase II chromatin interaction analysis by paired-end tag (ChIA-PET) reveals that rhythmic BMAL1 target gene expression correlates with rhythmic chromatin interactions. These data thus support that much of BMAL1 target gene transcription depends on BMAL1 capacity to rhythmically regulate a network of enhancers.
Circadian clocks are found ubiquitously across all kingdoms of life and provide a time-keeping mechanism for organisms to anticipate rhythmic environmental changes. In mammals, virtually every cell harbors the same circadian clockwork that regulates rhythmic gene expression along with temporal cues and systemic signals, such that biological functions occur at the most appropriate time of day. The mammalian circadian clock relies on transcriptional feedback loops initiated by the heterodimeric transcription factor CLOCK:BMAL1 (Partch et al. 2014; Takahashi 2017). CLOCK:BMAL1 rhythmically binds DNA to drive the rhythmic transcription of the genes Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2), which, upon translation, form a repressive complex that feedbacks to inhibit CLOCK:BMAL1-mediated transcription. CLOCK:BMAL1 binds to E-boxes and activates transcription more potently during the middle of the day, and maximal repression occurs during the middle of the night (Rey et al. 2011; Koike et al. 2012). CLOCK:BMAL1 is also responsible for the transcriptional regulation of many clock-controlled genes, which allows for rhythms in biochemistry, physiology, and behavior (Panda 2016).
Characterization of the rhythmic transcriptional outputs driven by the circadian clock indicates that genes expressed in a rhythmic manner vary greatly between tissues in the mouse (Panda et al. 2002; Storch et al. 2002; Zhang et al. 2014). Similar findings in plants, insects, and primates also revealed that circadian gene expression is largely tissue-specific (Endo et al. 2014; Meireles-Filho et al. 2014; Mure et al. 2018). However, it is still unknown how the same circadian clock mechanism can generate tissue-specific rhythmic gene expression. Previous work in Drosophila revealed that CLOCK:CYC, the CLOCK:BMAL1 homolog, exhibits tissue-specific DNA binding between the body and head, and that much of this tissue specificity is due to the cooperation between CLOCK:CYC and tissue-specific transcription factors (ts-TFs) (Meireles-Filho et al. 2014). Tissue-specific binding of another mammalian clock component, Rev-erbα, has also been described, but the underlying mechanisms have not yet been characterized (Zhang et al. 2015).
To address the mechanisms by which the mammalian circadian clock generates tissue-specific circadian transcriptional programs, we performed BMAL1 chromatin immunoprecipitation (ChIP) at the genome-wide level (ChIP-seq) in the mouse liver, kidney, and heart. Our data revealed that the majority of BMAL1 DNA binding is tissue-specific and suggest that accessibility of the chromatin (as defined by DNase I-hypersensitive sites [DHSs]) (Thurman et al. 2012) and that cobinding of ts-TFs accounts for much of this tissue specificity. In addition, we found large discrepancies between BMAL1 DNA binding and rhythmic gene expression with, for example, many genes targeted by BMAL1 in all three tissues only exhibiting rhythmicity in a single tissue. Characterization of the underlying mechanism suggests that the ability of BMAL1 to drive tissue-specific rhythmic gene expression depends on how BMAL1 promotes not only the activity of the DHSs to which it binds but also the activity of other neighboring DHSs. Together, our data suggest that BMAL1 transcriptional output is controlled through enhancer–enhancer interactions and that BMAL1-driven rhythmic transcription depends on the capacity of BMAL1 to rhythmically regulate a network of DHSs.
Results
BMAL1 DNA-binding signal is largely tissue-specific
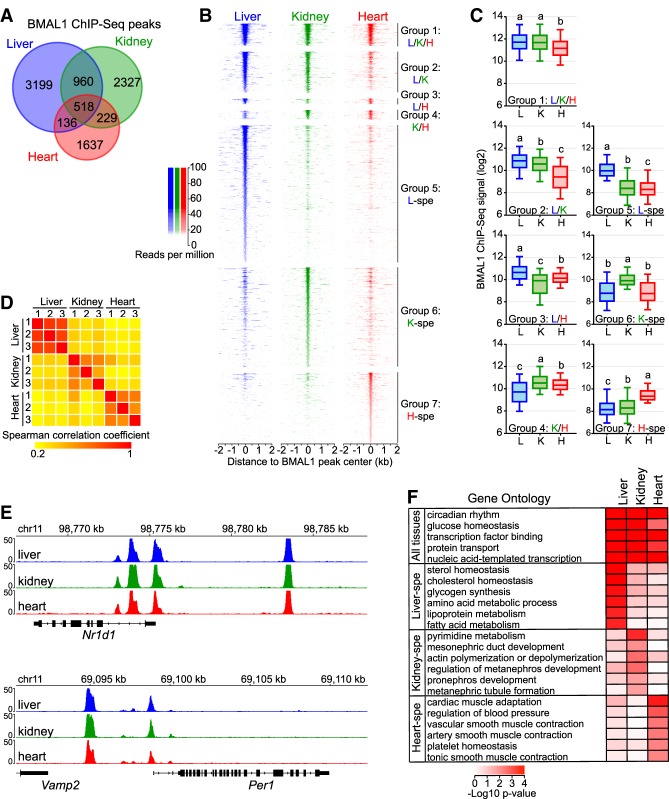
To determine whether BMAL1 cistrome differs between tissues, we performed BMAL1 ChIP-seq in the mouse liver, kidney, and heart with three biological replicates per tissue. Because CLOCK:BMAL1 DNA-binding sites in the liver are virtually identical over the course of the day and only exhibit homogenous differences in DNA-binding strength (Rey et al. 2011; Koike et al. 2012), we performed BMAL1 ChIP at the time of maximal binding; i.e., Zeitgeber time 6 (ZT6; ZT0 is defined at the light on signal) (Supplemental Fig. S1A). Identification of BMAL1 DNA-binding sites using HOMER software (Heinz et al. 2010) revealed that most BMAL1 peaks were tissue-specific, with <6% of BMAL1 peaks common to the liver, kidney, and heart (Fig. 1A,B; Supplemental Table S1). Closer inspection revealed that tissue-specific BMAL1 peaks were more often associated with BMAL1 signal below peak detection threshold in the other tissues rather than a complete absence of signal (Fig. 1B; Supplemental Fig. S1B; see below). In addition, quantification of BMAL1 ChIP-seq signal revealed that stronger BMAL1 binding is associated with peaks common to more than one tissue (Fig. 1C; see below).
Figure 1.
BMAL1 cistromes are largely tissue specific. (A) Overlap of BMAL1 ChIP-seq peaks in the mouse liver, kidney, and heart. (B) BMAL1 ChIP-seq signal at BMAL1 peak center ±2 kb in the mouse liver, kidney, and heart, parsed based on tissues in which BMAL1 peaks were detected. (Group 1) Peaks common to all three tissues; (group 2) peaks common to the liver and kidney; (group 3) peaks common to the liver and heart; (group 4) peaks common to the kidney and heart; (group 5) liver-specific peaks; (group 6) kidney-specific peaks; (group 7) heart-specific peaks. (C) BMAL1 ChIP-seq signal calculated at peak center ±250 base pairs (bp) in the mouse liver, kidney, and heart. Groups with different letters are statistically different. P < 0.05, Kruskal-Wallis test. (D) Spearman correlation coefficients of BMAL1 ChIP-seq signal between each biological replicate (n = 3 per tissue), calculated at all 9006 BMAL1 ChIP-seq peaks. (E) Genome browser view of BMAL1 ChIP-seq signal at Nr1d1 and Per1 gene loci. (F) Gene ontology analysis performed using BMAL1 peaks detected in the liver, kidney, or heart, and reporting functions enriched across the three tissues or in only one tissue. P-value < 0.05.
To ensure that differences in BMAL1 ChIP-seq signal between tissues represent true biological variation, we compared BMAL1 ChIP-seq signal between each of the nine data sets and found a high degree of correlation between samples originating from the same tissue but not between tissues (Fig. 1D). Interreplicates variation between replicates was higher for the kidney than for the liver and heart; however, this might reflect the higher cell type heterogeneity and the variability in the chromatin accessibility landscape that are observed in the kidney when compared with the heart and liver (Cusanovich et al. 2018). We also found that BMAL1 ChIP-seq signal at core clock genes is nearly identical between all three tissues, further indicating that differences in BMAL1-binding signal between tissues stem from biological variations (Fig. 1E). Consistent with this finding, gene ontology analysis of BMAL1 target genes for each tissue revealed that while circadian rhythm and other general terms are enriched in all three tissues, tissue-specific biological processes are enriched in a tissue-specific manner (Fig. 1F). Many of these tissue-specific processes have been shown to be regulated by the circadian clock, such as glycogen synthesis, lipid metabolism, and cholesterol homeostasis in the liver (Doi et al. 2010; Gnocchi et al. 2015; Ma et al. 2015) and blood pressure and vascular smooth muscle contraction in the heart (Wang et al. 2008; Xie et al. 2015). Together, these results indicate that BMAL1 predominantly binds DNA in a tissue-specific manner and targets genes involved in the regulation of tissue-specific biological functions.
Chromatin accessibility contributes to tissue-specific BMAL1 DNA binding
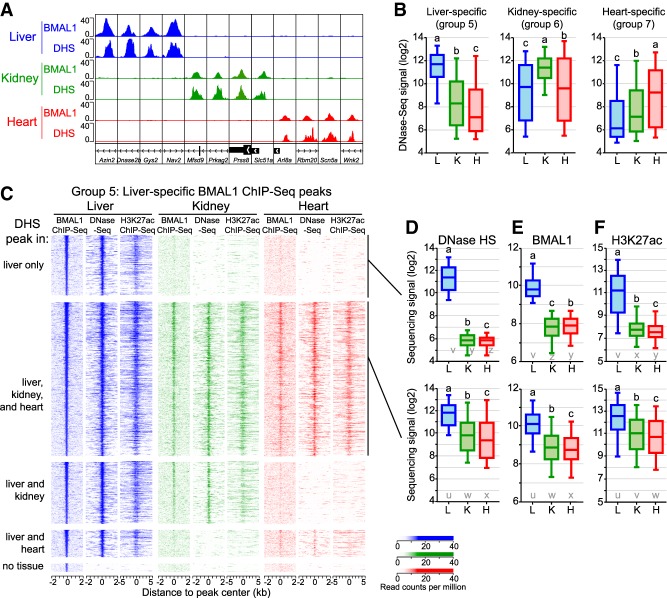
To understand the large differences in BMAL1 ChIP-seq signal between tissues, we set out to define the mechanisms that enable BMAL1 to bind DNA in a tissue-specific manner. Characterization of the chromatin landscape by the ENCODE consortium and others revealed that 94.4% of TF ChIP-seq peaks are located in an accessible chromatin region; i.e., a region that is hypersensitive to DNase I (Thurman et al. 2012; Yue et al. 2014). Because mouse liver CLOCK:BMAL1 DNA-binding sites are located predominantly in DHSs (Sobel et al. 2017; Trott and Menet 2018), we hypothesized that tissue-specific BMAL1 binding may be, at least in part, caused by differences in DNA accessibility between the liver, kidney, and heart. To test our hypothesis, we used DNase sequencing (DNase-seq) data sets from the mouse ENCODE project (Vierstra et al. 2014; Yue et al. 2014) and found that many tissue-specific BMAL1 peaks are located in a chromatin region that is accessible only in the corresponding tissue (Fig. 2A). Quantification of DNase-seq signal at liver-, kidney-, and heart-specific BMAL1 peaks (named groups 5, 6, and 7 here, respectively) confirmed that tissue-specific BMAL1 peaks are associated with higher DNase-seq signal (Fig. 2B). However, some DNase-seq signal could be detected above background level in all three tissues, suggesting that tissue-specific BMAL1 peaks may not be always associated with a complete absence of DNase-seq signal in other tissues.
Figure 2.
The chromatin environment shapes tissue-specific BMAL1 binding. (A) Genome browser view of BMAL1 ChIP-seq and DNase-seq signals in the mouse liver, kidney, and heart at 12 BMAL1 tissue-specific peaks. (B) DNase-seq signal calculated at BMAL1 peak center ±250 bp in the mouse liver, kidney, and heart for tissue-specific BMAL1 peaks. Groups with different letters are statistically different. P < 0.05, Kruskal-Wallis test. (C) BMAL1 ChIP-seq, DNase-seq, and H3K27ac ChIP-seq signal at liver-specific BMAL1 peaks, parsed based on the presence of a DHS peak in the liver, kidney, and heart. BMAL1 ChIP-seq and DNase-seq signals are displayed with a window of ±2 kb, whereas H3K27ac ChIP-seq signal is displayed with a window of ±5 kb. (D–F) Quantification of DNase-seq (D), BMAL1 ChIP-seq (E), and H3K27ac ChIP-seq (F) signals for liver-specific BMAL1 peaks located at (group 5A; top) liver-specific DHS or (group 5B; bottom) DHS peaks common to the liver, kidney, and heart. Groups with different letters are statistically different. P < 0.05, Kruskal-Wallis test. u–z denote the outcome of the statistical analysis performed using groups 5A and 5B together.
To investigate whether tissue-specific BMAL1 binding occurs at DHSs common to several tissues, we mapped the genomic location of DHSs in the liver, kidney, and heart (Supplemental Fig. S2A), and determined the fraction of tissue-specific BMAL1 peaks found at tissue-specific DHSs or at DHSs common to more than one tissue, focusing primarily on liver-specific BMAL1 peaks. As expected, almost all mouse liver BMAL1 peaks (97.7%) are located in chromatin regions that are accessible in the mouse liver (Fig. 2C). However, only ∼20% of liver-specific BMAL1 peaks are located at a liver-specific DHSs; i.e., most liver-specific BMAL1 peaks are located within chromatin regions that are also accessible in other tissues (Fig. 2C). Importantly, we found that while no BMAL1 signal is detectable in the kidney and heart at liver-specific DHSs, some BMAL1 signal is visible at common DHSs even if it is not defined as significant BMAL1 peaks by HOMER (Fig. 2C–E). Similar results were observed for kidney- and heart-specific BMAL1 peaks (Supplemental Fig. S2B,C). Because DNase I hypersensitivity does not assess the activity of accessible chromatin regions, we also analyzed public ChIP-seq data sets for histone modifications found at transcriptionally active/primed DHSs; i.e, acetylated Lys27 of histone 3 (H3K27ac) and monomethylated Lys4 of histone 3 (H3K4me1). Remarkably, both H3K27ac and H3K4me1 ChIP-seq signals mirrored the DNase-seq signal and were detected in particular at kidney and heart DHSs exhibiting liver-specific BMAL1 peaks (Fig. 2C,F; Supplemental Fig. S2D).
In summary, our data indicate that only a minority of BMAL1-binding sites are located at tissue-specific DHSs. For most tissue-specific BMAL1 peaks, chromatin is accessible in other tissues and harbors histone modifications that mark transcriptionally active DHSs, albeit to lower levels. Importantly, some BMAL1 DNA binding can be detected at those sites, although at levels too low to be categorized as ChIP-seq peaks by peak-calling algorithms.
Tissue-specific BMAL1 peaks are enriched for ts-TF DNA-binding motifs
Several mechanisms may contribute to tissue-specific BMAL1 binding. First, BMAL1 DNA binding may only rely on the presence of its binding motif in an accessible DHS. Under this scenario, BMAL1-binding signal would correlate with DNase-seq signal, and differences between tissues would mostly depend on the proportion of cells harboring an accessible DHS and/or the extent to which the DHS is accessible. Alternatively, tissue-specific BMAL1 binding may be, as suggested for some other TFs (Hu and Gallo 2010; Gertz et al. 2013; Meireles-Filho et al. 2014), facilitated by ts-TFs that bind at sites nearby BMAL1. Here, BMAL1 DNA-binding signal would not correlate well with DNase-seq signal, and motifs for ts-TFs would be enriched at tissue-specific BMAL1 peaks.
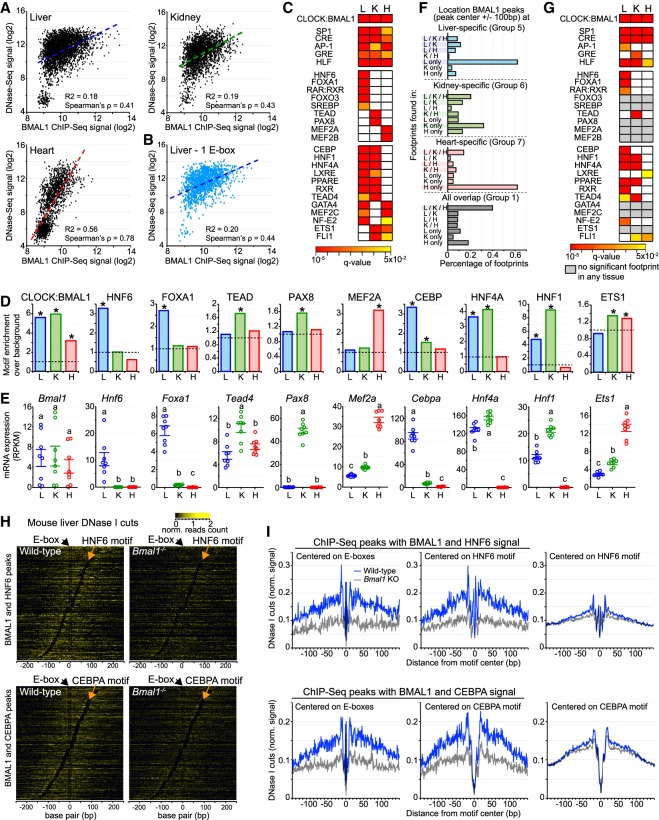
To assess whether either or both of these mechanisms promote tissue-specific BMAL1 binding, we first compared BMAL1 ChIP-seq signal with DNase-seq signal at liver-, kidney-, and heart-specific BMAL1 peaks and found that both signals were correlated in the heart but not in the liver and kidney (Fig. 3A). Differences in BMAL1 ChIP-seq signal of >16-fold are detected at liver DHSs displaying similar DNase-seq signals, and similar BMAL1 ChIP-seq signals were observed at DHSs harboring differences of DNase-seq signal of >30-fold in both the liver and kidney (Fig. 3A). Importantly, these differences are independent of the number of E-boxes within each DHS (Fig. 3B; Supplemental Fig. S3A). These results thus indicate that, at least for the liver and kidney, the extent to which the chromatin is accessible cannot solely explain the differences in tissue-specific BMAL1 binding.
Figure 3.
Motifs and footprints for ts-TFs are enriched at tissue-specific BMAL1 enhancers. (A) Correlation between DNase-seq and BMAL1 ChIP-seq signals (calculated at BMAL1 peak center ±250 bp) at liver-, kidney-, and heart-specific BMAL1 peaks. (B) Correlation between DNase-seq and BMAL1 ChIP-seq signal for liver-specific BMAL1 peaks harboring one E-box only. (C) Enrichment of TF motifs at liver-, kidney-, and heart-specific BMAL1 peaks, performed using HOMER at BMAL1 peak center ±100 bp. Enrichments are shown if q-value was <0.05. (D) Fold-enrichment of TF motif over background at tissue-specific BMAL1 peaks (BMAL1 peak center ±100 bp; q-value < 0.05). (E) mRNA expression in the mouse liver, kidney, and heart of Bmal1 and TFs whose motifs were enriched at BMAL1 ChIP-seq peaks. (RNA-seq data sets from Zhang et al. 2014). Groups with different letters are significantly different. P-value < 0.05, one-way ANOVA. (F) Distribution of DNase I footprints identified using pyDNase and detected at BMAL1 peak center ±100 bp, and parsed based on the tissues in which they were found. (G) Motif enrichment of TFs performed at DNase I footprints identified in liver-, kidney-, and heart-specific BMAL1 peaks (footprint center ±15 bp). Enrichments are displayed if q-value was <0.05, and colored in gray if no significant footprint is detected in any of the three tissues. (H) Heat map representing DNase I cuts at BMAL1 peaks containing an E-box and an HNF6 motif (top) or a CEBP (bottom). DNase I cut signal is centered on the E-box and sorted based on the distance between the E-box and the liver-specific TF motif. (I) Quantification of DNase I cut signal at BMAL1 peaks containing an E-box and an HNF6 motif (top) or a CEBP (bottom) in the liver of wild-type (left) or Bmal1−/− (right) mice. Quantification was performed using signal centered on E-boxes (left) or on the ts-TF motif (middle). As control, quantification was also performed at HNF6 or CEBPA peaks that do not harbor a BMAL1 ChIP-seq peak (right; signal corresponds to the average of 1000 decile-normalized iterations).
To determine whether ts-TFs may be involved, we performed a motif enrichment analysis at tissue-specific BMAL1 peaks using HOMER. As expected, E-boxes are enriched at tissue-specific BMAL1 peaks for all three tissues, along with a few additional motifs for ubiquitously expressed TFs (u-TFs) (Fig. 3C,D; Supplemental Table S2). However, the majority of the motifs were significantly enriched in only one or two tissues, with many of them corresponding to binding sites of well-known ts-TFs like Foxa1 and Mef2a/b (Fig. 3C,D). To verify that these TFs are legitimate ts-TFs, we analyzed their levels of expression in the liver, kidney, and heart using public mouse RNA-seq data sets (Zhang et al. 2014) and the Genotype Tissue Expression (GTEx) portal for human tissues (The GTEx Consortium 2013). For almost all of the >20 TFs that we examined, mRNA levels were specific to one or two tissues, confirming that these TFs are true ts-TFs (Fig 3E; Supplemental Fig. S3B,C). Importantly, the tissue-specific pattern of expression matched particularly well with the tissue specificity of the motif enrichment, suggesting that these TFs bind DNA in a tissue-specific manner.
Genomic footprints for ts-TFs are specifically enriched at tissue-specific BMAL1 peaks
The concordance between ts-TF motif enrichment and expression pattern (Fig. 3C–E; Supplemental Fig. S3B,C) does not directly show that ts-TFs bind at BMAL1 DHSs in a tissue-specific manner. To further investigate this possibility, we took advantage of the genomic footprints left by chromatin-associated proteins following treatment with DNase I (Hesselberth et al. 2009). This analysis, which has been used successfully to define the genome-wide DNA-binding profiles of dozens of TFs (Neph et al. 2012; Sherwood et al. 2014; Stergachis et al. 2014), exploits DNase-seq data sets to uncover regions within DHSs that are protected from DNase I activity by proteins bound to DNA (Supplemental Fig. S3D,E). To this end, we analyzed the mouse liver, kidney, and heart DNase-seq data sets using pyDNase (Piper et al. 2015) and identified the genome-wide location of TF footprints.
This analysis revealed that most footprints found at tissue-specific BMAL1 peaks are significantly enriched in the corresponding tissue. For example, 60% of the footprints found at liver-specific and heart-specific BMAL1 peaks are only detected in the liver and heart, respectively (Fig. 3F; Supplemental Fig. S3F). Conversely, a large fraction of the footprints identified at common BMAL1 peaks are detected in all three tissues (Fig. 3F; Supplemental Fig. S3F). To identify the proteins generating these footprints, we performed a motif analysis restricted to the footprint genomic locations found at liver-, kidney- and heart-specific BMAL1 peaks. While motifs for CLOCK:BMAL1 and some u-TFs are enriched in all three tissues, most significantly enriched motifs correspond to ts-TFs binding sites (Fig. 3G; Supplemental Fig. S3G; Supplemental Table S3). For example, footprints are enriched at motifs for Hnf6, Foxa1, Cebp, Hnf1, and Hnf4a in the mouse liver, and for Hnf1, Hnf4a, and Tead in the kidney. We noticed that several ts-TFs whose motifs are enriched at BMAL1 DHSs (Fig. 3C) do not exhibit a significant footprint (e.g., Foxo3, Srebp, Pax8, Gata4, and Ets1) (Fig. 3G), suggesting that they do not bind to BMAL1 DHSs despite the presence of a cognate motif. However, the absence of footprints may also reflect the inability of these TFs to generate distinctive footprints because of DNA residence time and DNase I cut bias, as shown previously for a few other TFs (He et al. 2014; Sung et al. 2014).
Using heat maps, we then examined DNase I cuts at BMAL1 peaks and found that DHSs exhibit footprints at E-box and motifs for ts-TFs, suggesting that ts-TFs may be bound to DNA concurrently with CLOCK:BMAL1 (Fig. 3H; Supplemental Fig. S3H). Interestingly, quantification of DNase I cut signal in wild-type versus Bmal1−/− mice revealed that Bmal1 knockout decreases footprint signal for the ts-TFs HNF6 and CEPBA more potently at BMAL1 peaks than at control peaks (i.e., HNF6 and CEPBA peaks without BMAL1 ChIP-seq signal), suggesting that BMAL1 may increase the binding of ts-TFs at tissue-specific DHSs (Fig. 3I).
Tissue-specific and common BMAL1 peaks are cobound by different classes of TFs
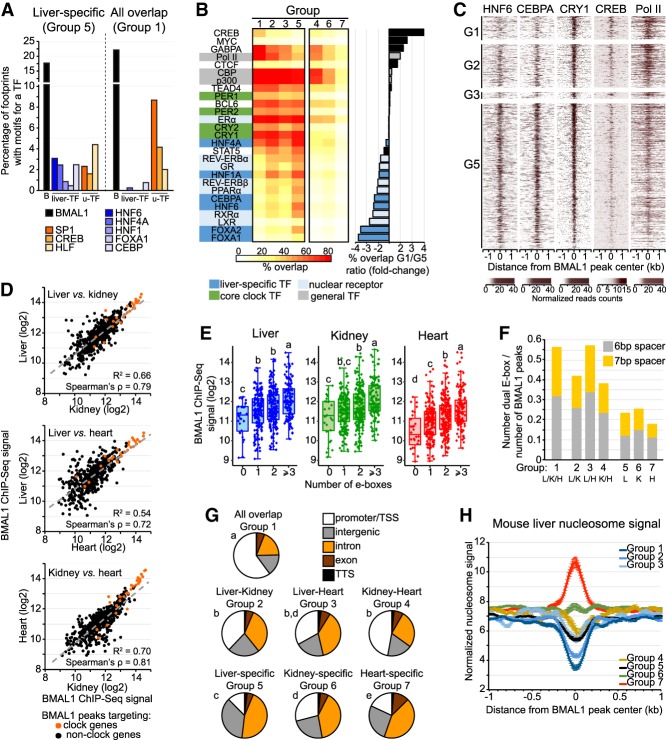
To further assess whether ts-TFs may contribute to tissue-specific BMAL1 binding, we determined the relative enrichment of footprints for several ts-TFs and u-TFs at BMAL1 peaks either specific to the liver (group 5) or common to all three tissues (group 1). We found that footprints for ts-TFs were preferentially detected at liver-specific BMAL1 peaks when compared with common BMAL1 peaks (Fig. 4A; Supplemental Fig. S4A). Conversely, footprints for several u-TFs were enriched at BMAL1 peaks common to all three tissues, when compared with liver-, kidney-, or heart-specific BMAL1 peaks (Fig. 4A; Supplemental Fig. S4A). Differences in footprint enrichment between ts-TFs and u-TFs were also observed at liver-specific BMAL1 peaks based on whether DHS are liver-specific or ubiquitous, respectively (Supplemental Fig. S4B). These results thus suggest that ts-TFs are more effectively recruited to tissue-specific BMAL1 peaks, whereas u-TFs may be more effectively recruited to BMAL1 peaks common to different tissues.
Figure 4.
BMAL1 DNA-binding sites common to the mouse liver, kidney, and heart exhibit unique features. (A) Percentage of footprints for BMAL1 (black), liver-specific TFs (blue), and u-TFs (orange) identified at liver-specific BMAL1 peaks (group 5) or at BMAL1 peaks common to all three tissues (group 1). (B) Percentage overlap between BMAL1 peaks and ChIP-seq peaks for various TFs and Pol II in the mouse liver. Results are parsed based on tissues in which BMAL1 peaks were detected. (G1) Peaks common to all three tissues; (G2) peaks common to the liver and kidney; (G3) peaks common to the liver and heart; (G4) peaks common to the kidney and heart; (G5) liver-specific peaks; (G6) kidney-specific peaks; (G7) heart-specific peaks. (C) Heat map representation of mouse liver ChIP-seq signal for HNF6 (data set from Faure et al. 2012), CEBPA (data set from Faure et al. 2012) CRY1 (CT04) (data set from Koike et al. 2012), CREB (from Everett et al. 2013), and Pol II (ZT06) (data set from Sobel et al. 2017) at BMAL1 peaks ordered based on BMAL1 ChIP-seq signal (as in Fig. 1B) for groups 1–3, and 5. (D) Correlation of BMAL1 ChIP-seq signal between the mouse liver, kidney, and heart for BMAL1 peaks common to all three tissues (group 1). (E) BMAL1 ChIP-seq signal for BMAL1 peaks common to all tissues parsed based on the number of E-boxes (canonical E-box CACGTG and degenerated E-boxes CACGTT and CACGNG). (F) Fraction of BMAL1 peaks harboring a dual E-box motif (two E-boxes separated with 6 or 7 bp). (G) Location of BMAL1 peaks for each of the seven groups. Statistical analysis was performed using a χ2 test, and post-hoc analysis was performed with Fisher's exact test. Groups with different letters are statistically different. P < 0.01. (H) Nucleosome signal at BMAL1 DNA-binding sites parsed based on tissues in which BMAL1 peaks were detected (groups 1–7; see above). Nucleosome signal was retrieved from mouse liver MNase-seq (micrococcal nuclease [MNase] digestion of chromatin followed by deep sequencing) data sets (Menet et al. 2014), which consists of six time points, each separated by 4 h with n = 4 mice for each time point, and is displayed as the average of the 24 data sets ± S.E.M.
To address this possibility more directly, we determined the relative distribution of TF ChIP-seq peaks between tissue-specific and common BMAL1 peaks using public mouse liver ChIP-seq data sets for >25 TFs. Results largely confirmed the footprint analysis, as ts-TF ChIP-seq peaks were more potently found at liver-specific BMAL1 peaks than at peaks common to all three tissues (Fig. 4B,C; Supplemental Fig. S4C). Moreover, ChIP-seq peaks for RNA polymerase II (Pol II) and the u-TFs CREB and GABPA were preferentially observed at common BMAL1 peaks. Intriguingly, we noticed that liver-specific BMAL1 peaks were also more frequently associated with ChIP-seq for nuclear receptors (Fig. 4B,C). Taken together, these results thus indicate that BMAL1 peaks harbor a remarkable dissimilarity of TF bound near BMAL1, with ts-TFs and nuclear receptors being enriched at tissue-specific BMAL1 peaks while Pol II and some u-TFs being enriched at BMAL1 peaks common to different tissues.
BMAL1 peaks common to all three tissues exhibit stronger BMAL1 signal, harbor more E-boxes, are enriched at promoters, and display less nucleosome signal
More than 500 BMAL1 ChIP-seq peaks targeting 294 genes are common to the liver, kidney, and heart (Fig. 1A). These genes comprise all expected core clock genes and regulate biological processes that are ubiquitous to most mammalian tissues (Supplemental Fig. S4D). Consistent with the hypothesis that BMAL1 binding at peaks common to several tissues is less dependent on ts-TFs than tissue-specific BMAL1 peaks, we found that BMAL1 ChIP-seq signal at common BMAL1 peaks is well correlated with DNase-seq signal (Supplemental Fig. S4E) and between tissues (Fig. 4D). Analysis of the DNA-binding motifs at common BMAL1 peaks revealed that E-boxes were enriched along with motifs for the clock genes Rev-erbα/β and Rorα/β/γ (RORE) and E4bp4/Dbp/Tef/Hlf (D-box) as well as motifs for well-characterized u-TFs such as CREB, SP1, and HLF (Supplemental Fig. S4F,G). The reasons why REV-ERBα/β DNA binding is lower at common BMAL1 peaks (Fig. 4B,C; Supplemental Fig. S4C) in spite of RORE motif enrichment are unknown but may include the recently described recruitment of REV-ERBα to DNA-bound ts-TFs like HNF6 (Zhang et al. 2015). To investigate whether the higher enrichment for E-boxes at common BMAL1 peaks (Supplemental Fig. S4F) may contribute to the increased BMAL1 ChIP-seq signal at those sites (Fig. 1C), we quantified BMAL1 ChIP-seq signal based on the number of E-boxes and found that BMAL1 signal was positively correlated with an increasing number of E-boxes for all three tissues (Fig. 4E). Moreover, dual E-boxes—i.e., two E-box motifs separated by 6 or 7 nucleotides and which have been proposed to be a preferred CLOCK:BMAL1-binding site (Paquet et al. 2008)—are also more enriched at common BMAL1 peaks (Fig. 4F). Characterization of the location of BMAL1 peaks revealed that peaks common to all three tissues were predominantly located at promoter/transcription start site (TSS), whereas tissue-specific BMAL1 peaks were more consistently located within introns and intergenic regions (Fig. 4G). Finally, nucleosome signal at common BMAL1 peaks was lower compared with other groups (Fig. 4H; Supplemental Fig. S4H). This result, which is likely due to differences in BMAL1 peak location between groups, suggests that DNA is more accessible at peaks common to all three tissues.
Together, these results indicate that BMAL1 peaks common to all three tissues, which target genes that are involved in generic biological processes, are enriched for E-boxes, are predominantly localized at promoter regions/TSS, and exhibit less nucleosome signal. These features likely contribute to the higher BMAL1 ChIP-seq signal observed at those sites.
BMAL1 DNA binding contributes only partially to rhythmic gene expression
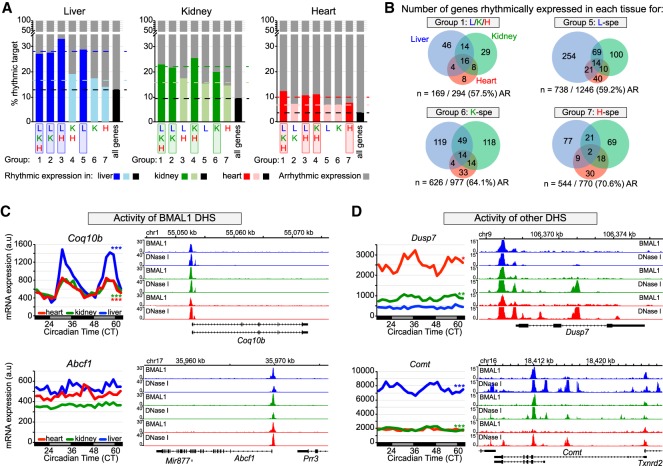
To determine whether tissue-specific BMAL1 binding contributes to generating tissue-specific circadian transcription, we analyzed BMAL1 target gene expression in the mouse liver, kidney, and heart using public data sets that assessed the rhythmic transcriptome in those tissues (Zhang et al. 2014). In agreement with the literature (Rey et al. 2011; Menet et al. 2012), genes targeted by BMAL1 exhibit a higher proportion of rhythmic mRNA expression than those not targeted by BMAL1. Specifically, rhythmic expression of BMAL1 targets in the liver, kidney, and heart is increased by approximately twofold compared with all genes and by ∼1.3-fold to 1.5-fold compared with genes targeted by BMAL1 in other tissues (Fig. 5A). Surprisingly, genes targeted by BMAL1 in all three tissues (group 1) do not exhibit substantially more rhythmic expression despite increased BMAL1 ChIP-seq signal, increased number of E-boxes, and preferential peak location at promoter/TSS (Fig. 5A). Moreover, the majority of BMAL1 target genes are not rhythmically expressed in any of the three tissues (Fig. 5A). These results thus suggest that although BMAL1 DNA binding contributes to increased rhythmic expression, it is not sufficient to drive rhythmic transcription.
Figure 5.
Transcriptional activities of BMAL1 DHS and other DHS contribute to BMAL1-mediated rhythmic transcription. (A) Percentage of rhythmically expressed gene for the seven groups of BMAL1-binding sites. Gene expression data originate from public microarray data sets (Zhang et al. 2014) and is considered rhythmic if q-value was <0.05 using JTK-cycle. (B) Overlap of rhythmic expression for peaks common to all three tissues (group 1), and for liver-, kidney-, and heart-specific BMAL1 peaks (groups 5, 6, and 7, respectively). The number of genes that are rhythmically expressed for the mouse liver (blue), kidney (green), and heart (red) is displayed, along with the number and percentage of nonrhythmically expressed genes (AR). (C,D) mRNA expression (left) and genome browser view (right) of BMAL1 ChIP-seq and DNase-seq signals in the mouse liver (blue), kidney (green), and heart (red). Rhythmic expression determined by JTK cycle is defined as three asterisks if q-value was <0.001, two asterisks if q-value was <0.01, and one asterisk if q-value was <0.05. The genes Coq10b and Abcf1 (C) represent two examples for which the activity of BMAL1 DHS likely contributes to the differences in mRNA expression, where the genes Dusp7 and Comt (D) represent two examples for which the activity of other DHS likely contributes to BMAL1-mediated rhythmic transcription.
Consistent with this notion, BMAL1 targets common to all three tissues display a widely heterogeneous rhythmic output, with most genes being rhythmic in only one tissue (83 genes out of 125 rhythmically expressed genes) (Fig. 5B). In fact, only 16 genes targeted by common BMAL1 peaks are rhythmically expressed in all three tissues, and most of them are core clock genes. We also found that many genes targeted by BMAL1 in one tissue were rhythmic only in another tissue (Fig. 5B). To characterize the mechanisms underlying this poor overlap between BMAL1 binding and rhythmic gene expression, we examined the profiles of DNase-seq and BMAL1 ChIP-seq data sets in the liver, kidney, and heart. While we were unable to detect a unified mechanism that may explain the variability in BMAL1 transcriptional output, we identified two important features that likely contribute to rhythmic transcription. First, similar BMAL1 ChIP-seq signal between tissues can result in significantly different levels and/or rhythmicity of target gene expression (Fig. 5C; Supplemental Fig. S5A). This observation was made at genes targeted by a unique DHS, suggesting that factors other than just BMAL1 contribute to the activity of BMAL1-bound DHSs and to rhythmic BMAL1 target gene expression. Second, we found that rhythmicity of BMAL1 target genes was often associated with higher levels of expression and more DHSs, thus suggesting that rhythmic gene expression results from functional interactions between BMAL1 DHSs and other DHSs (Fig. 5D; Supplemental Fig. S5B). We also found a few instances where more DHSs were associated with decreased expression, suggesting that interactions between multiple DHSs may also result in decreased transcription activation (Supplemental Fig. S6C).
In summary, our results indicate that, as reported recently (Trott and Menet 2018), BMAL1 DNA binding is not sufficient to drive rhythmic transcription. They also suggest that rhythmic BMAL1 target gene expression likely results from (1) interactions between DHSs bound by BMAL1 and other DHSs and (2) the activity of each DHS, including those bound by BMAL1.
BMAL1-bound DHSs physically interact with other DHSs
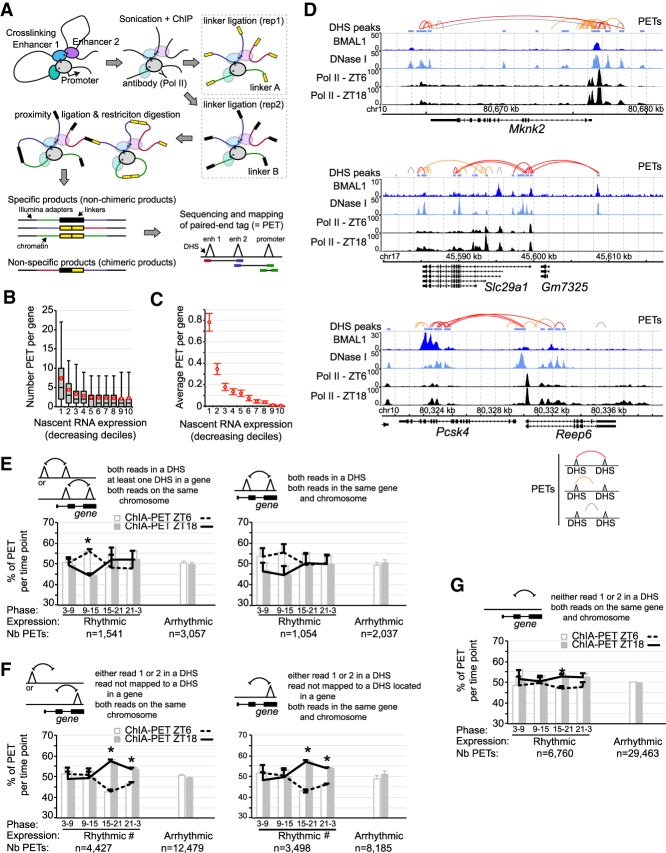
To determine the extent to which BMAL1 target gene transcription relies on physical interactions between DHSs, we performed RNA Pol II chromatin interaction analysis by paired-end tag (ChIA-PET) sequencing in the mouse liver at ZT6 and ZT18 with three biological replicates per time point. By incorporating a Pol II ChIP step, this technique identifies interactions between enhancers and/or TSS of genes that are being transcribed (Fig. 6A; Li et al. 2012; Kieffer-Kwon et al. 2013).
Figure 6.
Analysis of chromatin interactions by RNA Pol II ChIA-PET in the mouse liver. (A) Illustration of the ChIA-PET technique. (B) Number of PETs with both reads mapped to the same gene and parsed based on gene nascent RNA expression in the mouse liver. Red circles represent the average PET number for each decile ±95% confidence intervals. (C) Average number of PETs per gene in the mouse liver with both reads located in different DHS and mapped to the same gene, parsed based on gene nascent RNA expression. Error bars represent the 95% confidence intervals. (D) Genome browser view of mouse liver BMAL1 ChIP-seq (this study), DNase-seq (ENCODE), and Pol II ChIP-seq (Sobel et al. 2017). PETs with both reads mapped to DHS are in red, while PETs with one read mapped to a DHS are in orange and those not mapped to a DHS are in gray. (E–G) Percentage of PETs detected at ZT6 (white bar and dashed black line) or ZT18 (gray bar and solid black line), displayed as the average ± S.E.M. of three independent experiments based on the type of PETs, the rhythmicity of gene expression, and the phase of expression. (*) P < 0.05 between ZT6 and ZT18; (#) an interaction found in the two-way ANOVA (P < 0.05) between the phase of expression and the time of the ChIA-PET experiment (ZT6 or ZT18). Triangles represent DHSs and may be located within a gene or not.
To assess the efficiency of our mouse liver Pol II ChIA-PET in detecting chromatin interactions at transcriptionally active genes, we determined the number of PETs mapped to a single gene based on gene transcription using public mouse liver Nascent-seq (genome-wide sequencing of nascent RNA) data sets (Menet et al. 2012). We found that most PETs mapped to transcriptionally active genes, with 23.4% of all PETs located within the top 10% of genes transcribed in the mouse liver (Fig. 6B; Supplemental Fig. S7A; Supplemental Table S4). Analysis of PETs with reads located in two different DHSs and visualization of chromatin interactions at mouse liver BMAL1 target genes further confirmed that actively transcribed genes harbor many PETs mapping to different DHSs, including some bound by BMAL1 (Fig. 6C,D). These data indicate that DHSs located within the same gene physically interact via gene looping, and suggest that transcriptional activation results from the synergistic interaction between DHSs. To assay whether rhythmic gene expression is associated with changes in chromatin interactions, we determined the number of PETs between DHSs based on the phase of rhythmic gene expression. We found small day versus night differences in chromatin interactions in the mouse liver, with ∼10% more PETs between two DHSs at ZT6 for genes with a peak of expression at ZT9-15 (Fig. 6E). In addition, analysis of interactions between a DHS and a non-DHS site; e.g., a site with elongating Pol II that is expected to better reflect transcriptional activity revealed that DHS–non-DHS interactions exhibit day versus night differences, with more interactions at ZT18 for genes expressed from ZT15 to ZT3 (Fig. 6F,G).
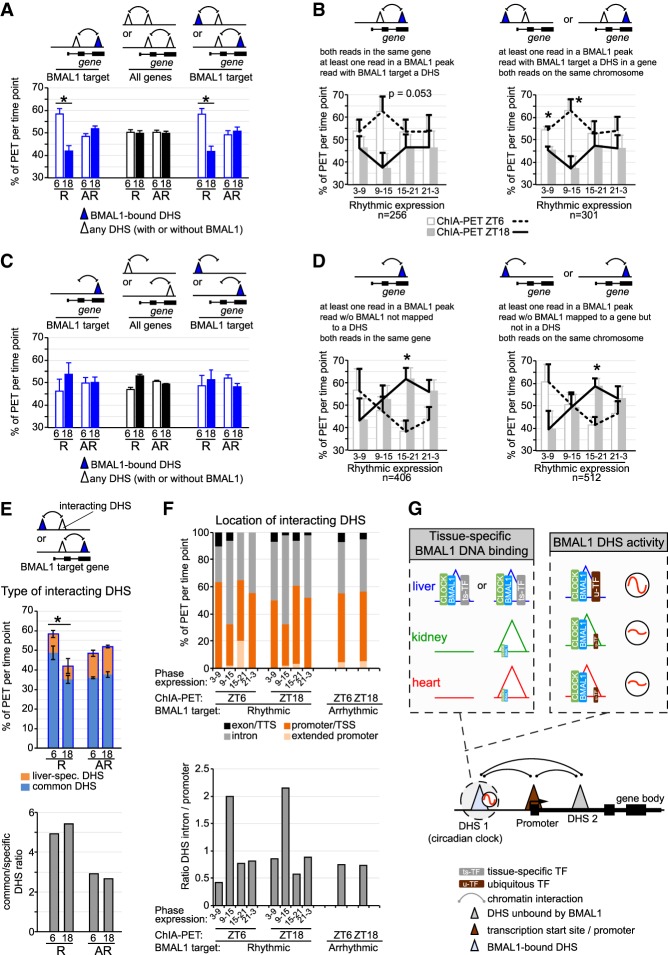
A few recent reports suggested that rhythmic interactions between BMAL1 DHSs and other DHSs underlie the rhythmic transcription of BMAL1 target genes (Aguilar-Arnal et al. 2013; Mermet et al. 2018; Yeung et al. 2018). To investigate this possibility, we quantified the number of PETs mapped to BMAL1 target genes. We found that at rhythmically expressed BMAL1 target genes the number of chromatin interactions between two DHSs containing at least one BMAL1 peak is higher at ZT6 when BMAL1 binding is maximal (Fig. 7A,B). This higher number of PETs at ZT6 versus ZT18 was specifically observed for rhythmically expressed targets peaking at ZT3–ZT15 (i.e., at times corresponding relatively well with BMAL1 DNA binding) and after incorporation of chromatin interactions involving distal BMAL1-bound DHSs, which account for ∼15%–20% of DHS–DHS interactions (Fig. 7A,B). In addition, analysis of interactions between a BMAL1 peak and a non-DHS site revealed that while the proportion of PETs between ZT6 and ZT18 was similar for rhythmically and arrhythmically expressed targets, more PETs were found at ZT18 for the target genes rhythmically expressed at a similar phase (Fig. 7C,D).
Figure 7.
Rhythmic chromatin interactions are more prevalent for rhythmically expressed genes. (A–D) Percentage of PETs detected at ZT6 (empty/white bar and dashed black line) or ZT18 (solid bar and solid black line) are displayed as the average ± S.E.M. of three independent experiments based on the type of PET, the rhythmicity of gene expression, the presence of a BMAL1-bound DHS, and the phase of gene expression. R = rhythmic expression; AR = arrhythmic expression. Triangles represent DHSs, and may be located within a gene or not. (*) P < 0.05 between ZT6 and ZT18. (E) Type of DHS (liver-specific DHS or DHS common to the liver, kidney, and heart) interacting with a BMAL1 DHS is displayed based on the transcriptional output of BMAL1 target genes (rhythmic or nonrhythmic) and the time at which the ChIA-PET experiment was performed (ZT6 or ZT18). Results are shown as percentage of PETs per ChIA-PET time point (as in A; top) or as the ratio of common DHS over liver-specific DHS (bottom). (F) Genomic location of DHSs interacting with BMAL1-bound DHS is displayed based on the transcriptional output of BMAL1 target genes (phase of expression, and rhythmic or nonrhythmic) and the time at which the ChIA-PET experiment was performed (ZT6 or ZT18). The locations of DHSs consist of exon and transcription termination site (TTS; in black), introns (in gray), promoter and TSS (−1 kb to +100 bp from TSS; in dark orange), and extended promoter (−10 kb to −1 kb from TSS; in light orange). (G) Hypothetical model illustrating how CLOCK:BMAL1 generates tissue-specific rhythmic transcriptional programs. This model incorporates mechanisms on how BMAL1 binds to DNA in a tissue-specific manner (chromatin accessibility and cobinding with ts-TFs) and how u-TFs might contribute to BMAL1 DHS rhythmic transcriptional activity. It also illustrates how functional interaction between BMAL1 DHSs and other DHSs (including tissue-specific DHS) may contribute to rhythmic transcription.
To further characterize chromatin interactions between BMAL1-bound DHSs and other DHSs, we analyzed the type of DHSs with which BMAL1-bound DHSs interact (i.e., liver-specific DHS or DHS common to the kidney and/or heart) as well as their genomic location (i.e., promoter, intron, etc.). We found that interactions between BMAL1-bound DHSs and DHSs common to several tissues were enriched for rhythmically expressed genes (Fig. 7E), and that interactions with DHSs located in introns were enriched for rhythmically expressed genes with a peak of expression at ZT9–ZT15; i.e., a few hours after maximal BMAL1 DNA binding (Fig. 7F). Future experiments will be required to determine whether these differences in the type and location of chromatin interactions are relevant for BMAL1-mediated rhythmic gene expression.
Taken together, these results indicate that BMAL1-bound DHSs interacts with other DHSs, and that these interactions are more prevalent when BMAL1 is bound to DNA at ZT6 for rhythmic targets expressed at ZT3–ZT15. Because the number of interactions is not rhythmic for arrhythmically expressed targets, this suggests that rhythmic chromatin interactions between BMAL1-bound DHSs and other DHS may contribute to rhythmic gene expression.
Discussion
Although the same clockwork mechanism is found in essentially all tissues, clock-controlled gene rhythmic expression is mainly tissue-specific, reflecting the large number of biological functions that are clock controlled (Panda et al. 2002; Storch et al. 2002; Zhang et al. 2014; Mure et al. 2018). By characterizing BMAL1 cistromes in three mammalian tissues, we show that tissue-specific circadian transcriptional programs can be directly initiated by the core circadian clock. We provide evidence that BMAL1 DNA-binding profiles are largely tissue-specific, which coincides with tissue-specific chromatin accessibility and cobinding with ts-TFs. Our analysis of BMAL1 target gene expression also indicates that rhythmic CLOCK:BMAL1 transcriptional output relies, at least in part, on the rhythmic interaction between BMAL1-bound DHSs and other DHSs. Given that some of these other DHSs are either tissue-specific or more accessible in specific tissues (e.g., Fig. 5D; Supplemental Fig. S5B), our data suggest that enhancer–enhancer interactions can drive rhythmic BMAL1 target gene expression in a tissue-specific fashion (Fig. 7E). Taken together, chromatin accessibility and enhancer–enhancer interactions may explain how the circadian clock generates tissue-specific circadian transcriptional programs, thereby regulating biological functions in a tissue-specific manner. We anticipate that these mechanisms apply to other TFs that regulate gene expression in a tissue-specific manner.
Recent characterization of the chromatin-accessible landscape revealed that most TFs bind to regions that are hypersensitive to nuclease digestion (Thurman et al. 2012; Yue et al. 2014). Because many of these DHSs are tissue specific (Supplemental Fig. S2A), it is not surprising that differences in the openness of the chromatin between tissues can promote tissue-specific BMAL1 binding. However, the majority of tissue-specific BMAL1 peaks are located in chromatin regions accessible in other tissues (Fig. 2C; Supplemental Fig. S2C,D). DHSs are clusters of TF-binding motifs, and TF recruitment to DHSs often results from cooperative binding between TFs. In particular, ts-TFs can cooperate with u-TFs to facilitate DNA binding: CEBPB cooperates with glucocorticoid receptor in the mouse liver (Grøntved et al. 2013), FOXA1 and GATA3 with the estrogen receptor in primary breasts tumors (Hurtado et al. 2011; Theodorou et al. 2013), and OPA and SERPENT with Drosophila CLK:CYC (Meireles-Filho et al. 2014). Our finding that binding motifs, genomic footprints, and ChIP-seq signal for liver-specific TFs are enriched at liver-specific BMAL1 peaks, but not at common peaks strongly suggests that this ts-TF/u-TF cooperation applies to BMAL1 and that ts-TFs facilitate tissue-specific CLOCK:BMAL1 binding (Fig. 7C).
By analyzing the genome-wide nucleosome positioning in the mouse liver over the course of the day, it has recently been shown that CLOCK:BMAL1 promotes rhythmic nucleosome removal at its binding sites and proposed that CLOCK:BMAL1 is a pioneer-like TF (Menet et al. 2014). Our findings that BMAL1 binds in a tissue-specific manner even at DHSs common to several tissues seems contradictory to BMAL1 pioneering-like function. Instead of generating new DHSs (an expected feature from pioneer TFs) (Zaret and Carroll 2011), our data rather suggest that BMAL1 DNA binding prevents the folding of DNA around histones and thus nucleosome formation. As a result, BMAL1 DNA binding would increase the exposure of naked DNA for other TFs and DNase I and facilitate their binding on DNA. This scenario, which is consistent with the analysis of nucleosome signal at tissue-specific BMAL1 peaks in Bmal1−/− mice (Supplemental Fig. S4H), would explain why DNase I signal is decreased in Bmal1−/− mice around not only E-boxes but also ts-TFs motifs (Fig. 3H,I). Conversely, ts-TFs may play a similar role at tissue-specific DHSs; i.e., their binding to DNA may facilitate the access of CLOCK:BMAL1 to exposed E-boxes by preventing nucleosome formation. This mechanism, which has been proposed previously to illustrate cooperative TF binding to DHSs (Mirny 2010), may explain at least in part tissue-specific BMAL1 binding at DHSs common to several tissues. Future experiments will be required to experimentally validate this hypothesis and eventually clarify BMAL1 pioneering like function.
Genes targeted by BMAL1 in all three tissues do not exhibit a higher rate of rhythmic expression and display a remarkable heterogeneity in the rhythmic output (Fig. 5A,B), indicating that even robust BMAL1 binding is not sufficient to drive rhythmic transcription. As recently suggested (Trott and Menet 2018), this may be explained by the contribution of u-TFs in regulating the transcriptional activity of BMAL1 DHSs and promoting gene expression. However, our analysis suggests an additional level of regulation that includes DHSs unbound by BMAL1, and through which interactions between BMAL1-bound DHSs and other DHSs would contribute to the regulation of rhythmic gene expression (Fig. 5D). To characterize whether DHSs not bound by BMAL1 might contribute to rhythmic BMAL1 transcriptional output, we conducted mouse liver Pol II ChIA-PET to identify physical interactions between DHSs within genes engaged in active transcription (Fig. 6). In agreement with the literature, we found that the number of DHS–DHS interactions correlates with transcriptional levels. Relevant to how the circadian clock might regulate rhythmic gene expression, we identified a large number of interactions between BMAL1-bound DHSs and other DHSs, suggesting that BMAL1 target gene transcription relies more on the functional interaction between DHSs rather than just BMAL1-bound DHSs. In addition, and consistent with previous reports that used the 4C technique (Aguilar-Arnal et al. 2013; Mermet et al. 2018; Yeung et al. 2018), we found that the number of interactions between BMAL1 DHSs and other DHSs is higher at ZT6; i.e., at the time of maximal BMAL1 DNA binding. Importantly, this higher number of chromatin interactions at ZT6 was observed for rhythmically expressed target genes that peak during the day and not at arrhythmically expressed targets, suggesting that BMAL1-mediated rhythmic transcription relies on the capacity of BMAL1 to regulate rhythmic enhancer–enhancer interactions. These results are concordant with recent reports showing that rhythmic transcription in mammals is associated with rhythmic long-range interactions between cis-regulatory elements and rhythmic gene looping (Aguilar-Arnal et al. 2013; Kim et al. 2018; Mermet et al. 2018; Yeung et al. 2018). While the mechanisms underlying rhythmic enhancer–enhancer interactions remain vastly unknown, they likely involve the Mediator complex and transcriptional regulators recognizing histone modifications associated with transcriptional activation (Kim et al. 2018). In addition, our and others’ findings raise the hypothesis that the expression of BMAL1 target genes relies on the relative contribution of BMAL1-bound DHSs versus other DHSs, and that changes in the activity of other DHSs can impair the rhythmic expression of BMAL1 targets without affecting the molecular clockwork mechanism. Such a mechanism may well explain why circadian transcriptional programs can be reprogrammed by environmental challenges without affecting much of the clockwork mechanism (Eckel-Mahan et al. 2013; Murakami et al. 2016; Sato et al. 2017).
In summary, our results provide novel insights into how BMAL1 regulates rhythmic gene expression in a tissue-specific manner and shed light on the role of enhancer–enhancer interactions in generating circadian transcriptional programs. We anticipate that these findings will be relevant for our understanding of how the circadian clock regulates a wide array of biological functions under normal and diseased states and will apply more generally to how other TFs regulate tissue-specific gene expression.
Materials and methods
Animals
Male C57BL/6J and Bmal1−/− mice were housed under 12 h light:12 h dark (LD12:12) with food and water available ad libitum. Bmal1−/− were kindly provided by Christopher Bradfield (Bunger et al. 2000). The age of animals collected was between 3 and 6 mo old. All experiments were approved by the Texas A&M University Institutional Animal Care and Use Committee.
BMAL1 ChIP-seq
BMAL1 ChIPs were performed as described previously (Menet et al. 2014) with modifications. Formaldehyde crosslinked nuclei were purified by centrifugation on a 24% sucrose cushion at 20,000g, washed, and sonicated to obtain chromatin fragments of about 100–600 base pairs (bp) in length. Immunoprecipitation was performed using 1 µL of BMAL1 antibody (chicken anti-BMAL1) and BMAL1 ChIP efficiency was verified by qPCR. ChIP-seq libraries (n = 3 mice per tissue) were generated using NEBNext ChIP-seq library preparation master mix set as per the manufacturer's instructions and sequenced using an Illumina NextSeq.
Sequencing data sets and alignment to the mouse genome
All public data sets were downloaded as fastq or Short Read Archive (SRA) file formats (accession numbers are provided in the Supplemental Material). To avoid issues due to the utilization of different protocols/procedures for each tissue, each of the liver, kidney, and heart ChIP-seq, DNase-seq and mRNA expression data sets used in cross-comparisons were generated from the same research laboratory. ChIP-seq and DNase-seq data sets were aligned to the mouse genome (version mm10) using bowtie2 with the parameters: -x and --end-to-end. Uniquely mapped reads were only considered for analysis, and up to three duplicated sequences were kept for each BMAL1 ChIP-seq data set. Visualization files were generated using bedtools and normalized to 10,000,000 reads. Input files were processed individually and then merged as bam files using samtools.
For DNase-seq data sets, bam files from all technical and biological replicates were merged and no downsampling was performed. Reads from the RNA-seq data set were trimmed using fastx_trimmer with the following parameters -f 1 -l 100 -Q 33, and aligned to the genome using STAR with the default parameters and the option: –outFilterIntronMotifs RemoveNoncanonical. Gene expression data were retrieved using cufflinks and the genome version GRCm38.p5_M14 and default parameters.
Sequencing data set analysis
Peak calling for both BMAL1 ChIP-seq and DNase-seq data was performed with findPeaks from the HOMER suite. A minimum local enrichment of fourfold was set up as necessary, and the following parameters were used: -style factor and -i (for BMAL1 ChIP-seq) or -style dnase and -region (for DNase-seq). Overlap between ChIP-seq or DNase-seq peaks was determined using the function intersectBed from Bedtools suite with default parameters. Heat maps were using the Rscript pheatmap.R.
BMAL1 peaks were assigned to their target genes using the Perl script annotatePeaks.pl from the HOMER suite. The HOMER gene annotation script outputs each peak into the following categories: (1) promoter–TSS, corresponding to TSS −10 kb to TSS +1 kb; (2) transcription termination site (TTS), corresponding to TTS −100 bp to TTS +1 kb; (3) exons; (4) introns; and (5) intergenic, which corresponds to peaks located upstream of the TSS by >10 kb and downstream from the TTS by >1 kb. Intron and exon assignments were not adjusted from the output of HOMER annotatepeaks.pl. BMAL1 peaks labeled as intergenic were not assigned to a target gene.
ChIP-seq and DNase-seq signal was calculated using scripts from Bedtools and the uniquely mapped reads and the genomic coordinates of BMAL1 ChIP-seq peaks (Supplemental Table S1) or mouse liver DHS peaks (Supplemental Table S5). Signal was calculated in a ±250-bp region from the peak center for DNase-seq as well as for BMAL1 and other TF ChIP-seq signal and in a ±1-kb region from the peak center for H3K27ac ChIP-seq signal.
Detection of footprints was performed using the python script wellington_footprints.py from the pyDNase suite. All parameters were set to default, and a P-value of −20 was used along with a false discovery rate of 0.01. Motif analysis was performed at BMAL1 DNA-binding sites using the perl script findMotifsGenome.pl from the HOMER suite using the parameter -size 200. Motifs were considered as significantly enriched if the q-value was <0.05. Motif enrichment was calculated based on the background from the output of findMotifsGenome.pl.
The genomic location of DNase I cuts was retrieved from DNase-seq data sets by reporting the position and strandness of the first nucleotide of each read in a bam file. Average DNase I cut signal was calculated at E-boxes and other TF-binding motifs using scripts from Bedtools. Quantification of the average DNase I cut signal at control peaks was performed by randomly selecting an equivalent number of TF ChIP-seq peaks without BMAL1 to the number of peaks with BMAL1. Because most TF ChIP-seq peaks with BMAL1 signal exhibit strong DNase-seq signal (i.e., most of them are within the strongest DHSs), the random selection was performed by matching the number of peaks within each DNase-seq signal decile. This process was repeated 1000 times, and the average of 1000 iterations calculated.
Generation of motifs for E-boxes and dual E-boxes was performed with the perl script seq2profile.pl from the HOMER suite. The E-boxes considered for analysis were CACGTG, CACGNG, and CACGTT, and the dual E-box motif tolerated up to two mismatches between the two E-boxes and contained a spacer of either six or seven base pairs. Gene ontology was performed using the perl script annotatePeaks.pl from the HOMER suite with the parameter -go and mm10 genome.
Rhythmic expression of BMAL1 target genes was determined using public microarray data sets performed in the same research laboratory (Zhang et al. 2014). Files containing expression values for each microarray probe were downloaded from the NCBI Web site (GSE54652), and no analysis of the original files was performed. Rhythmic gene expression was determined using JTK-cycle with the following parameters: time points 18–64, and all other parameters were left to default and considered significant if q-value <0.05. The output regarding phase of expression, which is reported for every gene, was used in the ChIA-PET analysis. For the results presented in Figure 5, A and B, genes targeted by two or more BMAL1 ChIP-seq peaks assigned to different categories (e.g., a gene targeted by two BMAL1 peaks: one common to all three tissues, and one specific to the liver) were not considered.
Graphs in Supplemental Fig. S3C displaying the RPKM (reads per kilobase per million mapped reads) values of different TFs in human livers, kidneys, and hearts (atrial appendage and left ventricle) were retrieved from GTEx portal in April 2016.
Mouse liver Pol II ChIA-PET
Mouse liver Pol II ChIA-PET experiments were performed at ZT6 and ZT18 with three biological replicates according to published protocols (Li et al. 2010) with some modifications. Liver nuclei were either dual cross-linked with 1.5 mM EGS and 1% formaldehyde (two experiments) or single cross-linked with 1% formaldehyde (one experiment). Sonicated chromatin was immunoprecipitated with an anti-RNA Pol II 8WG16 antibody. ChIA-PET libraries were sequenced on a HiSeq 2000 or a MiSeq. Additional information is in the Supplemental Material.
Sequencing and computational analysis of Pol II ChIA-PET libraries
ChIA-PET reads from the fastq files were processed to extract the tags 1 and 2 along with their accompanying half-linker code using a custom-made Python script and to generate two fastq files (R1 and R2 files) containing the sequence identifier, the raw sequence, and the sequence quality values for each tag. These two files were then aligned to the mouse genome (mm10 version) as paired-end reads using bowtie2. Only paired tags with both reads mapping uniquely to the mouse genome were considered in our analysis. PETs were parsed based on the half-linker barcodes into nonchimeric PETs (specific products) or chimeric PETs (nonspecific products) and duplicated PETs were removed for both chimeric and nonchimeric products. PETs with a tag location shifted by 1 bp compared with an existing PET were also considered as PCR duplicates and removed.
PETs with both reads on the same chromosome and with a distance between reads ≥500 bp were only considered in our analysis. For all PETs, each of the two tags was extended to 200 bp (tag location ±100 bp) and this tag genomic location (chr:start-end) was used to map tags to a gene, and a DHS, using intersectBed from bedtools. Mapping to DHS was performed using a more stringent analysis of the mouse liver DNase-seq data sets from ENCODE (Supplemental Table S5) and which mostly reports stronger DHSs (see Fig. 6D). DHSs harboring a mouse liver BMAL1 peak were identified with intersectBed between the DHS peak list described above and the list of mouse liver ChIP-seq peaks generated in this manuscript (Supplemental Table S1). To validate that mouse liver PETs contribute to gene transcription (Fig. 6B,C), we used public mouse liver Nascent-seq data sets, and averaged values for each of the 12 independent samples. Finally, we considered genes to be rhythmically expressed based on the analysis of the microarray data sets from Zhang et al. (2014) as described above.
Statistical analysis
Statistical analysis was carried out in JMP version 12.0.1. ChIP-seq and DNase-seq signals were analyzed using a Kruskal-Wallis test and post-hoc analysis with a Wilcoxon each pair test. Analysis of TF mRNA expression between the three tissues was performed using a one-way ANOVA. Analysis of the differences in BMAL1 peaks genomic locations was performed using a χ2 test, and differences in the number of BMAL1 peaks per genomic locations we analyzed by a Fisher's exact test. Spearman correlation was used to determine the degree of correlation between signals (e.g., ChIP-seq with DNA-seq) or signal between tissues. Results were considered significant if P-value was <0.05 for the Kruskal-Wallis, Student's t-test, and ANOVA tests, and P-value was <0.01 for Fisher's exact test.
Data availability
The sequencing data sets generated in this study (BMAL1 ChIP-seq and Pol II ChIA-PET) have been deposited to Gene Expression Omnibus under the accession code GSE110604.
Supplementary Material
Acknowledgments
We thank Christopher Bradfield for kindly providing the Bmal1−/− mouse; Craig Kaplan, Christine Merlin, and Aldrin Lugena for insightful suggestions at various stages of the project; Christine Merlin, Deborah Bell-Pedersen, and Paul Hardin for comments on the manuscript; the Texas A&M Institute for Genome Sciences and Society for providing access and maintenance to their high-performance computing cluster; Michael Rosbash, Kate Abruzzi, and Ryanne Spann for helping with sequencing BMAL1 ChIP-seq libraries; and Charles Johnson and Richard Metz for helping with sequencing the ChIA-PET libraries. We are also grateful to Paul Hardin and Matthew Sachs laboratories for technical support. This work was supported by startup funds from Texas A&M University. The laboratory of S.-H.Y. is supported by the National Institute of General Medical Sciences (R01GM114424), and the laboratory of Z.C. is supported by the Robert A. Welch Foundation (AU-1731-20160319) and the National Institute on Aging (R01AG045828). J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Author contributions: J.R.B. and J.S.M. conceived and designed the research. J.R.B. performed most of the experiments and the bioinformatics analysis. J.S.M. performed the ChIA-PET experiments with the help of A.J.T. and J.R.B. B.G., C.A.O., and J.S.M. helped with the bioinformatics analysis. J.S. and H.V. performed ChIPs at an early stage of the project. S.-H.Y., Z.C., and J.S.T. contributed to the BMAL1 antibody. N.G. helped with the ChIA-PET bioinformatics analysis. J.R.B. and J.S.M. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.322198.118.
References
- Aguilar-Arnal L, Hakim O, Patel VR, Baldi P, Hager GL, Sassone-Corsi P. 2013. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat Struct Mol Biol 20: 1206–1213. 10.1038/nsmb.2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103: 1009–1017. 10.1016/S0092-8674(00)00205-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. 2018. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174: 1309–1324.e18. 10.1016/j.cell.2018.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi R, Oishi K, Ishida N. 2010. CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. J. Biol Chem 285: 22114–22121. 10.1074/jbc.M110.110361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. 2013. Reprogramming of the circadian clock by nutritional challenge. Cell 155: 1464–1478. 10.1016/j.cell.2013.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Shimizu H, Nohales MA, Araki T, Kay SA. 2014. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422. 10.1038/nature13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett LJ, Le Lay J, Lukovac S, Bernstein D, Steger DJ, Lazar MA, Kaestner KH. 2013. Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver. BMC Genomics 14: 337 10.1186/1471-2164-14-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odom DT, Flicek P. 2012. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res 22: 2163–2175. 10.1101/gr.136507.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Savic D, Varley KE, Partridge EC, Safi A, Jain P, Cooper GM, Reddy TE, Crawford GE, Myers RM. 2013. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell 52: 25–36. 10.1016/j.molcel.2013.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnocchi D, Pedrelli M, Hurt-Camejo E, Parini P. 2015. Lipids around the clock: focus on circadian rhythms and lipid metabolism. Biology (Basel) 4: 104–132. 10.3390/biology4010104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grøntved L, John S, Baek S, Liu Y, Buckley JR, Vinson C, Aguilera G, Hager GL. 2013. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J 32: 1568–1583. 10.1038/emboj.2013.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2013. The genotype-tissue expression (GTEx) project. Nat Genet 45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, Rao PK, Fei T, Xu H, Long H, et al. 2014. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods 11: 73–78. 10.1038/nmeth.2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. 2009. Global mapping of protein–DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289. 10.1038/nmeth.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Gallo SM. 2010. Identification of interacting transcription factors regulating tissue gene expression in human. BMC Genomics 11: 49 10.1186/1471-2164-11-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. 2011. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–33. 10.1038/ng.730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grøntved L, et al. 2013. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155: 1507–1520. 10.1016/j.cell.2013.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Marhon SA, Zhang Y, Steger DJ, Won KJ, Lazar MA. 2018. Rev-erbα dynamically modulates chromatin looping to control circadian gene transcription. Science 359: 1274–1277. 10.1126/science.aao6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354. 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, et al. 2010. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol 11: R22 10.1186/gb-2010-11-2-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98. 10.1016/j.cell.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Liu T, Chang L, Rui C, Xiao Y, Li S, Hogenesch JB, Chen YE, Lin JD. 2015. The liver clock controls cholesterol homeostasis through Trib1 protein-mediated regulation of PCSK9/low density lipoprotein receptor (LDLR) axis. J Biol Chem 290: 31003–31012. 10.1074/jbc.M115.685982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles-Filho AC, Bardet AF, Yáñez-Cuna JO, Stampfel G, Stark A. 2014. cis-regulatory requirements for tissue-specific programs of the circadian clock. Curr Biol 24: 1–10. 10.1016/j.cub.2013.11.017 [DOI] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. 2012. Nascent-seq reveals novel features of mouse circadian transcriptional regulation. eLife 1: e00011 10.7554/eLife.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Pescatore S, Rosbash M. 2014. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev 28: 8–13. 10.1101/gad.228536.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermet J, Yeung J, Hurni C, Mauvoisin D, Gustafson K, Jouffe C, Nicolas D, Emmenegger Y, Gobet C, Franken P, et al. 2018. Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev 32: 347–358. 10.1101/gad.312397.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. 2010. Nucleosome-mediated cooperativity between transcription factors. Proc Natl Acad Sci 107: 22534–22539. 10.1073/pnas.0913805107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Tognini P, Liu Y, Eckel-Mahan KL, Baldi P, Sassone-Corsi P. 2016. Gut microbiota directs PPARγ-driven reprogramming of the liver circadian clock by nutritional challenge. EMBO Rep 17: 1292–1303. 10.15252/embr.201642463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, et al. 2018. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359: eaao0318 10.1126/science.aao0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. 2012. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489: 83–90. 10.1038/nature11212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S. 2016. Circadian physiology of metabolism. Science 354: 1008–1015. 10.1126/science.aah4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320. 10.1016/S0092-8674(02)00722-5 [DOI] [PubMed] [Google Scholar]
- Paquet ER, Rey G, Naef F. 2008. Modeling an evolutionary conserved circadian cis-element. PLoS Comput Biol 4: e38 10.1371/journal.pcbi.0040038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. 2014. Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24: 90–99. 10.1016/j.tcb.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J, Assi SA, Cauchy P, Ladroue C, Cockerill PN, Bonifer C, Ott S. 2015. Wellington-bootstrap: differential DNase-seq footprinting identifies cell-type determining transcription factors. BMC Genomics 16: 1000 10.1186/s12864-015-2081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. 2011. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9: e1000595 10.1371/journal.pbio.1000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Solanas G, Peixoto FO, Bee L, Symeonidi A, Schmidt MS, Brenner C, Masri S, Benitah SA, Sassone-Corsi P. 2017. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 170: 664–677 e611. 10.1016/j.cell.2017.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Hashimoto T, O'Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. 2014. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol 32: 171–178. 10.1038/nbt.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JA, Krier I, Andersin T, Raghav S, Canella D, Gilardi F, Kalantzi AS, Rey G, Weger B, Gachon F, et al. 2017. Transcriptional regulatory logic of the diurnal cycle in the mouse liver. PLoS Biol 15: e2001069 10.1371/journal.pbio.2001069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Sandstrom R, Haugen E, Reynolds AP, Zhang M, Byron R, Canfield T, Stelhing-Sun S, Lee K, et al. 2014. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature 515: 365–370. 10.1038/nature13972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417: 78–83. 10.1038/nature744 [DOI] [PubMed] [Google Scholar]
- Sung MH, Guertin MJ, Baek S, Hager GL. 2014. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol Cell 56: 275–285. 10.1016/j.molcel.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS. 2017. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18: 164–179. 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou V, Stark R, Menon S, Carroll JS. 2013. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res 23: 12–22. 10.1101/gr.139469.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott AJ, Menet JS. 2018. Regulation of circadian clock transcriptional output by CLOCK:BMAL1. PLoS Genet 14: e1007156 10.1371/journal.pgen.1007156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346: 1007–1012. 10.1126/science.1246426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Yang G, Jia Z, Zhang H, Aoyagi T, Soodvilai S, Symons JD, Schnermann JB, Gonzalez FJ, Litwin SE, et al. 2008. Vascular PPARγ controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab 8: 482–491. 10.1016/j.cmet.2008.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Su W, Liu S, Zhao G, Esser K, Schroder EA, Lefta M, Stauss HM, Guo Z, Gong MC. 2015. Smooth-muscle BMAL1 participates in blood pressure circadian rhythm regulation. J Clin Invest 125: 324–336. 10.1172/JCI76881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung J, Mermet J, Jouffe C, Marquis J, Charpagne A, Gachon F, Naef F. 2018. Transcription factor activity rhythms and tissue-specific chromatin interactions explain circadian gene expression across organs. Genome Res 28: 182–191. 10.1101/gr.222430.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci 111: 16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, et al. 2015. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348: 1488–1492. 10.1126/science.aab3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data sets generated in this study (BMAL1 ChIP-seq and Pol II ChIA-PET) have been deposited to Gene Expression Omnibus under the accession code GSE110604.