This Outlook discusses Beytebiere et al.’s finding that the ability of BMAL1 to drive tissue-specific rhythmic transcription is associated with not only the activity of BMAL1-bound enhancers but also the activity of neighboring enhancers.
Keywords: circadian clock, enhancer–enhancer interactions, tissue-specific cistromes, transcription
Abstract
The circadian clock in the suprachiasmatic nucleus (SCN) of mammals drives 24-h rhythms of sleep/wake cycles. Peripheral clocks present in other organs coordinate local and global physiology according to rhythmic signals from the SCN and via metabolic cues. The core circadian clockwork is identical in all cells. However, there is only a small amount of overlap of the circadian transcriptomes in different organs and tissues. A novel study by Beytebiere and colleagues (pp. 294–309) indicates that the regulation of tissue-specific rhythmic gene expression involves the cooperation of the circadian transcription factor (TF) BMAL1:CLOCK with tissue-specific TFs (ts-TFs) and correlates with the potential of BMAL1:CLOCK to facilitate rhythmic enhancer–enhancer interactions.
The core circadian oscillator consists of a handful of transcription factors (TFs) interlocked in autoregulatory feedback loops. In mammals, the core clock components BMAL1 and CLOCK form heterodimers with the potential to activate thousands of genes, including those of the negative regulators Period (Per) and Cryptochrome (Cry) and the transcription repressors Rev-Erbα and Rev-Erbβ (Partch et al. 2014). Progressive accumulation and nuclear import of PERs and CRYs results in delayed repression and subsequent removal of BMAL1:CLOCK from DNA. REV-ERBα and REV-ERBβ repress Bmal1 transcription and establish an interconnected negative feedback loop.
Rhythmic transcription of genes controlled by BMAL1:CLOCK constitutes the primary output of the core circadian oscillator. Subsequent transcriptional and posttranscriptional processes orchestrate complex rhythms in mRNA and protein abundance. While the circadian clock has the potential to regulate ∼80% of the protein-coding transcripts, only a subset of genes (∼10%) is rhythmically expressed in any particular tissue, and there is surprisingly modest overlap between the circadian transcriptomes of different organs (Mure et al. 2018). How the same molecular oscillator controls distinct sets of rhythmically expressed genes in different organs remains enigmatic.
In this issue of Genes & Development, Beytebiere et al. (2019) analyzed the primary transcriptional output of the core circadian oscillator in different organs. To assess mechanisms responsible for tissue-specific circadian transcription controlled by BMAL1, they combined systematic analysis of novel and available genomic data. Using ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) analysis in the mouse liver, heart, and kidney, Beytebiere et al. (2019) demonstrate that BMAL1 binding is tissue-specific and exhibits only minor overlaps between different organs. The overlap includes mostly genes involved in circadian rhythms and housekeeping processes. Similar observations were reported previously for CYC:CLK, the core transcription factor of the Drosophila clock (Meireles-Filho et al. 2014).
These findings raise questions concerning the mechanisms underlying tissue specificity of DNA binding. Beytebiere et al. (2019) show that DNA recruitment of BMAL1 in the liver coincides almost entirely with DNase-hypersensitive sites (DHSs). However, liver-specific open chromatin regions (DHSs) account for only a small fraction of the liver-specific BMAL1-binding sites, whereas the chromatin around the majority of liver-specific BMAL1-binding sites remains DNase-hypersensitive in other tissues where the sites are not bound by BMAL1. Analysis of MNase-seq (micrococcal nuclease [MNase] sequencing) data revealed that hepatic BMAL1-binding sites are in fact nucleosome-depleted in the liver but display rather high nucleosome occupancy in the kidney and heart (despite DNase sensitivity), suggesting that tissue-specific TFs (ts-TFs) may somehow facilitate nucleosome depletion and BMAL1 recruitment. In agreement, Beytebiere et al. (2019) show that hepatic BMAL1 peaks overlap with footprints and binding motifs of liver-specific TFs, such as HNF6 or CEBPA, whereas BMAL1 peaks common in all three tissues were cobound by ubiquitously expressed TFs (u-TF). Hence, binding of ts-TFs or u-TFs to their cognate sites may open and remodel the chromatin and thereby expose E-boxes that are otherwise covered by nucleosomes. This process seems to be mutual, since binding sites of hepatic TFs (HNF6 and CEBPA) showed significantly reduced DNase accessibility and TF-specific footprints in Bmal1 knockout mice.
It therefore seems conceivable that BMAL1 and other TFs may mutually facilitate their recruitment by depletion of nucleosomes from their binding sites. A similar mechanism was described recently for the glucocorticoid receptor and CEBP (Grøntved et al. 2013). Synergism of TF recruitment has been previously proposed and modeled, and the mechanism was termed nucleosome-mediated cooperativity (Mirny 2010).
Earlier data from the Menet laboratory (Trott and Menet 2018) had suggested that BMAL1 may facilitate oscillations in the permissive state of the chromatin, thereby allowing rhythmic access and transcription activation by other, noncircadian TFs. RNA polymerase II (Pol II) ChIA-PET (chromatin interaction analysis by paired-end tag) analysis by Beytebiere et al. (2019) revealed that both high expression levels and high-amplitude circadian rhythms correlated with rhythmic interaction of BMAL1-specific DHSs with other enhancer sites (DHSs) that were not directly targeted by BMAL1.
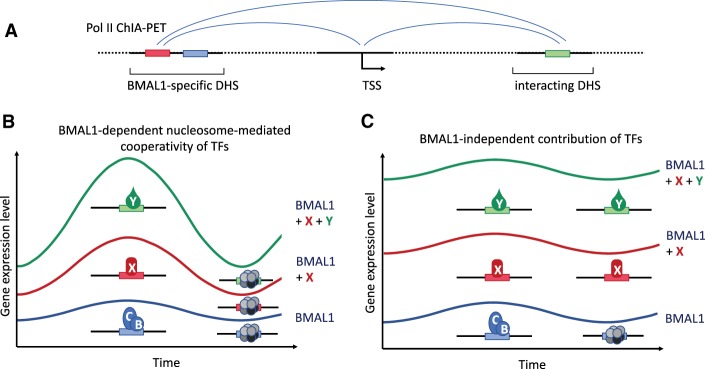
How can these observations be integrated into a working model? TFs support multiple functions (Spitz and Furlong 2012), including recruitment of chromatin modifiers and remodelers to generate nucleosome-depleted regions (NDRs) at gene promoters and enhancers, recruitment of Mediator to promote assembly of preinitiation complexes (PICs) at the NDRs, and recruitment of machinery facilitating the transition of promotor-proximally paused Pol II into a productive elongation complex. A single TF is generally not sufficient to support all of these functions in an efficient manner. Hence, multiple TFs recruited via the same or different enhancers often cooperate to activate transcription. It is therefore not surprising that efficient transcription of many BMAL1 target genes is also associated with the physical interaction of multiple DHSs and seems to depend on the activity of multiple TFs. Nucleosome-mediated cooperativity (Mirny 2010) of BMAL1:CLOCK with noncircadian TFs could account for the synergistic activity that is required to support high-amplitude expression levels and rhythms. The relative contribution of BMAL1 versus other TFs to generate NDRs (at promoters and enhancers) may determine whether a gene is rhythmically transcribed (Fig. 1). If BMAL1 is limiting to shift the nucleosome–TF competition toward nucleosome depletion, its contribution is synergistic and transcription will be rhythmic (Fig. 1B, left panel). On the other hand, if the noncircadian TFs are sufficient to remodel the chromatin and expose enough TF-binding sites, additional clock-dependent binding of BMAL1 will contribute in an additive manner and the transcription rhythms (peak:trough ratio) might be severely blunted (Fig. 1B, right panel). In fact, the Menet group (Beytebiere et al. 2019) found many genes with similar BMAL1-binding signals in different tissues that were rhythmically expressed in only one tissue, suggesting that factors in addition to BMAL1 determine expression levels and rhythms. Furthermore, recent studies by the Naef and Lazar laboratories (Kim et al. 2018; Mermet et al. 2018; Yeung et al. 2018) demonstrated the importance of rhythmic chromatin interactions.
Figure 1.
Model of interaction of BMAL1:CLOCK with noncircadian transcription factors. (A) Schematic of Pol II ChIA-PET. A DHS with binding sites for BMAL1:CLOCK (blue box) and the hypothetical noncircadian transcription factor X (red box) interacts with the transcription start site (TSS) of the gene, and both interact in addition with a gene-specific DHS with a binding site for the noncircadian transcription factor Y (green box). (B) Nucleosome-mediated cooperativity of BMAL1/CLOCK (BMAL1) with X and Y. BMAL1, X, and Y are required to mutually facilitate their recruitment by depleting nucleosomes (gray octamers) from their binding sites. Rhythmic binding of BMAL1 synergistically supports rhythmic recruitment of the noncircadian TFs X and Y and thereby supports high expression levels and high-amplitude rhythms. (C) X and Y bind independently of BMAL1. The rhythmic contribution of BMAL1 is blunted in presence of X and Y.
In summary, the data by Beytebiere et al. (2019) suggest that tissue-specific versus ubiquitous transcription levels and rhythms may depend on nucleosome-mediated cooperativity of BMAL1:CLOCK with ts-TFs or u-TFs via enhancer–enhancer interactions.
Acknowledgments
M.B. is an investigator of CellNetworks and is supported by the Deutsche Forschungsgemeinschaft (SFB1036).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.324046.119.
References
- Beytebiere JR, Trott AJ, Greenwell B, Osborne CA, Vitet H, Spence J, Yoo S-H, Chen Z, Takahashi JS, Ghaffari N, et al. 2019. Tissue-specific BMAL1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer–enhancer interactions. Genes Dev (this issue). 10.1101/gad.322198.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grøntved L, John S, Baek S, Liu Y, Buckley JR, Vinson C, Aguilera G, Hager GL. 2013. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J 32: 1568–1583. 10.1038/emboj.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Marhon SA, Zhang Y, Steger DJ, Won KJ, Lazar MA. 2018. Rev-erbα dynamically modulates chromatin looping to control circadian gene transcription. Science 359: 1274–1277. 10.1126/science.aao6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles-Filho ACA, Bardet AF, Yáñez-Cuna JO, Stampfel G, Stark A. 2014. Cis-regulatory requirements for tissue-specific programs of the circadian clock. Curr Biol 24: 1–10. 10.1016/j.cub.2013.11.017. [DOI] [PubMed] [Google Scholar]
- Mermet J, Yeung J, Hurni C, Mauvoisin D, Gustafson K, Jouffe C, Nicolas D, Emmenegger Y, Gobet C, Franken P, et al. 2018. Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev 32: 347–358. 10.1101/gad.312397.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. 2010. Nucleosome-mediated cooperativity between transcription factors. Proc Natl Acad Sci 107: 22534–22539. 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, et al. 2018. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359: eaao0318 10.1126/science.aao0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. 2014. Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24: 90–99. 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13: 613–626. 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Trott AJ, Menet JS. 2018. Regulation of circadian clock transcriptional output by CLOCK:BMAL1. PLoS Genet 14: e1007156 10.1371/journal.pgen.1007156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung J, Mermet J, Jouffe C, Marquis J, Charpagne A, Gachon F, Naef F. 2018. Transcription factor activity rhythms and tissue-specific chromatin interactions explain circadian gene expression across organs. Genome Res 28: 182–191. 10.1101/gr.222430.117. [DOI] [PMC free article] [PubMed] [Google Scholar]