Intelligence is highly heritable1 and a major determinant of human health and well-being2. Recent genome-wide meta-analyses have identified 24 genomic loci linked to variation in intelligence3–7, but much about its genetic underpinnings remains to be discovered. Here, we present the largest genetic association study of intelligence to date (N=269,867), identifying 205 associated genomic loci (190 novel) and 1,016 genes (939 novel) via positional mapping, expression quantitative trait locus (eQTL) mapping, chromatin interaction mapping, and gene-based association analysis. We find enrichment of genetic effects in conserved and coding regions and associations with 146 nonsynonymous exonic variants. Associated genes are strongly expressed in the brain, specifically in striatal medium spiny neurons and hippocampal pyramidal neurons. Gene-set analyses implicate pathways related to nervous system development and synaptic structure. We confirm previous strong genetic correlations with multiple health-related outcomes, and Mendelian randomization results suggest protective effects of intelligence for Alzheimer’s disease and ADHD, and bidirectional causation with pleiotropic effects for schizophrenia. These results are a major step forward in understanding the neurobiology of cognitive function as well as genetically related neurological and psychiatric disorders.
We performed a genome-wide association (GWAS) meta-analysis of 14 independent epidemiological cohorts of European ancestry and 9,295,118 genetic variants passing quality control (Table 1; Supplementary Table 1; Supplementary Figure 1). A flowchart of the study methodology is presented in Supplementary Figure 2 and additional details of the methods and results are presented in the Supplementary Note.
Table 1.
Overview of cohorts included in a GWAS meta-analysis of general intelligence.
| Cohort | N | Age | Phenotype |
|---|---|---|---|
| 1. UKB | 195,653 | 39–72 | Verbal and mathematical reasoning |
| 2. COGENT | 35,289 | 8–96 | One or more neuropsychological tests from three or more domains of cognitive performance |
| 3. RS | 6,182 | 45–98 | Letter-digit substitution, Stroop, verbal fluency, delayed recall |
| 4. GENR | 1,929 | 5–9 | SON-R (spatial visualization and abstract reasoning subsets) |
| 5. STR | 3,215 | 18 | Logical, verbal, spatial, and technical ability subtests |
| 6. S4S | 2,818 | 17–18 | SAT test scores |
| 7. HiQ / HRS | 9,410 | * | High IQ cases / unselected population controls |
| 8. TEDS | 3,414 | 12 | WISC-III verbal and nonverbal reasoning; Raven's progressive matrices |
| 9a. DTR - MADT | 737 | 55–80 | Verbal fluency, digit span, immediate and delayed recall tests |
| 9b. DTR - LSADT | 253 | 73–94 | Verbal fluency, digit span, immediate and delayed recall tests |
| 10. IMAGEN | 1,343 | 14 | WISC-IV, CANTAB factor score |
| 11a. BLTS - Children | 530 | 12–13 | VSRT-C factor score |
| 11b. BLTS - Adolescents | 2,598 | 15–30 | MAB-II IQ score |
| 12. NESCOG | 252 | 18–79 | WAIS IQ score |
| 13. GfG | 5,084 | 15–91 | ICAR verbal reasoning test |
| 14a. STSA - SATSA+GENDER | 703 | 50–94 | Verbal, spatial, episodic memory, and processing speed tests |
| 14b. STSA - HARMONY | 448 | 65–96 | Verbal, spatial, episodic memory, and processing speed tests |
HiQ/HRS sample used a case-control design rather than a cognitive test score ascertained at a specific age; see Online Methods and Supplementary information 1.1.
Intelligence was assessed using various neurocognitive tests, primarily gauging fluid domains of cognitive functioning (Supplementary Information 1.1–1.2). Despite variation in form and content, cognitive test scores display a positive manifold of correlations, a robust empirical phenomenon that is observed in multiple populations8. Statistically, the variance common across cognitive tasks can be modeled as a latent factor denoted as g (the general factor of intelligence)9,10. In addition, twin- and family studies show strong genetic correlations across diverse cognitive domains11, suggesting pleiotropy, and across levels of ability11, substantiating the view of general intelligence as an aetiological continuum (with rare syndromic forms of severe intellectual disability being the exception12). Additionally, g-factors extracted from different sets of cognitive tests correlate very strongly (>.9813,14), supporting the universality of g15,16. In meta-analyzing cognitive scores obtained using a variety of tests, we aim to boost the statistical power to detect genetic variants underlying g, which are likely to have pleiotropic effects across multiple domains of cognitive functioning.
Despite sample and methodological variations, genetic correlations (rg) between cohorts were considerable (mean=0.67), and there was no evidence of heterogeneity between cohorts in the single nucleotide polymorphism (SNP) associations (Supplementary Table 2; Supplementary Results 2.1). Age-stratified meta-analyses indicated high genetic correlations (rg>0.62), and comparable heritability across age, as captured by the SNPs included in the analysis (h2SNP=0.19–0.22) (Supplementary Table 3; Supplementary Results 2.2). The full sample h2SNP was 0.19 (SE=0.01), in line with previous findings4,5, and an LD score intercept17 of 1.08 (SE=0.02) indicated that most of the inflation (λGC=1.92) could be explained by polygenic signal6 (Supplementary Table 4; Supplementary Figure 3).
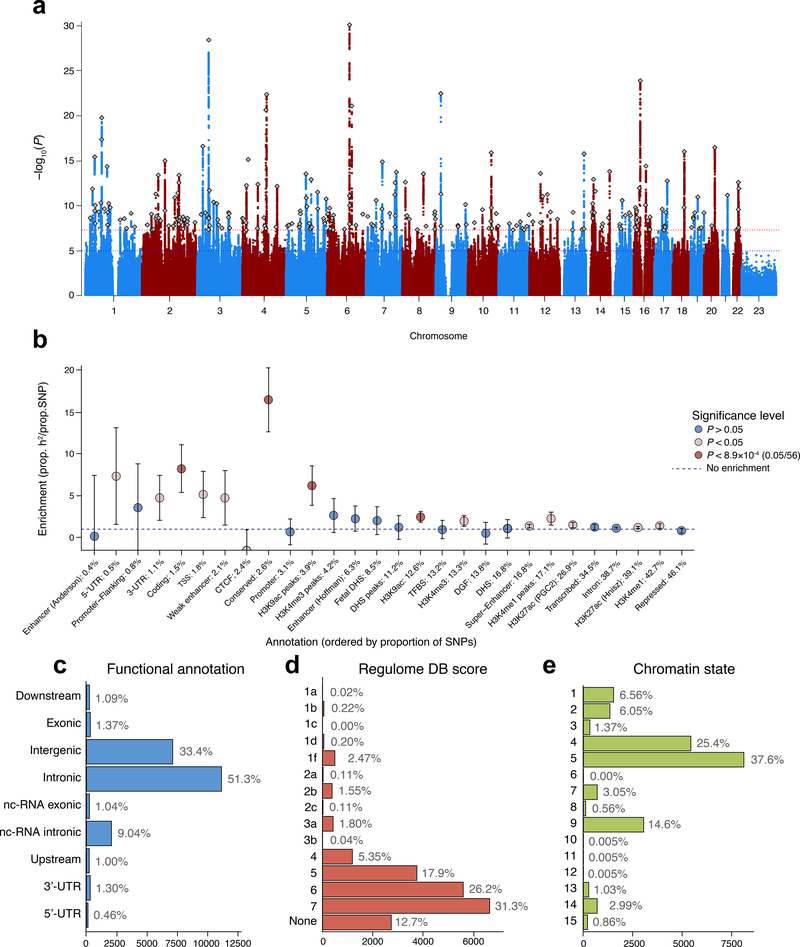
In the meta-analysis, 12,110 variants indexed by 242 lead SNPs in approximate linkage equilibrium (r2<0.1) reached genome-wide significance (GWS; P<5×10−8) (Figure 1a; Supplementary Tables 5–7; Supplementary Figures 4–5). These were located in 205 distinct genomic loci (Supplementary Results 2.3.1). We tested for replication using the proxy phenotype of educational attainment, which is correlated phenotypically (r=~0.40)18 and genetically (r=~0.70)19 with intelligence. We confirmed this high genetic correlation (rg=0.73) and observed sign concordance with educational attainment for 93% of GWS SNPs (P<1×10−300), with replication for 48 loci (Supplementary Results 2.3.2; Supplementary Table 8). Using polygenic score prediction20,21, the current results explain up to 5.2% of the variance in intelligence in four independent samples (Supplementary Table 9, Supplementary Results 2.3.3).
Figure 1. SNP-based associations with intelligence in the GWAS meta-analysis of N=269,867 independent individuals.
(a) Manhattan plot showing the −log10 transformed two-tailed P-value of each SNP from the GWAS meta-analysis (of linear and logistic regression statistics) on the y-axis and base pair positions along the chromosomes on the x-axis. The dotted red line indicates Bonferroni-corrected genome-wide significance (P<5×10−8); the blue line the threshold for suggestive associations (P<1×10−5). Independent lead SNPs are indicated by a diamond. (b) Heritability enrichment of 28 functional annotation categories for SNPs in the meta-analysis, calculated with stratified LD score regression. Error bars show 95% confidence intervals around the enrichment estimate. The dashed horizontal line indicates no enrichment of the annotation category. Red dots indicate significant Bonferroni-corrected two-tailed P-values and beige dots indicate suggestive (P<.05) values. UTR=untranslated region; TSS=transcription start site; CTCF=CCCTC-binding factor; DHS=DNaseI Hypersensitive Site; TFBS=transcription factor binding site; DGF=DNaseI digital genomic footprint. (c) Distribution of functional consequences of SNPs in genomic risk loci in the meta-analysis. (d) Distribution of RegulomeDB score for SNPs in genomic risk loci, with a low score indicating a higher likelihood of having a regulatory function (Online methods). (e) The minimum chromatin state across 127 tissue and cell types for SNPs in genomic risk loci, with lower states indicating higher accessibility and states 1–7 referring to open chromatin states (Online Methods).
We observed enrichment for heritability of SNPs in conserved regions (P=2.01×10−12), coding regions (P=1.67×10−6), and H3K9ac histone regions/peaks (P<6.26×10−5), and among common (minor allele frequency > 0.3) variants (Figure 1b; Supplementary Results 2.3.4; Supplementary Table 10; Supplementary Figures 6–7). Conserved and regulatory regions have previously been implicated in cognitive functioning22 but coding regions have not.
Functional annotation of all candidate SNPs in the associated loci (SNPs with an r2≧0.6 with one of the independent significant SNPs, a suggestive P-value (P<1×10−5) and a MAF>0.0001; n=21,368) showed that these were mostly intronic/intergenic (Supplementary Table 6; Figure 1), yet 146 (81 GWS) SNPs were exonic non-synonymous (ExNS) (Supplementary Table 11, Supplementary Results 2.3.5). Convergent evidence of strong association (Z=9.49) and the highest observed probability of a deleterious protein effect (CADD23 score=34) was found for rs13107325. This missense mutation (MAF=0.065, P=2.23×10−21) in SLC39A8 was the lead SNP in locus 71 and the ancestral allele C was associated with higher scores on intelligence measures. The effect sizes for ExNS were individually small, with each effect allele accounting for a difference of 0.01 to 0.08 standard deviations. Supplementary Tables 6 and 11 and Supplementary Results 2.3.5 present a detailed catalog of variants in the associated genomic loci.
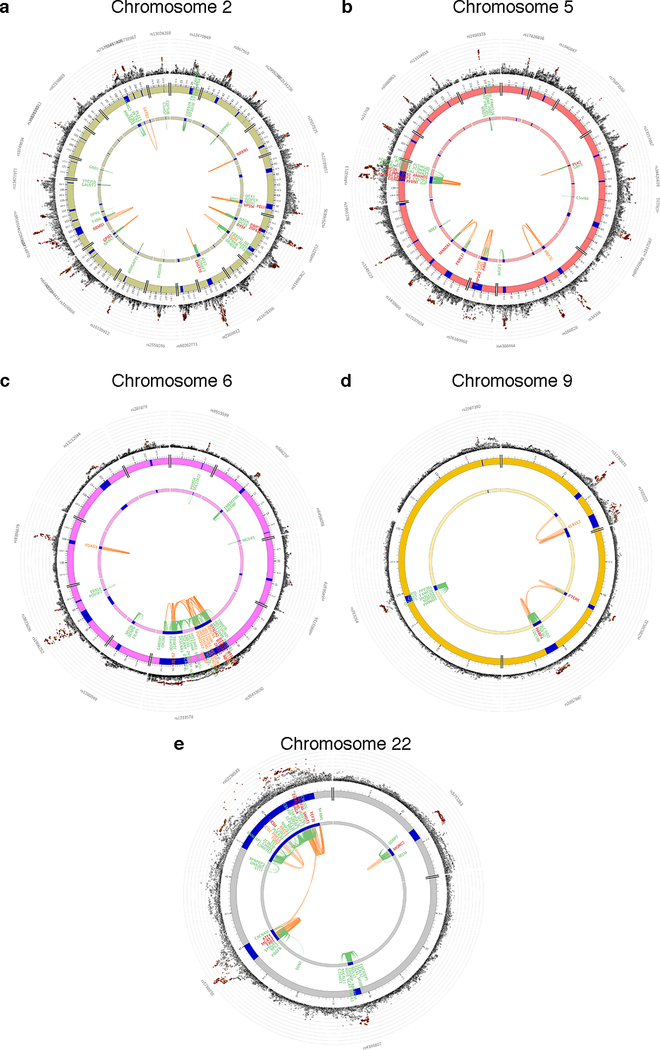
To link the associated variants to genes, we applied three gene-mapping strategies implemented in FUMA24. Positional gene-mapping aligned SNPs to 522 genes by genomic location, eQTL (expression quantitative trait loci) gene-mapping matched cis-eQTL SNPs to 684 genes whose expression levels they influence, and chromatin interaction mapping annotated SNPs to 227 genes based on three-dimensional DNA-DNA interactions (Figure 2; Supplementary Results 2.3.6; Supplementary Figures 8–9; Supplementary Tables 12–14). This resulted in 859 unique mapped genes, 435 of which were implicated by at least two mapping strategies and 139 by all three (Figure 3). Although not all of these genes are certain to have a role in intelligence, they point to potential functional links for the GWAS associated variants and give higher credibility to genes with convergent evidence of association from multiple sources. The FUMA-mapped genes were enriched for brain tissue expression and several regulatory biological gene-sets (Supplementary Results 2.3.6). Fifteen genes are particularly notable as they are implicated via chromatin interactions between two independent genomic risk loci (Figure 2; Supplementary Results 2.3.6). Cross-locus interactions implicated ELAVL2, PTCH1, ATF4, FBXL17, and MAN2A1 in left ventricle of the heart tissue, SATB2 in liver tissue, and MEF2C in 5 tissues. Multiple interactions in multiple tissue types were seen for a cluster of 8 genes on chromosome 6 encoding zinc finger proteins and histones.
Figure 2. Cross-locus interactions for genomic regions associated with intelligence in 269,867 independent individuals.
Circos plots showing genes on chromosomes 2 (a), 5 (b), 6 (c), 9 (d), and 22 (e) that were linked to genomic risk loci in the GWAS meta-analysis (blue regions) by eQTL mapping (green lines connecting an eQTL SNP to its associated gene), and/or chromatin interactions (orange lines connecting two interacting regions) and showed evidence of interaction across two independent genomic risk loci. Genes implicated by eQTL are in green, by chromatin interactions in orange, and by both eQTL and chromatin interactions mapping in red. The outer layer shows a Manhattan plot containing the −log10 transformed two-tailed P-value of each SNP from the GWAS meta-analysis (of linear and logistic regression statistics), with genome-wide significant SNPs colored according to linkage disequilibrium patterns with the lead SNP. Circos plots for all chromosomes are provided in Supplementary Fig. 8.
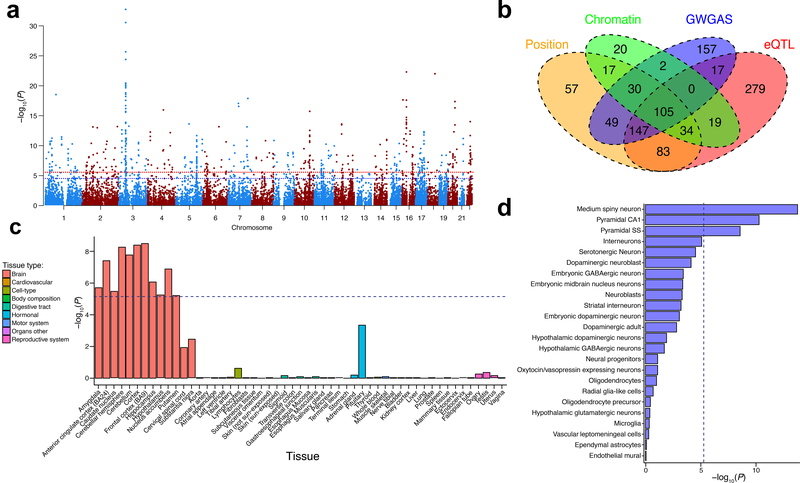
Figure 3. Implicated genes, pathways, and tissue- and cell- expression profiles for intelligence in 269,867 independent individuals.
(a) Manhattan plot of the genome-wide gene-based association analysis (GWGAS). The y-axis shows the −log10 transformed two-tailed P-value of each gene from a linear model, and the chromosomal position on the x-axis. The red dotted line indicates the Bonferroni-corrected threshold for genome-wide significance of the gene-based test (P<2.76×10−6; 0.05/18,128 genes), and the blue line indicates the suggestive threshold (P<2.76×10−5; 0.5/18,128 genes) (b) Venn diagram showing overlap of genes implicated by positional mapping, eQTL mapping, chromatin interaction mapping, and GWGAS. (c) Gene expression profiles of associated genes in 53 tissue types. The y-axis shows the −log10 transformed two-tailed P-value of association of GWGAS test statistics with tissue-specific gene expression levels in a linear model. Expression data were extracted from the Genotype-Tissue Expression (GTEx) database. Expression values (RPKM) were log2 transformed with pseudocount 1 after winsorization at 50 and averaged per tissue. The dotted blue line indicates the Bonferroni-corrected significance threshold (P=0.05/7,323 gene-sets=6.83×10−6). (d) Single-cell gene-expression analysis of genes related to intelligence in 24 cell-types. The x-axis shows the −log10 transformed two-tailed P-value of association of GWGAS test statistics with cell-specific gene expression levels in a linear model. The dotted blue line indicates the Bonferroni-corrected significance threshold (P=0.05/7,323 gene-sets=6.83×10−6).
We performed genome-wide gene-based association analysis (GWGAS) using MAGMA25 to estimate aggregate associations based on all SNPs in a gene (whereas FUMA annotates individually significant SNPs to genes). GWGAS identified 507 associated genes (Figure 3a; Supplementary Results 2.4.1; Supplementary Table 15), of which 350 were also mapped by FUMA (Figure 3b). In total, 105 genes were implicated by all four strategies (Supplementary Table 16).
In gene-set analysis, six Gene Ontology26 gene-sets were significantly associated with intelligence: neurogenesis (P=4.78×10−7), neuron differentiation (P=4.82×10−6), central nervous system neuron differentiation (P=3.31×10−6), regulation of nervous system development (P=9.30×10−7), positive regulation of nervous system development (P=1.00×10−6), and regulation of synapse structure or activity (P=5.42×10−6) (Supplementary Results 2.4.2; Supplementary Tables 17–18). Conditional analysis indicated that there were three independent associations, regulation of nervous system development, central nervous system neuron differentiation, and regulation of synapse structure or activity, which together accounted for the associations of the other sets.
Linking gene-based P-values to tissue-specific gene-sets, we observed strong associations with gene expression across multiple brain areas (Figure 3c; Supplementary Results 2.4.2; Supplementary Table 19), particularly the frontal cortex (P=3.10×10−9). In brain single-cell expression gene-set analyses, we found significant associations of striatal medium spiny neurons (P=2.02×10−14) and pyramidal neurons in the CA1 hippocampal (P=5.67×10−11) and cortical somatosensory regions (P=2.72×10−9) (Figure 3d; Supplementary Results 2.4.2; Supplementary Table 20). Conditional analysis showed that the independent association signal in brain cells was driven by medium spiny neurons, neuroblasts, and pyramidal CA1 neurons.
Intelligence has been associated with a wide variety of human behaviors15 and brain anatomy27. Confirming previous reports5,6, we observed negative genetic correlations with ADHD (rg=−0.36, P=4.58×10−23), depressive symptoms (rg=−0.27, P=6.20×10−10), Alzheimer’s disease (rg=−0.27, P=2.03×10−5), and schizophrenia (rg=−0.21, P=3.82×10−17), and positive correlations with longevity (rg=0.43, P=7.96×10−8) and autism (rg=0.25, P=3.14×10−7), among others (Supplementary Table 21; Supplementary Figure 10). Comparison with previous GWAS28 supported these correlations, showing numerous shared genetic variants across phenotypes (Supplementary Results 2.5; Supplementary Tables 22–23). Low enrichment (87 of 1,518 genes, P=0.05) was found for genes previously linked to intellectual disability or developmental delay, indicating largely distinct biological processes. However, our results extend previous genetic research on normal variation in general intelligence, as catalogued in Supplementary Tables 24–25.
We used Generalized Summary-statistic-based Mendelian Randomization29 to test for potential credible causal associations between intelligence and genetically correlated traits (Supplementary Results 2.5.3; Supplementary Figures 11–12; Supplementary Table 26). We observed a strong bidirectional effect of cognitive ability on educational attainment (bxy=0.549, P<1×10−320) and of educational attainment on intelligence (byx=0.480, P=6.85×10−82). Such findings are consistent with previous studies implicating bidirectional causal effects30,31. There was also a bidirectional association showing a strong protective effect of intelligence on schizophrenia (OR=0.50, bxy=−0.685, P=2.02×10−57) and a relatively smaller reverse effect (byx= −0.214, P=4.19×10−52), with additional evidence for pleiotropy (Supplementary Results 2.5.3). A number of previous reports support both a causal link and genetic overlap between these phenotypes32,33. Our results also suggested that higher intelligence had a protective effect on ADHD (OR=0.48, bxy=−0.734, P=2.57×10−46) and Alzheimer’s disease (OR=0.65, bxy=−0.435, P=3.59×10−14), but was associated with higher risk of autism (OR=1.38, bxy=0.321, P=1.12×10−3).
In the present study, we have affirmed and expanded existing knowledge of the genetics of general intelligence, identifying 190 novel loci and 939 novel associated genes and replicating previous associations with 15 loci and 77 genes. The combined strategies of functional annotation and gene-mapping using biological data resources provide extensive information on the likely consequences of relevant genetic variants and put forward a rich set of plausible gene targets and biological mechanisms for functional follow-up. Gene-set analyses contribute novel insight into underlying neurobiological pathways, confirming the importance of brain-expressed genes and neurodevelopmental processes in fluid domains of intelligence and pointing towards the involvement of specific cell types. Our results indicate overlap in the genetic processes involved in both cognitive functioning and neurological and psychiatric traits and provide suggestive evidence of causal associations that may drive these correlations. These results are important for understanding the biological underpinnings of cognitive functioning and contribute to our understanding of related neurological and psychiatric disorders.
Online Methods
Study Cohorts
The meta-analysis included new and previously reported GWAS summary statistics from 14 cohorts: UK Biobank (UKB), Cognitive Genomics Consortium (COGENT), Rotterdam Study (RS), Generation R Study (GENR), Swedish Twin Registry (STR), Spit for Science (S4S), High-IQ/Health and Retirement Study (HiQ/HRS), Twins Early Development Study (TEDS), Danish Twin Registry (DTR), IMAGEN, Brisbane Longitudinal Twin Study (BLTS), Netherlands Study of Cognition, Environment and Genes (NESCOG), Genes for Good (GfG), and the Swedish Twin Studies of Aging (STSA). All samples were obtained from epidemiological cohorts ascertained for research on a variety of physical and psychological outcomes. Participants ranged from children to older adults, with older samples being screened for cognitive decline to exclude the possibility of dementia affecting performance on cognitive tests.
Different measures of intelligence were assessed in each cohort but were all operationalized to index a common latent g factor underlying multiple dimensions of cognitive functioning. With the exception of HiQ/HRS, all cohorts extracted a single sum score, mean score, or factor score from a multidimensional set of cognitive performance tests and used this normally-distributed score as the phenotype in a covariate-adjusted (e.g. age, sex, ancestry principal components) GWAS using linear regression methods. For HiQ/HRS, a logistic regression GWAS was run with “case” status reflecting whether participants were drawn from an extreme-sampled population of very high intelligence (i.e. at the upper ~0.03% of the tail of the normal distribution) versus an epidemiological sample of unselected population “controls”. Detailed descriptions of the samples, measures, genotyping, quality control, and analysis procedures for each cohort are provided in the Supplementary Note (Supplementary Information 1.1–1.2), Supplementary Table 1, and in the Life Sciences Reporting Summary.
Meta-analysis
Stringent quality control measures were applied to the summary statistics for each GWAS cohort before combining. All files were checked for data integrity and accuracy. SNPs were filtered from further analysis if they met any of the following criteria: imputation quality (INFO/R2) score < 0.6, Hardy-Weinberg equilibrium (HWE) P < 5×10−6, study-specific minor allele frequency (MAF) corresponding to a minor allele count (MAC) < 100, and mismatch of alleles or allele frequency difference greater than 20% from the Haplotype Reference Consortium (HRC) genome reference panel16. Some cohorts used more stringent criteria (see Supplementary Information 1.1). Indels and SNPs that were duplicated, multi-allelic, monomorphic, or ambiguous (A/T or C/G with a MAF >0.4) were also excluded. Visual inspection of the distribution of the summary statistics was completed, and Manhattan plots and QQ plots were created for the cleaned summary statistics from each cohort (Supplementary Figure 1).
The SNP association P-values from the GWAS cohorts were meta-analyzed with METAL34 (see URLs) in two phases. First, we meta-analyzed all cohorts with quantitative phenotypes (all except HiQ/HRS) using a sample-size weighted scheme. In the second phase, we added the HiQ/HRS study results to the first phase results, weighting each set of summary statistics by their respective non-centrality parameter (NCP). This method improves power when using an extreme case sampling design such as HiQ35 and provides a comparable metric with which to combine information from different analytic designs while accounting for their differences in power/effective sample size. NCPs were estimated using the Genetic Power Calculator36, as described by Coleman et al.37. After combining all data, meta-analysis results were further filtered to exclude any variants with N < 50,000. We additionally included a random-effects meta-analysis for each phase, as implemented in METAL, to evaluate potential heterogeneity in the SNP association statistics between cohorts.
The X chromosome was treated separately in the meta-analysis because imputed genotypes were not available for the X chromosome in the largest cohort (UKB), and there was little overlap between the UKB called genotypes and imputed data from other cohorts (NSNPs < 500). We therefore included only the called X chromosome variants in UKB for these analyses after performing X-specific quality control steps38.
We conducted a series of meta-analyses on subsets of the full sample using the same methods as above. Age group-specific meta-analyses were run in the cohorts of children (age < 17; GENR, TEDS, IMAGEN, BLTS; N=9,814), young adults (age ~17–18; S4S, STR; N=6,033), adults (age > 18, primarily middle-aged or older: UKB, RS, DTR, NESCOG, STSA; N=204,228), and older adults (mean age > 60, RS, DTR, STSA; N=8,323), excluding studies whose samples overlapped child/young adult and adult groups (COGENT, HiQ/HRS, GfG; N=49,792). To create independent discovery samples for use in polygenic score validation, we also conducted meta-analyses with a “leave-one-out” strategy in which summary statistics from four validation datasets were, respectively, excluded from the meta-analysis (see Polygenic Scoring, below).
Cohort Heritability and Genetic Correlation
LD score regression17 was used to estimate genomic inflation and heritability of the intelligence phenotypes in each of the 14 cohorts using their post-quality control summary statistics, and to estimate the cross-cohort genetic correlations39. Pre-calculated LD scores from the 1000 Genomes European reference population were obtained online (see URLs). Genetic correlations were calculated on HapMap3 SNPs only. LD score regression was also used on the age subgroup meta-analyses to estimate heritability and cross-age genetic correlations.
Genomic Risk Loci Definition
Independently associated loci from the meta-analysis were defined using FUMA24 (see URLs), an online platform for functional mapping of genetic variants. We first identified independent significant SNPs which had a Bonferroni-corrected genome-wide significant two-tailed P-value (P<5×10−8) and represented signals that were independent from each other at r2<0.6. These SNPs were further represented by lead SNPs, which are a subset of the independent significant SNPs that are in approximate linkage equilibrium with each other at r2<0.1. We then defined associated genomic loci by merging any physically overlapping lead SNPs (linkage disequilibrium [LD] blocks <250kb apart). Borders of the associated genomic loci were defined by identifying all SNPs in LD (r2≧0.6) with one of the independent significant SNPs in the locus, and the region containing all of these candidate SNPs was considered to be a single independent genomic locus. All LD information was calculated from UK Biobank genotype data.
Proxy-replication with Educational Attainment
We conducted GWAS of educational attainment, an outcome with a high genetic correlation with intelligence5, in a non-overlapping European subset of the UKB sample (N=188,435) who did not complete the intelligence measure. Educational attainment was coded as maximum years of education completed, using the same methods as earlier analyses40 and GWAS was conducted using the same quality control and analytic procedures as described for the UKB intelligence phenotype (Supplementary Information 1.1.1). To test replication of the SNPs with this proxy phenotype, we performed a sign concordance test for all GWS SNPs from the meta-analysis using the two-tailed exact binomial test. For each independent genomic locus, we considered it to be evidence for replication if the lead SNP or another correlated SNP in the region was sign concordant with the corresponding SNP in the intelligence meta-analysis and had a two-tailed P-value of association with educational attainment smaller than 0.05/242 independent tests=0.0002.
Polygenic Scoring
We calculated polygenic scores (PGS) based on the SNP effect sizes of the leave-one-out meta-analyses, from which four cohorts were (separately) excluded and reserved for score validation. These included a child (GENR), young adult (S4S), and adult sample (RS). We also included the UKB-wb sample to test for validation in a very large (N = 53,576) cohort with the greatest phenotypic similarity to the largest contributor to the meta-analysis statistics (UKB-ts), in order to maximize potential predictive power. PGS were calculated on the genotype data using LDpred21, a Bayesian PGS method that utilizes a prior on effect size distribution to remodel the SNP effect size and account for LD, and PRSice20, a PLINK41-based program that automates optimization of the set of SNPs included in the PGS based on a high-resolution filtering of the GWAS P-value threshold. LDpred PGS were applied to the called, cleaned, genotyped variants in each of the validation cohorts with UK Biobank as the LD reference panel. PRSice PGS were calculated on hard-called imputed genotypes using P-value thresholds from 0.0 to 0.5 in steps of 0.001. The explained variance (ΔR2) was derived from a linear model in which the GWAS intelligence phenotype was regressed on each PGS while controlling for the same covariates as in each cohort-specific GWAS, compared to a linear model with GWAS covariates only.
Stratified Heritability
We partitioned SNP heritability using stratified LD Score regression42 in three ways: 1) by functional annotation category, 2) by minor allele frequency (MAF) in six percentile bins, and 3) by chromosome. Annotations for 28 binary categories of putative functional genomic characteristics (e.g. coding or regulatory regions) were obtained from the LD score website (see URLs). With this method, enrichment/depletion of heritability in each category is calculated as the proportion of heritability attributable to SNPs in the specified category divided by the proportion of total SNPs annotated to that category. The Bonferroni-corrected significance threshold was .05/56 annotations=.0009.
Functional Annotation of SNPs
Functional annotation of SNPs implicated in the meta-analysis was performed using FUMA24 (see URLs). We selected all candidate SNPs in associated genomic loci having an r2≧0.6 with one of the independent significant SNPs (see above), a suggestive P-value (P<1e-5) and a MAF>0.0001 for annotations. Predicted functional consequences for these SNPs were obtained by matching SNPs’ chromosome, base-pair position, and reference and alternate alleles to databases containing known functional annotations, including ANNOVAR43 categories, Combined Annotation Dependent Depletion (CADD) scores23, RegulomeDB44 (RDB) scores, and chromatin states45,46. ANNOVAR categories identify the SNP’s genic position (e.g. intron, exon, intergenic) and associated function. CADD scores predict how deleterious the effect of a SNP is likely to be for a protein structure/function, with higher scores referring to higher deleteriousness. A CADD score above 12.37 is the threshold to be potentially pathogenic23. The RegulomeDB score is a categorical score based on information from expression quantitative trait loci (eQTLs) and chromatin marks, ranging from 1a to 7 with lower scores indicating an increased likelihood of having a regulatory function. Scores are as follows: 1a=eQTL + Transcription Factor (TF) binding + matched TF motif + matched DNase Footprint + DNase peak; 1b=eQTL + TF binding + any motif + DNase Footprint + DNase peak; 1c=eQTL + TF binding + matched TF motif + DNase peak; 1d=eQTL + TF binding + any motif + DNase peak; 1e=eQTL + TF binding + matched TF motif; 1f=eQTL + TF binding / DNase peak; 2a=TF binding + matched TF motif + matched DNase Footprint + DNase peak; 2b=TF binding + any motif + DNase Footprint + DNase peak; 2c=TF binding + matched TF motif + DNase peak; 3a=TF binding + any motif + DNase peak; 3b=TF binding + matched TF motif; 4=TF binding + DNase peak; 5=TF binding or DNase peak; 6=other; 7=Not available. The chromatin state represents the accessibility of genomic regions (every 200bp) with 15 categorical states predicted by a hidden Markov model based on 5 chromatin marks for 127 epigenomes in the Roadmap Epigenomics Project46. A lower state indicates higher accessibility, with states 1–7 referring to open chromatin states. We annotated the minimum chromatin state across tissues to SNPs. The 15-core chromatin states as suggested by Roadmap are as follows: 1=Active Transcription Start Site (TSS); 2=Flanking Active TSS; 3=Transcription at gene 5’ and 3’; 4=Strong transcription; 5= Weak Transcription; 6=Genic enhancers; 7=Enhancers; 8=Zinc finger genes & repeats; 9=Heterochromatic; 10=Bivalent/Poised TSS; 11=Flanking Bivalent/Poised TSS/Enhancer; 12=Bivalent Enhancer; 13=Repressed PolyComb; 14=Weak Repressed PolyComb; 15=Quiescent/Low. Standardized SNP effect sizes were calculated for the most impactful SNPs by transforming the sample size-weighted meta-analysis Z score, as described by Zhu et al.47.
Gene-mapping
Genome-wide significant loci obtained by the GWAS meta-analysis were mapped to genes in FUMA24 using three strategies:
Positional mapping maps SNPs to genes based on physical distance (within a 10kb window) from known protein coding genes in the human reference assembly (GRCh37/hg19).
eQTL mapping maps SNPs to genes with which they show a significant eQTL association (i.e. allelic variation at the SNP is associated with the expression level of that gene). eQTL mapping uses information from 45 tissue types in 3 data repositories (GTEx48, Blood eQTL browser49, BIOS QTL browser50), and is based on cis-eQTLs which can map SNPs to genes up to 1Mb apart. We used a false discovery rate (FDR) of 0.05 to define significant eQTL associations.
Chromatin interaction mapping was performed to map SNPs to genes when there is a three-dimensional DNA-DNA interaction between the SNP region and a gene region. Chromatin interaction mapping can involve long-range interactions as it does not have a distance boundary. FUMA currently contains Hi-C data of 14 tissue types from the study of Schmitt et al51. Since chromatin interactions are often defined in a certain resolution, such as 40kb, an interacting region can span multiple genes. If a SNPs is located in a region that interacts with a region containing multiple genes, it will be mapped to each of those genes. To further prioritize candidate genes, we selected only interaction-mapped genes in which one region involved in the interaction overlaps with a predicted enhancer region in any of the 111 tissue/cell types from the Roadmap Epigenomics Project46 and the other region is located in a gene promoter region (250bp up and 500bp downstream of the transcription start site and also predicted by Roadmap to be a promoter region). This reduces the number of genes mapped but increases the likelihood that those identified will have a plausible biological function. We used a FDR of 1×10−5 to define significant interactions, based on previous recommendations51 modified to account for the differences in cell lines used here.
Functional annotation of mapped genes
Genes implicated by mapping of significant GWAS SNPs were further investigated using the GENE2FUNC procedure in FUMA24, which provides hypergeometric tests of enrichment of the list of mapped genes in 53 GTEx48 tissue-specific gene expression sets, 7,246 MSigDB gene-sets52, and 2,195 GWAS catalog gene-sets28. The Bonferroni-corrected significance threshold was 0.05/9,494 gene-sets=5.27×10-6.
Gene-based analysis
SNP-based P-values from the meta-analysis were used as input for the gene-based genome-wide association analysis (GWGAS). 18,128 protein-coding genes (each containing at least 1 GWAS SNP) from the NCBI 37.3 gene definitions were used as basis for GWGAS in MAGMA25 (see URLs). The Bonferroni-corrected genome-wide significance threshold was .05/18,128 genes=2.76×10-6.
Gene-set analysis
Results from the GWGAS analyses were used to test for association in three types of predefined gene-sets:
7,246 curated gene-sets representing known biological and metabolic pathways were derived from 9 data resources, catalogued by and obtained from the MsigDB version 5.229 (see URLs)
gene expression values from 53 tissues obtained from GTEx48, log2 transformed with pseudocount 1 after winsorization at 50 and averaged per tissue
cell-type specific gene expression in 24 types of brain cells, which were calculated following the method described in Skene et al.53 and Coleman et al.37 Briefly, brain cell-type expression data was drawn from single-cell RNA sequencing data from mouse brains. For each gene, the value for each cell-type was calculated by dividing the mean Unique Molecular Identifier (UMI) counts for the given cell type by the summed mean UMI counts across all cell types. Single-cell gene-sets were derived by grouping genes into 40 equal bins by specificity of expression.
These gene-sets were tested for association with the GWGAS gene-based test statistics using MAGMA. We computed competitive P-values, which represent the test of association for a specific gene-set compared to other gene-sets. This method is more robust to Type I error than self-contained tests that only test for association of a gene-set against the null hypothesis of no association25. The Bonferroni-corrected significance threshold was 0.05/7,323 gene-sets=6.83×10-6. Conditional analyses were performed as a follow-up using MAGMA to test whether each significant association observed was independent of all others. The association between each gene-set was tested conditional on the most strongly associated set, and then - if any substantial (p<.05/number of gene-sets) associations remained - by conditioning on the first and second most strongly associated set, and so on until no associations remained. Gene-sets that retained their association after correcting for other sets were considered to be independent signals. We note that this is not a test of association per se, but rather a strategy to identify, among gene-sets with known significant associations whose defining genes may overlap, which set(s) are responsible for driving the observed association.
Cross-Trait Genetic Correlation
Genetic correlations (rg) between intelligence and 38 phenotypes were computed using LD score regression39, as described above, based on GWAS summary statistics obtained from publicly available databases (see URLs; Supplementary Table 18). The Bonferroni-corrected significance threshold was 0.05/38 traits=1.32×10-3.
GWAS catalog lookup
We used FUMA to identify SNPs with previously reported (P < 5×10−5) phenotypic associations in published GWAS listed in the NHGRI-EBI catalog28 which overlapped with the genomic risk loci identified in the meta-analysis. As an additional relevant phenotype of interest, we examined whether the genes associated with intelligence in this study (by FUMA mapping or GWGAS) were overrepresented in a set of 1,518 genes linked to intellectual disability and/or developmental delay, as compiled by RegionAnnotater (see URLs). Many of these have been identified by non-GWAS sources and are not represented in the NHGRI catalog. We tested for enrichment using a hypergeometric test with a background set of 19,283 genomic protein-coding genes, as in FUMA. Manual lookups were also performed to identify overlapping loci/genes with known previous GWAS of intelligence.
Mendelian Randomization
To infer credible causal associations between intelligence and traits that are genetically correlated with intelligence, we performed Generalized Summary-data based Mendelian Randomization29 (GSMR; see URLs). This method utilizes summary-level data to test for causal associations between a putative risk factor (exposure) and an outcome by using independent genome-wide significant SNPs as instrumental variables. HEIDI-outlier detection was used to filter genetic instruments that show clear pleiotropic effects on both the exposure phenotype and the outcome phenotype. We used a threshold p-value of 0.01 for the outlier detection analysis in HEIDI which removes 1% of SNPs by chance if there is no pleiotropic effect. To test for a potential causal effect of intelligence on various outcomes, we selected traits in non-overlapping samples that showed significant genetic correlations (rg) with intelligence. We tested for bi-directional causation by repeating the analyses while switching the role of each correlated phenotype as an exposure and intelligence as the outcome. For each trait, we selected independent (r2=<0.1), GWS lead SNPs as instrumental variables in the analyses. For traits with less than 10 GWS lead SNPs (i.e. the minimum number of SNPs on which GSMR can perform a reliable analysis), the GWS threshold was lowered to 1×10−5, allowing a sufficient number of SNPs to conduct the reverse GSMR analysis for former smoker status, autism, and intracranial volume.
The method estimates a putative causal effect of the exposure on the outcome (bxy) as a function of the relationship between the SNPs’ effects on the exposure (bzx) and the SNPs’ effects on the outcome (bzy), given the assumption that the effect of non-pleiotropic SNPs on an exposure (x) should be related to their effect on the outcome (y) in an independent sample only via mediation through the phenotypic causal pathway (bxy). The estimated causal effect coefficients (bxy) are approximately equal to the natural log odds ratio (OR) for a case-control trait29. An OR of 2 can be interpreted as a doubled risk compared to the population prevalence of a binary trait for every SD increase in the exposure trait. For quantitative traits the bxy can be interpreted as a one standard deviation increase explained in the outcome trait for every SD increase in the exposure trait. This method can help differentiate the likely causal direction of association between two traits but cannot make any statement about the intermediate mechanisms involved in any potential causal process.
Supplementary Material
Acknowledgments
This work was funded by The Netherlands Organization for Scientific Research (NWO VICI 453–14-005 and NWO VIDI 452–12-014 to D.P.) and the Sophia Foundation for Scientific Research (SSWO grant S-1427 to P.R.J.). The analyses were carried out on the Genetic Cluster Computer, which is financed by the Netherlands Scientific Organization (NWO: 480–05-003), Vrije Universiteit, Amsterdam, The Netherlands, and the Dutch Brain Foundation, and is hosted by the Dutch National Computing and Networking Services SurfSARA. Support for data analysis was also provided by the Swiss National Science Foundation (to J.B.). This research has been conducted using the UK Biobank resource under application number 16406. We thank the numerous participants, researchers, and staff from many studies who collected and contributed to the data. Additional acknowledgments can be found in the Supplementary Information file.
Footnotes
Competing Financial Interests Statement
PF Sullivan reports the following potentially competing financial interests: Lundbeck (advisory committee), Pfizer (Scientific Advisory Board member), and Roche (grant recipient, speaker reimbursement). G Breen reports consultancy and speaker fees from Eli Lilly and Illumina and grant funding from Eli Lilly. J Hjerling-Leffler reports interests from Cartana (Scientific Advisor) and Roche (grant recipient). TD Cannon is a consultant to Boehringer Ingelheim Pharmaceuticals and Lundbeck A/S. All other authors declare no financial interests or potential conflicts of interest.
URLs:
UK Biobank website: http://ukbiobank.ac.uk
UK Biobank genotyping: http://www.biorxiv.org/content/early/2017/07/20/166298
Health and Retirement study: http://hrsonline.isr.umich.edu
Genes for Good study: http://genesforgood.org
International Cognitive Ability Resource measure (Genes for Good): https://icar-project.com/
Functional Mapping and Annotation (FUMA) software: http://fuma.ctglab.nl
Multi-marker Analysis of GenoMic Annotation (MAGMA) software: http://ctg.cncr.nl/software/magma
METAL software: http://genome.sph.umich.edu/wiki/METAL_Program
LD Score Regression software: https://github.com/bulik/ldsc
LD Hub (GWAS summary statistics): http://ldsc.broadinstitute.org/
LD scores: https://data.broadinstitute.org/alkesgroup/LDSCORE/
GeneCards: http://www.genecards.org
Psychiatric Genomics Consortium (GWAS summary statistics): http://www.med.unc.edu/pgc/results-and-downloads
MSigDB curated gene-set database: http://software.broadinstitute.org/gsea/msigdb/collections.jsp
NHGRI GWAS catalog: https://www.ebi.ac.uk/gwas/
RegionAnnotator: https://github.com/ivankosmos/RegionAnnotator
Generalized Summary-data-based Mendelian Randomization software: http://cnsgenomics.com/software/gsmr/
Data Availability Statement
Summary statistics will be available for download upon publication from https://ctg.cncr.nl.
References
- 1.Polderman TJ et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47, 702–709, doi: 10.1038/ng.3285 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wraw C, Deary IJ, Gale CR & Der G Intelligence in youth and health at age 50. Intelligence 53, 23–32, doi: 10.1016/j.intell.2015.08.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies G et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949). Mol Psychiatry 20, 183–192, doi: 10.1038/mp.2014.188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies G et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151). Mol Psychiatry 21, 758–767, doi: 10.1038/mp.2016.45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sniekers S et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat Genet 49, 1107–1112, doi: 10.1038/ng.3869 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trampush JW et al. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol Psychiatry 22, 336–345, doi: 10.1038/mp.2016.244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zabaneh D et al. A genome-wide association study for extremely high intelligence. Mol Psychiatry, doi: 10.1038/mp.2017.121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen AR The G Factor: The Science of Mental Ability. (Praeger, 1998). [Google Scholar]
- 9.Carroll JB Human Cognitive Abilities: A Survey of Factor-Analytic Studies. (Cambridge University Press, 1993). [Google Scholar]
- 10.Spearman C “General Intelligence,” Objectively Determined and Measured. The American Journal of Psychology 15, 201–292, doi: 10.2307/1412107 (1904). [DOI] [Google Scholar]
- 11.Plomin R & Kovas Y Generalist genes and learning disabilities. Psychol Bull 131, 592–617, doi: 10.1037/0033-2909.131.4.592 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Plomin R & von Stumm S The new genetics of intelligence. Nat Rev Genet, doi: 10.1038/nrg.2017.104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson W, Bouchard TJ, Krueger RF, McGue M & Gottesman II Just one g: consistent results from three test batteries. Intelligence 32, 95–107, doi: 10.1016/S0160-2896(03)00062-X (2004). [DOI] [Google Scholar]
- 14.Johnson W, Nijenhuis J. t. & Bouchard TJ Still just 1 g: Consistent results from five test batteries. Intelligence 36, 81–95, doi: 10.1016/j.intell.2007.06.001 (2008). [DOI] [Google Scholar]
- 15.Deary IJ, Penke L & Johnson W The neuroscience of human intelligence differences. Nat Rev Neurosci 11, 201–211, doi: 10.1038/nrn2793 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Deary IJ Intelligence. Annu Rev Psychol 63, 453–482, doi: 10.1146/annurev-psych-120710-100353 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–295, doi: 10.1038/ng.3211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deary IJ, Strand S, Smith P & Fernandes C Intelligence and educational achievement. Intelligence 35, 13–21, doi: 10.1016/j.intell.2006.02.001 (2007). [DOI] [Google Scholar]
- 19.Rietveld CA et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340, 1467–1471, doi: 10.1126/science.1235488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Euesden J, Lewis CM & O’Reilly PF PRSice: Polygenic Risk Score software. Bioinformatics 31, 1466–1468, doi: 10.1093/bioinformatics/btu848 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilhjalmsson BJ et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet 97, 576–592, doi: 10.1016/j.ajhg.2015.09.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill WD et al. Molecular genetic aetiology of general cognitive function is enriched in evolutionarily conserved regions. Transl Psychiatry 6, e980, doi: 10.1038/tp.2016.246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kircher M et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46, 310–315, doi: 10.1038/ng.2892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe K, Taskesen E, van Bochoven A & Posthuma D FUMA: Functional mapping and annotation of genetic associations. Nat Commun 8, 1826. doi: 10.1038/s41467-017-01261-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11, e1004219, doi: 10.1371/journal.pcbi.1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29, doi: 10.1038/75556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posthuma D et al. The association between brain volume and intelligence is of genetic origin. Nat Neurosci 5, 83–84, doi: 10.1038/nn0202-83 (2002). [DOI] [PubMed] [Google Scholar]
- 28.MacArthur J et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45, D896–d901, doi: 10.1093/nar/gkw1133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Z et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 9, 224, doi: 10.1038/s41467-017-02317-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson W, Deary IJ & Iacono WG Genetic and environmental transactions underlying educational attainment. Intelligence 37, 466–478, doi: 10.1016/j.intell.2009.05.006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards M & Sacker A Is education causal? Yes. Int J Epidemiol 40, 516–518, doi: 10.1093/ije/dyq166 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Kendler KS, Ohlsson H, Sundquist J & Sundquist K IQ and Schizophrenia in a Swedish National Sample: Their Causal Relationship and the Interaction of IQ with Genetic Risk. Am J Psychiatry 172, 259–265, doi: 10.1176/appi.ajp.2014.14040516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Hellard S et al. Identification of Gene Loci That Overlap Between Schizophrenia and Educational Attainment. Schizophr Bull 43, 654–664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-Only References
- 34.Willer CJ, Li Y & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191, doi: 10.1093/bioinformatics/btq340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peloso GM et al. Phenotypic extremes in rare variant study designs. Eur J Hum Genet 24, 924–930, doi: 10.1038/ejhg.2015.197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S, Cherny SS & Sham PC Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19, 149–150 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Coleman J et al. Biological annotation of genetic loci associated with intelligence in a meta-analysis of 87,740 individuals. Mol Psychiatry, doi: 10.1038/s41380-018-0040-6 (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konig IR, Loley C, Erdmann J & Ziegler A How to include chromosome X in your genome-wide association study. Genet Epidemiol 38, 97–103, doi: 10.1002/gepi.21782 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236–1241, doi: 10.1038/ng.3406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okbay A et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542, doi: 10.1038/nature17671 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7, doi: 10.1186/s13742-015-0047-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–1235, doi: 10.1038/ng.3404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Li M & Hakonarson H ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164, doi: 10.1093/nar/gkq603 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle AP et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22, 1790–1797, doi: 10.1101/gr.137323.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernst J & Kellis M ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9, 215–216, doi: 10.1038/nmeth.1906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330, doi: 10.1038/nature14248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Z et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48, 481–487, doi:10.1038/ng.3538 10.1038/ng.3538http://www.nature.com/ng/journal/v48/n5/abs/ng.3538.html#supplementary-informationhttp://www.nature.com/ng/journal/v48/n5/abs/ng.3538.html#supplementary-information (2016). [DOI] [PubMed] [Google Scholar]
- 48.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660, doi: 10.1126/science.1262110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westra HJ et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45, 1238–1243, doi: 10.1038/ng.2756 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhernakova DV et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat Genet 49, 139–145, doi: 10.1038/ng.3737 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Schmitt AD et al. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell reports 17, 2042–2059, doi: 10.1016/j.celrep.2016.10.061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liberzon A et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740, doi: 10.1093/bioinformatics/btr260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skene NG et al. Genetic Identification Of Brain Cell Types Underlying Schizophrenia. bioRxiv, doi: 10.1101/145466 (2017). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.