Summary
Planarians are flatworms capable of regenerating any missing body part in a process requiring stem cells and positional information. Muscle is a major source of planarian positional information and consists of several types of fibers with distinct regulatory roles in regeneration. The transcriptional regulatory programs used to specify different muscle fibers are poorly characterized. Using single-cell RNA sequencing, we define the transcriptomes of planarian dorsal-ventral muscle (DVM), intestinal muscle (IM), and pharynx muscle. This analysis identifies foxF-1, which encodes a broadly conserved Fox-family transcription factor, as a master transcriptional regulator of all non-body wall muscle. The transcription factor genes nk4 and gata4/5/6–2 specify two different subsets of DVM, lateral and medial, respectively, whereas gata4/5/6–3 specifies IM. These muscle types all express planarian patterning genes. Both lateral and medial DVM are required for medial-lateral patterning in regeneration whereas medial DVM and IM have a role in maintaining and regenerating intestine morphology. In addition to the role in muscle, foxF-1 is required for the specification of multiple cell types with transcriptome similarities, including high expression levels of cathepsin genes. These cells include pigment cells, glia, and several other cells with unknown function. cathepsin+ cells phagocytose E. coli, suggesting these are phagocytic cells. In conclusion, we describe a regulatory program for planarian muscle cell subsets and phagocytic cells both driven by foxF-1. FoxF proteins specify different mesoderm-derived tissues in other organisms, suggesting that FoxF regulates formation of an ancient and broadly conserved subset of mesoderm derivatives in the Bilateria.
eTOC Blurb
Planarian muscle provides positional information. Scimone et al. describe the transcriptome of major muscle subsets, identify the transcription factors required for their specification, and analyze their regenerative function. Besides a role in muscle, foxF-1 is also required for specification of previously unknown planarian phagocytic cells.
Introduction
Planarian regeneration and tissue turnover involve stem cells called neoblasts and positional information, which involves signaling molecules that pattern the planarian body plan. Genes proposed to encode positional information in planarians, often called position control genes (PCGs), are expressed predominantly in muscle cells in a regionally-restricted manner across body axes [1, 2].
Planarians have multiple muscle types (Figure 1A; [3]). Body-wall muscle (BWM) exists subepidermally and contains circular, diagonal, and longitudinal fibers. Dorsal-ventral muscle (DVM) connects dorsal and ventral surfaces. Intestinal muscle (IM) surrounds intestine branches. Finally, pharynx muscle consists of circular and longitudinal fibers associated with the elaborate movements of this feeding organ. In many animals, muscle has been classified as skeletal/somatic, cardiac, or visceral/intestinal. Based on ultrastructure, muscle is classified into striated or smooth. In vertebrates, skeletal and cardiac muscle cells are striated, but IM is smooth. In Drosophila and C. elegans most muscles, including IM, are striated [4–6]. Therefore, understanding the evolutionary relationship of different muscle types in bilaterians requires study of additional organisms. Annelids (Platynereis) have both smooth and striated muscles, which express conserved transcription factors (TFs) associated with muscle specification in other organisms [7, 8]. Planarian muscles resemble smooth muscles from vertebrates, although they express effector genes typically found in striated muscles (e.g., troponin) [3].
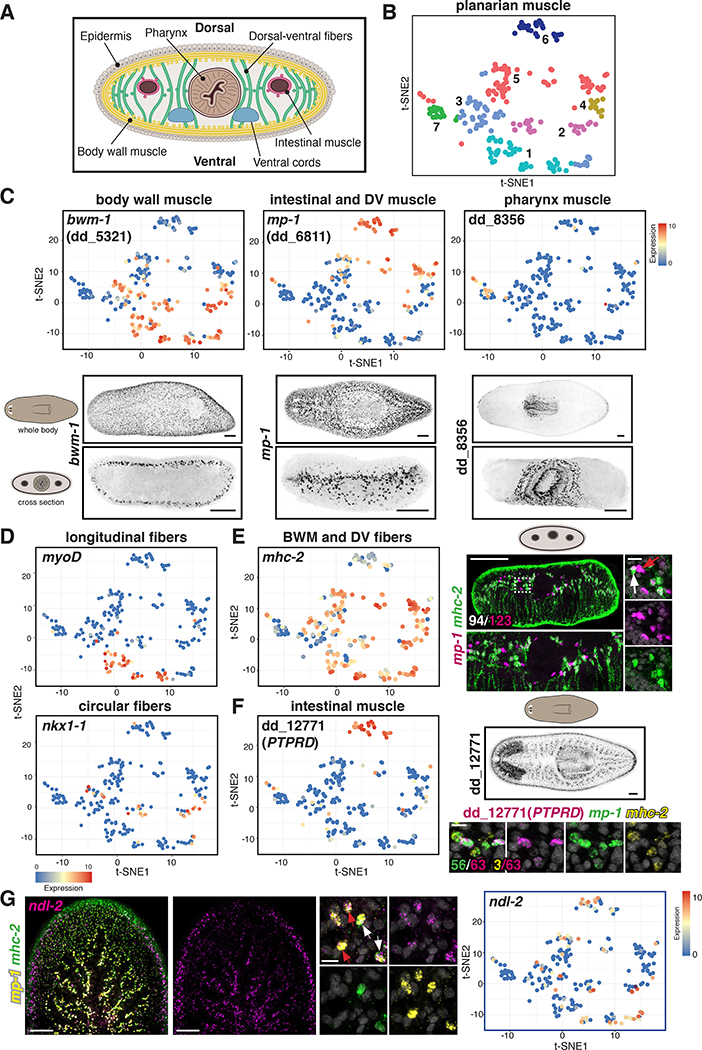
Figure 1. Single-muscle-cell RNA sequencing identifies distinct muscle subset transcriptomes.
(A) Diagram of a planarian cross section. (B) t-SNE representation of clustered muscle cells (dots) colored according to their planarian muscle-cluster-SSC assignment. (C) Top: t-SNE plots colored by gene expression of muscle genes. Bottom: Expression pattern of those genes. (D) t-SNE plots colored according to TF gene expression in longitudinal (top) and circular (bottom) muscle fibers. (E) Left: t-SNE plot colored according to mhc-2 gene expression. Right: Expression of mhc-2+/mp-1+ DVM cells and mhc-2-/mp-1+ IM cells. White arrow, DVM (mhc-2+/mp-1+); red arrow, IM (mhc-2-/ mp-1+). In white, number of mp-1+/mhc-2+ cells out of total mp-1+ cells. (F) Left: t-SNE plot colored according to IM dd_12771(PTPRD) gene expression. Right top: Expression pattern of those genes. Green, number of mp-1+/ dd_12771 (PTPRD)+; yellow, number of mhc-2+ dd_12771(PTPRD)+ out of total dd_12771(PTPRD)+ cells. (G) Left: ndl-2 expression in DVM (white arrows) and IM (red arrows) cells. Right: t-SNE plot colored according to ndl-2 expression. t-SNE plots: blue-to-red represents low-to-high expression (log2 CPM). Images are maximal intensity projections of the entire DV axis in C, or of planes around intestinal branches in E-G. Cartoons depict location of image shown. Bars: FISH panels, 100 μm; zoom-ins, 10 μm. See also Figures S1, S2, S3; Tables S1, S2, and Data S1.
Planarians provide an attractive model system to study muscle function and evolution because of their phylogenetic position within the Spiralia [9], because of the role of muscle in planarian patterning, and because of the ease of performing functional assays to study the roles of TF genes in planarian cell-fate specification. Planarian muscle specification does not appear to be controlled by a single transcriptional regulatory program. Instead, we previously found that myoD specifies BW longitudinal fibers and nkx1–1 specifies BW circular fibers [10]. Importantly, BW longitudinal and circular muscle subsets have distinct regulatory roles during planarian regeneration [10], raising the question of how the multiple other planarian muscle types are specified and whether they have specific regenerative functions. We utilized single-cell RNA sequencing and RNA interference (RNAi) to identify and study roles of TF genes in the specification of all major planarian muscle classes and to identify regeneration roles for these cells. Our findings unexpectedly revealed a role for a visceral muscle-specifying TF gene, foxF-1, in specification of previously unknown planarian phagocytic cells.
Results
Single-muscle-cell sequencing reveals transcriptomes for different muscle types
We combined previously published Smart-Seq single-cell RNA sequencing (Smart-Seq SCS) datasets of planarian muscle cells [2, 11] with an additional dataset of pre-pharyngeal muscle (Figure S1A,B; Table S1). 240 muscle cells were analysed using Seurat [12] and data were visualized using t-stochastic neighbour embedding (t-SNE; Methods). We identified seven muscle cell clusters (Figure 1B), which expressed well-characterized muscle markers (Figure S1C; [1, 11, 13]). Differential expression analysis between clusters identified cluster-enriched markers (Table S2). Neoblasts, planarian dividing cells, express smedwi-1 [14]. During neoblast specification into particular cell types, smedwi-1 expression is reduced concurrently with the tissue-specific marker expression. smedwi-1 expression levels were higher in cells on the left half of the t-SNE map (Figure S1D), suggesting that these were muscle progenitors.
Gene expression analysis by Smart-Seq SCS and fluorescence in situ hybridization (FISH) showed that previously described muscle markers [3, 11, 13] were expressed in specific muscle cell clusters. For example, body wall muscle-1 (dd_5321, bwm-1) (Figure S1E; [11]) was expressed in all BWM cells (by FISH) and its expression was highly enriched in clusters 1, 2, 3, and 4 but excluded from clusters 5, 6, and 7 (Figure 1C). Furthermore, known BWM cell types were found in cluster 1 (myoD+ longitudinal muscle cells) and cluster 2 (nkx1–1+ circular muscle cells), further classifying clusters 1–4 as BWM (Figure 1D). The previously published candidate IM marker multiplexin-1 (dd_6811, mp-1) (Figure S1E; [10, 11]) was expressed in clusters 5 and 6 and was excluded from BWM (Figure 1C). myosin heavy chain-2 (dd_432, mhc-2), which is expressed in BWM and DVM (Figure S1E; [3]), was expressed in cluster 5 but not 6 (Figure 1E), suggesting that cluster 5 represents DVM. FISH for mp-1 and mhc-2 showed that all DVM co-expressed both markers; however, some cells around intestinal branches expressed only mp-1 (Figure 1E; Figure S1E,F). Moreover, the recently described marker dd_12771(PTPRD) [13] had enriched expression in cluster 6 (Figure 1F), and FISH showed that dd_12771(PTPRD)+ cells also expressed mp-1 but not mhc-2, indicating that cluster 6 represents IM (Figure 1F; Figure S1E,F). Finally, pharynx muscle marker (dd_8356) expression [13, 15] was restricted to cluster 7, which was therefore considered pharynx muscle (Figure 1C). In summary, we identified transcriptomes for four main classes of muscle: BWM (including longitudinal and circular fibers), DVM, IM, and pharynx muscle.
To validate these findings, we performed a clustering analysis of muscle cells from a recently described Drop-Seq SCS (in short, Drop-Seq) dataset [13], generating 13 cell clusters (Figure S2A,B; Data S1). Similar to the Smart-Seq SCS analysis described above, cells with high smedwi-1 expression levels were clustered together with cells with low or no smedwi-1 expression but higher expression levels of differentiated markers, suggesting these cells represent transient stages of muscle differentiation (Figure S2C). Based on the expression of smedwi-1 and the muscle markers described above, all clusters from the Drop-Seq data, except cluster 10 (which contains a heterogeneous progenitor mixture), could be assigned to one of the four main muscle classes (Figure S2D-K; Data S1). Therefore, two independent SCS strategies identified similar muscle cell clusters. Pharynx muscle in the Drop-Seq data clustered into several subclusters (Figure S2L). Genes with enriched expression in cluster 7 of the Smart-Seq SCS data (Table S2) or in clusters 3, 4, 5, 8, 9, and 11 of the Drop-Seq data (Data S1) included several genes encoding broadly conserved TFs, many belonging to the bHLH TF family, and cofactors (Figure S2M-V).
The transcriptomes for all major muscle cell classes allow assessment of regeneration-regulatory genes in any muscle cell type of interest. PCGs are expressed in a constitutive and regional manner and are associated with planarian patterning pathways. Many PCGs are prominently expressed in BWM [1, 2]. Utilizing DVM and IM markers from the SCS analyses, we found that multiple PCGs, including ndl-2 (Figure 1G), ndl-3, several wnt genes, and slit, among others, were also expressed in DVM, IM, or in both muscle types (Figure S3). Interestingly, PCGs with AP-restricted BWM-expression domains displayed similar AP-restricted expression domains within DVM and IM (Figure S3C). These findings suggest that positional information is provided by different muscle subsets and that expression of these signalling molecules throughout the DV axis might be associated with patterning.
Distinct transcription factors are expressed in dorsal-ventral and intestinal muscle fibers
Differential expression analysis between the different muscle clusters identified genes encoding conserved TFs with enriched expression in the DVM and IM clusters, including nk4, gata4/5/6–2, gata4/5/6–3, and foxF-1 (Figure 2A; Figure S4; Table S2; Data S1). To understand the specification and regeneration roles of planarian DVM and IM, we focused study on these TF genes.
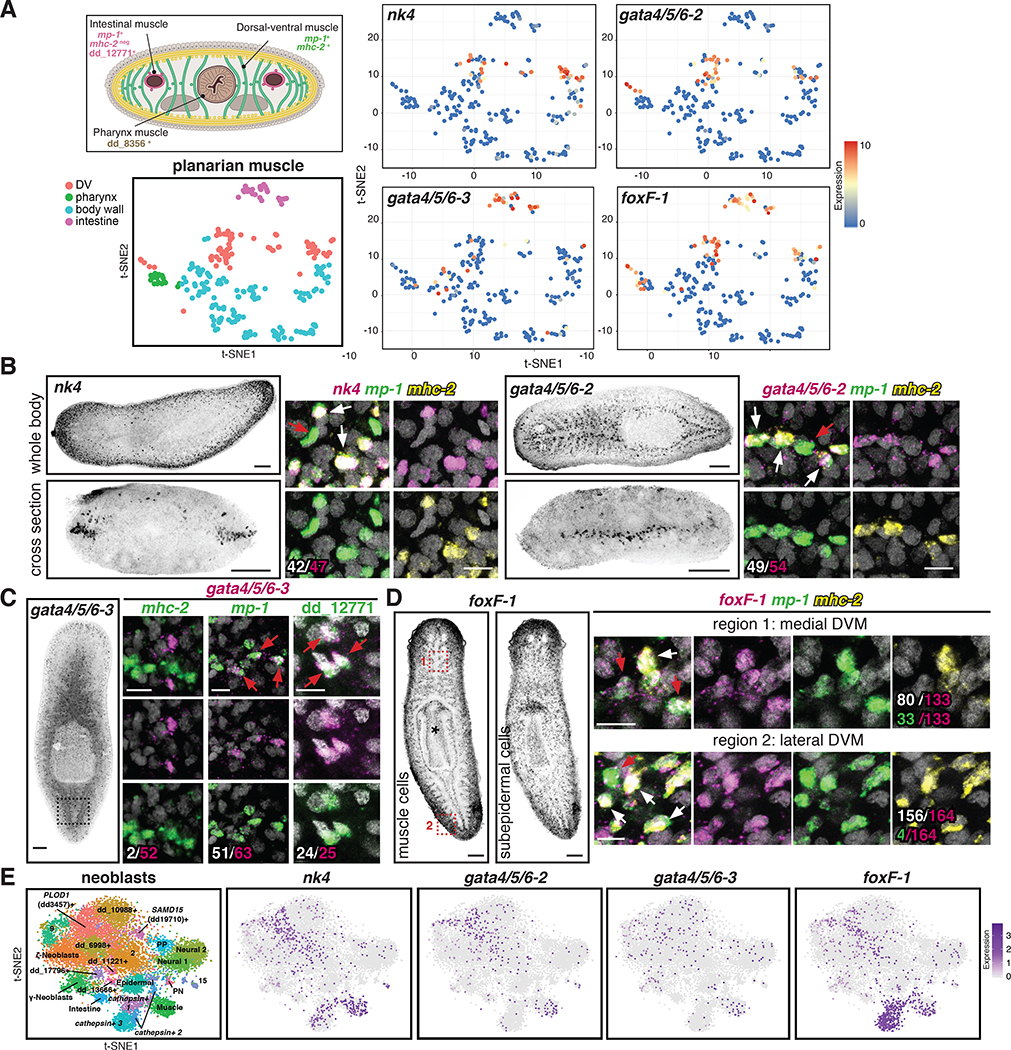
Figure 2. Conserved transcription factors are expressed in each muscle cluster.
(A) Top: Cross section diagram showing different muscle markers. Bottom: t-SNE representation of major planarian muscle classes. Right: t-SNE plots colored according to muscle-cluster TF gene expression. t-SNE plots: blue-to-red represents low-to-high expression (log2 CPM). (B) Left: nk4 (lateral DVM) and gata4/5/6–2 (medial DVM) expression patterns. Right: higher magnification of one confocal plane. White: number of TF+/ mp-1+/mhc-2+ cells out of total cells expressing the TF. (C) Left: gata4/5/6–3 (IM) expression. Right: higher magnification of one confocal plane showing gata4/5/6–3+/mhc-2-, mp-1+, and dd_12771(PTPRD)+ cells. White: number of muscle marker+/gata4/5/6–3+ out of total gata4/5/6–3+ cells. (D) Left: foxF-1 expression patterns. * pharynx muscle. Right: higher magnification of one confocal plane. White: number of mp-1+ mhc-2+foxF-1+ cells out of total foxF-1+ cells. Green: number of mp-1+/foxF-1+ cells out of total foxF-1+ cells. B-D: White arrows, DVM cells; red arrows, IM cells. (E) Left: t-SNE representation of subclusters from smedwi-1+ neoblasts [13]. Right: t-SNE plots showing nk4, gata4/5/6–2, gata4/5/6–3, and foxF-1 expression in neoblasts. Images are maximal intensity projections of the entire DV axis in B,C, or of planes around intestinal branches in D left. Bars: FISH panels, 100 μm; zoom in pictures, 10 μm. See also Figure S4; Table S2, and Data S1.
nk4 encodes a homeodomain TF that clusters in phylogenetic analyses with the NK4/Tinman class of NK homeobox genes (Figure S4A). Support for this homology assignment is modest; however, no other identified S. mediterranea gene had better support to represent the planarian ortholog of this widely conserved NK-family TF class (Figure S4A). nk4/Tinman genes have prominent roles in cardiac development in both Drosophila and vertebrates [16–18]. nk4 was expressed in planarian DVM (Figure 2A; Figure S4D,E). gata4/5/6–2 and gata4/5/6–3 encode homologs of the conserved zinc-finger DNA-binding-family of TFs (Figure S4B; [19]). GATA4/5/6-family members have prominent roles in cardiac development [20–22] and planarian gata4/5/6–2 was also expressed in DVM (Figure 2A; Figure S4D,E). gata4/5/6–3 was previously identified as gata1/2/3b [19], however, phylogenetic analysis suggested that this gene is better classified to the GATA4/5/6 family ([23]; Figure S4B). gata4/5/6–3 was expressed in planarian IM (Figure 2A; Figure S4D,E). Finally, planarian foxF-1 encodes a homolog of Drosophila Biniou and vertebrate FoxF (Figure S4C), a forkhead homeodomain-family with broad myogenesis roles. foxF-1 was expressed in DVM (cluster 5), IM (cluster 6), and part of the pharynx muscle (cluster 7), but its expression was excluded from BWM (clusters 1, 2, 3, and 4) (Figure 2A; Figure S4D,E).
FISH experiments were consistent with the sequencing data and further revealed that the DVM has two domains of muscle cells. Specifically, nk4 was highly expressed laterally, around the animal periphery, whereas gata4/5/6–2 was expressed more internally around intestinal branches (Figure 2B). As expected, both nk4+ and gata4/5/6–2+ cells co-expressed the DVM markers mhc-2 and mp-1 (Figure 2B; Figure S4E). gata4/5/6–3 was not exclusively expressed in muscle cells (Figure S4F), but gata4/5/6–3+ muscle cells were found sparsely around the intestinal branches, were mp-1+/ mhc-2-, and expressed the IM marker dd_12771(PTPRD), demonstrating that gata4/5/6–3 is indeed expressed in IM cells (Figure 2C; Figure S4E). gata4/5/6–3 was also expressed in pharynx muscle (Figure S4D,G). foxF-1 had several expression domains (Figure 2D; Figure S4D,E,G): it was expressed in lateral and medial DVM (mp-1+/ mhc-2+), in IM cells (mp-1+/ mhc-2-), and in pharynx muscle. In addition, foxF-1 was also expressed broadly in a non-muscle subepidermal layer (Figure 2D; [24]). SCS data showed co-expression offoxF-1 with nk4 and gata4/5/6–2 in DVM cells, and with gata4/5/6–3 in IM cells (Figure S4H). nk4 and gata4/5/6–2 were also co-expressed in some DVM cells (Figure S4H). However, none of these genes were substantially expressed in myoD+ BWM cells (Figure S4H). Taken together, these data identified molecular signatures, including TF genes, for two populations of DVM cells (lateral DVM: nk4+/ foxF-1+ and medial DVM: gata4/5/6–2+/foxF-1+) and IM cells (gata4/5/6–3+/foxF-1+).
nk4, gata4/5/6–2, and gata4/5/6–3 are required for the specification and maintenance of muscle subsets
TF-encoding genes required for the specification of differentiated cell types in planarians are often expressed in those differentiated cells and in neoblast subsets. The four muscle-TF genes identified above were also expressed in smedwi-1+ neoblasts, including muscle-progenitor subclusters (Figure 2E; [13]). Moreover, in the Smart-Seq SCS data, foxF-1 was co-expressed with nk4, gata4/5/6–2, or gata4/5/6–3 genes in smedwi-1+ cells (Figure S4I), consistent with a possible role in specifying different muscle cell subsets.
To determine whether nk4, gata4/5/6–2, gata4/5/6–3, and foxF-1 were important for muscle cell specification, we inhibited their expression with RNAi (Figure S5). Inhibition of nk4 and gata4/5/6–3 did not result in gross morphological defects during tissue turnover (Figure 3A). However, gata4/5/6–2 RNAi caused animals to expel their pharynges, which subsequently regrew (Figure 3A, B). Using RNA sequencing (Data S2) and FISH on RNAi animals, we determined that nk4 was required for detection of mhc-2+, col4–5+, mhc-3+ lateral DVM cells, the region where nk4+ cells were predominantly found (Figure 3C; Figure S5F-H), but was not essential for medial DVM (Figure S5G), dd_12771(PTPRD)+ IM (Figure S5H; Figure 3E), or BWM cells (Figure S5H). By contrast, gata4/5/6–2 RNAi animals had reduced numbers of medial DVM (mp-1+/ mhc-2+) but not IM (detected as mp-1+l mhc-2” or as dd_12771(PTPRD)+) or BWM cells (Figure 3D,E; Figure S5I). Lateral DVM cells (nk4+) were not detectably affected in gata4/5/6–2 RNAi animals (Figure S5J), indicating that this TF is mostly required to specify and/or maintain medial DVM cells. Finally, gata4/5/6–3 RNAi animals had reduced numbers of IM (detected as mp- 1+/ mhc-2” cells and as dd_12771(PTPRD)+ cells; Figure 3D,E) but not DVM or BWM cells (Figure 3D; Figure S5I), indicating a role for this gene in IM specification. In summary, these data indicate that nk4 is required for lateral DVM, gata4/5/6–2 is required for medial DVM, and gata4/5/6–3 is required for IM.
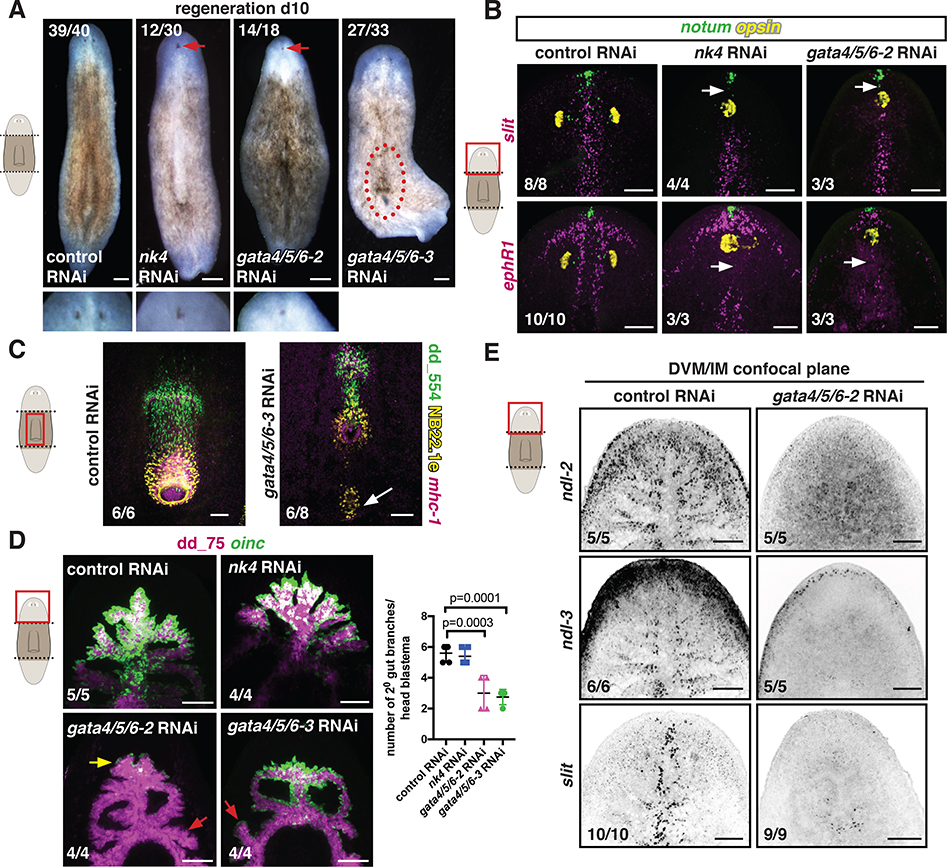
Figure 3. nk4, gata4/5/6–2, and gata4/5/6–3 are required for the specification of distinct muscle subsets.
(A) Uninjured RNAi animals. * missing pharynx. (B) New pharynx formation in gata4/5/6–2 RNAi animals. (C) Reduced lateral DVM cells (red arrow) in nk4 RNAi animals. Right, quantification. Mean ± SD, unpaired Student’s t-test. (D) Reduced medial DVM and IM cells in gata4/5/6–2 and gata4/5/6–3 RNAi animals, respectively. Bottom: quantification. Mean ± SD. One-way ANOVA, post Dunnetts’ test. (E) Reduced IM cells (red arrow) in a gata4/5/6–3 RNAi animal. Bottom: Reduced dd_12771(PTPRD)+ IM cells in a cross section. Below, quantification. Mean ± SD. One-way ANOVA, post Dunnetts’ test. 6G10 Ab labels muscle fibers. *intestinal lumen. (F) Intestinal defects (yellow arrows) in gata4/5/6–2 and gata4/5/6–3 RNAi animals. Bottom left: proportion of intestinal-branch fusion in RNAi conditions. Cartoon: primary and secondary intestinal branches. Bottom right: secondary branches numbers. Mean ± SD. One-way ANOVA, post Dunnetts’ test. Cartoons depict location of image shown or region where quantification was performed. Number of representative animals is shown. Images are maximal intensity projections of the entire DV axis in F, or of planes around intestinal branches in C-E. Bars: FISH and live images, 100 μm; except D,10 μm. See also Figure S5 and Data S2.
gata4/5/6–2 and gata4/5/6–3 are essential for gut morphogenesis
We used RNA sequencing (RNA-Seq) to assess changes in gene expression following muscle-TF RNAi (Data S2). Some muscle genes showed significantly reduced expression in gata4/5/6–2 RNAi animals (Figure S5F) and unexpectedly, several genes with intestine-enriched expression were also significantly downregulated (Figure S5K). Gene expression changes could reflect direct or indirect regulation by the Gata4/5/6–2 TF. During development of many vertebrates and Drosophila, visceral mesoderm (which generates intestinal muscle) and endoderm (which generates gut) are involved in reciprocal interactions important for gut formation [25]. Therefore, given this RNA- Seq data, we examined whether reduction of DVM or IM cells affected the intestine. RNAi of nk4 (lateral DVM) did not impact gut morphology (Figure S5H; Figure 3F). However, inhibition of gata4/5/6–2 (medial DVM) or gata4/5/6–3 (IM) resulted in intestine-structure defects, with intestine-branch fusion frequently observed (Figure 3F). Furthermore, the outer intestinal cell marker, oinc (dd_115+) expression was significantly reduced (FDR<0.05, Figure S5K) and fewer oinc+ cells were observed by FISH in gata4/5/6–2 RNAi animals (Figure 3F). gata4/5/6–2 was not expressed in intestinal cells and FISH experiments showed no co-expression of this TF and oinc (Figure S5L). gata4/5/6–3 RNAi animals also showed defects in intestinal branching, with secondary and tertiary branches severely reduced (Figure 3F). These data indicate that medial DVM has a role in formation/maintenance of outer intestine cells and that both medial DVM and IM cells are required to preserve normal intestinal branching morphology.
DVM cells are required for normal medial-lateral regeneration
The ability to ablate different muscle fibers presented the opportunity to assess their roles in planarian regeneration. nk4 (lateral DVM) or gata4/5/6–2 (medial DVM) inhibition did not affect wound closure but resulted in patterning defects affecting the medial-lateral (ML) axis during head and tail regeneration (Figure 4A; [19]). During regeneration, an unpigmented outgrowth (blastema) in which new tissues form is generated. nk4 and gata4/5/6–2 RNAi head blastemas were cyclopic and had perturbed expression domains of midline genes (Figure 4B; Figure S6A,B). This ML-patterning phenotype was similar to that of regenerating animals lacking the anterior pole, a group of notum-expressing cells that function as an organizer of the anterior and midline blastema regions [26, 27]. However, anterior-pole cells were still present in these RNAi animals (Figure 4B). Consistent with the phenotypes observed during tissue turnover, regenerating nk4 RNAi animals had reduced lateral but roughly normal medial DVM; regenerating gata4/5/6–2 RNAi animals had reduced medial but unaffected lateral DVM; and regenerating gata4/5/6–3 RNAi animals had reduced IM but no change in DVM cells (Figure S6C). Unexpectedly, some gata4/5/6–3 regenerating trunk fragments displayed ectopic mouths and pharynges (Figure 4C). Moreover, gata4/5/6–2 and gata4/5/6–3 RNAi animals also showed gut defects similar to their homeostatic RNAi phenotypes (Figure 4D).
Figure 4. nk4 and gata4/5/6–2 are required for normal medial-lateral patterning during regeneration.
(A) Regenerating RNAi animals. Higher magnification shows cyclopia. Red-dotted line, ectopic pharynx. (B) Abnormal expression patterns (white arrows) of midline genes (slit, ephR41) by FISH. opsin, photoreceptor neurons; notum, anterior pole. (C) Ectopic pharynx and mouth (white arrow) in a gata4/5/6–3 RNAi animal. (D) Reduced oinc+ cells (yellow arrow) in regenerating gata4/5/6–2 RNAi animals and abnormal intestinal branches in gata4/5/6–2 and gata4/5/6–3 RNAi animals (red arrows). Right, secondary branch numbers. Mean ± SD. One-way ANOVA, post Dunnetts’ test. (E) Reduced DVM expression of PCGs in a gata4/5/6–2 RNAi animal. Images are maximal intensity projections of the entire DV axis in B-D, or of planes around intestinal branches in E. Cartoons indicate image location. Bars, 100 μm. See also Figure S6.
The ML-patterning defect observed in nk4 and gata4/5/6–2 RNAi animals was also reminiscent of the slit RNAi phenotype. slit is a conserved midline-regulatory gene [1, 28]. Because gata4/5/6–2 RNAi animals had reduced numbers of medial DVM cells, which are the most abundant muscle cells around the gut branches, the expression of multiple PCGs, including slit, was severely reduced in the DVM (BWM PCG expression was normal) in regenerating fragments (Figure 4E). These results indicate that DVM and IM contribute patterning information that influences body-plan regeneration.
foxF-1 is required for DVM, IM, and pharynx muscle fibers
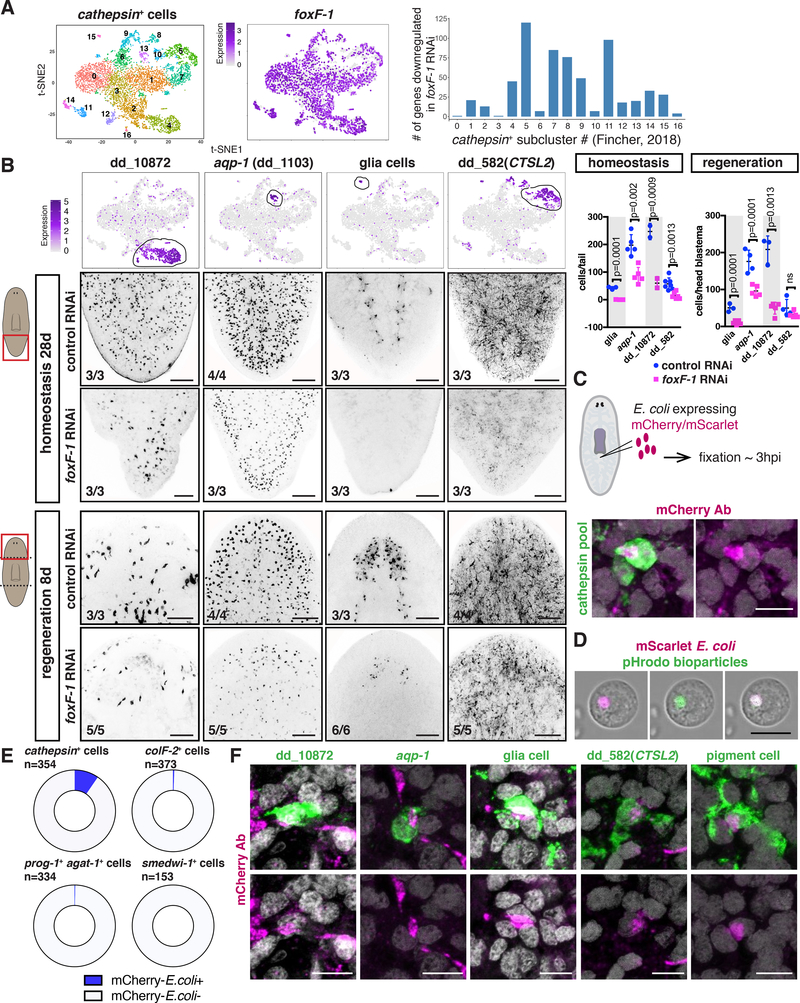
foxF-1 was unique among the identified muscle-associated TFs. First, foxF-1 was expressed in multiple muscle cell types, including lateral DVM, medial DVM, IM, and pharynx muscle cells, but not in BWM (Figure 2A). Second, foxF-1 was also expressed in a broad, non-muscle, subepidermal cell layer (Figure 2D; Figure 5A; Figure S7A). These non-muscle foxF-1-expressing cells were recently identified [13, 15] and belong to a large heterogenous group of cells that express a cathepsin gene similar to human CTSL2 (dd_175) and therefore have been referred to as “ cathepsin+ cells”. The cathepsin+ cluster contains several cell-type classes, including pigment cells, glia cells, and numerous cells of unknown function [13, 15]. foxF-1 was recently shown to be required for the specification of planarian pigment cells [24]. Accordingly, foxF-1 RNAi animals were depigmented (Figure 5B; Figure S7B; [24]).
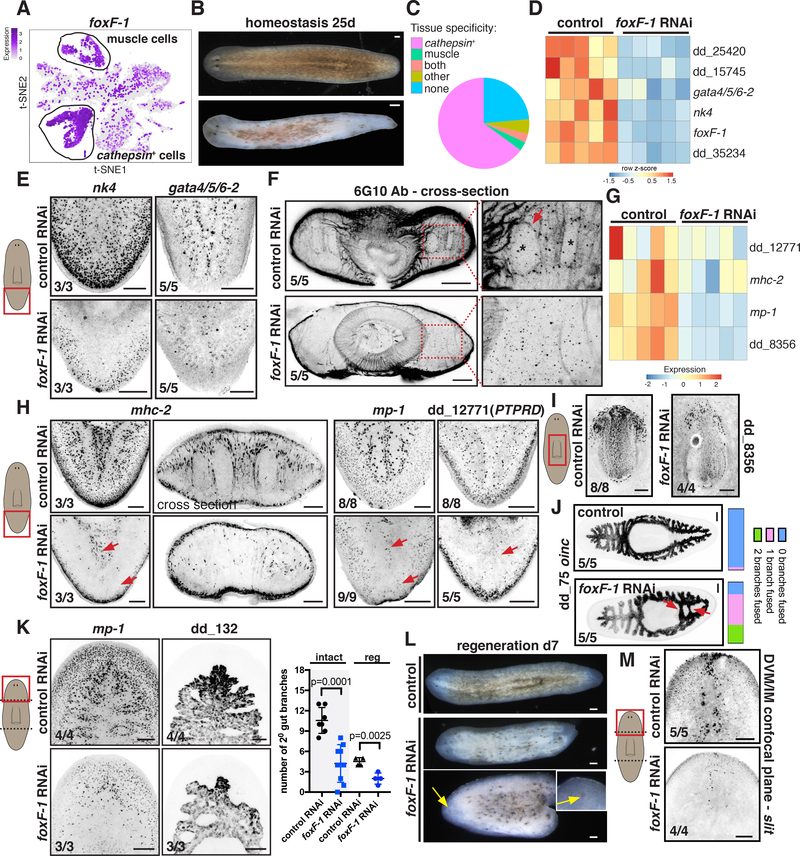
Figure 5. foxF-1 is required for the specification of all non-BWM in planarians.
(A). t-SNE plot: foxF-1 expression in muscle and non-muscle cathepsin+ cells. (B) Depigmentation of an uninjured foxF-1 RNAi animal. (C) Pie chart shows fraction of tissue-specific genes significantly downregulated (FDR < 0.05) in uninjured foxF-1 RNAi animals. (D) Heatmap of TF expression downregulation (FDR<0.001) in uninjured foxF-1 RNAi animals. Gene expression z-scores were calculated per row. (E) Reduced expression of nk4 and gata4/5/6–2 in uninjured foxF-1 RNAi animals. (F) Reduced DVM and IM fibers in an uninjured foxF-1 RNAi animal by immunostaining. (G, H) Reduced muscle-marker expression (red arrows) in uninjured foxF-1 RNAi animals (G) Heatmap, (H) FISH. (I) Reduced pharynx muscle numbers in a foxF-1 RNAi animal. (J) Abnormal intestinal structure in a foxF-1 RNAi animal (red arrows). Right: intestine-branch fusion quantification. (K) Left: Reduced mp-1+ cells and intestine-branching defects in regenerating foxF-1 RNAi animals. Right: secondary intestinal branch numbers in intact (tail region) and regenerating (head blastema) animals. Mean ± SD. Unpaired Student’s t-test. (L) Depigmentation and cyclopia (yellow arrows) of regenerating foxF-1 RNAi animals. (M) Reduced DVM cells expressing the PCG slit in a regenerating foxF-1 RNAi animal. Images are maximal intensity projections of planes around intestinal branches. Cartoons depict image location. Bars, 100 mm. See also Figure S7 and Data S2.
In contrast to nk4, gata4/5/6–2, or gata4/5/6–3 RNAi animals, which all were viable during RNAi (> 84 days), foxF-1 RNAi animals died between 20 and 30 days following RNAi initiation. Gene expression analysis of these animals (Data S2) showed significant downregulation (FDR<0.05) of 534 transcripts that had enriched expression in cathepsin+ cells and 183 muscle genes (Figure 5C; Figure S7C). foxF-1 RNAi animals also had significantly decreased expression of the DVM TFs nk4 and gata4/5/6–2 (FDR = 6.40E-28 and 1.14E-5, respectively, Figure 5D,E). Immunostainings of foxF-1 RNAi animals showed a severe decrease of DVM and IM fibers (Figure 5F), but no detectable changes in BWM fibers (Figure S7D). Pharynx muscle regeneration was also impaired (Figure S7E). Consistent with these findings, expression of DVM, IM, and pharynx muscle markers (mhc- 2, mp-1 (FDR=1.86E-28), dd_12771(PTPRD), dd_8356 (FDR=2.55E-21)) was severely reduced in foxF-1 RNAi animals, whereas BWM genes myoD and nkx1–1 were unaffected (Figure 5G-I; Figure S7E; Data S2).
Reduced presence of cells expressing DVM and IM muscle markers was also observed during regeneration offoxF-1 RNAi animals (Figure 5K). Some foxF-1 RNAi animals (7/29 trunk fragments) regenerated with ML defects (Figure 5L), similar to those observed in gata4/5/6–2 and nk4 RNAi animals, and had reduced slit (midline PCG) expression (Figure 5M). Some foxF-1 fragments failed to regenerate (4/29 trunk pieces). Regenerating and uninjured foxF-1 RNAi animals also showed abnormal intestine phenotypes, similarly to gata4/5/6–2 and gata4/5/6– 3 RNAi animals: reduced outer intestinal cells (dd_135+ and oinc+; FDR=9.51E-7 and 0.0002, respectively) and intestine-branching defects (Figure 5J,K; Figure S7F). These data together indicate that the TF-gene foxF-1 promotes specification and/or maintenance of medial and lateral DVM, IM, and pharynx muscle.
foxF-1 is essential for the specification of cathepsin+ cells
foxF-1 was broadly expressed in cells of the cathepsin+ cluster, despite the seemingly disparate functions of cell types in this group (Figure 6A; [13]). foxF-1 RNAi animals showed significantly decreased expression of cathepsin+-cell subcluster markers, which represent putative different cell types (Figure 6A, Data S2). foxF-1 thus not only regulates pigment cells [24] but also many other cathepsin+ cell types. FISH validated these findings in both uninjured and regenerating animals: foxF-1 RNAi animals had significantly fewer cathepsin+ cell types expressing the markers dd_10872, acq-1, and dd_582(CTSL2), as well as pigment and glia cells (Figure 6B; Figure 5B,L). These animals formed blastemas, indicating that regeneration capacity does not overtly require cathepsin+ cells. Lethality following foxF-1 RNAi indicates that reduction of IM and DVM together with cathepsin+ cells was incompatible with viability.
Figure 6. foxF-1 is required for the specification of the cathepsin+ cells.
(A) Left: t-SNE representation of all cathepsin+ subclusters [13]. Middle: t-SNE plot colored by foxF-1 expression. Graph: number of genes significantly downregulated in each cathepsin+ subcluster after foxF-1 RNAi. (B) t-SNE plots colored by gene expression for genes in different cathepsin subclusters. FISH: Reduced expression of those genes in uninjured (middle panels) and regenerating (bottom panels) foxF-1 RNAi animals. Graphs: quantification. Mean ± SD. Unpaired Student’s t-test. (C) Top: cartoon, mCherry/mScarlet-E.coli injection site. Bottom: co-labelling of mCherry and a pool of cathepsin+ cell marker probes (n=16 animals). (D) Co-labelling of mScarlet-expressing E.coli and pHrodo green bioparticles in live cells. (E) Higher proportion (p<1E-6, empirical test) of cathepsin+ cells labelled with the mCherry Ab compared to muscle (colF-2), epidermal progenitors (prog-1/agat-1), or neoblasts (smedwi-1). (F) Co-labelling of mCherry with different cathepsin+ subcluster markers. Cartoons indicate location of image shown. Number of representative animals is shown. Scale bars in B, 100 mm; in C, D, and F, 10 mm. See also Figure S7 and Data S2.
Most cathepsin+ cells are found throughout the planarian body and have elaborate morphologies with long processes [13]. Cathepsins are proteases mostly active at low pHs and enriched within lysosomes [29]. Lysosomes help degrade and process extracellular material taken up by endocytosis or intracellular material through autophagy. Based on the enriched lysosomal enzyme expression [13, 15] and their specialized morphology, we hypothesized that at least some cathepsin+-cluster cells are phagocytic. We tested this hypothesis by injecting planarians with mCherry-expressing E. coli and fixing the animals 2–3 hours later (Figure 6C). FISH and immunostaining experiments showed cathepsin+ cells containing mCherry protein (Figure 6C; Figure S7G). To further assess whether the bacterial particles were inside cells, we co-injected animals with mScarlet-expressing E. coli together with pHrodo green bioparticles™. These bioparticles contain E.coli fragments conjugated to a pH-sensitive fluorophore (fluorescent at low pH). mScarlet E.coli and green fluorescence overlapped tightly in live cells (Figure 6D; Figure S7H). To examine the specificity of the mCherry E.coli and cathepsin+ cell co-labeling, we utilized FISH. The fraction of cathepsin+ cells containing mCherry was significantly higher (p<1E-6, empirical test) than for any other neighbouring cell type tested (Figure 6E; Figure S7I). Occasionally, mCherry E.coli was also observed associated with protonephridia (Figure S7J). After animal dissociation, cells associated with mCherry/mScarlet-labelled E. coli or with bioparticles had similar complex cytoplasmic appearances, with numerous intracellular bodies observable by Nomarski microscopy (Figure S7H-L). Using an endogenous bacterial-level assay [30], foxF-1 RNAi animals had significantly more bacteria than controls (Figure S7M). This could reflect intestinal and/or cathepsin+ cell dysfunction. Cells labelled with the markers dd_10872, dd_582(CTSL2),and acq-1, as well as glia and pigment cells, were all associated with uptake of mCherry E. coli (Figure 6F). Taken together, these data suggest that many different cathepsin+ cell types are able to perform phagocytosis and they all required foxF-1 for their specification.
Discussion
Muscle is a fundamental tissue existing widely in animals. Mesoderm-derived muscle emerged with the Bilateria >550 million years ago. Planarian muscle is an attractive target for molecular study because planarians are placed within the Spiralia [9] and because of the positional information role of planarian muscle. Single-cell-RNA sequencing allowed us to identify different muscle subsets, to examine their molecular signatures, and to identify the genes that are required for specifying them. Planarian muscle is not specified by a single transcriptional program (Figure 7A). Instead, we previously described that myoD specified longitudinal fibers whereas nkx1–1 specified circular fibers of the BWM [10]. Interestingly, myoD is expressed specifically in longitudinal muscle in the annelid Platynereis dumerilii [7], raising the possibility that the ancestral role of myoD is for specification of longitudinal skeletal muscle-like cells. Here, we identified transcriptomes for planarian non-BWM, including DVM, IM, and pharynx muscle. We found two distinct DVM subsets, lateral and medial, that differ in their location and in the TFs involved in their specification. Both DVM subsets were required for normal ML patterning during regeneration whereas medial DVM and IM affect intestinal branching and morphology. Like BWM, all non-BWM subsets also expressed PCGs. Distributing PCG expression across different muscle types might be a mechanism by which signaling molecules expressed predominantly in muscle can regulate neoblast biology, and/or other differentiated tissues, throughout the DV/ML axes.
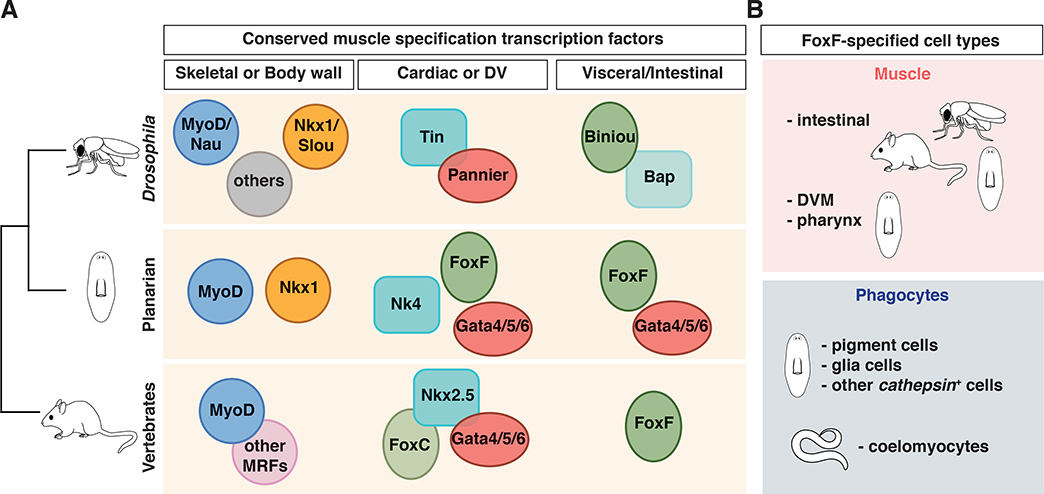
Figure 7. Conserved transcription factors in bilaterian muscle specification.
(A) Conserved TFs required for muscle specification in different model organisms. For skeletal/somatic/BW muscle: myoD for all skeletal muscle in vertebrates, for only a subset in Drosophila, and for longitudinal muscle in planarians. myoD is also specifically expressed in Platynereis longitudinal muscle [7]. Other TFs, such as nkx1/ slouch are required for the specification of other somatic/BWM subsets. For cardiac muscle: tinman (tin)/nk4/nkx2.5 andgata4/5/6 gene families are widely used. In many organisms (including vertebrates and C. intestinalis [50]), a member of the foxF/C class of TF genes is also required. In planarians, foxF-1 is expressed in and specifies DVM, which also express nk4 and/or gata4/5/6–2. Moreover, foxF homologs (biniou in Drosophila) are essential for visceral/IM specification. In Drosophila, a nkx3/bap (bagpipe) TF is also required. In planarians, another GATA4/5/6-family member is also required for IM specification. (B) foxF homologs are involved in visceral/IM specification in a wide group of model organisms including several vertebrates, flies, and planarians, and are also required for the specification of phagocytes in C. elegans and planarians, suggesting a broader role in specifying mesoderm derivatives.
Whereas lateral-DVM specification depends on foxF-1 and nk4, medial DVM requires both foxF-1 and gata4/5/6–2. In addition, IM required both foxF-1 and gata4/5/6–3. Homologs offoxF, nk4, and gata4/5/6 are involved in the specification of different muscle types (visceral/intestinal and cardiac) in many organisms. Homologs of the nk4/tinman and gata4/5/6 genes have roles in the specification and differentiation of cardiac muscle in Drosophila, the ascidian Ciona intestinalis, and several vertebrates [31, 32]. Certain gata4/5/6-family proteins have also roles in endoderm biology [33], and indeed, gata4/5/6–1 in planarians is involved in intestine differentiation [19, 34, 35]. Interestingly, only one GATA gene has been detected in the cnidarian Nematostella vectensis, and this is expressed around the gastrovascular cavity [36]. This suggests that in the ancestor of the Bilateria (in which mesoderm arose) duplication and divergence of an ancestral gata gene led to some family members with roles in muscle formation and others with roles in endoderm development. The requirements of some gata4/5/6-family genes in IM and DVM in planarians, and another gata4/5/6 gene in planarian intestine supports this model.
FoxF genes are widely conserved and essential for the specification and differentiation of visceral mesoderm into intestinal muscle in Drosophila, mouse, and Xenopus [37–39]. Moreover, FoxF and gata4/5/6 are also expressed in annelid intestinal muscle [7], suggesting broad utilization of FoxF genes in visceral muscle. Interestingly, even though vertebrates and Drosophila utilize similar TFs to specify visceral muscle, the ultrastructure of IM in these two animal groups is different (smooth in vertebrates and striated in Drosophila), suggesting that a molecular profile of core regulatory genes might be sufficient to determine cell type homology [40]. Recent ancestral state reconstruction from ultrastructural data suggests that cardiac and visceral muscle might both be derived from ancestral smooth muscle [7]. Even though each subset of planarian muscle fibers studied here utilized distinct TFs, all non-BWM required foxF-1 (Figure 7A). RNA-Seq and FISH experiments demonstrated diminished DVM subsets, IM cells, and pharynx muscle in foxF-1 RNAi animals. These results suggest that a unique gene regulatory network specifies each muscle type in planarians, with foxF-1 being a common component for all of the major non-BWM types. The existence of a similar function of FoxF in specifying intestinal muscle in Drosophila, vertebrates, and in planarians supports a model that FoxF had an ancient role in specification of visceral-like mesoderm, and that this molecular program has been conserved throughout bilaterian evolution.
In addition to the role of FoxF in muscle, we found that foxF-1 was essential for the specification and/or maintenance of a variety of non-muscle cell types, including pigment [24] and glia cells (Figure 7B). These different cell types highly express cathepsin genes [13, 15], lysosomal enzymes essential for processing of extracellular or intracellular particles. Different cathepsin+ cell types also have a dendritic-like shape reminiscent of phagocytic cells in other systems. Phagocytic cells suppress pathogen spread and can also remove apoptotic cells and cellular debris. Planarians are exposed to a wide range of microbes not only through their diet but also at wounds [30, 41]. Ingested bacteria can be phagocytosed by intestinal cells [42, 43], however, it is unclear if those cells are the only cells capable of phagocytosis. We found that several cathepsin+ cell types are able to phagocytose E. coli. The planarian cathepsin+ cluster contains cell types with some similarity to phagocytic cells in other organisms. For example, microglia, a type of glia cells of mesodermal origin [44], functions in the surveillance of the central nervous system [45]. Blastocoelar and pigment cells in larval sea urchins, which are specified from non-skeletogenic mesoderm [46], can phagocytose microbes [47]. Little evidence connects FoxF to the specification of phagocytic cells in other organisms; a C. elegans FoxF/FoxC-family gene, let-381, is expressed in the mesodermal M-lineage and is required for the formation of coelomocytes [48], which are scavenger cells in the animal pseudocoelom cavity [49].
In conclusion, we described here the molecular signature of all major muscle subsets in the planarian Schmidtea mediterranea, which is a member of the Spiralian superphylum, and studied their impact during regeneration. myoD and nkxl TF genes have restricted roles in different BWM subsets [10] and gata4/5/6, nk4, and foxF-1 TF genes have important roles for non-BWM muscle. Prominently in this gene set, foxF-1 is associated with all non-BWM muscle classes. Like BWM, our results indicate roles for non-BWM planarian muscle types beyond contractility, as a regulatory source of patterning information in regeneration. The broad role of foxF-1 in visceral muscle supports a view that this TF has an ancient and conserved role in specification of this class of muscle. Our findings also raise the possibility that FoxF might have a broad, but presently largely unrecognized, role in specifying glia, pigment cells, and/or other phagocytic cells in animals.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by, the Lead Contact, Dr.Peter W. Reddien (reddien@wi.mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Schmidtea mediterranea clonal strain CIW4 animals, starved for 7–14 days prior experimentation, were used for all experiments. Asexual animals were used and have indeterminate age because they are asexual. All animals utilized were of normal health, not used in previous procedures, and were of wild-type genotype. Animals were cultured in plastic containers, petri dishes for experiments, in 1x Montjuic water (1.6 mmol/l NaCl, 1.0 mmol/l CaCl2, 1.0 mmol/l MgSO4, 0.1 mmol/l MgCl2, 0.1 mmol/l KCl and 1.2 mmol/l NaHCO3 prepared in Milli-Q water) at 20°C in the dark. Animals were fed blended calf liver.
METHOD DETAILS
Replication, size estimation and randomization
At least two independent FISH and immunostaining experiments with a minimum of three animals/experiment were performed for the characterization of muscle markers, muscle TFs, and PCGs in DVM/IM (Figures 1, 2, S1, S3, S4). For RNAi phenotype characterization, numbers of animals used in each staining are indicated in each panel. No sample size estimation was performed. Animals for all experiments were randomly selected from a large collection of clonal animals for all experiments. All animals have been included in the statistical analysis, no exclusions have been done. Images were randomized before quantification.
Gene nomenclature
Genes that encode proteins with a clear domain structure have been assigned a name accordingly. For example, dd_6811 encodes a Multiplexin protein, and therefore has been named multiplexin-1 or mp-1. Similarly, dd_2500 encodes a Collagen 4 protein, and is therefore named col4–5. dd_1434 encodes a Myosin heavy chain and is named mhc-3. Genes that encode proteins with no clear domain structure, or clear homology to a vertebrate protein were identified using a transcriptome contig id number. Sequences with a clear human best blast hit were labelled with a transcriptome contig id and with the name of the human best blast hit in parentheses.
Muscle single cell isolation
Animals were dissected and the prepharyngeal region was isolated (Figure S1A). Muscle cells were isolated from the dorsal, ventral and lateral regions of this prepharyngeal region. Fragments were dissociated into single cells in a solution of CMFB and 1 mg/ml of collagenase [2]. Single cell suspensions for each region were labelled with Hoechst, and non-dividing single cells were sorted by flow cytometry into 96 well plates containing 5 ml of total cell lysis buffer (Qiagen, Germany) with 1% β-mercaptoethanol. Subsequently, amplified cDNA libraries were prepared from each single cell using the SmartSeq2 method [11, 51] and tested by qRT-PCR for the expression of the muscle markers collagen and troponin (colF-2 Fw: GGTGTACTTGGAGACGTTGGTTTA, colF-2 Rv: GGTCTACCTTCTCTTCCTGGAAC; troponin Fw: ACAGGGCCTTGCAACTATTTTCATC, troponin Rv: GAAGCTCGACGTCGACAGGA). Cells expressing either or both of these muscle-specific genes (~5 in 96 cells) were used for library preparation using the Nextera XT method (Illumina, Inc) [11]. Libraries were sequenced using Illumina HiSeq.
Smart-Seq sequencing analysis
SCS data from three sources were merged with SCS data of planarian neoblasts [11]. The merged SCS expression data were analyzed with the Seurat v1.4 [12]. Cells expressing less than 1,000 or over 10,000 transcripts were removed from further analysis. Cells that were identified as belonging to the epidermal lineage based on expression (> 5 log2 CPM) of prog-1 or prog-2 (dd_920 and dd_899, respectively) were removed from further analysis, and reads mapped to contigs dd_Smed_v4_10881_0_1 and dd_Smed_v4_5614_0_1 were excluded, as they represented misalignments of primer amplification sequences [11]. Seurat object was initiated using the setup function [min.cells = 3, min.genes = 1000, max.genes = 10000, is.expr= 0.1]. Genes were selected for initial PCA using params [y.cutoff = 2, x.low.cutoff = 6, fxn.y = logVarDivMean] and were supplemented by genes known to be enriched in muscle, neoblasts, and epidermis [2, 10, 11]: dd_1021, dd_10216, dd_10673, dd_11500, dd_11840, dd_12035, dd_12634, dd_13343, dd_13518, dd_14391, dd_14783, dd_16209, dd_1694, dd_17951, dd_19327, dd_1985, dd_21801, dd_2373, dd_2592, dd_26182, dd_306, dd_3098, dd_31435, dd_323, dd_3244, dd_332, dd_364, dd_402, dd_4075, dd_432, dd_436, dd_4877, dd_5014, dd_54, dd_579, dd_659, dd_6746, dd_6811, dd_69, dd_6910, dd_6999, dd_702, dd_7038, dd_7326, dd_7371, dd_7837, dd_8833, dd_899, dd_920, dd_9259, dd_9910. t-SNE was performed with parameters [dims.use = c(1:13), perplexity = 14, do.fast = T], and differentially expressed genes were called using the bimodal test implemented in Seurat [12].
Drop-Seq clustering analysis
Cells assigned a muscle identity from [13] were re-clustered from the existing muscle Seurat object [13] using Seurat package, v2.2 [52]. Briefly, the Seurat function FindVariableGenes [y.cutoff = 1, x.low.cutoff=.2, x.high.cutoff=5, mean.function = expMean, dispersion.functin = LogVMR] was used to identify genes with high variance and high expression. These genes were used as input for principal component analysis using the function RunPCA [pcs.compute = 10]. Clustering was performed using the function FindClusters [reduction.type = “pca”, dims.use = c(1: 10), resolution = 1] and cells were plotted in 2 dimensions by t-SNE. 13 clusters were generated and a cell identity was assigned based on expression of highly enriched transcripts (Data S1). Specifically, clusters 0 and 7 were defined and renamed as DVM; clusters 1, 2, and 6 were defined and renamed as BWM; clusters 3, 4, 5, 8, 9, and 11 were defined and renamed as pharynx muscle; cluster 12 was defined and renamed as IM; and cluster 10 contained a heterogeneous mix of cells isolated from both inside and outside the pharynx and was labelled nd (not determined). Cluster assignment were highly consistent with the original clustering from [13], as demonstrated in the table below, though re-clustering yielded better separation between smedwi-1+ BWM and DVM progenitors. The apparent discrepancy between the expression of the differentiated marker mp-1 in the pharynx muscle cluster between the Smart-Seq SCS analysis (Figure 1C) and the Drop-Seq analysis (Figure S2E) is explained by the fact that the former analysis did not include pharyngeal cells and most of the cells in cluster 7 represented pharynx muscle progenitors (higher levels of smedwi-1 expression, Figure S1D), whereas the Drop-Seq dataset did contain pharyngeal cells at several differentiation stages and therefore expressed mp-1.
| Cluster assignment from this manuscript | Cluster assignment from [13] |
|---|---|
| 0 | 6 |
| 1 | 1 |
| 2 | 0 and 5 |
| 3 | 2 |
| 4 | 8 |
| 5 | 8 |
| 6 | 7 |
| 7 | 0, 3, and 5 |
| 8 | 9 |
| 9 | 10 |
| 10 | None |
| 11 | 11 |
| 12 | 13 |
Gene cloning
Two contigs dd_9259 and dd_25344 spanned fragments of nk4 gene sequence (http://planmine.mpi-cbg.de, [53]). Both contigs were amplified and used combined for RNAi experiments. dd_9259 was amplified_using the following primers: forward 5’ATATTAGCTTGATACCGTGTCAC; reverse 5’AATCTGCTGTGGGAGGTGTT, and for dd_25344 forward 5 ‘ AATTAT CT AAT GCCT CAAGT GCA and reverse 5’TACTTGTTGTGGAGTCATTTTCA; gata4/5/6–2 was amplified using the following primers: forward 5’CACCAGCAACAATCACCAGA; reverse 5’CGAACAATGAAGACCCCTCC; gata4/5/6–3 was amplified using forward 5’ ACCAAATCGACACTTAAAACCG and reverse 5’ GTACAATTTCTCGGGTGATCGTG; foxF-1 was amplified using forward 5’ GTCCTATTTCCAGCACACAGC and reverse 5’ TCCGGAATCGTGCTGAGG. All constructs were cloned from cDNA into the pGEM vector (Promega). These constructs were used to synthesized RNA probes and dsRNA for RNAi experiments.
RNAi
For RNAi experiments, dsRNA was synthesized by in vitro transcription reactions (Promega) using PCR-generated templates with flanking T7 promoters, followed by ethanol precipitation, and annealed after resuspension in water. The concentration of dsRNA varied in each prep between 4 and 7 mg/ml. dsRNA was then mixed with planarian food (liver) [2] and 2 ml of this mixture per animal (liver containing dsRNA) was used for feedings. For nk4 RNAi, a 1:1 mixture of dsRNA from dd_9259 and dd_25344 was used. nk4 and gata4/5/6–3 RNAi animals had eight to ten feedings. gata4/5/6–2 RNAi were fed six to eight times, after which they ejected their pharynx and stop eating. foxF-1 RNAi animals were fed four to six times, after which they lysed. Feedings were performed every other three days. In all cases, animals were fixed seven days after the last feeding. For regeneration experiments, animals were amputated into three pieces (head, trunk and tail pieces) one week after the last RNAi feeding. Seven or nine days after amputation, trunk pieces were scored, and fixed for further analysis. For RNA-Seq experiments in uninjured animals, control, nk4, gata4/5/6–2, and gata4/5/6–3 RNAi animals were fed eight times; foxF-1 RNAi animals and their controls were fed six times.
Fluorescence in situ hybridizations and immunostainings
RNA probes were synthesized and whole-mount FISH was performed [2]. Briefly, animals were killed in 5% NAC and treated with proteinase K (2 μg/ml). Following overnight hybridizations, samples were washed twice in each of pre-hybridization buffer, 1: 1 pre-hybridization-2X SSC, 2X SSC, 0.2X SSC, PBS with Triton-X (PBST). Subsequently, blocking was performed in 1% Western Blocking Reagent (Roche, 11921673001) PBST solution and anti-DIG, anti-DNP, or anti-FITC antibodies were used. Antibody washes were then performed for three hours followed by tyramide development. Peroxidase inactivation with 1% sodium azide was done for 90 minutes at room temperature. Brightfield images were taken with a Zeiss Discovery Microscope. Fluorescent images were taken with a Zeiss LSM700 Confocal Microscope using ZEN software or with a Leica SP8 Confocal Microscope. Colocalization analyses of FISH signals were performed using Fiji/ImageJ. For each channel, histograms of fluorescence intensity were used to determine the cut-off between signal and background. All FISH images are representative of all images taken in each condition. For immunostainings, animals were fixed as for in situ hybridizations, blocked in 1% Western Blocking Reagent (Roche, 11921673001) PBST solution for one hour and then stained with the antibody of interest. An anti-muscle mouse monoclonal antibody 6G10 [54] was used in a 1:1,000 dilution, and an anti-mouse Alexa conjugated antibody (Life Tech) was used in a 1:500 dilution. Probes used for labelling glia cells were if-1 and cali; for pigment cells,pbgd-1. The pool of probes used for labelling cathepsin+ cells included: dd_1161 1, pbgd-1, if-1, cali, dd_582 (CTSL2), dd_9, dd_10872, dd_1831, dd_5690, aqp-1 (dd_1103), dd_7593, dd_6149, and dd_1260.
RNA-Seq experiments
Total RNA was isolated using Trizol (Life Technologies) from single animals. Libraries were prepared using the Kapa Stranded mRNA-Seq Kit Illumina Platform (KapaBiosystems). Libraries were sequenced on an Illumina Hi-Seq. Libraries were mapped to the dd_Smed_v4 transcriptome (http://planmine.mpi-cbg.de; [53] using bowtie v1.1.2 [55] with -best alignment parameter. The number of mapped reads per contig in every cell was quantified using the coverageBed utility from the bedtools v2.26.0 suite [56] and reads from the same isotig were summed to generate raw read counts for each transcript. Reads mapped to contigs dd_Smed_v4_9259_0_1 and dd_Smed_v4_25344_0_1, which are fragments of the gene nk4, were summed together for downstream analysis. Pairwise differential expression analysis was performed using DESeq [57]. Expression values from DESeq normalization were scaled, row-wise, to generate z-scores for heatmaps [11]. Pheatmap was used to generate scaled heatmaps. Differentially expressed genes were determined using edgeR [58] with threshold of FDR < 0.05 or otherwise indicated in the figure. Cell type specific expression of differentially expressed genes was annotated using a recently published Drop-Seq analysis of planarian cells [13].
Preparation and injection of red fluorescent bacterial cultures
E. coli cultures expressing red fluorescent proteins were prepared by transformation of BL21(DE3) cells with either pET His6 mCherry (Addgene 29722) or pmScarlet (Addgene 85063) [59]. Cultures were grown at 37C in LB media to an OD600 of 0.5. Protein production was induced by addition of IPTG to a final concentration of 0.5 mM. After 16–20h, 25mL cultures were collected by centrifugation and resuspended in calcium and magnesium free planarian water (CMF) at a ratio of 1 mL of CMF to 2 mL of culture. Cultures were then loaded into needles backfilled with mineral oil. Needles were prepared from Drummond 3.5” borosilicate glass capillaries pulled on a Narishge (model PN-30) needle puller set at heater level 95 and magnet level 45. Needles were filled and injected using a Drummond Nanoject III (model 3–000-207, Drummond Scientific company) mounted on a micromanipulator. Planarians were injected on the ventral side immediately posterior to the pharyngeal cavity. Planarians were placed on wetted filter paper on a cold plate to limit their movement during injection. After initial puncture, each injection delivered 25 nL over 2s. Injections were repeated 2–3 times until subtle inflation of the injected posterior was observed. Needles were cleaned or trimmed with forceps to eliminate clogging between injections. Animals were injected in batches over the course of 20–30 minutes for each subsequent time point of interest. Visual inspection of injected worms under a fluorescence dissecting scope was performed to confirm and spot check injection quality. Mocked injected animals received CMF only. Similar injections were performed with a suspension of E.coli (K12 strain) BioParticles™ conjugated with Alexa Fluor 488 resuspended following manufacturer specifications (Molecular Probes, Thermofisher) and diluted 1:5 in CMF, and with a suspension of pHrodo™ Green E.coli BioParticles™ conjugate (Molecular Probes, Thermofisher) resuspended in CMF and injected at 1 mg/ml.
Cell dissociation
Animals were cut into small fragments and resuspended in CMFB and 1mg/ml collagenase and incubated for ten minutes at room temperature gently pipetting up and down. Dissociated single cells were then pipetted through a 40 microns filter, centrifuged at 300g for five minutes, resuspended in CMFB and mounted to image. Brightfield and fluorescent images were taken with a Leica SP8 Confocal Microscope.
Bacterial colony assays
Control or foxF-1 RNAi animals were washed three times in fresh planarian water. Three or four animals were pooled and homogenized in 100 μl of deionized water. 25 μl was then plated on LB agar without any antibiotics and incubated in the dark at room temperature [30]. A water control was also plated, but no bacterial colonies grew on those plates at the time of bacterial colony counting. One day after plating, bacterial colonies were counted, and total numbers of bacterial colony forming units (CFU) per animal were shown in graph (Figure S7M).
Ink feeding
Animals were fed with a 1:1 mix of liver and ink solution. The ink solution was prepared by mixing one-part black india ink and one-part planarian water. Animals were fed for 1–2 hours and were immediately imaged after using a Zeiss Discovery Microscope.
Phylogenetic analysis
The NKX tree shows 49 homeobox and the Fox tree shows 95 homeobox proteins from diverse organisms. Protein sequences were aligned using MUSCLE [60] with default settings and trimmed with Gblocks [61]. Maximum likelihood analyses were run using PhyML with 100 or 1,000 bootstrap replicates, the WAG model of amino acid substitution, four substitution rate categories and the proportion of invariable sites estimated from the dataset. Trees were visualized in FigTree. All ML bootstrap values are shown. For the NKX tree, we used the homeobox containing domain found in the cDNA contig PL06018A1E07 which contains parts of both dd_9529 and dd_25344 contigs. The GATA DNA-binding domain amino acids of 76 diverse GATA family members were aligned by MUSCLE [60] and trimmed by Gblocks [61] with the settings ‘b3=15 -b4=2 -b5=a -e=.gb -p=t -g’ to select 92 conserved amino acids, the same strategy and sequences used in [23]. Maximum likelihood analyses were run using PhyML, the WAG model of amino acid substitution ‘-m WAG -f e -v e -c 4 -b 1000’ and evaluated using the SH- like approximate Likelihood Ratio Test. Similar to [23] our results support placement of dd_8208 (previously published as Smed-gata1/2/3b) as a member of the GATA4/5/6 family (aLTR 0.846), and therefore we renamed the gene to Smed-gata4/5/6–3. This assignment is consistent with a tree generated that included for all four planarian GATA transcription factors in [23]. All aLTR values are shown. Hs,Homo sapiens; Mm,Mus musculus; Dm, Drosophila melanogaster; Smed, Schmidtea mediterranea; Xl, Xenopus laevis; Sd; Suberites domuncula; Bf, Branchiostoma floridae; Ci, Ciona intestinalis; Hv, Hydra vulgaris; Nv, Nematostella vectensis; Dj, Dugesia japonica; Cs, Ciona selvatgi; Ml, Mnemiopsis leidyi, Ct, Capitella teleta.
QUANTIFICATIONS AND STATISTICAL ANALYSIS
Numbers of mhc-2+ and col4–5+ cells (lateral DVM) were counted within a 0.02 mm2 rectangle in the tail tip of each RNAi animal (Figure 3C). Numbers of mp-1+ and mhc-2+ cells were counted in each animal within the tail stripe region (Figure 3D, S5G) or in the head blastema (Figure S6C). Ratios (double-positive cells (DVM) to single mp-1+ cells (IM)) were calculated per animal as indicated. In addition, lateral DVM (mp-1+mhc-2+) cells were also counted within a 0.02 mm2 rectangle of the head blastema (Figure S6C). Total number of IM cells (dd_12771(PTPRD)+) in the animal tail stripe were counted in different RNAi animals (animals have comparable sizes, Figure 3E). Intestinal branches fusion (no fusion, fused in one point, fused at two different points) and secondary gut branches were counted in the tail of RNAi animals (Figures 3F; 5J,K) or in the head blastema (Figure 4D; 5K). Total numbers of glia, aqp-1+, dd_10872+, and dd_582(CTSL2)+ cells were counted either in tails (homeostasis) or head blastemas (regeneration) (Figure 6B). Cells with mCherry+ signal associated to the nucleus (DAPI staining) were counted as positive for phagocytosis (Figure 6E). One-way ANOVA test followed by Dunnett’s multiple comparison test was used when analyzing more than two conditions. Unpaired Student’s t-test was used when comparing two conditions. Mean ± SD is shown in all graphs. Empirical p-value for enrichment of mCherry+ in cathepsin+ cells was calculated by randomly sampling cells (n=37, the number of detected positive cells) and marking them as positive in a mock population of cells representing all counted cell types (i.e., cathepsin+, muscle, epidermal progenitors, and neoblasts) according to the total cell number counted of each type. Then, the number of cathepsin+ cells that were randomly assigned as mCherry+ was documented. The process was repeated 1,000,000 times, and finally the results of the random sampling (i.e. expected results) were compared with the observed results.
DATA AVAILABILITY
Sequence of gata4/5/6–2 has been previously deposited in Genbank with the accession number KX827244. Sequences of nk4, gata4/5/6–3, andfoxF-1 have been deposited in Genbank with the accession numbers MH392337–39. Single cell sequencing data and mRNA-Seq data have been deposited in Genbank under PRJNA471168. The accession numbers of reported data used in this study are PRJNA353867 (from [11]), GSE74360 (from [2]), and GSE111764 (from [13]).
Supplementary Material
Data S1. List of enriched gene expression in distinct muscle clusters by Drop-Seq analysis. Related to Figures 1, 2.
Data S2. RNA-sequencing analyses of uninjured nk4, gata4/5/6-2, gata4/5/6-3, and foxF-1 RNAi animals. Related to Figures 3, 5, 6.
Table S1. Description of single Smart-Seq libraries of muscle cells. Related to Figure 1.
Table S2. Cluster enriched genes in single-muscle-cell Smart-Seq analysis. Related to Figures 1, 2.
Highlights.
foxF-1 specifies planarian non-body wall muscle
nk4, gata4/5/6–2, and gata4/5/6–3 specify different muscle subsets
DVM and IM are required for normal patterning in planarians
Phagocytic cells in planarians are specified by foxF-1
Acknowledgments
We thank Lauren Cote for plasmids and members of the Reddien lab for discussions and comments on the manuscript. We acknowledge support by NIH R01GM080639. PWR is an investigator of HHMI and an associate member of the Broad Institute. We thank the Eleanor Schwartz Charitable Foundation for support.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Witchley JN, Mayer M, Wagner DE, Owen JH, and Reddien PW (2013). Muscle cells provide instructions for planarian regeneration. Cell Reports 4, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scimone ML, Cote LE, Rogers T, and Reddien PW (2016). Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cebriá F (2016). Planarian Body-Wall Muscle: Regeneration and Function beyond a Simple Skeletal Support. Front Cell Dev Biol 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein MA, and Burdette WJ (1971). Striated visceral muscle of Drosophila melanogaster. J Morphol 134, 315–334. [DOI] [PubMed] [Google Scholar]
- 5.White J (1988). 4 the anatomy. Nematode Caenorhabditis elegans. Cold Spring Harbor Perspectives in Biology 17, 81–122. [Google Scholar]
- 6.Paniagua R, Royuela M, García-Anchuelo RM, and Fraile B (1996). Ultrastructure of invertebrate muscle cell types. Histol Histopathol 11, 181–201. [PubMed] [Google Scholar]
- 7.Brunet T, Fischer AH, Steinmetz PR, Lauri A, Bertucci P, and Arendt D (2016). The evolutionary origin of bilaterian smooth and striated myocytes. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saudemont A, Dray N, Hudry B, Le Gouar M, Vervoort M, and Balavoine G (2008). Complementary striped expression patterns of NK homeobox genes during segment formation in the annelid Platynereis. Dev Biol 317, 430–443. [DOI] [PubMed] [Google Scholar]
- 9.Laumer CE, Bekkouche N, Kerbl A, Goetz F, Neves RC, Sorensen MV, Kristensen RM, Hejnol A, Dunn CW, Giribet G, et al. (2015). Spiralian phylogeny informs the evolution of microscopic lineages. Curr Biol 25, 2000–2006. [DOI] [PubMed] [Google Scholar]
- 10.Scimone ML, Cote LE, and Reddien PW (2017). Orthogonal muscle fibres have different instructive roles in planarian regeneration. Nature 551, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wurtzel O, Cote LE, Poirier A, Satija R, Regev A, and Reddien PW (2015). A Generic and Cell-Type-Specific Wound Response Precedes Regeneration in Planarians. Dev Cell 35, 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, and Reddien PW (2018). Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, and Sánchez Alvarado A (2005). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330. [DOI] [PubMed] [Google Scholar]
- 15.Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glazar P, Obermayer B, Theis FJ, Kocks C, and Rajewsky N (2018). Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360. [DOI] [PubMed] [Google Scholar]
- 16.Bodmer R (1993). The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118, 719–729. [DOI] [PubMed] [Google Scholar]
- 17.Azpiazu N, and Frasch M (1993). tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev 7, 1325–1340. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka M, Kasahara H, Bartunkova S, Schinke M, Komuro I, Inagaki H, Lee Y, Lyons GE, and Izumo S (1998). Vertebrate homologs of tinman and bagpipe: roles of the homeobox genes in cardiovascular development. Dev Genet 22, 239–249. [DOI] [PubMed] [Google Scholar]
- 19.González-Sastre A, De Sousa N, Adell T, and Saló E (2017). The pioneer factor Smed-gata456–1 is required for gut cell differentiation and maintenance in planarians. Int J Dev Biol 61, 53–63. [DOI] [PubMed] [Google Scholar]
- 20.Gajewski K, Fossett N, Molkentin JD, and Schulz RA (1999). The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila. Development 126, 5679–5688. [DOI] [PubMed] [Google Scholar]
- 21.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, and Leiden JM (1997). GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev 11, 1048–1060. [DOI] [PubMed] [Google Scholar]
- 22.Molkentin JD, Lin Q, Duncan SA, and Olson EN (1997). Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev 11, 1061–1072. [DOI] [PubMed] [Google Scholar]
- 23.Gillis WQ, Bowerman BA, and Schneider SQ (2008). The evolution of protostome GATA factors: molecular phylogenetics, synteny, and intron/exon structure reveal orthologous relationships. BMC Evol Biol 8, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He X, Lindsay-Mosher N, Li Y, Molinaro AM, Pellettieri J, and Pearson BJ (2017). FOX and ETS family transcription factors regulate the pigment cell lineage in planarians. Development 144, 4540–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kedinger M, Simon-Assmann PM, Lacroix B, Marxer A, Hauri HP, and Haffen K (1986). Fetal gut mesenchyme induces differentiation of cultured intestinal endodermal and crypt cells. Dev Biol 113, 474–483. [DOI] [PubMed] [Google Scholar]
- 26.Scimone ML, Lapan SW, and Reddien PW (2014). A forkhead transcription factor is wound-induced at the planarian midline and required for anterior pole regeneration. PLoS genetics 10, e1003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oderberg IM, Li DJ, Scimone ML, Gavino MA, and Reddien PW (2017). Landmarks in Existing Tissue at Wounds Are Utilized to Generate Pattern in Regenerating Tissue. Curr Biol 27, 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cebriá F, Guo T, Jopek J, and Newmark PA (2007). Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev Biol 307, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turk V (2012). Special issue: Proteolysis 50 years after the discovery of lysosome in honor of Christian de Duve. Biochim Biophys Acta 1824, 1–2. [DOI] [PubMed] [Google Scholar]
- 30.Arnold CP, Merryman MS, Harris-Arnold A, McKinney SA, Seidel CW, Loethen S, Proctor KN, Guo L, and Sanchez Alvarado A (2016). Pathogenic shifts in endogenous microbiota impede tissue regeneration via distinct activation of TAK1/MKK/p38. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cripps RM, and Olson EN (2002). Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol 246, 14–28. [DOI] [PubMed] [Google Scholar]
- 32.Tolkin T, and Christiaen L (2012). Development and evolution of the ascidian cardiogenic mesoderm. Curr Top Dev Biol 100, 107–142. [DOI] [PubMed] [Google Scholar]
- 33.Aronson BE, Stapleton KA, and Krasinski SD (2014). Role of GATA factors in development, differentiation, and homeostasis of the small intestinal epithelium. Am J Physiol Gastrointest Liver Physiol 306, G474–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner DE, Wang IE, and Reddien PW (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flores NM, Oviedo NJ, and Sage J (2016). Essential role for the planarian intestinal GATA transcription factor in stem cells and regeneration. Dev Biol 418, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martindale MQ, Pang K, and Finnerty JR (2004). Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa). Development 131, 2463–2474. [DOI] [PubMed] [Google Scholar]
- 37.Zaffran S, Kuchler A, Lee HH, and Frasch M (2001). biniou (FoxF), a central component in a regulatory network controlling visceral mesoderm development and midgut morphogenesis in Drosophila. Genes Dev 15, 2900–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahlapuu M, Ormestad M, Enerback S, and Carlsson P (2001). The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development 128, 155–166. [DOI] [PubMed] [Google Scholar]
- 39.Tseng HT, Shah R, and Jamrich M (2004). Function and regulation of FoxF1 duringXenopus gut development. Development 131, 3637–3647. [DOI] [PubMed] [Google Scholar]
- 40.Arendt D (2008). The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet 9, 868–882. [DOI] [PubMed] [Google Scholar]
- 41.Peiris TH, Hoyer KK, and Oviedo NJ (2014). Innate immune system and tissue regeneration in planarians: an area ripe for exploration. Semin Immunol 26, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morita M (1991). Phagocytic response of planarian reticular cells to heat-killed bacteria. Hydrobiologia 227, 193–199. [Google Scholar]
- 43.Abnave P, Mottola G, Gimenez G, Boucherit N, Trouplin V, Torre C, Conti F, Ben Amara A, Lepolard C, Djian B, et al. (2014). Screening in planarians identifies MORN2 as a key component in LC3-associated phagocytosis and resistance to bacterial infection. Cell Host Microbe 16, 338–350. [DOI] [PubMed] [Google Scholar]
- 44.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frost JL, and Schafer DP (2016). Microglia: Architects of the Developing Nervous System. Trends Cell Biol 26, 587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruffins SW, and Ettensohn CA (1993). A clonal analysis of secondary mesenchyme cell fates in the sea urchin embryo. Dev Biol 160, 285–288. [DOI] [PubMed] [Google Scholar]
- 47.Ho EC, Buckley KM, Schrankel CS, Schuh NW, Hibino T, Solek CM, Bae K, Wang G, and Rast JP (2017). Perturbation of gut bacteria induces a coordinated cellular immune response in the purple sea urchin larva. Immunol Cell Biol 95, 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amin NM, Shi H, and Liu J (2010). The FoxF/FoxC factor LET-381 directly regulates both cell fate specification and cell differentiation in C. elegans mesoderm development. Development 137, 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fares H, and Greenwald I (2001). Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics 159, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beh J, Shi W, Levine M, Davidson B, and Christiaen L (2007). FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development 134, 3297–3305. [DOI] [PubMed] [Google Scholar]
- 51.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, and Sandberg R (2013). Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10, 1096–1098. [DOI] [PubMed] [Google Scholar]
- 52.Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, et al. (2014). Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu SY, Selck C, Friedrich B, Lutz R, Vila-Farre M, Dahl A, Brandl H, Lakshmanaperumal N, Henry I, and Rink JC (2013). Reactivating head regrowth in a regeneration-deficient planarian species. Nature 500, 81–84. [DOI] [PubMed] [Google Scholar]
- 54.Ross KG, Omuro KC, Taylor MR, Munday RK, Hubert A, King RS, and Zayas RM (2015). Novel monoclonal antibodies to study tissue regeneration in planarians. BMC Dev Biol 15, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, et al. (2017). mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods 14, 53–56. [DOI] [PubMed] [Google Scholar]
- 60.Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Talavera G, and Castresana J (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56, 564–577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. List of enriched gene expression in distinct muscle clusters by Drop-Seq analysis. Related to Figures 1, 2.
Data S2. RNA-sequencing analyses of uninjured nk4, gata4/5/6-2, gata4/5/6-3, and foxF-1 RNAi animals. Related to Figures 3, 5, 6.
Table S1. Description of single Smart-Seq libraries of muscle cells. Related to Figure 1.
Table S2. Cluster enriched genes in single-muscle-cell Smart-Seq analysis. Related to Figures 1, 2.
Data Availability Statement
Sequence of gata4/5/6–2 has been previously deposited in Genbank with the accession number KX827244. Sequences of nk4, gata4/5/6–3, andfoxF-1 have been deposited in Genbank with the accession numbers MH392337–39. Single cell sequencing data and mRNA-Seq data have been deposited in Genbank under PRJNA471168. The accession numbers of reported data used in this study are PRJNA353867 (from [11]), GSE74360 (from [2]), and GSE111764 (from [13]).