Synopsis:
Hereditary alpha tryptasemia is an autosomal dominant genetic trait caused by increased germline copies of TPSAB1 encoding alpha-tryptase. Individuals with this trait have elevated basal serum tryptase, and may present with associated multisystem complaints, including systemic immediate hypersensitivity reactions, cutaneous flushing and pruritus, functional gastrointestinal diseases, connective tissue abnormalities, and symptoms suggestive of autonomic dysfunction. Both basal serum tryptase levels and severity of clinical symptoms display a gene dose relationship with TPSAB1, whereby higher tryptase levels and greater symptom severity are correlated with increasing numbers of alpha-encoding TPSAB1 copies. Complex structural variation at the tryptase locus limits accurate quantification of TPSAB1 copy number by methods other than droplet digital PCR. As the functional effects of increased basal serum tryptase and/or altered tryptase gene expression are elucidated, greater insights will be gained into the symptoms associated with hereditary alpha tryptasemia and their potential therapy.
Keywords: mast cell activation, hypertryptasemia, autosomal dominant, genotyping
Introduction
Tryptase is a protein expressed by mast cells and basophils (1, 2). Mature, enzymatically active tryptases are tetrameric serine proteases that are stored in mast cell secretory granules and contribute to allergic inflammation (3). Experiments inhibiting mature tryptases have demonstrated their role in promoting inflammatory cell recruitment, vascular permeability, and airway hypersensitivity and remodeling, in animal models. However, the specific contribution of mature tryptases to allergic reactions in humans is less clear (4). Pro-tryptases, which have not undergone enzymatic conversion into mature tetrameric tryptases, are constitutively secreted into serum in their monomeric form, and provide the vast majority of measured basal serum tryptase (BST) in healthy individuals (5, 6) (Fig. 1). Pro-tryptases are also the predominant forms of tryptase present in the serum from patients with systemic mastocytosis (7, 8). During mast cell degranulation, as occurs during IgE-mediated immediate hypersensitivity reactions, mature tryptases are released with other mast cell mediators and contribute to symptoms of type I allergic reactions. Thus, serum tryptase in this setting is a useful biomarker for the clinical diagnosis of anaphylaxis (9).
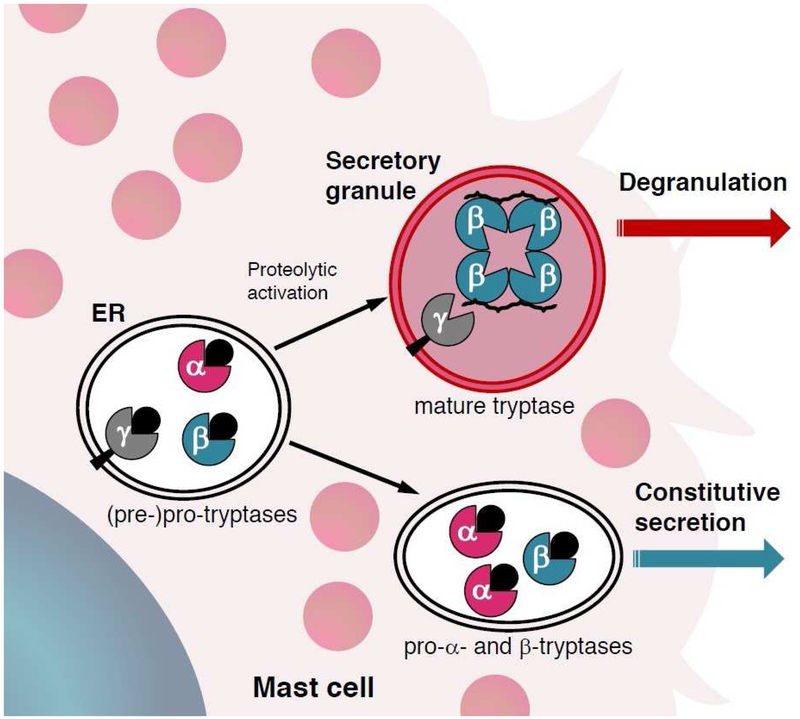
Figure 1. Schematic of tryptase secretion from human mast cells.
Pro-tryptases generated in mast cells undergo sequential proteolytic cleavage to become mature tetrameric tryptase, stabilized by heparin, and stored in secretory granules (top) awaiting appropriate stimuli to induce degranulation. Alternatively, pro-tryptases can be secreted constitutively into serum as enzymatically inactive pro-peptides (bottom).
Adapted from Caughey GH. Tryptase genetics and anaphylaxis. J Allergy Clin Immunol 2006;117(6):1412; with permission.
However, elevated basal serum tryptase (BST) – currently defined clinically as > 11.4 ng/mL – appears to be quite common, being reported in 4-6% of the general population (10, 11). While in some individuals reported increases may be due to end stage renal disease or clonal expansion of myeloid or mast cells, including mastocytosis (12-14), it has recently been discovered that a number of individuals with elevated BST inherit this trait (15-18). Further, in the small cohorts studied thus far, the data suggest that this trait may also be relatively common, and frequently the cause for elevated BST in the general population (16, 18). The focus of this review is to discuss the details of this genetic trait and the complexities surrounding genotyping patients, as well as the associated clinical features and management approaches for patients with the multisystem complaints associated with hereditary alpha tryptasemia.
Tryptase locus and isotype differences
The tryptase locus contains four tryptase encoding genes (TPSG1, TPSB2, TPSAB1, and TPSD1) and is present on the distal portion of the short arm of chromosome 16 at position p13.3 (Fig. 2). One additional human tryptase (epsilon) encoded by PRSS22 also exists on 16p just outside of this cluster. While all of these genes encode tryptases, only TPSB2 and TPSAB1 encode the secreted isoforms of tryptase that are measured and reported as serum tryptase by clinical laboratories (19).
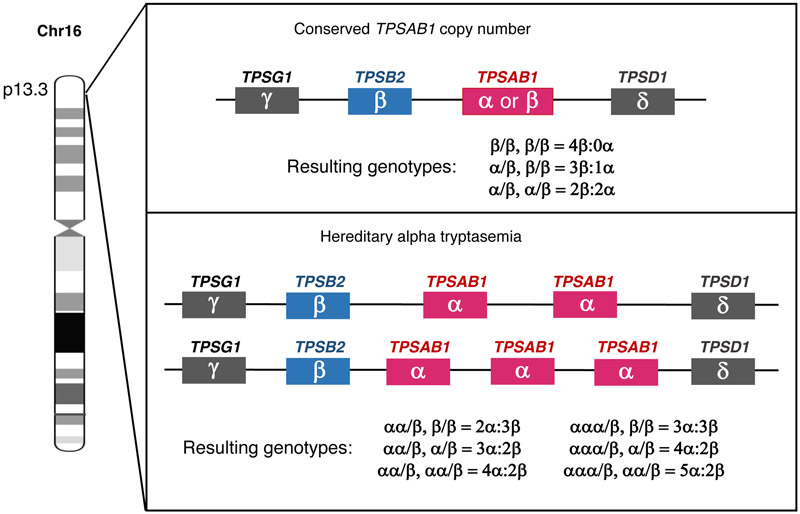
Figure 2. Identified tryptase genotypes encoded at TPSAB1 and TPSB2.
Canonical alpha- and beta-tryptase genotypes based upon conserved copy number (top) and those identified resulting from increased TPSAB1 copy number encoding one or two additional alpha-tryptase copies on single alleles (bottom). Additional genotypes are likely to be identified, and other variant beta isoforms that have already been identified (19) are excluded for simplicity.
While TPSB2 is believed to encode only beta-tryptase isoforms, the TPSAB1 locus encodes either alpha or beta isoforms (20) (Fig. 2). Each of these isoforms is remarkably similar – being at least 97% identical – making detection of distinct tryptase isoforms extremely difficult. Recent publications have clarified a number of misconceptions about the biology of alpha-tryptase isoforms encoded at TPSAB1. We now know that alpha pro-tryptase can be processed into mature tryptase (21). Further, the monoclonal tryptase antibody (G5 clone) which has been reported to distinguish alpha- from beta- tryptases, is now know to recognize any tryptase – including alpha-tryptase – which has been processed to maturity. This includes destabilized monomers which had previously been tetrameric enzyme (6). There is still no antibody that can differentiate alpha- or beta-tryptase protein. Despite our limitations in detecting protein differences in vivo, a number of important functional distinctions have been identified between alpha- and beta-tryptase isoforms in vitro. Importantly, alpha-tryptase sequences contain two isoform-defining variants: one which may promote constitutive secretion, and a second at the enzymatic active site which appears to eliminate functional activity of homo-tetrameric alpha tryptases (22-24).
Until recently, it was believed that individuals have one copy each of TPSB2 and TPSAB1. Based upon the tryptase isoform expression at these two genetic loci, there have been three canonical genotypes described in the literature with a total tryptase gene number of four: 0α:4β, 1α:3β, and 2α:2β (Fig. 2). Importantly, among individuals with a 2α:2β genotype, the two alpha-tryptase copies are on opposite alleles (one being inherited from each parent), and do not represent increased TPSAB1 copy number. However, we now know that increased TPSAB1 copy number encoding alpha-tryptase can occur on a single allele, and when present leads to elevated BST; duplications and triplications have thus far been identified (16) (Fig. 2). Altered TPSB2 or TPSAB1 copy number encoding beta-tryptase has not been reported, but may also occur, and any associated biochemical findings or clinical phenotypes have yet to be described.
Several individuals have been reported with inherited TPSAB1 duplications on both alleles (i.e. both parents carried duplications and passed them to these individuals) suggesting that hereditary alpha tryptasemia may be relatively common. Complete segregation of elevated BST with increased TPSAB1 copy number in the two unselected populations that have been studied (16) and the relatively high prevalence of elevated BST in multiple populations (10, 11) also suggest this trait may be a common cause for elevated BST. Additional population-based studies are required to establish the prevalence of increased TPSAB1 copy as the cause for elevated BST in the general population.
How increased mono-allelic TPSAB1 copy number leads to increased BST and the clinical features associated with hereditary alpha tryptasemia remains unknown. However, it appears that mast cells and basophils over-express and secrete pro-tryptase(s) in excess when increased alpha-encoding TPSAB1 copies are present. A greater number of extra copies, leads to higher BST and more reported symptoms – or an observed gene dosage effect. However, total tryptase within basophils and mast cells does not appear to be increased, though this has not been studied in detail. While alpha-tryptase tetramers do not exhibit the trypsin-like protease activity of their beta-tryptase counterparts (22), the potential effects of alpha-tryptase over-expression on mature tryptases has not previously been studied and is an area of active investigation. There is limited evidence to support non-enzymatic activity of tryptases through unknown mechanisms; enzymatically inactivated human tryptases can exhibit mitogenic activity on human lung fibroblasts (25). Ongoing investigations into the effects of altered tryptase gene expression and increased BST levels may provide new insights into the mechanisms underlying the clinical symptoms associated with hereditary alpha tryptasemia.
Genotyping strategies
Complex structural variation at the tryptase locus has thus far precluded its direct sequencing. The few published human genome assemblies, derived from single molecule real time sequencing (SMRT) sequencing – a technology which allows very large contiguous reads – have demonstrated remarkably structural diversity at this locus, even among ostensibly healthy individuals (26). Because of the high degree of structural variation present and conservation of sequence between tryptase isoforms, clinically available exome or genome sequencing data do not identify increased TPSAB1 copy number. Further limiting conventional mapping and interpretation of genetic sequence is copy number variation of wild-type TPSAB1, where unaffected individuals may have 0, 1, or even 2 copies of alpha-encoding sequence on opposing alleles. There are several published approaches that have been applied to overcome these obstacles and establish alpha-encoding TPSAB1 copy number in silico or in vitro (16, 20, 27).
Relative tryptase gene quantitation by Sanger sequencing and modified Southern blotting
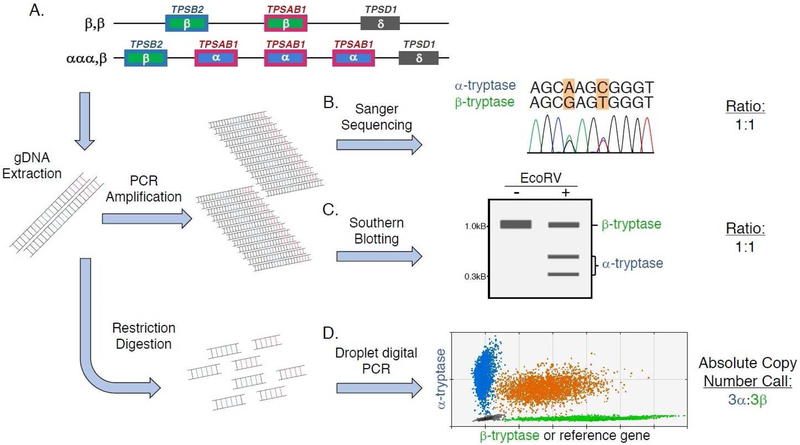
Two of the original methods described for tryptase genotyping (Fig. 3) provide relative quantification of alpha- and beta-tryptase encoding copies at TPSAB1 and TPSB2 (Fig. 1A-C). Both approaches rely upon the calculated ratio between alpha- and beta-tryptase genomic sequences. The first method employs Sanger gene sequencing of the tryptase locus (Fig. 1B). To accomplish this method corresponding regions of TPSAB1 and TPSB2 are amplified in a single reaction, and sequenced using the chain termination (Sanger) method. Peak heights observed in the resulting chromatograms at nucleic acid residues that are only present in either alpha- or beta-tryptase sequences are directly measured, and the ratios of the two different nucleic acid signals are determined to provide relative quantitation of alpha- and beta-tryptases (20).
Figure 3. Strategies for tryptase genotyping.
(A) Two alleles in an individual with hereditary alpha tryptasemia – one allele with two beta-tryptase sequences at TPSAB1 and TPSB2 (top), and the second trait-associated allele with three alpha-tryptase encoding TPSAB1 copies and a single beta-tryptase encoding TPSB2 copy (bottom). Genomic DNA (gDNA) extracted from cells from this individual, containing these six different alpha- or beta-tryptase sequences are either amplified or restriction digested. The amplified gDNA can then either be (B) Sanger Sequenced, or (C) treated with the restriction enzyme EcoRV and Southern blotted to perform relative quantitation of alpha- and beta-tryptases. In both cases the ratio of alpha- to beta-tryptases calculated would be 1:1. (D) Unamplified digested gDNA is assayed by droplet digital PCR (ddPCR), which allows for absolute copy number detection of alpha- and beta-tryptase sequences, yielding the genotype determination 3α:3β.
The second method employs a similar strategy in which homologous genomic DNA from both TPSAB1 and TPSB2 is amplified. However, to determine the relative prevalence of sequences, the investigators took advantage of a conserved alpha-specific variant which results in the introduction of new restriction site. Treating the amplified DNA from TPSAB1 and TPSB2 with the restriction enzyme EcoRV results in cleavage of only alpha-tryptase amplicons. Gel electrophoresis and Southern blotting is then performed on the digested DNA (or undigested DNA in the case of beta-sequences) (Fig. 1C). Alpha-sequences result in a smaller-sized band, while beta-sequences remain as an uncut larger band. Band signal intensities are then quantified to determine a relative quantity of alpha- and beta-tryptases (27).
A major drawback of both of these methods, is that they rely upon conservation of both TPSAB1 and TPSB2 copy number to determine genotype. Therefore, with these methods an individual with a TPSAB1 triplication and an ααα/β:β/β genotype would not be distinguished from an individual without increased TPSAB1 copy number and alpha-sequences on both alleles (or the α/β:α/β genotype); both individuals would have equal quantities of alpha- and beta-tryptase sequences (Fig. 1). Moreover, both methods rely upon amplification of genomic DNA, and subtle differences in efficiency resulting from unique alpha- and beta-tryptase sequences potentially limit the precision required to distinguish individuals with TPSAB1 duplications and an αα/β:β/β genotype from those with an α/β:α/β genotype.
Bioinformatic re-alignment and copy number determination
Conventional exome or genome sequencing relies upon sequencing of small fragments of DNA in massive parallel (sizes typically range from 50-500 base pairs in length). Because the sequences at TPSAB1 and TPSB2 have a very high degree of homology, and TPSD1 sequences also have areas of significant homology with these genes, small fragments from all 6 loci, or more in the case of hereditary alpha tryptasemia, are frequently misaligned to any of these three reference genes. There are a number of publicly available algorithms designed to resolve highly complex loci that can be adapted to circumvent this issue (28). The strategy we developed employed de novo assembly of unselected sequences (from genome sequencing data) that mapped to the tryptase locus. We defined a ~500 base-pair tryptase “consensus” sequence with a number of unique identifiers that could distinguish alpha1/2-, beta1/3-, beta2-, and delta-tryptases from one another; gamma-tryptase was not homologous enough to require deconvolution.
To perform genotyping, all reads from exome and genome sequencing that map to TPSAB1, TPSB2, or TPSD1 are realigned to this “consensus” sequence using an algorithm and the number of reads assigned to each isoform (coverage) is used to estimate relative copy number (16). There are several major limitations to this and similar approaches. First, this process is highly dependent upon the quality, and quantity (or depth of coverage) at the locus. Poor capture is particularly problematic with older exon capture kits, where probes designed to extract the exome (coding DNA sequence) do not fully cover the tryptase locus leaving gaps in the genes. Genome sequencing avoids this issue, but the locus is also highly repetitive and GC-rich which can cause dimerization and stem-loop formation of DNA, both of which can hinder DNA amplification and/or extension still leading to poor coverage. Second, this method is still only a relative quantitation of copy. While coverage can be normalized to the average genomic coverage in the areas around the locus, the precision observed with these methods remains moderate. Because of this, the problem remains that an individual with an αα/β:β/β genotype can be difficult to distinguish from an individual with an α/β:α/β using this method.
TPSAB1 and TPSB2 allele-specific genotyping by droplet digital PCR
In order to overcome the issues of TPSAB1 copy number variation and reliance upon relative quantitation, we developed a droplet digital PCR (ddPCR) based assay (16). This platform allows for more precise and absolute quantification of alpha- and beta-tryptase specific sequences. Within the identified tryptase “consensus” sequence we identified a region in exon 3 of tryptase-encoding genes which allowed us to distinguish alpha1/2-, beta1/3-, beta2, and delta-tryptases from one another, and designed probes for each.
To accomplish genotyping, unamplified genomic DNA is restriction digested to separate each copy of tryptase sequence and then partitioned into droplets with two multiplexed primer/probe sets: one specific for alpha-tryptase and the other for beta-tryptase or a copy number reference gene. The samples are then placed on a thermal cycler in a process very similar to real-time PCR. However, rather than determining cycle time, all reactions are taken to completion (maximal fluorescence) and then run through a flow-based detector system much like a flow cytometer. Based upon the number of positive and negative droplets, a Poisson distribution-based calculation is used to absolutely quantify copy number, yielding highly reproducible and accurate quantification of tryptase genotype (Fig. 1D).
Manipulation of DNA digestion strategies can further resolve monoallelic copy number changes. Two copies of alpha- or beta-tryptase sequence on the same allele are in close proximity, and without DNA shearing or digestion, do not randomly segregate into droplets. Therefore, the resulting copy number call for alpha- or beta-tryptase is suppressed when the allelic copy number is increased; this is normalized with digestion. By comparing the results of the assay using digested DNA, to those results obtained without restriction digestion, allows for confirmation that alpha- or beta-tryptase sequences exist on the same allele (allele specificity). While there are some limitations to this assay, including the inability to resolve certain allelic genotypes displaying increased copy number without sequencing of additional family members (e.g. individuals with 4α:2β genotypes cannot be further resolved as ααα/β,α/β or αα/β,αα/β), it accurately determines absolute TPSAB1 and TPSB2 alpha- and beta-tryptase encoding copy number, including theoretic copy number loss, and accurately determine most allelic genotypes. Efforts are underway to make this assay available clinically.
Associated clinical features
All individuals identified to date with increased alpha-encoding TPSAB1 copy number have basal serum tryptases above 8 ng/mL (average BST caused by a duplication is 15 ± 5 ng/mL, and a triplication is 24 ± 6 ng/mL); on this basis hereditary alpha tryptasemia is currently believed to be a fully penetrant genetic trait. The expressivity of associated clinical phenotypes reported has been more variable, with some individuals reporting few if any symptoms. However, a number of symptoms are frequently reported by individuals with hereditary alpha tryptasemia (Table 1). Many of these phenotypes have also been reported in association with elevated BST in unselected cohorts, strengthening the clinical association between clinical features and increased TPSAB1 copy number (10, 16, 17, 29-31).
Table 1.
Clinical features reported in association with hereditary alpha tryptasemia
| Manifestation | Reported Prevalence* | Association Supported in an Unselected Cohort‡ |
|---|---|---|
| Basal serum tryptase >8ng/mL | 100% | Yes |
| Chronic gastroesophageal reflux symptoms | 56-77% | No |
| Arthralgia | 44-45% | No |
| Body pain/Headache | 33-47% | No |
| Flushing/Pruritus | 32-55% | Yes |
| Irritable bowel syndrome (Rome III) | 28-49% | Yes |
| Sleep disruption | 22-39% | No |
| Systemic immediate hypersensitivity reaction | 21-28% | No |
| Retained primary dentition | 20-33% | Yes |
| Systemic venom reaction | 14-22% | Yes |
| Congenital skeletal abnormality | 11-26% | No |
| Joint Hypermobility | 0-28% | No |
| Positive Tilt-table test | 0-11% | No |
Among the most commonly reported clinical symptoms among individuals with hereditary alpha tryptasemia are functional gastrointestinal complaints. A number of these, such as dyspepsia and odynophagia without observable pathology can be hard to characterize or quantify. However, irritable bowel syndrome (IBS) which has a number of validated measures, has been reported in approximately half of affected individuals within highly symptomatic families, and in a third of unselected individuals, using the Rome III criteria. This prevalence is approximately 2-5 times the estimated population prevalence in North America (32).
Half of individuals in selected and unselected populations with TPSAB1 duplications were reported with recurrent cutaneous symptoms that include flushing and pruritus; induration, angioedema, and urticaria were less commonly present. In some individuals these symptoms were spontaneous; however, vibration or minor trauma such as hand clapping, were frequently reported triggers. While these symptoms are suggestive of mast cell mediator release, few of these patients had identifiable evidence of chronic mast cell mediator release, and symptomatic events were not sufficiently studied to confirm these symptomatic episodes were mast cell-related.
Systemic reactions consistent with IgE-mediated immediate hypersensitivity to stinging insects (e.g. hymenoptera or honey bee) have been reported in approximately 20% of patients with hereditary alpha tryptasemia. This prevalence is 3-4 fold that of what has been reported in a similar population (33). While the established association between elevated BST and severe anaphylaxis to stinging insects has been largely attributed to clonal mast cell disease (34), these independent findings of an association between elevated BST caused by hereditary alpha tryptasemia and stinging insect allergy suggest some of this signal may come from increased TPSAB1 copy number, and requires further study.
A number of additional clinical manifestations have been reported in association with hereditary alpha tryptasemia including: connective tissue abnormalities such as joint hypermobility; retained primary dentition, and congenital abnormalities; symptoms suggestive of autonomic dysfunction, such as orthostatic hypotension, palpitations, tachycardia, presyncope and syncope; and constitutional symptoms, such as chronic pain and fatigue. Many of these symptoms are difficult to characterize or quantify and require additional validation. Finally, eosinophilic gastrointestinal disease, multiple food intolerances, failure to thrive, and IgE-mediated allergy have been observed in a small number of highly symptomatic families with increased TPSAB1 copy number. Whether these findings are generalizable to all individuals with hereditary alpha tryptasemia remains to be determined.
Despite the fact that tryptase genotype has not been evaluated extensively, a number of independent studies have examined the relationship between elevated BST, ostensibly in the absence of clonal mast cell disease or mastocytosis, and many of the clinical phenotypes observed in association with hereditary alpha tryptasemia. In addition to the well documented association of elevated BST and severe anaphylaxis to stinging insects (10, 30, 34-36), a recent publication demonstrated an association between elevated BST and anaphylaxis in children with food allergy (31). In this study, a BST > 14.5 ng/mL had a 90% positive predictive value for severe anaphylaxis.
In a separate, case control study of patients in a clinical practice in Austria, 100 individuals with elevated BST and 100 individuals with BST <11.4 ng/mL (mean BST 3.7 ng/mL) were administered a questionnaire which evaluated a number of the symptoms later reported in individuals with hereditary alpha tryptasemia. Significant associations were observed between elevated BST and cutaneous symptoms (flushing, angioedema), gastrointestinal symptoms (abdominal pain, nausea, meteorism, diarrhea), symptoms suggestive of autonomic dysfunction (tachycardia, palpitations, vertigo) and collapse of unclear eitiology; fatigue, pain symptoms, and mood alterations were also reported to be associated with increased BST (10).
In a third study examining patients with chronic idiopathic urticaria (CIU) who experience systemic symptoms during disease flares (n = 155), BST >8.2 ng/mL was not observed in patients without systemic complaints. The complaints reported included: gastrointestinal symptoms, flushing, joint pain or swelling, cardiovascular manifestations, respiratory symptoms, and other constitutional complaints. Overall, significantly higher BST were observed in patients with systemic symptoms, and were associated with significantly more severe urticaria (determined by several validated measures). Further, examining patients with BST ≥10 ng/mL in this cohort, the mean itch score was nearly twice what was reported in the remainder of individuals (29).
In a final report, a three-generation Belgian family was described with dominantly inherited BST elevations associated with severe episodic gastrointestinal cramping and diarrhea. Serum tryptase levels were observed to rise in association with symptomatic events. In one affected individual, evidence of clonal mast cell disease was also reported [hepatosplenomegaly and the missense KIT p.(D816V) in bone marrow] consistent with the diagnosis of mono-clonal mast cell activation syndrome (MMAS) (17).
Management approaches
Clinical treatment approaches for patients with hereditary alpha tryptasemia are currently personalized based upon symptoms. Like other patients with the kinds of multisystem complaints reported by individuals with hereditary alpha tryptasemia, responses to therapy are often mixed and variable from person-to-person. As there is little data to support a particular strategy, and no prospective studies on which to rely, the following management approaches are based solely upon the limited clinical experience at the NIH Clinical Center, and focus on the commonest symptoms.
For cutaneous and gastrointestinal symptoms, we recommend trials of maximal antihistamine therapy targeting both H1 and H2 receptors twice daily – as used in CIU or indolent forms of mastocytosis (37, 38). We also prescribe oral cromolyn sodium if gastrointestinal symptoms are severe. While our center does not have access to oral ketotifen, a number of patients have also reported some improvement with this medication. However, the overall clinical responses to these mast cell-directed therapies have been disappointing. While one patient reported complete resolution of diarrhea, abdominal pain, dysphagia, and migraine headaches with addition of oral ketotifen to maximal twice daily anti-histamines, most others have reported only modest benefit of mast cell stabilizers, and even responses to anti-histamines have been mixed. Anecdotally, a few patients who have received omalizumab for allergic asthma or CIU, have reported improvement in some additional symptoms.
For individuals with recurrent severe systemic symptoms and/or anaphylaxis, triggers should be identified and avoided, and epi-pens should be provided – as is the standard of care. However, many systemic events in these patients are difficult to characterize, and in some cases may not represent immediate hypersensitivity reactions. Regardless of etiology, particularly when anxiety plays a large contributing role, biofeedback – as frequently employed in management of patients with mastocytosis – has proven beneficial in a number of patients.
A number of other medications that have been anecdotally observed as beneficial for symptoms in some patients include: tricyclic antidepressants (TCAs), clemastine fumarate, and gabapentin. More effective medications are greatly needed for the symptoms associated with hereditary alpha tryptasemia, and prospective clinical trials are critically lacking in order to evaluate the efficacy of current treatment approaches in these difficult to treat patients.
Future Considerations / Summary
In summary, hereditary alpha tryptasemia is a relatively common genetic trait caused by increased copies of TPSAB1 encoding alpha-tryptase on a single allele, and is inherited in an autosomal dominant manner. Affected individuals have elevated BST and may present with multisystem complaints, both of which positively correlate with the number of additional TPSAB1 copies, in a gene dose manner. A number of questions remain around the clinical phenotypes associated with hereditary alpha tryptasemia and whether this trait may play a causative or modifying role in the findings that have been reported. A common haplotype reported to be co-inherited with increased TPSAB1 copy number in approximately 2/3 of individuals was recently characterized (18). Additional variants in TPSG1 encoding gamma-tryptase and partial gain-of-function variants in CACNA1H encoding a T-type voltage gated calcium channel were identified. While no effect on clinical phenotype could be observed, the CACNA1H variants are quite intriguing as this channel has been implicated in nociception and IBS in animal models (39, 40).
Given that hereditary alpha tryptasemia appears to be a common cause for elevated BST, TPSAB1 copy number should be considered in the clinical evaluation of patients with BST >8 ng/mL. Once clinically available, tryptase genotyping of patients will likely be a useful tool for evaluation of patients with suspected clonal mast cell disease and other myeloid abnormalities that can also be associated with elevated BST. The normal reference range for BST also bears reevaluation in this context. Currently the upper limit of normal was assigned as 11.4 ng/mL, somewhat arbitrarily. Now that a genetic basis has been established which appears to explain a large number of individuals with BST > 10ng/mL, a rationally identified upper limit of normal at 8-10 ng/mL should be considered, and has been suggested in the literature (30).
As more individuals and larger cohorts are genotyped, and the association between elevated BST and increased TPSAB1 copy number at the population level is clarified, the strength of association between hereditary alpha tryptasemia and the multiple clinical phenotypes that have been reported will no doubt evolve. Further, as the functional effects of elevated BST and/or altered tryptase gene expression are elucidated, potential mechanisms for causation and/or modification of clinical disease are likely to be elucidated. Once identified, these findings will provide a rationale for future therapeutic intervention in these patients, and potentially others with similar clinical presentations.
Key points:
Hereditary alpha tryptasemia is a genetic trait which leads to elevated basal serum tryptase.
Some individuals with hereditary alpha tryptasemia present with a syndrome comprised of multisystem complaints.
Increased TPSAB1 copy number encoding alpha-tryptase on a single allele is the cause of hereditary alpha tryptasemia.
A gene dosage effect exists between number of additional TPSAB1 copies, basal serum tryptase levels, and severity of clinical symptoms in affected individuals.
Complex structural variation at the tryptase locus prevents identification of increased TPSAB1 copy number by conventional exome or genome sequencing.
Acknowledgements:
The author is grateful to Kelly Stone M.D. Ph.D. for his thoughtful comments on the manuscript.
Funding: This research was supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
Disclosure statement: The author declares no competing or conflicting interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Theoharides TC, Valent P, Akin C. Mast Cells, Mastocytosis, and Related Disorders. N Engl J Med. 2015;373(2):163–72. [DOI] [PubMed] [Google Scholar]
- 2.Foster B, Schwartz LB, Devouassoux G, Metcalfe DD, Prussin C. Characterization of mast-cell tryptase-expressing peripheral blood cells as basophils. J Allergy Clin Immunol. 2002;109(2):287–93. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz LB, Lewis RA, Seldin D, Austen KF. Acid hydrolases and tryptase from secretory granules of dispersed human lung mast cells. J Immunol. 1981;126(4):1290–4. [PubMed] [Google Scholar]
- 4.Caughey GH. Mast cell tryptases and chymases in inflammation and host defense. Immunol Rev. 2007;217:141–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz LB, Sakai K, Bradford TR, Ren S, Zweiman B, Worobec AS, et al. The alpha form of human tryptase is the predominant type present in blood at baseline in normal subjects and is elevated in those with systemic mastocytosis. J Clin Invest. 1995;96(6):2702–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz LB, Min HK, Ren S, Xia HZ, Hu J, Zhao W, et al. Tryptase precursors are preferentially and spontaneously released, whereas mature tryptase is retained by HMC-1 cells, Mono-Mac-6 cells, and human skin-derived mast cells. J Immunol. 2003;170(11):5667–73. [DOI] [PubMed] [Google Scholar]
- 7.Kanthawatana S, Carias K, Arnaout R, Hu J, Irani AM, Schwartz LB. The potential clinical utility of serum alpha-protryptase levels. J Allergy Clin Immunol. 1999;103(6):1092–9. [DOI] [PubMed] [Google Scholar]
- 8.Akin C, Soto D, Brittain E, Chhabra A, Schwartz LB, Caughey GH, et al. Tryptase haplotype in mastocytosis: relationship to disease variant and diagnostic utility of total tryptase levels. Clin Immunol. 2007;123(3):268–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz LB. Diagnostic value of tryptase in anaphylaxis and mastocytosis. Immunol Allergy Clin North Am. 2006;26(3):451–63. [DOI] [PubMed] [Google Scholar]
- 10.Fellinger C, Hemmer W, Wohrl S, Sesztak-Greinecker G, Jarisch R, Wantke F. Clinical characteristics and risk profile of patients with elevated baseline serum tryptase. Allergol Immunopathol (Madr). 2014;42(6):544–52. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Quintela A, Vizcaino L, Gude F, Rey J, Meijide L, Fernandez-Merino C, et al. Factors influencing serum total tryptase concentrations in a general adult population. Clin Chem Lab Med. 2010;48(5):701–6. [DOI] [PubMed] [Google Scholar]
- 12.Sirvent AE, Gonzalez C, Enriquez R, Fernandez J, Millan I, Barber X, et al. Serum tryptase levels and markers of renal dysfunction in a population with chronic kidney disease. J Nephrol. 2010;23(3):282–90. [PubMed] [Google Scholar]
- 13.Valent P, Akin C, Metcalfe DD. Mastocytosis: 2016 updated WHO classification and novel emerging treatment concepts. Blood. 2017;129(11):1420–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valent P, Sperr WR, Sotlar K, Reiter A, Akin C, Gotlib J, et al. The serum tryptase test: an emerging robust biomarker in clinical hematology. Expert Rev Hematol. 2014;7(5):683–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyons JJ, Sun G, Stone KD, Nelson C, Wisch L, O'Brien M, et al. Mendelian inheritance of elevated serum tryptase associated with atopy and connective tissue abnormalities. J Allergy Clin Immunol. 2014;133(5):1471–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyons JJ, Yu X, Hughes JD, Le QT, Jamil A, Bai Y, et al. Elevated basal serum tryptase identifies a multisystem disorder associated with increased TPSAB1 copy number. Nat Genet. 2016;48(12):1564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabato V, Van De Vijver E, Hagendorens M, Vrelust I, Reyniers E, Fransen E, et al. Familial hypertryptasemia with associated mast cell activation syndrome. J Allergy Clin Immunol. 2014;134(6):1448–50 e3. [DOI] [PubMed] [Google Scholar]
- 18.Lyons JJ, Stotz SC, Chovanec J, Liu Y, Lewis KL, Nelson C, et al. A common haplotype containing functional CACNA1H variants is frequently coinherited with increased TPSAB1 copy number. Genet Med. 2017. [DOI] [PubMed] [Google Scholar]
- 19.Caughey GH. Tryptase genetics and anaphylaxis. J Allergy Clin Immunol. 2006;117(6):1411–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trivedi NN, Tamraz B, Chu C, Kwok PY, Caughey GH. Human subjects are protected from mast cell tryptase deficiency despite frequent inheritance of loss-of-function mutations. J Allergy Clin Immunol. 2009;124(5):1099–105 e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le QT, Min HK, Xia HZ, Fukuoka Y, Katunuma N, Schwartz LB. Promiscuous processing of human alphabeta-protryptases by cathepsins L, B, and C. J Immunol. 2011;186(12):7136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C, Li L, Krilis SA, Chanasyk K, Tang Y, Li Z, et al. Human tryptases alpha and beta/II are functionally distinct due, in part, to a single amino acid difference in one of the surface loops that forms the substrate-binding cleft. J Biol Chem. 1999;274(28):19670–6. [DOI] [PubMed] [Google Scholar]
- 23.Marquardt U, Zettl F, Huber R, Bode W, Sommerhoff C. The crystal structure of human alpha1-tryptase reveals a blocked substrate-binding region. J Mol Biol. 2002;321(3):491–502. [DOI] [PubMed] [Google Scholar]
- 24.Selwood T, Wang ZM, McCaslin DR, Schechter NM. Diverse stability and catalytic properties of human tryptase alpha and beta isoforms are mediated by residue differences at the S1 pocket. Biochemistry. 2002;41(10):3329–40. [DOI] [PubMed] [Google Scholar]
- 25.Brown JK, Jones CA, Rooney LA, Caughey GH, Hall IP. Tryptase's potent mitogenic effects in human airway smooth muscle cells are via nonproteolytic actions. Am J Physiol Lung Cell Mol Physiol. 2002;282(2):L197–206. [DOI] [PubMed] [Google Scholar]
- 26.Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517(7536):608–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le QT, Lotfi-Emran S, Min HK, Schwartz LB. A simple, sensitive and safe method to determine the human alpha/beta-tryptase genotype. PLoS One. 2014;9(12):e114944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handsaker RE, Van Doren V, Berman JR, Genovese G, Kashin S, Boettger LM, et al. Large multiallelic copy number variations in humans. Nat Genet. 2015;47(3):296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doong JC, Chichester K, Oliver ET, Schwartz LB, Saini SS. Chronic Idiopathic Urticaria: Systemic Complaints and Their Relationship with Disease and Immune Measures. J Allergy Clin Immunol Pract. 2017;5(5):1314–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kucharewicz I, Bodzenta-Lukaszyk A, Szymanski W, Mroczko B, Szmitkowski M. Basal serum tryptase level correlates with severity of hymenoptera sting and age. J Investig Allergol Clin Immunol. 2007;17(2):65–9. [PubMed] [Google Scholar]
- 31.Sahiner UM, Yavuz ST, Buyuktiryaki B, Cavkaytar O, Yilmaz EA, Tuncer A, et al. Serum basal tryptase may be a good marker for predicting the risk of anaphylaxis in children with food allergy. Allergy. 2014;69(2):265–8. [DOI] [PubMed] [Google Scholar]
- 32.Saito YA, Schoenfeld P, Locke GR, 3rd. The epidemiology of irritable bowel syndrome in North America: a systematic review. Am J Gastroenterol. 2002;97(8):1910–5. [DOI] [PubMed] [Google Scholar]
- 33.Sturm GJ, Kranzelbinder B, Schuster C, Sturm EM, Bokanovic D, Vollmann J, et al. Sensitization to Hymenoptera venoms is common, but systemic sting reactions are rare. J Allergy Clin Immunol. 2014;133(6):1635–43 e1. [DOI] [PubMed] [Google Scholar]
- 34.Bonadonna P, Zanotti R, Muller U. Mastocytosis and insect venom allergy. Curr Opin Allergy Clin Immunol. 2010;10(4):347–53. [DOI] [PubMed] [Google Scholar]
- 35.Haeberli G, Bronnimann M, Hunziker T, Muller U. Elevated basal serum tryptase and hymenoptera venom allergy: relation to severity of sting reactions and to safety and efficacy of venom immunotherapy. Clin Exp Allergy. 2003;33(9):1216–20. [DOI] [PubMed] [Google Scholar]
- 36.Rueff F, Przybilla B, Bilo MB, Muller U, Scheipl F, Aberer W, et al. Predictors of severe systemic anaphylactic reactions in patients with Hymenoptera venom allergy: importance of baseline serum tryptase-a study of the European Academy of Allergology and Clinical Immunology Interest Group on Insect Venom Hypersensitivity. J Allergy Clin Immunol. 2009;124(5):1047–54. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein JA, Lang DM, Khan DA, Craig T, Dreyfus D, Hsieh F, et al. The diagnosis and management of acute and chronic urticaria: 2014 update. J Allergy Clin Immunol. 2014;133(5):1270–7. [DOI] [PubMed] [Google Scholar]
- 38.Frieri M, Alling DW, Metcalfe DD. Comparison of the therapeutic efficacy of cromolyn sodium with that of combined chlorpheniramine and cimetidine in systemic mastocytosis. Results of a double-blind clinical trial. Am J Med. 1985;78(1):9–14. [DOI] [PubMed] [Google Scholar]
- 39.Choi S, Na HS, Kim J, Lee J, Lee S, Kim D, et al. Attenuated pain responses in mice lacking Ca(V)3.2 T-type channels. Genes Brain Behav. 2007;6(5):425–31. [DOI] [PubMed] [Google Scholar]
- 40.Marger F, Gelot A, Alloui A, Matricon J, Ferrer JF, Barrere C, et al. T-type calcium channels contribute to colonic hypersensitivity in a rat model of irritable bowel syndrome. Proc Natl Acad Sci U S A. 2011;108(27):11268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]