Abstract
Dual-axis confocal (DAC) microscopy is an optical imaging modality that utilizes simple low-numerical aperture (NA) lenses to achieve effective optical sectioning and superior image contrast in biological tissues. The unique architecture of DAC microscopy also provides some advantages for miniaturization, facilitating the development of endoscopic and handheld DAC systems for in vivo imaging. This article reviews the principles of DAC microscopy, including its differences from conventional confocal microscopy, and surveys several variations of DAC microscopy that have been developed and investigated as non-invasive real-time alternatives to conventional biopsy and histopathology.
Keywords: confocal microscopy, point-of-care pathology, miniature imaging system
I. Introduction
The microscopic evaluation of surgical and biopsy specimens with tabletop microscopes, through a process known as histology, is currently regarded by the medical community as the “gold standard” for the diagnosis of diseases. However, this core technology for clinical pathology has certain limitations that can lead to poor inter-observer concordance and limited diagnostic accuracy for prognostication and prediction of treatment response. For example, the invasive physical resection of tissues is often not desired by patients, especially in cases where the tissues are of functional importance (e.g. brain), cosmetic value (e.g. skin), and in cases where there is a low probability of malignancy (e.g. biopsies of suspicious lesions in the oral cavity). In addition, dynamic information (e.g. blood flow) and physiological parameters (e.g. pH, oxygenation, electrolyte concentration, etc.) are often lost or altered during ex vivo tissue processing, which can reduce diagnostic accuracy. Once tissues are excised, the standard procedure of sample preparation - involving fixation, dehydration, wax embedding, sectioning, mounting of tissue sections on glass slides, and staining - is labor-intensive, complex, and time-consuming, potentially resulting in treatment delays and process-induced errors. Moreover, the multi-step procedure is known to introduce artifacts (e.g. shrinkage and cracking due to dehydration) as well as sampling errors since only a small fraction of most tissue specimens are processed onto glass slides for imaging. Therefore, there has been a long-standing interest in developing in vivo microscopes for real-time non-invasive microscopic examination of vital tissues. Such in situ evaluation can circumvent some of the drawbacks of ex vivo tissue processing and can provide immediate (and potentially more accurate) feedback to the clinicians, thus accelerating and improving diagnoses and treatments.
Over the past few decades, various portable research microscopes, and commercialized in vivo microscopy systems have been developed to address a host of clinical needs [1–28]. Many of these devices have been based on the technology of confocal microscopy [29], which provides cross-sectional images of intact specimens (i.e. optical sectioning) with high resolution and contrast (i.e. signal-to-background ratio, SBR). In confocal microscopy, a spatial filter (e.g. a pinhole or a slit) is placed at a conjugate image plane so that only the signal originating from a localized focal volume within the tissue is efficiently transmitted onto a detector, while out-of-focus and multiply scattered photons (from tissue regions away from the focal volume of interest) are largely blocked by the pinhole (or slit). In order for a conventional confocal microscope (referred to as a single-axis confocal, SAC, microscope in this article) to achieve subcellular resolution while retaining adequate field-of-view (FOV) and working distance (WD), a bulky high numerical aperture (NA) objective lens, and a complex scanning mechanism are typically needed. A portable SAC microscope that images with high resolution and contrast is technically difficult to engineer but has been realized by several groups in academia and industry [1–11, 15–18]. As an alternative to conventional SAC microscopy, dual-axis confocal (DAC) microscopy relaxes, to some degree, the requirements for high-NA focusing in order to achieve efficient optical sectioning, and provides certain benefits (as described later in this article) for the miniaturization of confocal microscopes down to the scale of several millimeters. In addition, DAC microscopy has demonstrated superior contrast (i.e. SBR) and imaging depth compared to SAC microscopy, which can be of clinical value for non-invasive real-time in vivo pathology.
II. DAC ARCHITECTURE
Dual-axis confocal (DAC) microscopy, which was inspired by the concept of “theta” confocal microscopy [30–32], was developed in the early 2000s to address certain limitations of conventional SAC microscopes [33, 34]. DAC microscopy achieves optical sectioning using the same basic principle of confocal detection as conventional SAC microscopy, in which a spatial filter (i.e. a pinhole or slit) is used to reject out-of-focus light. The fundamental difference between SAC and DAC microscopy is how the focal volumes are generated and defined. In the SAC configuration, the illumination and detection optics share the same objective (Fig. 1a), which alone defines the focal volume of the system. However, in a DAC system (Fig. 1b), the illumination and collection beam paths (blue and green lines, respectively) do not overlap except at their foci. The effective focal volume of a DAC system is therefore defined by the region where the two individual foci intersect (black oval).
Fig. 1.
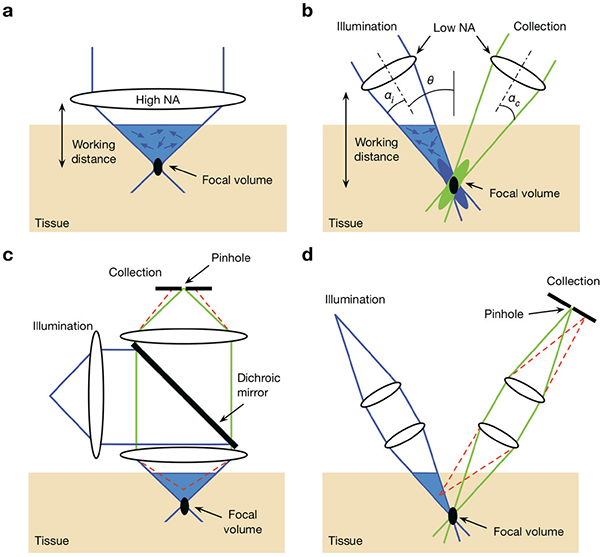
Comparison of the optical configurations for a conventional singleaxis confocal (SAC) microscope and a dual-axis confocal (DAC) microscope, (a) In order to achieve a tight focal volume (black oval), a SAC microscope requires a high-N A objective lens. This results in a short working distance that makes miniaturization and beam scanning more difficult, (b) A DAC microscope uses low-NA off-axis illumination and collection beams, in which the focal volume is defined by the overlapping foci of the two beams. The use of low-NA beams allows for a longer working distance, which provides advantages for miniaturization and beam scanning, (c) In SAC microscopy, out-of-focus light (an example beam path is shown with the dashed red lines) is not completely rejected by the pinhole, (d) In DAC microscopy, out-of-focus light is directed away from the pinhole and is more optimally rejected, thereby improving the signal-to-background ratio.
The spatially separated illumination and collection path of the DAC architecture provides certain advantages. First, in a DAC configuration, the effective axial resolution (optical-sectioning thickness) of the system is proportional to 1/NA, rather than 1/NA2 as in a SAC configuration, allowing a DAC system to achieve more effective optical sectioning using low-NA lenses (NA < 0.5), which are often lower in cost and more easily miniaturized. Second, the use of low-NA beams provides a longer working distance such that it is possible to place a scanning mirror between the focusing optics and the sample [35]. Such “post-objective scanning” allows the beams to maintain an on-axis alignment through the focusing optics regardless of the angle of the scanning mirror. By eliminating the possibility of off-axis aberrations, simple inexpensive low-NA lenses such as injection-molded spheres may be used rather than bulky compound objectives. Low-NA lenses are also more-easily scaled down to millimeter dimensions while maintaining a reasonable FOV and WD. Third, the DAC architecture has been shown to improve the imaging contrast (thus imaging depth) in highly scattering fresh tissues, mainly because the smaller acceptance cone of the low-NA collection lens, and the well-separated illumination and collection beam paths, are better for rejecting the multiply scattered photons [36, 37] that are the main contributors to the background for confocal microscopy within fresh tissues. Finally, the illumination and collection beam paths can be independently aligned to compensate for any chromatic effects due to the Stokes shift of fluorophores.
The point spread function (PSF) derived from diffraction theory predicts the theoretical response of an imaging system under ideal conditions (i.e. the diffraction-limited performance). In a typical DAC system (Fig. 2), two low-NA beams with focusing angles of αi, and αc, respectively, intersect at a half-angle of θ. Using diffraction theory under the paraxial approximation [33,38], the PSF of each beam can be calculated and is depicted by the blue and green cigar-shaped ovals in Fig. 2b. The overall PSF of the DAC microscope is defined by the product (the black oval) of the intersecting PSFs of the illumination and collection beams. For truncated circular Gaussian beams, the amplitude PSF, U, which describes the spatial distribution of the electric-field amplitude of a beam, is proportional to the Huygens-Fresnel diffraction integral [39]:
| (1) |
where W is the weighting function that accounts for the beam truncation, Jo is the zero-order Bessel function, ρ is a normalized spatial variable that describes the distance from the optical axis of the beam (i.e. the beam radius). The upper limit of the integral, a, is determined by the size of the aperture. Here we assume the use of pure Gaussian beams that are not truncated by apertures (i.e. a = ∞). For the case of apodized (truncated) beams, a more-detailed treatment may be found in a previous publication [38].
Fig. 2.
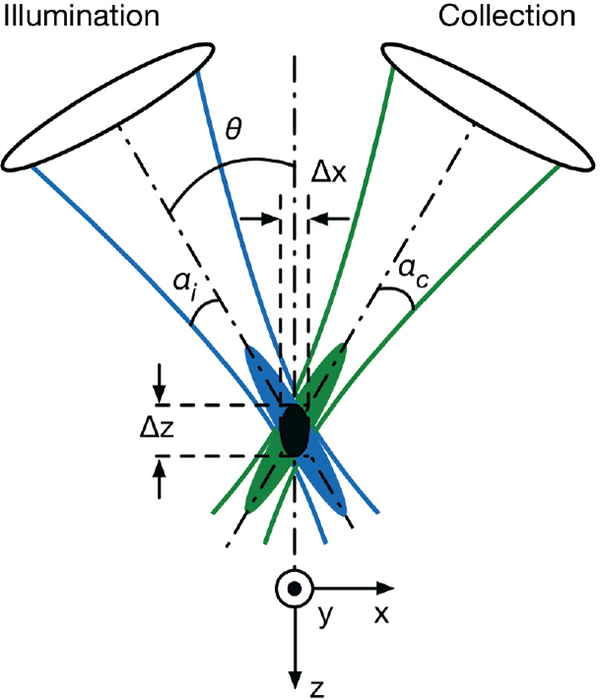
The DAC microscope architecture. Two low-NA beams (illumination and collection) with focusing angles of αi and αc, respectively, intersect at a half-angle of θ. The focal volume (black oval) of the system is defined by the product of the intersecting point spread functions (PSFs) of the illumination (blue) and collection (green) beams. The dimensions of the focal volume (ᐃx, ᐃy, and ᐃz) correspond to the spatial (lateral and axial) resolutions of the system.
The response of the DAC system to a delta-function point object (power received at the detector), Idac, is proportional to the square of the product of the illumination and the collection amplitude PSFs,Ui and Uc
| (2) |
The full-width at half-maximum (FWHM) extent of IDAC is often used to quantify the dimensions of the DAC focal volume, and provides an approximation of the spatial resolution of the imaging system. By assuming that the illumination and collection wavelengths are identical (λi = λc = λ), and that the focusing NA of the illumination and collection beams are also identical (αi, = αc = α), the FWHM spatial resolution along each dimension can be calculated as [12]:
| (3) |
where n is the refractive index of the medium, and 0 < θ < π/2.
In comparison, the theoretical resolution derived from diffraction theory for a SAC microscope system with uniform illumination are [34, 40–44]:
| (4) |
These results indicate that the spatial resolution of a DAC system in the x and z directions (the plane of intersection of the DAC beams) are θ-dependent, and that the axial response, Δz, of a DAC system is inversely proportional to the NA of the lenses, rather than to the square of the NA as with a SAC microscope. This suggests that a DAC system is able to provide effective optical sectioning even when low-NA lenses are used, especially when NA < 0.5, as is the case for DAC microscopy. In addition, as shown in Fig. 3 a, the axial response of a DAC microscope will be steeper compared with a SAC microscope that has an identical FWHM axial resolution.
Fig. 3.
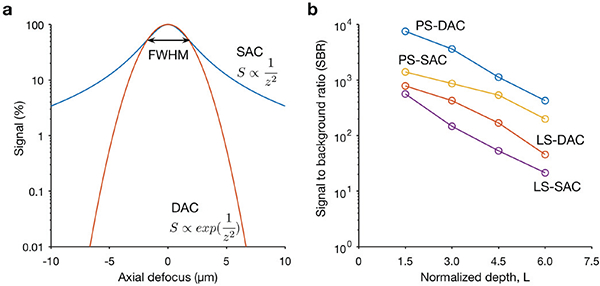
Quantitative comparison of the axial-sectioning response (a) and contrast (b) of typical SAC and DAC systems, (a) The theoretical axial response of a SAC and DAC microscope is shown, in which a point object is translated through the focus of the microscope in the z direction. In this case, the SAC and DAC microscopes have equivalent axial resolutions (FWHM). The signal rolls off more quickly in a DAC system (red) than in a SAC system (blue), showing that the axial sectioning performance (rejection of out-of-focus light) is superior for the DAC configuration. (b) Monte-Carlo scattering simulations to compare the contrast (signal-to-background ratio, SBR) of various microscope configurations when imaging an in-focus reflective object within highly scattering biological tissues, as a function of depth. The results show that both the point-scanned (PS) and the line-scanned (LS) versions of DAC microscopy provide superior image contrast in scattering media when compared with their SAC microscopy counterparts. The normalized depth refers to the number of mean free paths that ballistic photons would travel in a round-trip perpendicular path from the tissue surface to the focal volume, i.e. L=2μsd, where μs is the scattering coefficient, and d is the imaging depth in the direction normal to the tissue surface [46]
The PSF and axial response derived from diffraction theory cannot predict the effects of tissue scattering, which prevents ballistic photons (i.e. unscattered photons that are “diffraction-limited” in terms of their trajectory and focusing abilities) from penetrating deeply within tissues. Therefore, Monte-Carlo ray tracing models have previously been developed to simulate the performance of various DAC microscope configurations in scattering media [36, 37, 46] (Fig. 3b), including the role of θ and α on sectioning performance (contrast) [37, 45, 46] The simulation results showed that both the imaging contrast (signal to background ratio, SBR) and axial resolution consistently improve as θ is increased, suggesting that the crossing angle of the two beams in a DAC system should be maximized when possible. In general, the contrast is more sensitive to the crossing angle whereas the resolution is sensitive to both θ and α (the NA of the beams) [45] Maximizing both parameters typically gives the best performance, but also implies larger device sizes and/or shorter working distances, and potentially creates additional aberrations in the system.
III. DAC IMAGING SYSTEMS
A. Basic components
Geometric orientations
DAC microscope prototypes were initially developed as tabletop systems with primarily off-the-shelf optical components. For example, in several early systems [33, 35, 37, 38, 47–49], two low-NA (typically around 0.2) objective lenses were oriented at a half crossing angle of 30 deg. Note that the NA and the crossing angle has been varied in different DAC systems, and has often been determined by pragmatic concerns such as working distance and device size, as well as the position and size of the scanning mechanism (e.g. galvanometric or MEMS scanning mirrors).
Post-objective scanning
As discussed previously, the long working distance of low-NA beams provides room for a scanning mirror to be placed after the focusing optics, allowing the system to avoid the off-axis aberrations that must be compensated for with pre-objective scanning. This scanning scheme allows for large FOVs to be achieved even when small and simple focusing optics are used. Note that the same scanning mirror may be used to steer both the illumination and collection beams simultaneously (scanning the illumination beam while de-scanning the collection beam), which helps to ensure that the beams remain well-aligned and that they always intersect at their foci [12, 35].
Hemispherical solid immersion lens
The use of a hemispherical fused-silica solid immersion lens (SIL) has been a distinguishing feature of many previous DAC microscope prototypes [38, 47], as well as a related open-top light sheet microscope design described recently [56]. The SIL provides several advantages:
Minimizing off-axis aberrations: the curved surface of the hemisphere provides a normal interface for both the illumination and collection beams as they transition from air into the glass SIL, such that off-axis aberrations (e.g. coma and astigmatism) are minimized.
Minimizing spherical aberrations: the wave-front curvature of the focused beams is matched to the curved surface of the hemisphere, minimizing the spherical aberrations that would result from focusing a beam through a flat interface between two media with different refractive indices (e.g. from air to glass). Note that slight aberrations still occur while scanning the beams away from their ideal neutral positions.
Refractive index matching: since the refractive index of fused silica (n = 1.45) is similar to that of most biological tissues, aberrations are minimized as the beams travel across the interface between the tissue sample and the flat surface (distal surface) of the hemisphere.
Increasing the effective focusing NA: if a beam is being focused from air into a higher-index material, the NA is typically preserved due to Snell’s law (where ΝΑ = n sinα). However, the curved surface of the hemisphere, when well-aligned, preserves the ray angles of the focused beams, thus increasing the NA by a factor n. On the other hand, the curved surface of the hemisphere also acts to de-magnify the scanning range of the beams, to first order, by 1/n. For example, in the case of fused silica (n = 1.45), a 250-μm axial translation of the stage causes the focal volume to translate ∼150 μm within the sample. Similarly, lateral translations are also de-magnified by roughly 1 /n.
B. DAC microscopy variations
Early DAC microscopes utilized point-focused Gaussian illumination in conjunction with point-by-point confocal detection along a raster-scanned [28] or Lissajous-scanned trajectory [12]. While these systems successfully demonstrated the advantages of the DAC architecture, their speed was often limited due to the use of point scanning. In addition to a desire to improve upon imaging speeds, the ability to image deeper was also desired for certain in vivo imaging applications. To extend the potential of DAC microscopy, a wide range of DAC - variants have been explored. These approaches can be grouped into two main categories based on their aims: (1) improving the imaging speed by modifying either the scanning mechanism (e.g. with complex scanning mirrors [28, 50]) or the illumination patterns (e.g. line scanning [46, 47]); (2) improving the imaging depth (i.e. image contrast) by using different light sources (e.g. near infrared lasers [28, 35], Bessel beams [51]) or through advanced detection methods (e.g. temporal gating, lock-in detection [35, 38, 52]). Due to space constraints, this section highlights only the variants that have been developed within the past five years.
Line-scanned dual-axis confocal (LS-DAC) microscopy
In vivo imaging with a handheld device is subject to motion artifacts induced by the subjects and users. For example, early miniaturized point-scanned (PS) DAC systems (Fig. 4a) had a limited frame rate of < 5 Hz, which led to frequent motion artifacts (blurring and distortions) during in vivo use. Although it is possible for a PS-DAC system to acquire images at video rate [28, 41], the FOV is often limited or the scanning mechanisms are complicated, expensive, and difficult to scale down in size. As a result, a simpler line-scanned (LS) DAC microscopy (Fig. 4b) approach has been utilized in recent years to improve the imaging speed, with a trade-off in image contrast (SBR) due to the loss of confocality along one dimension (along the focal line). In brief, LS-DAC microscopes illuminate a focal line in the sample, instead of a localized point. That focal line is imaged by the collection optics onto a linear-array detector. Since an entire line of pixels is imaged and acquired simultaneously, the focal line (or tissue) only needs to be scanned in one direction to generate a 2D image. In addition to reducing the cost and complexity of the scanning mechanism, line-by-line data collection also has the potential to improve the signal-to-noise ratio (SNR) of the images because of the longer pixel integration times.
Fig. 4.
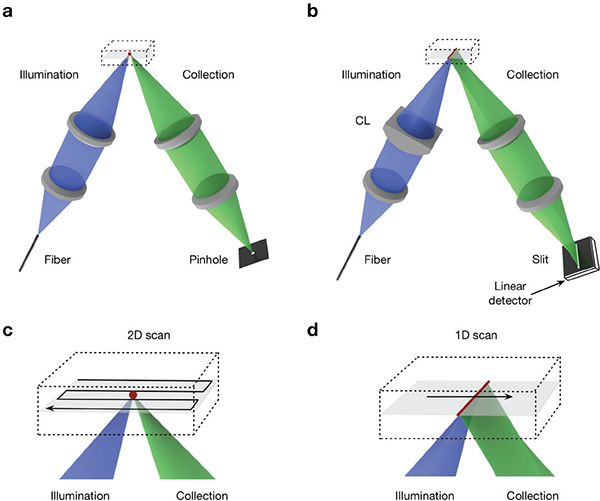
(a) In a point-scanned DAC system, light is tightly focused to a point within the sample, and a pinhole is used for confocal detection. To create an image, the point is scanned in two dimensions (e.g. following a raster- or Lissajous-scanned trajectory) and the image is generated point by point. (b) In the case of a line-scanned DAC system, the illumination objective lens is replaced with a cylindrical lens (CL) so that light is focused to a thin line within the sample, and a slit is used for confocal detection. The focal line only needs to be scanned in one dimension to create a 2D image. (c-d) Zoomed-in views of the scanning trajectories described in (a) and (b) are shown, respectively.
The first LS-DAC microscope utilized a cylindrical lens in the illumination path to generate a long focal line along the y axis of the imaging plane [47]. On the detection side, instead of using a single-mode fiber as a pinhole, as was done in previous PS-DAC systems, a digital slit (cropped region) was defined within a detector array to serve as a spatial filter for confocal detection. This configuration eliminated the need for a fast 2D scanning mirror to create an image, such that high-speed imaging could be achieved with a slow 1D galvanometric mirror [49, 53]. The frame rate was thus primarily limited by the sensitivity of the detector, rather than the mechanical scanning mechanism. It has been shown that LS-DAC and PS-DAC microscopy exhibit comparable imaging performance at shallow depths (<150 μm), but that the image quality (contrast) of LS-DAC microscopy deteriorates at deeper depths because of the reduced confocality of a line-scanned system [47].
Sheet-scanned dual-axis confocal (SS-DAC) microscopy
As discussed in the previous section, one of the drawbacks of LS-DAC microscopy is the deteriorated image contrast due to the loss of confocality along the focal line. Sheet-scanned (SS) DAC microscopy was developed to mitigate the reduced performance in LS-DAC microscopy by utilizing the additional spatial information provided by a 2D detector array [54]. The key principle of this approach is to partially mitigate the loss of confocality by utilizing the useful information contained in the slightly “out-of-focus” regions near the focal line that are typically rejected by a physical slit in a line-scanned confocal microscope. In particular, if the crossing angle of a DAC microscope approaches 90 deg, then the collection arm essentially images a “light sheet” generated by the illumination arm, from which additional information is provided that can be used to perform 3D deconvolution.
The SS-DAC concept was first demonstrated with a scientific complementary metal-oxide-semiconductor (sCMOS) detector array to image an oblique light sheet (effectively, an angled light sheet). By scanning the angled light sheet, a thin 3D volume of data was acquired, which could then be used for deconvolution of the main LS-DAC image if the PSF of the system was known (either measured or simulated). This simple technique was shown to improve the spatial resolution and contrast of the LS-DAC system [54].
Note that SS-DAC is slightly different from the recently developed technology of light-sheet microscopy (LSM), also known as selective plane illumination microscopy (SPIM) [55, 56]. In a SS-DAC microscope, the illumination beam is focused with a moderate NA to generate a fairly localized focal line, whereas LSM typically utilizes a lower NA to generate a thicker light sheet with a long depth of focus (in order to image a larger 3D volume by scanning the light sheet in only one direction). With LS-DAC and SS-DAC microscopy, the goal is to generate a 2D image by scanning in one direction, rather than a 3D volume of data. However, with SS-DAC, some 3D information is captured (similar to LSM), but for the purposes of 3D deconvolution to improve one 2D image, rather than to obtain volumetric information. LSM generates more data (in 3D) but sacrifices contrast due to the use of a very low-NA illumination beam whereas LS-DAC and SS-DAC generate 2D images, but with higher contrast [57]. In general, DAC microscopy is more ideal for imaging highly scattering fresh tissues (including in vivo) whereas LSM is ideal for rapid 3D microscopy of relatively transparent model organisms and optically cleared ex vivo tissues.
Modulated-alignment dual-axis (MAD) confocal microscopy
MAD confocal microscopy is a technology that combines the inherent strengths of focal-modulation microscopy [58] and PS-DAC microscopy, with the aim of improving image contrast and depth [52]. In a PS-DAC microscope, illumination and collection beams are spatially separated except at one single point (at the focus of the microscope). Optical sectioning with DAC microscopy relies on the precise alignment of the illumination-beam’s focus and detection-beam’s focus at the sub-micron level. For example, it has been shown that the confocal signal is reduced by an order of magnitude when the two beams are offset by only 1.4 times the beam radius (i.e. a distance on the order of a micrometer). This feature provides the opportunity to implement a “spatial overlap modulation” technique that was originally used in the context of nonlinear microscopy [59].
The first MAD confocal microscope utilized an acousto-optic deflector (AOD) in the illumination beam path to sinusoidally scan the illumination beam over a small range (+/− a few micrometers from the well-aligned condition) in the direction perpendicular to the plane defined by the dual-axis beams. This spatial modulation was performed at a frequency f resulting in a modulated signal at a frequency of 2f, which could be detected and distinguished from the static (non-modulated) background signal using 2flock-in detection. This strategy was shown to improve the image contrast (SBR) by ~6 dB in scattering media in comparison to standard PS-DAC microscopy [52].
There are a few limitations to the MAD confocal microscopy approach. First, the first-order diffracted light used as a spatially modulated illumination source in this system can vary in intensity over time due to the fact that the AOD diffraction efficiency typically varies with scanning angle, which can lead to a modulated background signal that competes with the MAD signal. Second, the acoustic wave within an AOD crystal has a limited propagation speed (~3.63 mm/μs in the early prototypes), which limits the modulation rate and thus the maximum frame rate for MAD imaging. Finally, the MAD technique may be limited in tissues with refractive heterogeneities, which can introduce aberrations and misalignments of the beams that will reduce the modulation depth of the MAD signals.
Bessel dual-axis confocal (DAC) microscopy
Bessel beams have been investigated as a means of improving deep-tissue microscopy in highly scattering and heterogeneous media [51, 60–63]. As discussed in the previous sections, DAC microscopy requires the precise intersection of two beams at their respective foci (micron scale). Therefore, the pointing accuracy of the beams and the quality of their foci are critical for optimal performance. A few recent studies have explored the adaptation of Bessel illumination for DAC microscopy and have shown that Bessel beams exhibit improved pointing accuracy and beam quality in samples with refractive heterogeneities, in comparison to conventional Gaussian beam [51, 63]. Consequently, spatial resolution is maintained more effectively with Bessel-DAC microscopy compared with standard Gaussian-DAC microscopy. One drawback of Bessel illumination is that the diffraction side lobes contain a significant amount of the beam energy, and contribute to an out-of-focus background that reduces image contrast [51]. Various approaches have been proposed to mitigate this effect for other imaging modalities, such as through the use of two-photon excitation and structured illumination, etc. [60, 62, 64, 65] Similar strategies are still under investigation for DAC microscopy systems.
Divided-pupil systems
As an alternative to using two separated objectives as discussed in the previous sections, off-axis illumination and collection has also been achieved using a single high-NA lens with a “divided pupil”, i.e. using one half of the lens for illumination and the other half for collection [66–70]. It should also be noted that a few light-sheet microscopy variants - for example, oblique plane microscopy [71] and swept confocally aligned planar excitation (SCAPE) microscopy [72] - have also utilized a similar configuration (off-axis illumination and collection beam paths that share one large objective lens) to achieve high-speed volumetric imaging.
IV. PORTABLE DAC SYSTEMS
Perhaps the greatest constraint for the design of in vivo microscopes is size. Although the DAC architecture has many unique properties that significantly simplify its miniaturization, smaller form factors are typically associated with reduced performance, as well as increased design complexity and cost. Each clinical device is designed for a specific biomedical application and careful deliberation is necessary to arrive at the most optimal design trade-offs. This section surveys some of the miniature DAC systems that have been built to address clinical applications such as gastrointestinal endoscopy, dermatopathology, neurosurgery, and the detection of head and neck cancers.
A. In vivo endoscopic microscopy of hollow organs
There is a clinical need for improved early detection and image-guided therapy of diseases in hollow organs such as the gastrointestinal (GI) tract. A critical requirement for such applications is that the device be small enough to fit within the instrument channel of a standard GI endoscope, which has a diameter of several millimeters. An endoscope-compatible DAC microscope with a diameter of 5.5-mm was developed at Stanford University for in vivo GI imaging at a frame rate of 5 Hz [73]. The device was deployed through the instrument channel of an endoscope, and was used to image the colonic mucosa of patients after topical application of an FDA-approved contrast agent (sodium fluorescein) [73]. Facilitated by the advancement of MEMS technology, a number of variants of this DAC endo-microscope have been developed with improved performance for a broad range of clinical applications [3, 50, 74, 80–81]. For example, a recent publication from the University of Michigan describes a similar DAC endomicroscope that incorporates a state-of-the-art 3D MEMS scanner that can alternate between a “tilting mode” (with actuation along the x and y axes) and a “pistoning mode” (with actuation along the y and z axes) to image either en face optical sections or vertical optical sections in real time (Fig. 5) [74, 79]. This specific topic of DAC endomicroscopy has been discussed in greater detail in a separate review [75].
Fig. 5.
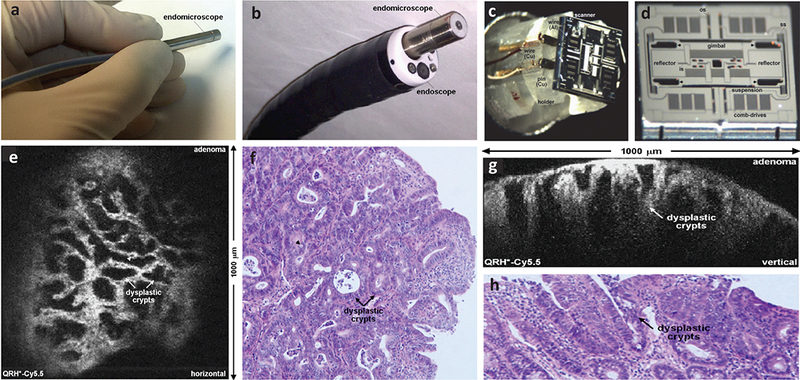
Photographs of a miniature DAC endo-microscope (a) that is fitted within a clinical GI endoscope (b). The head of the imaging probe has a diameter of 5.5 μm. (c-d) Photograph and scanning electron micrograph (SEM) of a custom-developed tri-axial MEMS scanner that enables the user to switch between two orthogonal imaging planes (either the en face plane or the vertical plane) in real time. (e) An example of en face optical sectioning of mouse colon after intravenous injection of a Cy5.5-labeled peptide, showing the dysplastic crypts (arrows) and goblet cells (arrowheads). (f) A corresponding H&E-stained histology section is shown of the mouse colon. (g) An example of vertical optical sectioning of the same mouse colon, showing EGFR expression from a region of adenoma up to 430 μm below the surface. (h) A corresponding H&E-stained histology section is shown of the mouse colon. Ref: [74]
B. Intraoperative neurosurgical guidance
Surgical resection (i.e. debulking) is the first step in the treatment of many brain tumors such as gliomas, in which a greater extent of resection has been associated with improved outcomes. However, the complete resection of gliomas is challenging because the tumor at the margins is often indistinguishable from the surrounding normal brain, and there is no quantitative metric (e.g. tumor-cell density) by which to optimize the extent of resection for these diffuse tumors that infiltrate far beyond the radiologically defined margins. While only a small set of untargeted fluorescence contrast agents (e.g. FITC, ICG, etc.) are approved for clinical use, a new compound, 5-ALA, has recently been approved by the U.S. FDA for neurosurgical guidance, in which a metabolic byproduct of the orally ingested agent is used to highlight the bulk tumor regions and to improve the extent of resection. However, image contrast is often still ambiguous and weak near the diffuse margins of the tumor. It has been suggested that intraoperative microscopy, which can provide images that approach the gold-standard of histopathology, may have value for neurosurgical oncologists to maximize the extent of resection while minimizing neurological damage. In particular, portable optical-sectioning microscopes provide sufficient resolution to detect and potentially quantify the sparse and disseminated tumor-cell populations at the margins of diffuse gliomas. Such sparse cell populations are often not visualized by other imaging technologies (e.g. low-power surgical microscopes, MRI, CT) since they lack the spatial resolution to detect individual disseminated cells, even if such isolated cells are effectively labeled by a contrast agent.
A pen-sized handheld DAC microscope with a distal diameter of 1.8 mm was described in 2010, in which images were obtained from the brains of living mice that were genetically engineered to develop medulloblastoma [12]. The device was able to achieve cellular resolution (4 μm laterally and 8 μm axially) and an imaging depth of up to 250 μm with the use of low-NA (∼0.075) beams focused with a parabolic mirror. A biaxial MEMS scanning mirror was used for postobjective scanning of the focal volume over a FOV of approximately 0.4 mm by 0.4 mm. The MEMS mirror was axially translated with a piezoelectric actuator to adjust the imaging depth by up to 250 μm. In addition, a customized gradient-index (GRIN) “needle lens” located at the distal tip was used, with a diameter of just 1.8 mm. A major limitation of this prototype was its slow frame rate (~4 Hz) due to the point-by-point Lissajous scanning pattern that was used, which made the device vulnerable to motion artifacts. In addition, the spatial resolution was not ideal.
A handheld LS-DAC microscope was developed (Fig. 6) recently with significantly improved frame rates and spatial resolution [27]. As discussed in Sec. III B, the LS-DAC architecture significantly simplifies the requirements of the scanning mechanism for high-speed imaging. A robust commercial MEMS mirror was used to scan the focal line in one dimension to create en face images at video rate (>16 fps). The improved resolution was achieved by using a custom-developed 1:3 de-magnifying relay objective at the distal end of the device. The relay lens effectively increased the NA of the beams as well as the crossing angles, at the cost of reducing the FOV. The system was able to achieve a lateral resolution of 1.1 μm with 2.0-μm axial resolution (optical sectioning thickness) over a FOV of roughly 350 μm by 350 μm. This high-speed LS-DAC microscope enabled depth-resolved imaging of red blood cells trafficking within the capillaries of a living mouse, as well as high-contrast imaging of ex vivo tissues stained with FDA-approved fluorophores (e.g. methylene blue). A trade-off of using line scanning in miniature devices is that confocal detection must be achieved with a digital line detector that is directly integrated within the device, instead of using a fiber-coupled point detector (in the case of a miniature PS-DAC microscope).
Fig. 6.
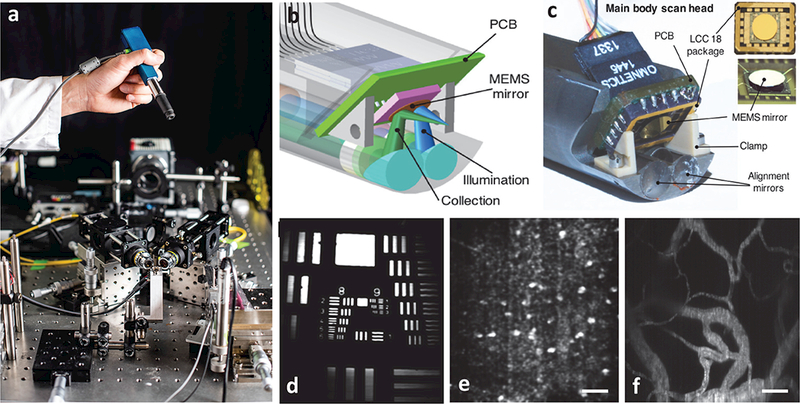
A handheld video-rate LS-DAC microscope for intraoperative guidance. (a) A miniature LS-DAC is held above a large tabletop LS-DAC prototype [photograph courtesy of Dennis Wise at the University of Washington]. (b-c) A design rendering and photograph, respectively, of the scan head of the handheld LS-DAC device. A single MEMS mirror is used to scan both beams in one dimension to rapidly generate 2D images. (d) An image of a 1951 USAF resolution target, showing the ability to resolve objects as small as ~1 μm. (e) A label-free in vivo image of hyper-reflective nuclei in human oral buccal mucosa. (f) A fluorescence in vivo image (maximum intensity projection from a depth range of 50 to 100 μm) of the vasculature of a mouse ear after retro-orbital injection of FITC-dextran. All scale bars represent 50 μm.
C. Label-free in vivo reflectance microscopy of skin and the oral cavity
Imaging the nuclear morphology and tissue architecture down to the dermal-epidermal junction (~100 μm below the skin surface) is valuable for the diagnosis and treatment of basal cell carcinoma, one of the most common cancers of the skin and oral cavity. As an alternative to invasive biopsy and histopathology, label-free reflectance confocal microscopy can provide a powerful method to allow suspicious lesions to be non-invasively and rapidly examined in real time. These clinical applications require the imaging device to be able to achieve (1) a resolution of 5 μm or less to distinguish nuclear morphology, (2) an imaging depth of at least 100 μm to reach the deeper layers of interest (dermal-epidermal junction), (3) a small imaging head that can fit within the oral cavity, and (4) a high frame rate to reduce motion artifacts during handheld use. A miniature reflectance LS-DAC was developed recently to address these clinical needs and design criteria. This device is similar to the fluorescence version that was discussed in the previous section, but is more compact owing to the use of a small and inexpensive (< $100) linear detector array integrated within the device. Although not as sensitive as the detector used in the fluorescence LS-DAC device, the low-cost linear array in the reflectance LS-DAC device has sufficient sensitivity for reflectance imaging, which generates much more signal than fluorescence imaging.
The portable divided-pupil line-scanned confocal microscope developed at Memorial Sloan Kettering Cancer Center is an additional example of a clinical device for label-free in vivo reflectance imaging of human skin (Fig. 7) [76]. In a divided-pupil system, the pupil of a high-NA (0.9, water-immersion) objective is divided into two halves, one for the illumination beam and the other half for the collection beam. An endoscopic relay lens was incorporated at the distal end so that the oral cavity could be accessed. With such reflectance-based devices, sub-cellular optical sectioning has been achieved at a depth of ~ 100 μm in human skin, while achieving a frame rate of 8 Hz over a FOV that is comparable to that of a standard 20X objective lens. Note that reflectance confocal microscopy has also been used as a complementary imaging modality with another common reflectance-mode optical imaging technique, optical coherent tomography (OCT), in which reflectance confocal microscopy provides high resolution images at a limited imaging depth while OCT provides lower resolution images at a greater imaging depth [82].
Fig. 7.
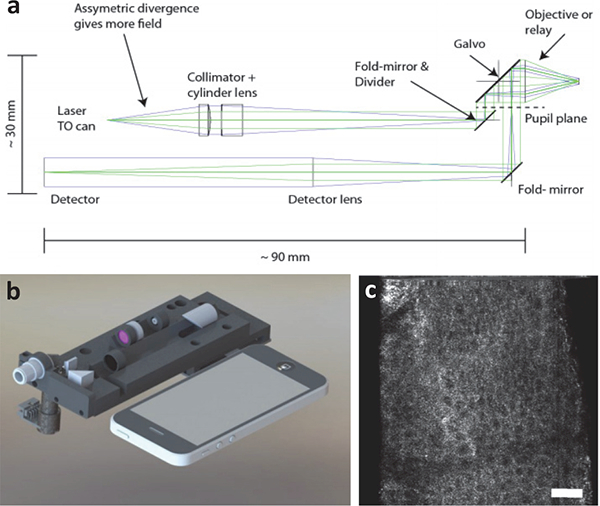
(a) Optical circuit of a compact line-scanned divided pupil confocal system. The pupil of the objective lens is physically divided into two halves, one to generate the illumination beam and the other for the collection path. A single galvanometric mirror is used to scan both beams to create an image. (b) A design rendering of the system is shown alongside a smartphone. (c) A label-free in vivo image is shown of human epidermis. Ref: [76]
V. Summary
This article provides a review of DAC microscope technology, an optical imaging modality that utilizes low-NA beams to achieve effective optical sectioning and superior image contrast in biological tissues. In contrast to the conventional confocal microscope invented by Minsky in 1957 [29], a DAC microscope utilizes spatially separated off-axis illumination and collection beam paths that only intersect at their foci - an optical architecture inspired by the initial works of Stelzer et al. [30, 32], and Webb et al. [34] in the 1990s - to improve image depth and to reduce the complexity of miniaturization. Since its first introduction in 2003 [35], DAC microscopy has been significantly improved in terms of imaging depth (>500 μm), speed (>30 fps), resolution (<1 μm laterally; <3 μm axially), and size (e.g. endoscope-compatible). Several portable DAC systems have been developed for a wide range of clinical applications such as for intraoperative guidance and for early disease detection, with some systems currently in the process of clinical testing.
In addition to confocal microscopy (SAC and DAC), there are many other portable optical sectioning microscopy systems such as multiphoton microscopy [19–22], OCT [23, 77], structured illumination microscopy [24–26], etc. These technologies are also being investigated as non-invasive and real-time alternatives to conventional biopsy and histopathology, and have been shown to be useful for certain clinical applications. Some of these systems have been discussed in a prior review [78]. Finally, it should be noted that the research and development of these miniature clinical devices has both facilitated and benefited from the advancement of miniature optical components such as MEMS scanners, ultra-small lenses, fiber-optics technologies, as well as compact detectors and other hardware for high-speed image acquisition and processing.
Acknowledgments
The authors acknowledge funding support from the NIH, including grants from the NIDCR (R01DE023497), the NCI (R01CA175391 ), and the NIBIB (K99/R00EB015016).
Biography
 Linpeng “Peter” Wei received a B.E. degree in biomedical engineering from Stony Brook University, Stony Brook, NY, USA, in 2015. After graduation, he joined Dr. Jonathan Liu’s research group at the University of Washington to work on the development of a handheld dual-axis confocal microscopy for fluorescence-guided neurosurgery. He is currently pursuing a Ph.D. degree in mechanical engineering and his research interests include biomedical optics and fluorescence imaging.
Linpeng “Peter” Wei received a B.E. degree in biomedical engineering from Stony Brook University, Stony Brook, NY, USA, in 2015. After graduation, he joined Dr. Jonathan Liu’s research group at the University of Washington to work on the development of a handheld dual-axis confocal microscopy for fluorescence-guided neurosurgery. He is currently pursuing a Ph.D. degree in mechanical engineering and his research interests include biomedical optics and fluorescence imaging.
 Chengbo Yin received a B.S. degree in aircraft manufacturing engineering from Beihang University, Beijing, China in 2012, and a M.S. degree in mechanical engineering from Northeastern University, Boston, MA, USA, in 2014. He is currently pursuing a Ph.D. degree in mechanical engineering at the University of Washington, Seattle, WA. His research aims to develop a miniature dual-axis confocal microscope for the early detection of oral cancers.
Chengbo Yin received a B.S. degree in aircraft manufacturing engineering from Beihang University, Beijing, China in 2012, and a M.S. degree in mechanical engineering from Northeastern University, Boston, MA, USA, in 2014. He is currently pursuing a Ph.D. degree in mechanical engineering at the University of Washington, Seattle, WA. His research aims to develop a miniature dual-axis confocal microscope for the early detection of oral cancers.
 Jonathan T.C. Liu received a B.S.E. degree in mechanical engineering from Princeton University in 1999, and an M.S. and Ph.D. degree from Stanford University in 2000 and 2005, respectively. He was a Postdoctoral Fellow in the Department of Electrical Engineering (Ginzton Labs) and the Molecular Imaging Program at Stanford (2005–2009), and was later appointed as an Instructor within the Stanford University School of Medicine (2009–2010). From 2010 to 2014, Jonathan was an Assistant Professor in the Biomedical Engineering Department at SUNY Stony Brook. He is currently an associate professor in the Mechanical Engineering Department, with an adjunct appointment in Pathology, at the University of Washington in Seattle, WA. His laboratory for molecular biophotonics develops optical strategies for improving the diagnosis and treatment of diseases.
Jonathan T.C. Liu received a B.S.E. degree in mechanical engineering from Princeton University in 1999, and an M.S. and Ph.D. degree from Stanford University in 2000 and 2005, respectively. He was a Postdoctoral Fellow in the Department of Electrical Engineering (Ginzton Labs) and the Molecular Imaging Program at Stanford (2005–2009), and was later appointed as an Instructor within the Stanford University School of Medicine (2009–2010). From 2010 to 2014, Jonathan was an Assistant Professor in the Biomedical Engineering Department at SUNY Stony Brook. He is currently an associate professor in the Mechanical Engineering Department, with an adjunct appointment in Pathology, at the University of Washington in Seattle, WA. His laboratory for molecular biophotonics develops optical strategies for improving the diagnosis and treatment of diseases.
References
- [1].Sung KB, Liang C, Descour M, Collier T, Follen M, Malpica A, et al. , “Near real time in vivo fibre optic confocal microscopy: sub-cellular structure resolved,” J Microsc, vol. 207, pp. 137–45, August 2002. [DOI] [PubMed] [Google Scholar]
- [2].Shin HJ, Pierce MC, Lee D, Ra H, Solgaard O, and Richards-Kortum R, “Fiber-optic confocal microscope using a MEMS scanner and miniature objective lens,” Optics Express, vol. 15, pp. 9113–9122, July 23 2007. [DOI] [PubMed] [Google Scholar]
- [3].Liu L, Wang EK, Zhang XY, Liang WX, Li XD, and Xie HK, “MEMS-based 3D confocal scanning microendoscope using MEMS scanners for both lateral and axial scan,” Sensors and Actuators a-Physical, vol. 215, pp. 89–95, August 15 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Carlson K, Chidley M, Sung KB, Descour M, Gillenwater A, Follen M, et al. , “In vivo fiber-optic confocal reflectance microscope with an injection-molded plastic miniature objective lens,” Applied Optics, vol. 44, pp. 1792–1797, April 1 2005. [DOI] [PubMed] [Google Scholar]
- [5].Olsovsky C, Hinsdale T, Cuenca R, Cheng YSL, Wright JM, Rees TD, et al. , “Handheld tunable focus confocal microscope utilizing a double-clad fiber coupler for in vivo imaging of oral epithelium,” Journal of Biomedical Optics, vol. 22, May 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Richards-Kortum R, Smithpeter CL, Bowman BS, and Descour MR, “Fiber-optic confocal imaging apparatus and methods of use,” ed: Google Patents, 2002. [Google Scholar]
- [7].BERIER F, BOURRIAUX S, Genet M, Viellerobe B, Loiseau A, and Abrat B, “Miniaturized focusing optical head in particular for endoscope,” ed: Google Patents, 2003. [Google Scholar]
- [8].Kanai M, “Condensing optical system, confocal optical system, and scanning confocal e ndoscope,” ed: Google Patents, 2005. [Google Scholar]
- [9].Kumar K, Avritscher R, Wang Y, Lane N, Madoff DC, Yu TK, et al. , “Handheld histology-equivalent sectioning laser-scanning confocal optical microscope for interventional imaging,” Biomed Microdevices, vol. 12, pp. 223–33, April 2010. [DOI] [PubMed] [Google Scholar]
- [10].Tanbakuchi AA, Rouse AR, Udovich JA, Hatch KD, and Gmitro AF, “Clinical confocal microlaparoscope for real-time in vivo optical biopsies,” J Biomed Opt, vol. 14, p. 044030, Jul-Aug 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang TD, Friedland S, Sahbaie P, Soetikno R, Hsiung PL, Liu JT, et al. , “Functional imaging of colonic mucosa with a fibered confocal microscope for real-time in vivo pathology,” Clin Gastroenterol Hepatol, vol. 5, pp. 1300–5, November 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu JT, Mandella MJ, Loewke NO, Haeberle H, Ra H, Piyawattanametha W, et al. , “Micromirror-scanned dual-axis confocal microscope utilizing a gradient-index relay lens for image guidance during brain surgery,” J Biomed Opt, vol. 15, p. 026029, Mar-Apr 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Seibel EJ, Brown CM, Dominitz JA, and Kimmey MB, “Scanning single fiber endoscopy: a new platform technology for integrated laser imaging, diagnosis, and future therapies,” Gastrointest Endosc Clin N Am, vol. 18, pp. 467–78, viii, July 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee CM, Engelbrecht CJ, Soper TD, Helmchen F, and Seibel EJ, “Scanning fiber endoscopy with highly flexible, 1 mm catheterscopes for wide-field, full-color imaging,” J Biophotonics, vol. 3, pp. 385–407, June 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kiesslich R, Burg J, Vieth M, Gnaendiger J, Enders M, Delaney P, et al. , “Confocal laser endoscopy for diagnosing intraepithelial neoplasias and colorectal cancer in vivo,” Gastroenterology, vol. 127, pp. 706–13, September 2004. [DOI] [PubMed] [Google Scholar]
- [16].Polglase AL, McLaren WJ, Skinner SA, Kiesslich R, Neurath MF, and Delaney PM, “A fluorescence confocal endomicroscope for in vivo microscopy of the upper- and the lower-GI tract,” Gastrointest Endosc, vol. 62, pp. 686–95, November 2005. [DOI] [PubMed] [Google Scholar]
- [17].Kumar K, Hoshino K, and Zhang X, “Handheld subcellular-resolution single-fiber confocal microscope using high-reflectivity two-axis vertical combdrive silicon microscanner,” Biomed Microdevices, vol. 10, pp. 653–60, October 2008. [DOI] [PubMed] [Google Scholar]
- [18].Tanbakuchi AA, Udovich JA, Rouse AR, Hatch KD, and Gmitro AF, “In vivo imaging of ovarian tissue using a novel confocal microlaparoscope,” Am J Obstet Gynecol, vol. 202, pp. 90 e1–9, January 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hoy CL, Durr NJ, Chen P, Piyawattanametha W, Ra H, Solgaard O, et al. , “Miniaturized probe for femtosecond laser microsurgery and two-photon imaging,” Opt Express, vol. 16, pp. 9996–10005, June 23 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Myaing MT, MacDonald DJ, and Li X, “Fiber-optic scanning two-photon fluorescence endoscope,” Opt Lett, vol. 31, pp. 1076–8, April 15 2006. [DOI] [PubMed] [Google Scholar]
- [21].Murari K, Zhang Y, Li S, Chen Y, Li MJ, and Li X, “Compensation-free, all-fiber-optic, two-photon endomicroscopy at 1.55 mum,” Opt Lett, vol. 36, pp. 1299–301, April 1 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Helmchen F, Fee MS, Tank DW, and Denk W, “A miniature head-mounted two-photon microscope. high-resolution brain imaging in freely moving animals,” Neuron, vol. 31, pp. 903–12, September 27 2001. [DOI] [PubMed] [Google Scholar]
- [23].Li X, Chudoba C, Ko T, Pitris C, and Fujimoto JG, “Imaging needle for optical coherence tomography,” Opt Lett, vol. 25, pp. 1520–2, October 15 2000. [DOI] [PubMed] [Google Scholar]
- [24].Bozinovic N, Ventalon C, Ford T, and Mertz J, “Fluorescence endomicroscopy with structured illumination,” Opt Express, vol. 16, pp. 8016–25, May 26 2008. [DOI] [PubMed] [Google Scholar]
- [25].Ford TN, Lim D, and Mertz J, “Fast optically sectioned fluorescence HiLo endomicroscopy,” J Biomed Opt, vol. 17, p. 021105, February 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kyrish M, Dobbs J, Jain S, Wang X, Yu D, Richards-Kortum R, et al. , “Needle-based fluorescence endomicroscopy via structured illumination with a plastic, achromatic objective,” J Biomed Opt, vol. 18, p. 096003, September 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yin C, Glaser AK, Leigh SY, Chen Y, Wei L, Pillai PC, et al. , “Miniature in vivo MEMS-based line-scanned dual-axis confocal microscope for point-of-care pathology,” Biomed Opt Express, vol. 7, pp. 251–63, February 1 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu JT, Mandella MJ, Ra H, Wong LK, Solgaard O, Kino GS, et al. , “Miniature near-infrared dual-axes confocal microscope utilizing a two-dimensional microelectromechanical systems scanner,” Opt Lett, vol. 32, pp. 256–8, February 1 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Minsky M, “Microscopy apparatus,” ed: Google Patents, 1961. [Google Scholar]
- [30].Stelzer EHK and Lindek S, “Fundamental Reduction of the Observation Volume in Far-Field Light-Microscopy by Detection Orthogonal to the Illumination Axis - Confocal Theta Microscopy,” Optics Communications, vol. 111, pp. 536–547, October 15 1994. [Google Scholar]
- [31].Cogswell CJ, Lindek S, Stelzer EH, and Carlsson K, “Confocal theta microscopy and 4Pi-confocal theta microscopy,” vol. 2184, pp. 188–194, 1994. [Google Scholar]
- [32].Lindek S and Stelzer EH, “Resolution improvement by nonconfocal theta microscopy,” Opt Lett, vol. 24, pp. 1505–7, November 1 1999. [DOI] [PubMed] [Google Scholar]
- [33].Wang TD, Mandella MJ, Contag CH, and Kino GS, “Dual-axis confocal microscope for high-resolution in vivo imaging,” Opt Lett, vol. 28, pp. 414–6, March 15 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Webb RH and Rogomentich F, “Confocal microscope with large field and working distance,” Appl Opt, vol. 38, pp. 4870–5, August 1 1999. [DOI] [PubMed] [Google Scholar]
- [35].Wang TD, Contag CH, Mandella MJ, Chan NY, and Kino GS, “Dual-axes confocal microscopy with post-objective scanning and low-coherence heterodyne detection,” Opt Lett, vol. 28, pp. 1915–7, October 15 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wong LK, Mandella MJ, Kino GS, and Wang TD, “Improved rejection of multiply scattered photons in confocal microscopy using dual-axes architecture,” Opt Lett, vol. 32, pp. 1674–6, June 15 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu JT, Mandella MJ, Crawford JM, Contag CH, Wang TD, and Kino GS, “Efficient rejection of scattered light enables deep optical sectioning in turbid media with low-numerical-aperture optics in a dual-axis confocal architecture,” J Biomed Opt, vol. 13, p. 034020, May-Jun 2008. [DOI] [PubMed] [Google Scholar]
- [38].Liu JT, Mandella MJ, Friedland S, Soetikno R, Crawford JM, Contag CH, et al. , “Dual-axes confocal reflectance microscope for distinguishing colonic neoplasia,” J Biomed Opt, vol. 11, p. 054019, Sep-Oct 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Born EWM, Principles of Optics, 7th ed.,. Cambridge, UK: Cambridgge Press, 1999. [Google Scholar]
- [40].Pawley J, Handbook of Biological Confocal Microscopy: Springer, 2006. [Google Scholar]
- [41].Rajadhyaksha M, Anderson RR, and Webb RH, “Video-rate confocal scanning laser microscope for imaging human tissues in vivo,” Appl Opt, vol. 38, pp. 2105–15, April 1 1999. [DOI] [PubMed] [Google Scholar]
- [42].Corle TR, Chou CH, and Kino GS, “Depth response of confocal optical microscopes,” Opt Lett, vol. 11, pp. 770–2, December 1 1986. [DOI] [PubMed] [Google Scholar]
- [43].Sheppard CJ and Wilson T, “Depth of field in the scanning microscope,” Opt Lett, vol. 3, p. 115, September 1 1978. [DOI] [PubMed] [Google Scholar]
- [44].Robert HW, “Confocal optical microscopy,” Reports on Progress in Physics, vol. 59, p. 427, 1996. [Google Scholar]
- [45].Chen Y and Liu JT, “Optimizing the performance of dual-axis confocal microscopes via Monte-Carlo scattering simulations and diffraction theory,” J Biomed Opt, vol. 18, p. 066006, June 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chen Y, Wang D, and Liu JT, “Assessing the tissue-imaging performance of confocal microscope architectures via Monte Carlo simulations,” Opt Lett, vol. 37, pp. 4495–7, November 1 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang D, Chen Y, Wang Y, and Liu JT, “Comparison of line-scanned and point-scanned dual-axis confocal microscope performance,” Opt Lett, vol. 38, pp. 5280–3, December 15 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Meza D, Wang D, Wang Y, Borwege S, Sanai N, and Liu JT, “Comparing high-resolution microscopy techniques for potential intraoperative use in guiding low-grade glioma resections,” Lasers Surg Med, vol. 47, pp. 289–95, April 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wei L, Chen Y, Yin C, Borwege S, Sanai N, and Liu JTC, “Optical-sectioning microscopy of protoporphyrin IX fluorescence in human gliomas: standardization and quantitative comparison with histology,” J Biomed Opt, vol. 22, p. 46005, April 1 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ra H, Piyawattanametha W, Taguchi Y, Lee D, Mandella MJ, and Solgaard O, “Two-dimensional MEMS scanner for dual-axes confocal microscopy,” Journal of Microelectromechanical Systems, vol. 16, pp. 969–976, August 2007. [Google Scholar]
- [51].Chen Y, Glaser A, and Liu JT, “Bessel-beam illumination in dual-axis confocal microscopy mitigates resolution degradation caused by refractive heterogeneities,” J Biophotonics, vol. 10, pp. 68–74, January 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Leigh SY, Chen Y, and Liu JT, “Modulated-alignment dual-axis (MAD) confocal microscopy for deep optical sectioning in tissues,” Biomed Opt Express, vol. 5, pp. 1709–20, June 1 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chen Y, Wang D, Khan A, Wang Y, Borwege S, Sanai N, et al. , “Video-rate in vivo fluorescence imaging with a line-scanned dual-axis confocal microscope,” J Biomed Opt, vol. 20, p. 106011, October 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang D, Meza D, Wang Y, Gao L, and Liu JT, “Sheet-scanned dual-axis confocal microscopy using Richardson-Lucy deconvolution,” Opt Lett, vol. 39, pp. 5431–4, September 15 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huisken J, Swoger J, Del Bene F, Wittbrodt J, and Stelzer EH, “Optical sectioning deep inside live embryos by selective plane illumination microscopy,” Science, vol. 305, pp. 1007–9, August 13 2004. [DOI] [PubMed] [Google Scholar]
- [56].Glaser AK, Reder NP, Chen Y, McCarty EF, Yin C, Wei L, et al. , “Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens,” Nature Biomedical Engineering, vol. 1, p. 0084, 06/26/online 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Glaser AK, Wang Y, and Liu JT, “Assessing the imaging performance of light sheet microscopies in highly scattering tissues,” Biomed Opt Express, vol. 7, pp. 454–66, February 1 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen N, Wong CH, and Sheppard CJ, “Focal modulation microscopy,” Opt Express, vol. 16, pp. 18764–9, November 10 2008. [DOI] [PubMed] [Google Scholar]
- [59].Isobe K, Kawano H, Takeda T, Suda A, Kumagai A, Mizuno H, et al. , “Background-free deep imaging by spatial overlap modulation nonlinear optical microscopy,” Biomed Opt Express, vol. 3, pp. 1594–608, July 1 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, et al. , “Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination,” Nat Methods, vol. 8, pp. 417–23, May 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Fahrbach FO, Simon P, and Rohrbach A, “Microscopy with self-reconstructing beams,” Nature Photonics, vol. 4, pp. 780–785, November 2010. [Google Scholar]
- [62].Lorenser D, Christian Singe C, Curatolo A, and ampson DD, “Energy-efficient low-Fresnel-number Bessel beams and their application in optical coherence tomography,” Opt Lett, vol. 39, pp. 548–51, February 1 2014. [DOI] [PubMed] [Google Scholar]
- [63].Chen Y and Liu JT, “Characterizing the beam steering and distortion of Gaussian and Bessel beams focused in tissues with microscopic heterogeneities,” Biomed Opt Express, vol. 6, pp. 1318–30, April 1 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Fahrbach FO, Gurchenkov V, Alessandri K, Nassoy P, and Rohrbach A, “Light-sheet microscopy in thick media using scanned Bessel beams and two-photon fluorescence excitation,” Opt Express, vol. 21, pp. 13824–39, June 3 2013. [DOI] [PubMed] [Google Scholar]
- [65].Zhao M, Zhang H, Li Y, Ashok A, Liang R, Zhou W, et al. , “Cellular imaging of deep organ using two-photon Bessel light-sheet nonlinear structured illumination microscopy,” Biomed Opt Express, vol. 5, pp. 1296–308, May 1 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Gareau DS, Abeytunge S, and Rajadhyaksha M, “Line-scanning reflectance confocal microscopy of human skin: comparison of full-pupil and divided-pupil configurations,” Opt Lett, vol. 34, pp. 3235–7, October 15 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Koester CJ, “Scanning mirror microscope with optical sectioning characteristics: applications in ophthalmology,” Appl Opt, vol. 19, pp. 1749–57, June 1 1980. [DOI] [PubMed] [Google Scholar]
- [68].Patel YG, Rajadhyaksha M, and Dimarzio CA, “Optimization of pupil design for point-scanning and line-scanning confocal microscopy,” Biomed Opt Express, vol. 2, pp. 2231–42, August 1 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Dwyer PJ, DiMarzio CA, Zavislan JM, Fox WJ, and Rajadhyaksha M, “Confocal reflectance theta line scanning microscope for imaging human skin in vivo,” Opt Lett, vol. 31, pp. 942–4, April 1 2006. [DOI] [PubMed] [Google Scholar]
- [70].Dwyer PJ, DiMarzio CA, and Rajadhyaksha M, “Confocal theta line-scanning microscope for imaging human tissues,” Appl Opt, vol. 46, pp. 1843–51, April 1 2007. [DOI] [PubMed] [Google Scholar]
- [71].Kumar S, Wilding D, Sikkel MB, Lyon AR, MacLeod KT, and Dunsby C, “High-speed 2D and 3D fluorescence microscopy of cardiac myocytes,” Optics Express, vol. 19, pp. 13839–13847, July 18 2011. [DOI] [PubMed] [Google Scholar]
- [72].Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, Mann RS, et al. , “Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms,” Nature Photonics, vol. 9, pp. 113–119, February 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Piyawattanametha W, Ra H, Qiu Z, Friedland S, Liu JT, Loewke K, et al. , “In vivo near-infrared dual-axis confocal microendoscopy in the human lower gastrointestinal tract,” J Biomed Opt, vol. 17, p. 021102, February 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Li G, Li H, Duan X, Zhou Q, Zhou J, Oldham KR, et al. , “Visualizing Epithelial Expression in Vertical and Horizontal Planes With Dual Axes Confocal Endomicroscope Using Compact Distal Scanner,” IEEE Trans Med Imaging, vol. 36, pp. 1482–1490, July 2017. [DOI] [PubMed] [Google Scholar]
- [75].Piyawattanametha W and Wang TD, “MEMS-Based Dual Axes Confocal Microendoscopy,” IEEE J Sel Top Quantum Electron, vol. 16, pp. 804–814, July 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Glazowski C, Kollias N, Peterson G, Choi B, Zeng H, Rajadhyaksha M, et al. , “Compact divided-pupil line-scanning confocal microscope for investigation of human tissues,” vol. 8565, p. 856523, 2013. [Google Scholar]
- [77].Liu X, Cobb MJ, Chen Y, Kimmey MB, and Li X, “Rapid-scanning forward-imaging miniature endoscope for real-time optical coherence tomography,” Opt Lett, vol. 29, pp. 1763–5, August 1 2004. [DOI] [PubMed] [Google Scholar]
- [78].Liu JT, Loewke NO, Mandella MJ, Levenson RM, Crawford JM, and Contag CH, “Point-of-care pathology with miniature microscopes,” Anal Cell Pathol (Amst), vol. 34, pp. 81–98, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Li HJ, Duan XY, Qiu Z, Zhou Q, Kurabayashi K, Oldham KR, et al. , “Integrated monolithic 3D MEMS scanner for switchable real time vertical/horizontal cross-sectional imaging,” Optics Express, vol. 24, pp. 2145–2155, February 8 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Qiu Z, Liu ZY, Duan XY, Khondee S, Joshi B, Mandella MJ, et al. , “Targeted vertical cross-sectional imaging with handheld near-infrared dual axes confocal fluorescence endomicroscope,” Biomedical Optics Express, vol. 4, pp. 322–330, February 1 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Gao Z, Li G, Li X, Zhou J, Duan X, Chen J, et al. , “In vivo near-infrared imaging of ErbB2 expressing breast tumors with dual-axes confocal endomicroscopy using a targeted peptide,” Sci Rep, vol. 7, p. 14404, October 31 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Iftimia N, Ferguson RD, Mujat M, Patel AH, Zhang EZ, Fox W, et al. , “Combined reflectance confocal microscopy/optical coherence tomography imaging for skin burn assessment,” Biomed Opt Express, vol. 4, pp. 680–95, May 1 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]


