Abstract
The functional cofactors derived from vitamin B3 are nicotinamide adenine dinucleotide (NAD+), its phosphorylated form, nicotinamide adenine dinucleotide phosphate (NADP+) and their reduced forms (NAD(P)H). These cofactors, together referred as the NAD(P)(H) pool, are intimately implicated in all essential bioenergetics, anabolic and catabolic pathways in all forms of life. This pool also contributes to post-translational protein modifications and second messenger generation. Since NAD+ seats at the cross-road between cell metabolism and cell signaling, manipulation of NAD+ bioavailability through vitamin B3 supplementation has become a valuable nutritional and therapeutic avenue. Yet, much remains unexplored regarding vitamin B3 metabolism. The present review highlights the chemical diversity of the vitamin B3-derived anabo-lites and catabolites of NAD+ and offers a chemical perspective on the approaches adopted to identify, modulate and measure the contribution of various precursors to the NAD(P)(H) pool.
Introduction
Niacin and niacinamide, also known as nicotinic acid (NA) and nicotinamide (Nam), are the better known forms of vitamin B3 [1,2]. Along with tryptophan (trp), they are biosynthetic precursors to nicotinamide adenine dinucleotide (NAD+), nicotinamide adenine dinucleotide phosphate (NADP+) and their respective reduced forms (NAD(P)H), altogether referred as the NAD(P)(H) pool. The vitamin B3 metabolome includes the biosynthetic precursors of NAD+ (anabolites; Table 1a), the cofactors derived from NAD+ (i.e. the NAD(P)(H) pool; Table 1a) and the derivatives generated through catabolic processes (catabolites; Table 1b) [3–8]. Altogether, the NAD+-derived cofactors are central to cellular homeostasis and growth through their roles in intermediary metabolism, mitochondrial respiration, the Krebs’ cycle, ATP production, reactive oxygen species generation and inhibition, and additional roles in post-translational protein modifications, protein regulation and second messengers’ generation [9–16]. Sub-optimal intracellular levels of these cofactors yield to cellular dysfunction, while acute vitamin B3 deficiency leads to pellagra [17,18], a debilitating and deadly disease still endemic in some regions of the world where malnutrition is common place. In more affluent countries, clinical vitamin B3 deficiency is due to poor food choices, adverse drug reactions, alcoholism and infectious or autoimmune diseases [19–22]. There are several additional excellent publications covering in detail the biological and physiological roles of the NAD(P)(H) pool and that of its biosynthetic precursors [23–30]. The present review covers the breadth of the vitamin B3 metabolome and presents an overview of the tools used to modulate the NAD(P)(H) pool and therefore the vitamin B3 metabolome in biological systems with a focus on mammalian systems. First, the known vitamin B3 metabolites (anabolites and catabolites) and the biosynthetic pathways to NAD(P)(H) will be summarized. A brief foray in the chemical and chemoenzymatic routes to NAD+ precursors will then follow along with an overview of isotopically labeled metabolic NAD+ intermediates, which have been used to report on the vitamin B3 metabolomic profiles..
Table 1a.
Chemical structures and abbreviations of the anabolites constituting the vitamin B3 metabolome
| Anabolites of the vitamin B3 metabolome, precursor to the NAD(P)(H) pool |
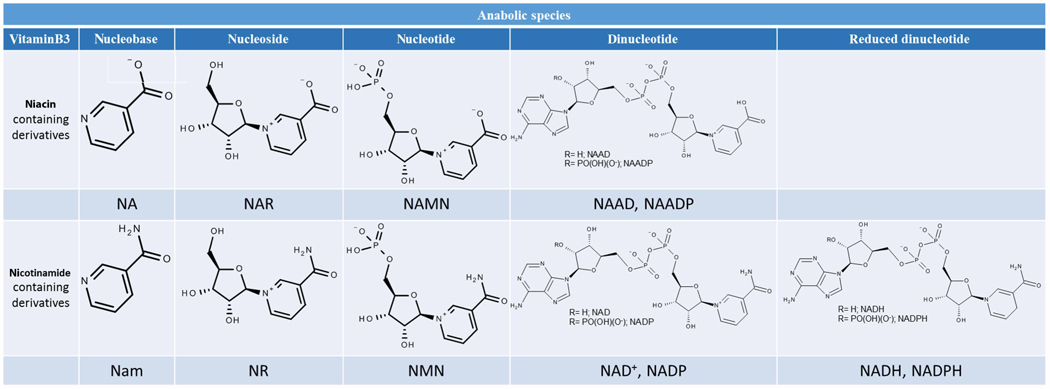 |
Abbreviations: NA, niacin/nicotinic acid; Nam, niacinamide/nicotinamide; NR, nicotinamide riboside; NAR, nicotinic acid riboside; NAMN, nicotinic acid mononucleotide; NMN, nicotinamide mononucleotide; NAAD, nicotinic acid adenine dinucleotide; NAADP*, nicotinic acid adenine dinucleotide phosphate; NAD+, nicotinamide adenine dinucleotide; NADP+, nicotinamide adenine dinucleotide phosphate; NADH, nicotinamide adenine dinucleotide reduced form; NADPH, nicotinamide adenine dinucleotide phosphate reduced form. *Generated via a yet unknown mechanism.
Table 1b.
Chemical structures and abbreviations of the catabolites constituting the vitamin B3 metabolome
| Catabolites of the vitamin B3 metabolome |
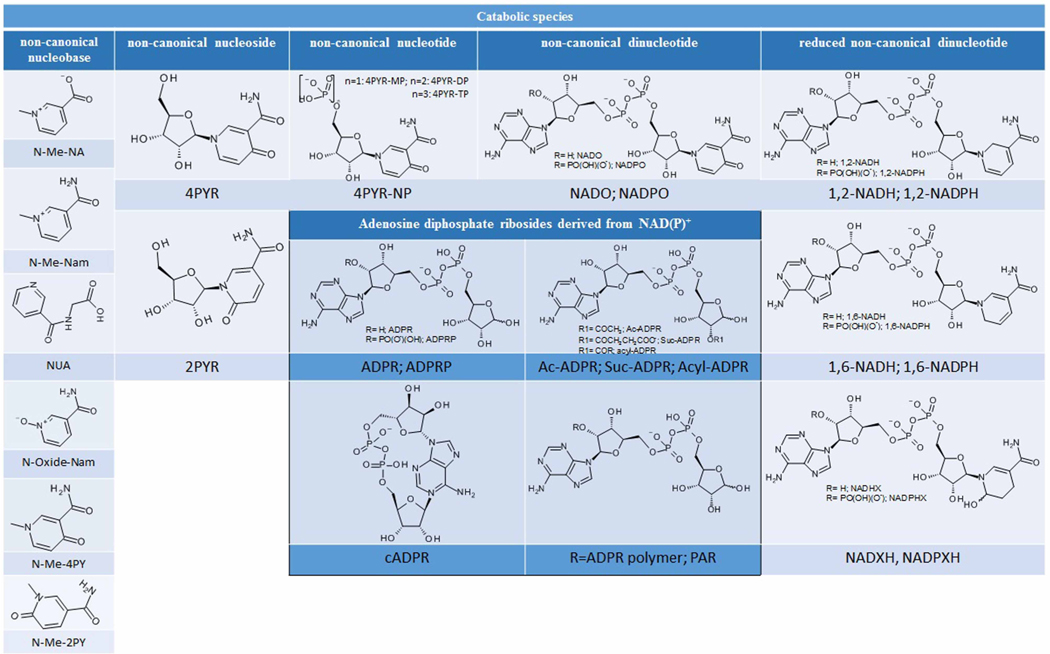 |
Abbreviations: N-Me-Nam, N-methyl nicotinamide or trigonellinamide; N-methyl-NA, N-methyl nicotinic acid or trigonelline; N-Oxide-Nam, N-oxide nicotinamide; N-Me-4PY, N-methyl-4-pyridone-3-carboxamide; N-Me-2PY, N-methyl-2-pyridone-4-carboxamide; 4PYR, 1-β-D-ribofuranosyl 4-pyridone-3-carboxamide; 4PYR-MP (n = 1), 1 -β-D-ribofuranosyl 4-pyridone-3-carboxamide monophosphate; 4PYR-DP (n = 2), 1 -β-D-ribofuranosyl 4-pyridone-3-carboxamide diphosphate; 4PYR-TP (n = 3), 1 -β-D-ribofuranosyl 4-pyridone-3-carboxamide triphosphate; NADO, 4-pyridone-3-carboxamide adenine dinucleotide; NADPO, 4-pyridone-3-carboxamide adenine dinucleotide phosphate; 1,2-NADH, 1,2-dihydronicotinamide adenine dinucleotide; 1,2-NADPH, 1,2-dihydronicotinamide adenine dinucleotide phosphate; 1,6-NADH, 1, 6-dihydronicotinamide adenine dinucleotide; 1,6-NADPH, 1,6-dihydronicotinamide adenine dinucleotide phosphate; NADHX, adenosine 5’-(trihydrogen diphosphate), -ester with 1,4,5,6-tetrahydro-6-hydroxy-1 -β-D-ribofuranosyl-3-pyridinecarboxamide also known as 6-hydroxylated nicotinamide adenine dinucleotide reduced form. NADPHX, adenosine 5’-(trihydrogen diphosphate), -ester with 1,4,5,6-tetrahydro-6-hydroxy-1-β-D-ribofuranosyl-3-pyridinecarboxamide phosphate also known as 6-hydroxylated nicotinamide adenine dinucleotide phosphate reduced form. cADPR, cyclic adenosine diphosphoriboside; ADPR, adenosine diphosphoribose; ADPRP, adenosine diphosphoribose phosphate; PAR, poly adenosine diphosphoriboside; Ac-ADPR, acetyl adenosine diphosphoribose; Suc-ADPR, succinyl adenosine diphosphoribose; acyl-ADPR, acyl adenosine diphosphoribose.
The chemistry of the NAD(P)(H) pool
The anabolites and catabolites of the vitamin B3 metabolome
Once generated from vitamin B3 derivatives via independent biosynthetic pathways, NAD+ can be converted to its reduced form NADH via redox processes or to its phosphorylated form NADP+, which, in turn, can enter redox processes to generate its reduced form, NADPH. Alternatively, NADH can be phosphorylated to NADPH. This constitutes the anabolic pathways to the NAD(P)(H) pool. Upon a range of biochemical and chemically driven processes, the components of the NAD(P)(H) pool are converted to nicotinamide or to catabolites which are either eliminated through excretion or recycled. The following describes these components in greater detail.
Vitamin B3 anabolites
Niacin (NA) and niacinamide (Nam) fall under the vitamin B3 denomination [1]. Intracellularly, NAD+ is generated from dietary vitamin B3 or trp (Figure 1) with the contribution made by the latter, known as the kynurenine pathway, varying greatly between species and organs [31–36]. Via the kynurenine pathway, biosynthetic precursors to NAD+ include kynurenine, 3-hydroxykinurenine, 3-hydroxyanthranilate and quinolinate, leading to nicotinic acid mononucleotide (NAMN) [37]. NAMN is also an NAD+ anabolite through the Preiss-Handler pathway, which uses NA [38], while nicotinamide mononucleotide (NMN) is generated in the salvage pathway, which uses Nam. Additional NAD+ anabolites include nicotinamide riboside (NR), nicotinic acid (NAR) and nicotinic acid adenine dinucleotide (NAAD) [8,39,40].
Figure 1. Precursors to NAD+.
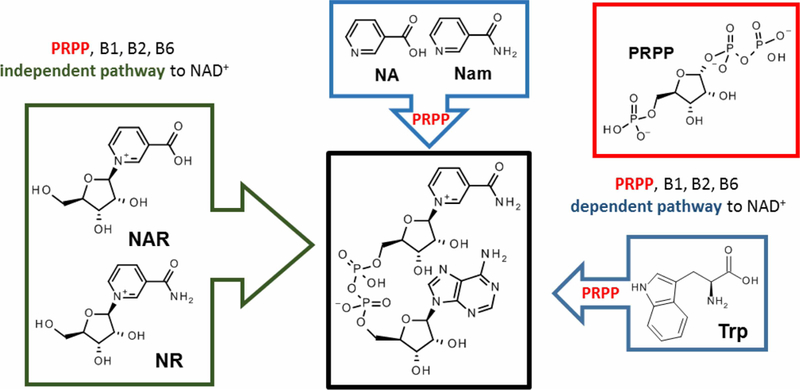
Blue box: PRPP-dependent NAD+ biosynthetic pathways; Green box: PRPP and vitamin B1, B2 and B6-independent pathways; PRPP, 5-phospho-1-pyrophosphoriboside; vitamin B1, thiamine; vitamin B2, riboflavin; vitamin B6, pyridoxine; NA, niacin/nicotinic acid; Nam, niacinamide/nicotinamide; NR, nicotinamide riboside; NAR, nicotinic acid riboside.
Vitamin B3 functional catabolites
NAD+ and NADP+ are substrates of enzymes capable of cleaving the glycosidic linkage between the northern ribose of the dinucleotide and nicotinamide, and replacing the latter with water, nucleophilic nucleobases or side chains of peptidic residues (e.g. hydroxyl or carboxylate) [41–49]. Unless chemical hydrolysis occurs, this cleavage is a finely orchestrated nucleophilic enzymatic process, leading to an exquisitely specific derivative. These derivatives are unique with regard to the biology they regulate [50]. NAADP is generated by an as-yet undiscovered biosynthetic pathway either from NAAD or from NADP+, regulating intracellular Ca2+ signaling processes [51]. The cyclic form of adenosine diphosphoribose, cADPR, produced by a cyclase [5], specifically mobilizes Ca2+ from ryanodine receptors [52], while its linear form, ADPR (adenosine diphosphoribose), generated from NAD+ by glycohydrolases, promotes Ca2+ cellular uptake [53]. Unlike ADPR itself, acylated forms of ADPR are products of NAD+- dependent post-translation modification catalyzed by sirtuins [10,26,54] hydrolyzed to ADPR by esterases [55]. Finally, the polymeric forms of ADPR, product of PARP enzymes, either covalently bound to proteins or free in solution, act as major complex recruiting agents in DNA repair [56–58].
Vitamin B3 catabolites
Many catabolic pathways are responsible for the loss of vitamin B3-derived cofactors and of their anabolites. Upon high NA intake, excess NA is converted to nicotinuric acid (NUA, Table 1b) in a phase 2 metabolic process when conjugated to glycine [59]. Excess Nam is readily oxidized to N-oxide-Nam ( Table 1b) by cytochrome P450 [60,61]. Yet, under standard dietary conditions, the bigger contributor to vitamin B3 catabolism in human physiology is the methylation of Nam, leading to N-methyl-Nam (N-Me-Nam; Table 1b). The formation of N-methyl-Nam requires S-adenosylmethionine. Therefore, in conjunction with homocysteine, N-methyl-Nam is a reporter of both the 1-carbon pathway efficacy and the vitamin B3 dietary status [62,63].
Trigonelline is N-methyl nicotinic acid, found abundantly in fenugreek and thought to be generated during coffee bean processing [64,65]. Trigonelline is also a catabolite found in tissues but less often measured [66] and for which the physiological properties remain unexplored. Oxidation of circulating N-methyl-Nam by aldehyde oxidase yields N-methyl-4-pyridone3-carboxamide (N-Me-4PY) and N-methyl-2-pyridone-5-carboxamide (N-Me-2PY) [6,67–69]. Much confusion exists in the literature as to the nomenclature of these two entities. The relative production of these catabolites is species-specific as well as driven by age and health status [70]. N-Me-2PY has been described as a uremic toxin because of the correlation between its abundance in blood and kidney disease states [71]. Critically, these two pyridones are produced systemically [72,73]. There, N-Me-2PY is thought to be an inhibitor of PARP function at physiologically relevant concentrations [67,74,75]. Another catabolite of vitamin B3 is N-ribosyl-3-carboxamide 4-pyridone (4PYR, Table 1b). This ribosylated pyridone is also found abundantly in circulation in uremic patients. Importantly, it is easily converted to its nucleotide forms (4PYR-MP, 4PYR-DP, 4PYR-TP, Table 1b) or adenylated to generate pyridone adenine dinucleotide species [NAD(P)O, Table 1b] [76–82]. Both the phosphorylated forms of 4PYR and its dinucleotide forms are endogenously generated. The synthesis of NADPO has been shown to occur as a side-reaction on NAD(P)+ catalyzed by flavin-dependent oxidases, such as ferrodoxin reductase [83–85]. In vitro,, the nucleotide forms show substantial ability to inhibit ATP-dependent kinases, while the dinucleotides are inhibitors of NAD(P)+-dependent metabolic redox enzymes at physiologically relevant concentrations [76,86]. A similar class of NAD(P)+ catabolites capable of inhibiting key metabolic enzymes are hydroxylated NAD(P)H (NAD(P)HX, Table 1b). The generation of these catabolites, which occurs chemically, is sufficiently critical to warrant a repair mechanism in all forms of life and the regeneration of NAD(P)H as accumulation of these catabolites causes central metabolomic perturbations [87,88]. Finally, other even less explored NAD(P)H catabolites are the 1,2-NAD(P)H and the 1,6-NAD(P)H [89]. These isomers can be mistaken for the α-anomeric forms of NAD(P)H [90–92] and are excellent inhibitors of isolated NAD(P)H-dependent redox enzymes. Renalase has been shown to re-oxidize these NAD(P)H isomers in vitro. It can then be viewed as a NAD(P)H repair enzyme directly affecting intracellular metabolism [93].
Overall, except for N-Me-Nam and N-Me-2PY, the catabolites of the NAD(P)(H) metabolome are rarely accounted for in metabolomic studies [94–96]. Furthermore, these compounds react readily under standard analytical conditions used for cell and tissues metabolomic measurements unless special care is applied and therefore go undetected. As such, an in-depth account of the detection protocols of the vitamin B3 metabolome is warranted but is beyond the scope of this review. Furthermore, mammalian cells have in place at least two known repair mechanisms to control dinucleotidic catabolite levels, renalase and NAD(P)HX dehydratase/ epimerase [87,88,93]. Dysregulation of such repair processes and accumulation of these catabolites surely impacts cellular homeostasis. Yet, the function, regulation and impact of the multiple vitamin B3 repair mechanisms have been vastly under-explored.
Biochemical pathways known to sustain the NAD(P)(H) pool
Notably, tryptophan, NA and Nam employ three convergent pathways which require molar equivalents of 5-phospho-1-pyrophosphoriboside (Scheme 1; PRPP) to convert quinolinic acid and NA to nicotinic acid adenine dinucleotide (NAAD) or Nam to NAD+ (Scheme 1) [34].
Scheme 1. Detailed biosynthetic pathways to the NAD(P)(H) pool components.
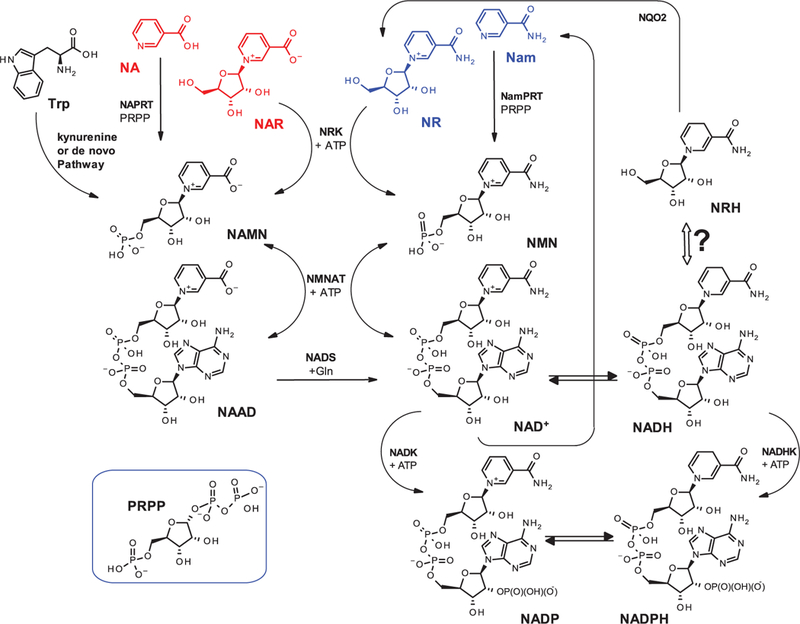
trp, tryptophan; Gln, glutamine; PRPP, 5-phosphoriboside pyrophosphate; NAPRT, nicotinic acid phosphoribosyl transferase; NamPRT, nicotinamide phosphoribosyl transferase; NMNAT, nicotinamide mononucleotide adenylyl transferase; NRK, nicotinamide riboside kinase; NADS, nicotinamide adenine dinucleotide synthase; NADK, nicotinamide adenine dinucleotide kinase; NADHK, NADH kinase; NQO2: N-ribosyldihydronicotinamide : quinone reductase 2.
While cofactors derived from riboflavin (vitamin B2) [97] and pyridoxine (vitamin B6) [98] are required by enzymes of the kynurenic pathway to generate quinolinic acid from tryptophan, NADP+ and thiamine (vitamin B1) diphosphate are cofactors required for the synthesis of PRPP from glucose 6-phosphate [36]. This highlights the dependency of the NAD(P)(H) biosynthetic pathways on the bioavailability of three other metabolic cofactors, all derived from water soluble B-vitamins.
Along with NA, Nam and trp, NAR and NR are also precursors of NAD+ (Scheme 1). Noticeably, QA, NA and Nam require phosphoribosylation as means of biosynthetic activation to NAMN [99] and NMN [100–102], while NR and NAR require phosphorylation by a specific kinase (Scheme 1) [8,39,103,104]. NAMN and NMN are biosynthetic intermediates to NAAD and NAD+, following an adenylyl transfer (Scheme 1) [105–109]. NAAD, acting as a pre-NAD+ storage pool [40], is converted to NAD+ by a ligase (NADS) (Scheme 1) [35,110]. NADP+ is generated from NAD+ by NAD+ kinase for which NADH is a weak substrate yielding NADPH [111]. It must be noted that while NR, NAR and NMN are PRPP-independent precursors to the NAD+, they are only molar equivalent precursors to NAD+. It is the generation of Nam through NAD+ consuming enzymes and its recycling to NAD+ which enables sustained NAD+ levels [112]. To sustain increased NAD+ levels through NR supplementation, NRK, NMNAT and NamPRT (nicotinamide phosphori- bosyl transferase; Scheme 1) must be functional, with turn-over in excess of that of Nam methylation by NNMT and cellular export mechanisms.
It is only recently that NR and NMN, both found in milk [113,114], have gained recognition as nutraceutical precursors of NAD+. NR supplementation in cell-based assays was evidenced to boost the NAD(P)(H) pool with a specific effect on the mitochondrial pool and function. Supplementation with NA and Nam, while critical in acute vitamin B3 deficiency, does not demonstrate the same physiological outcomes compared with that of supplementation with NR or NMN [7,15,34,94], indicative of additional controlling factors, such as intracellular biodistribution, expression of key biosynthetic enzymes and/or bioavailability of PRPP. To explore the parameters controlling functionalization and conversion of these NAD+ precursors and their biological endpoints in cell-based assays as well as in animals, an extensive synthetic program has been implemented over the past 50 years.
Accessing biosynthetic precursors of NAD+
NA and Nam can be readily obtained from bacterial broth, foodstuffs or generated from petroleum sources [1]. They are now widely available commercially along with some more clinically focused versions and formulations [115]. The ribosylated forms of NA or Nam have required the development of more substantial synthetic routes.
Enzymatic syntheses
NR may be prepared enzymatically from NAD+ and NMN by using snake venom phosphodiesterase and subsequent transformation of NMN to NR with prostatic monoesterase [116] or with 5’-nucleotidase [117]. Alternatively, NR can be generated from α-D-ribose-1-phosphate and Nam using purine nucleoside phosphor-ylase and sucrose phosphorylase [118]. There are only few reports in the literature describing the efficient chemical generation of NMN from NR [119–121]. In general, this process is often low yielding and associated with difficulties in removing phosphate contaminants. As such, enzymatic conversions with isolated NRK or whole cell production have been explored, but they too remain challenging. Accessing NAR has been even less explored. Yet, the generation of NAMN from NMN using a new cross-linked deamidase aggregate biocatalyst has been reported [122]. This offers new opportunities for a facile access to NAR via enzymatic routes using phosphatases such as 5’-nucleotidase [8].
Chemical syntheses of nicotinoyl ribosides and derivatives
Two main synthetic strategies have been developed to access NR salt forms (NR+X-) (Figure 2). One proceeds via a reaction between Nam or derivative A and a peracylated (halo)-D-ribofuranoside B resulting in acylated intermediate C that is subsequently converted into the desired NR+X– salt. This approach was also applied for the synthesis of NAR (NAR zwitterion). The other proceeds via the condensation of N-(2,4-dinitrophenyl)-3-carbamoylpyridinium salt D with derivatives of D-ribofuranosylamine E [120]. To date, the first approach has proved the most efficient in terms of overall yields and chemo-selectivity. We will summarize advances made with this first approach.
Figure 2.
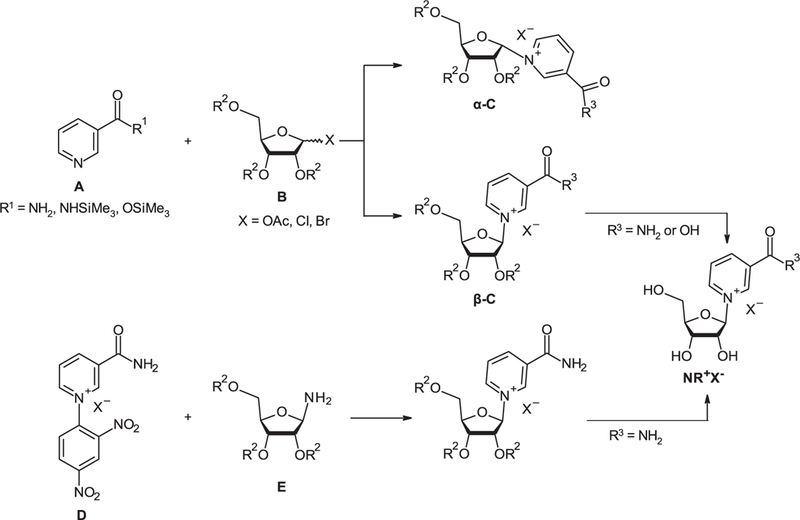
Synthetic routes to nicotinamide riboside (NR+X—).
Two anomeric α- and β-forms of NR (α-C and β-C; Figure 2) can be generated by glycosylation reactions with the stereochemical outcome of the synthesis being dependent on the nature and stereochemical position of the leaving group X, nature of the substituents at amide nitrogen atom in Nam and conditions of glycosylation, such as solvent and temperature. Because only the β-form of NR or NAR is of biochemical relevance, the most valuable synthetic methods offer β-stereoselectivity.
The first chemical syntheses of NR salts (NR+X-) was described by Todd and coworkers [123,124] and entailed the glycosylation of nicotinamide (Nam) 1a with either 1-bromo-2,3,5-tri-O-acetyl-D-ribofuranose to yield the bromide salt, 1-chloro-2,3,5-tri-O-acetyl-D-ribofuranose to yield the triacetylated chloride salt or 1-chloro-2,3,5-tri-O-benzoyl-D-ribofuranose to yield the tribenzoylated chloride salt. The halosugars were obtained from 1,2,3,5-tetra-O-acetyl-D-β-ribofuranose (2a) or 1-O-acetyl-2,3,5-tri-O-benzoyl-D-β-ribofuranose (2b) [125]. Such chemistry resulted in the generation of both pyridinium riboside anomers, with the best results in terms of β-/α-anomer stereoselectivity obtained when chlorosugars were used as precursors. Removal of the protecting groups in anhydrous methanol saturated with dry ammonia at 0°C yielded NR+Cl— as a 4: 1 mixture of β- and α-anomers [124]. Low temperature was required to minimize Nam release (Scheme 2).
Scheme 2.
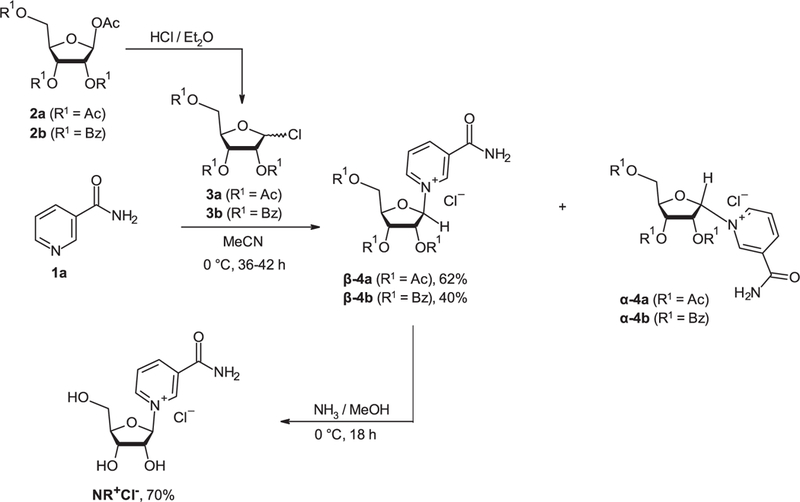
Synthetic sequence to 1-β-D-ribofuranoside nicotinamide chloride.
While several routes and optimization studies have been conducted [126] since the first synthetic route development, the most versatile uses tetra-acylated ribosides and TMSOTf as a catalyst [127,128]. Sauve and coworkers improved on the method and reported a very efficient one-pot procedure for the synthesis of β-NR from ethyl nicotinate [129,130]. Both routes generate the triflate salt forms of NR. The triflate salts, deemed unsuitable for pharmacological use, must be exchanged for pharmaceutically acceptable anions. Anion exchange either by liquid/liquid extraction [131] or by treatment with ion exchange resin such as using Amberlite IRA400-Cl have been successfully applied to generated NR+Cl— [132]. Oxidation of the reduced form of NR 7 on charcoal in the presence of protic salts, such as ammonium salts NH4X-, is an alternative method to anion exchange resulting in different salt forms of NR+X—. Acylated NR-triflate and acylated NAR prepared via mechanochemical methods, reduced to the acylated 1,4-dihydronicotinamide and dihydronicoti- noyl riboside, can be readily extracted in pure form in organic solvents (Scheme 3) [133]. The reduced forms of NR 6a-b and NAR are stable to Bronsted bases and, therefore, the acyl groups can be removed at room temperature at increased rates [134,135]. 1,4-Dihydronicotinamide riboside derivatives may be also oxidized with hexachloroacetone or cobalt(II) acetate in the presence of hydrogen peroxide. The later process requires removal of cobalt cations with QuadraSil AP resin [136].
Scheme 3.
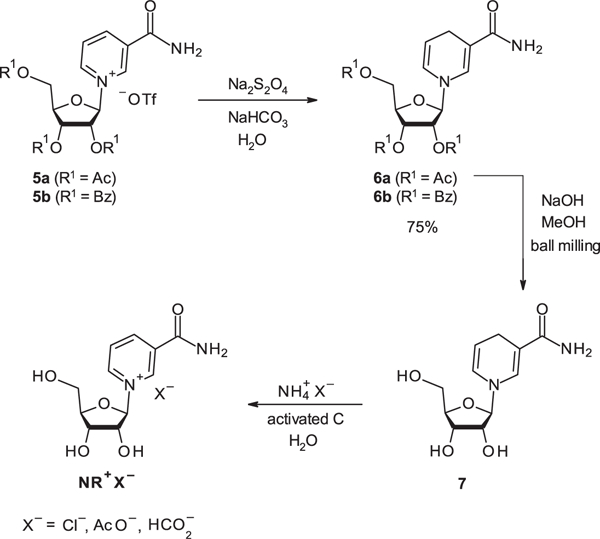
Synthesis of the reduced form of NR as a synthetic intermediate to NR.
Chemical reduction in N-substituted pyridinium salts results in three possible isomeric products: 1,2-, 1,4- and 1,6-dihydropyridines (DHP) as illustrated in Figure 3 for corresponding dihydro-1-P-D-ribofuranosyl-3- pyridinecarboxamides.
Figure 3.
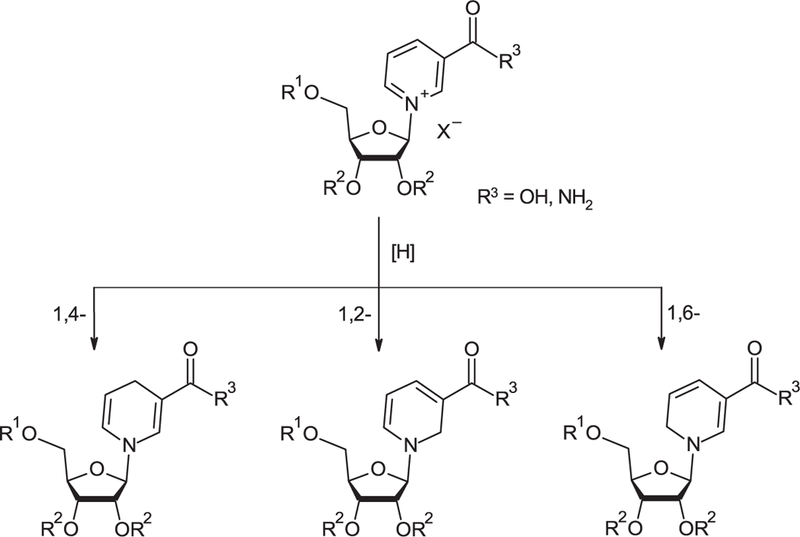
Reduction in derivatives of NR+X— into corresponding NRH derivatives.
Reduction in pyridinium salts to dihydropyridines has been extensively reviewed in the literature [137–140]. Sodium borohydride (NaBH4) and sodium dithionite (Na2S2O4) are the most commonly used reducing agents to reduce NAD(P), NMN and NR. However, these reagents are not equivalent. Na2S2O4 regioselectively reduces NAD+ to 1,4-dihydronicotinamide adenine dinucleotide (NADH), and NR to the 1,4–3-carboxamide dihydropyridinyl riboside, while reduction in NAD+ and NR+ with NaBH4 or milder hydride-based reducing agents results in a mixture of the 1,2-, 1,4- and 1,6-isomers.
Synthesis of pyridones
While the hydroxylated forms of 1,4–3-carboxamide dihydropyridinyl riboside derivatives (e.g. NAD(P)HX) are readily generated from the ribosylated species, the pyridone-derived catabolites (N-Me-2/4-PY, 4-PYR, NAD(P) O, 4-PYR-M/D/TP; Table 1b) require chemical syntheses. 4-Pyridone-3-carboxamide (4PY) and 2-pyridone-5- carboxamide (2PY) are generated from 4-chloro-3-carboxypyridine and 2-hydroxy-5-cyanopyridine, respectively [79]. These can then be used to prepare the nucleosides [79,141]. Critically, the nucleotide- and dinucleotide-derived pyridones are only prepared on analytical scale, generated as enzymatic side-reaction products [142].
Synthesis of isotope-labeled NAD+ precursors (isotopomers and isotopologues)
Decaying and stable isotope-labeled derivatives [143–146] have been used to study metabolic pathways and bio-distribution processes. Combining separation to detection and quantification allows for complex product distributions to be measured. To differentiate between biosynthetic components and pathways of the NAD(P)(H) pool, anabolites incorporating different profiles of stable isotopes can be used if their incorporation into NAD+ leads to versions of NAD+ which can be differentiated by mass (MS) or fragmentation patterns (MS2). Here, enter two critical definitions: that of isotopomers which are molecules which vary in the position of labeled atom, such as 2’−2H-NR versus 1’−2H-NR and isotopologues which are molecules which differ by containing different isotopes, such as 2H-NR versus 13C-NR. These isotopically labeled compounds will possess different exact molecular mass and/or fragments’ exact mass.
Presently, the rationale applied to selecting appropriate isotopologues is driven by the question being asked and the levels of the isotopically labeled derivatives to be detected. Bioavailability and biodistribution studies in animals are often addressed using decaying isotopomers. Combined to liquid chromatography, this highly sensitive method differentiates between the bio-transformed radio-isotopically labeled products, e.g. [147]. Furthermore, the uniformly labeled NAD+ is commercially available and amenable to chemoenzymatic transformations. For instance, radio-isotopically labeled NAD+ can be converted to its NADP+ parent by NAD+ kinase [148]. It is also reduced enzymatically to NADPH [148]. These can be used as starting materials in some of the enzymatic processes described below.
Non-decaying isotopomers provide the necessary versatility to interrogate fluxes if the building blocks and products can be traced with enough statistical confidence at levels above natural isotopic abundance. For instance, to establish in mammals whether NR was directly converted to NMN, or hydrolyzed to Nam prior to it being incorporated into NMN, the use of a doubly labeled isotopomer of NR was used [94]. This isotopomer incorporated one heavier isotope on the nicotinamide ring and one heavier isotope on the furanose. The glyco- sidic breakage led to mono-labeled NAD+, while the direct incorporation led to doubly labeled NAD+ being detected. In cell work study, this type of multi-site labeling informs on the regulation of the biosynthetic pathways and of the turn-over of NAD+ by consuming and biosynthetic enzymes [34,40,107].
Isotopically labeled forms of Nam, NA and trp are available commercially and can thus be selected at will. Similarly, ribosylated derivatives, for which isotope labels may be incorporated into the sugar residue (2H, 3H, 18O, 13C and 14C isotopes) and the nicotinamide or nicotinic core (18O, 13C, 14C and 15N isotopes), or both (Figure 4), require dedicated chemoenzymatic or chemical syntheses.
Figure 4.
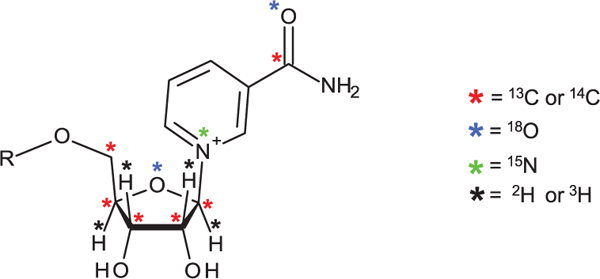
General representation of isotope-labeled NR derivative; illustrative labeled sites are shown by colored asterisks.
Decaying isotopically labeled NAD+ anabolites, such as synthetic tritiated nucleotides and 32P-containing ATP, have been combined to chemoenzymatic preparations to allow for the efficient generation of radiolabeled NAD(P)(H) pools [149–155]. The enzymatic methods allow preparation of not only NR derivatives labeled in the Nam core but in the ribosyl moiety as well when labeled phosphoribosyl pyrophosphate (PRPP) is used. An example of this methodology is found in work of Kašarov and Moat describing preparation of [carbonyl-14C]NR from corresponding 14C-labeled NAD+ catalyzed by enzymes from Proteus vulgaris OX-19 [156]. Saunders et al. [117] describe the preparation of [carbonyl-14C]NR and [4-3H]NR by the treatment of corresponding radiolabeled NMN with 5’-nucleotidase. Chemical synthesis of tritium-labeled NR and subsequent enzymatic synthesis of tritium-labeled NMN as well as corresponding [2’−3H]-NAD+ are described in work of Cen and Sauve [157], which also describes the synthesis of NAD+ containing 18O-label in the NR portion of the molecule and originating from [5-18O]glucose. Bull et al. [158] describes the chemical synthesis of deuterium-labeled [1’−2H]NR+Br— and [1’−2H]NMN. Once purified, this was used to prepare [1’−2H]NAD+ enzymatically, which was then enzymatically converted to [carbonyl-14C,1’−2H]NAD+ with [carbonyl-14C]Nam. In a series of papers by Schramm et al. dealing with the enzymatic synthesis of [3H,14C] NAD+ isotopomers, the authors used [2-3H]-, [5-3H]-, [6-3H]-, [2-14C]- and [6-14C]glucose and nicotinic acid to generate corresponding [1’−3H]-, [2’−3H]-, [4’−3H]-, [5’−3H]-, [1’−14C]-, [5’−14C]NAD+; they also describe preparation of 15N-labeled NAD+ isotopologues, such as [1’−14C,1-15N]NAD+ and [5’−14C,1-15N]NAD+ (primed numbers indicate atomic locations in the ribosyl residue of NR part of NAD+), using 15N-labeled NA as a source of the label [159–162]. The enzymatic synthesis of [14C]NR was achieved from unlabeled NAD+ and [carbonyl-14C] Nam in the presence of ADP-ribosylcyclase to give 14C-NAD+, followed by treatment with phosphodiesterase I and alkaline phosphatase [163].
While extremely sensitive, detection of radiation-emitting entities requires special laboratory set-up and therefore limits its use by the wider research community. Detection of non-decaying isotopic modifications are less sensitive but rely on more generally adopted protocols [145]. Yet, accurate measurements of the vitamin B3 metabolome in biological systems have been limited by the chemical availability of chemical standards and tailor-made vitamin B3 metabolites. However synthetic efforts have been undertaken towards achieving higher availability of labeled and non-labeled standards for an increased coverage, characterization and quantification of the metabolome. The use of isotopically labeled vitamin B3 metabolites combined to powerful targeted metabolomic analytical methods has proved particularly suited to improving our knowledge of vitamin B3 both at cellular and organismal levels, allowing rapid translational discoveries [34,164]. This has been enabled by dramatic advances in the field of mass spectroscopy, metabolomics and large data set management along with an increased access to molecules purposefully incorporating isotopes that enable their detection and quantification as well as inform of their modifications.
According to the approach described by Tran et al. [163], [13C,18O]NR can be generated from [U-13C] glucose and NA enzymatically converted to [13C]NAAD containing fully 13C-labeled ribosyl residue in NAR part of the NAAD molecule. This synthesis requires usage of 10 enzymes, along with ATP, phospho(enol)pyru-vate, NADP+ and α-ketoglutarate. In the second — again enzymatic — step, purified 13C-labeled NAAD was transformed to corresponding [13C]NAD+ by NAD+ synthetase. Then, purified [13C]NAD+ was incubated with [18O]Nam (prepared by chemical reaction of 3-cyanopyridine with 18O-water) in the presence of ADP-ribosylcyclase to give [13C,18O]NAD+ that was subsequently degraded by using phosphodiesterase I and alkaline phosphatase to quantitatively afford [13C,18O]NR. Furthermore, the enzymatic synthesis of 18O-labeled NAD+ from non-labeled NAD+ can be achieved using glycohydrolase/cyclase CD38 and 18O-nicotinamide (20-fold excess) [165]. Mills et al. [105] used double-labeled NMN prepared via a procedure based on the work of Lee et al., while Ratajczak et al. mention the synthesis of 18O-labeled NR from [18O]Nam and subsequent synthesis of 18O-labeled NNM by phosphorylation with NRK1 [94,107]. Chemical sequences described above were applied to generate [18O]NR from 18O-labeled Nam and [2’−2H,carbonyl-13C]NR generated from the 2’−2H-1,2,3,5-tetra-O-acetyl-β-D-ribofuranose and [carbonyl-13C]Nam to establish biodistribution and function [104]. Finally, [2’−2H,carbonyl-13C]NAR was synthesized and compared with [2’−2H, 18O]NR in metabolic fluxes and organelle transport experiments [34,40].
Conclusion
Overall, many chemical syntheses and chemoenzymatic syntheses have been developed to identify and trace the metabolites and precursors of NAD(P)(H) and quantify the metabolic distribution following supplementation. Current limitations associated with establishing a true representation of the vitamin B3 metabolome are associated with the breadth of molecules which this metabolome includes, the synthetic challenges associated with their individual preparation, the cost of the isotopically labeled reagents and the scale on which syntheses are carried out. However, these limitations appear to slowly fade as more efficient syntheses become available and enable cell-based kinetic studies and animal pharmacokinetics investigations. This review aimed to update our view of the vitamin B3 metabolome and the current chemical efforts undertaken in the field of NAD+ biology to better understand its role in cellular biology and physiology.
Perspectives.
Since NAD+ seats at the cross-road of metabolism and cellular signaling, there is an urgent need to acquire a greater evidence-based understanding of vitamin B3 metabolism and of its role in health, diseases and ageing.
Increased access to fit-for-purpose chemical entities and biosynthetic intermediates has greatly enabled the recent discoveries in the NAD+ field and facilitated translational research in ageing, metabolic diseases and nutrition.
As further analytical refinements are achieved, and analytical standards become more widely available, the cellular functions of endogenously generated vitamin B3 catabolites will come under greater scrutiny. Furthermore, as the functional co-dependence between the NAD(P)(H) pool and cofactors derived from vitamin B1 (thiamine), vitamin B2 (riboflavin), vitamin B5 ( pantothenate), vitamin B6 ( pyridoxine) and vitamin B9 (folate) becomes more apparent, vitamin B-targeted metabolomics will offer new functional perspectives on the B-vitaminome.
Acknowledgments
Funding
M.M. received support from National Center for Complementary and Integrative Health R21 AT009908–01.
Abbreviations
- 4PYR
N-ribosyl-3-carboxamide 4-pyridone
- ADPR
adenosine diphosphoribose
- ADPRP
adenosine diphosphoribose phosphate
- N-Me-2PY
N-methyl-2-pyridone-5-carboxamide
- N-Me-4PY
N-methyl-4-pyridone3-carboxamide
- NA
nicotinic acid
- NAAD
nicotinic acid adenine dinucleotide
- NAADP
nicotinic acid adenine dinucleotide phosphate
- NAD
adenine dinucleotide
- NADH
1,4-dihydronicotinamide adenine dinucleotide
- NADP
nicotinamide adenine dinucleotide phosphate
- NADS
nicotinamide Adenine dinucleotide synthase
- Nam
nicotinamide
- NAMN
nicotinic acid mononucleotide
- NAPRT
Nicotinic acid phosphoribosyl transferase
- NAR
nicotinic acid
- NMN
nicotinamide mononucleotide
- NMNAT
nicotinamide mononucleotide adenylyl transferase
- NR
nicotinamide riboside
- NRK
nicotinamide riboside kinase
- PRPP
5-phospho-1-pyrophosphoriboside
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Wolak N, Zawrotniak M, Gogol M, Kozik A and Rapala-Kozik M (2017) Vitamins B1, B2, B3 and B9-occurrence, biosynthesis pathways and functions in human nutrition. Mini. Rev. Med. Chem. 17, 1075–1111 [DOI] [PubMed] [Google Scholar]
- 2.Fricker RA, Green EL, Jenkins SI and Griffin SM (2018) The influence of nicotinamide on health and disease in the central nervous system. Int. J. Tiyptophan Res. 11, 1178646918776658 10.1177/1178646918776658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikiforov A, Kulikova V and Ziegler M (2015) The human NAD metabolome: functions, metabolism and compartmentalization. Crit. Rev. Biochem. Mol. Biol. 50, 284–297 10.3109/10409238.2015.1028612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warszta D, Nebel M, Fliegert R and Guse AH (2014) NAD derived second messengers: role in spontaneous diastolic Ca2+ transients in murine cardiac myocytes. DNA Repair 23, 69–78 10.1016/j.dnarep.2014.05.007 [DOI] [PubMed] [Google Scholar]
- 5.Ferrero E, Lo Buono N, Horenstein AL, Funaro A and Malavasi F (2014) The ADP-ribosyl cyclases-the current evolutionary state of the ARCs. Front. Biosci. 19, 986–1002 10.2741/4262 [DOI] [PubMed] [Google Scholar]
- 6.Maeta A, Sano M, Fukuwatari T and Shibata K (2014) Simultaneous measurement of nicotinamide and its catabolites, nicotinamide N-oxide, N1-methyl-2-pyridone-5-carboxamide, and N1-methyl-4-pyridone-3-carboxamide, in mice urine. Biosci. Biotechnol. Biochem. 78, 1306–1309 10.1080/09168451.2014.918495 [DOI] [PubMed] [Google Scholar]
- 7.Yoshino J, Baur JA and Imai SI (2018) NAD+ intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab. 27, 513–528 10.1016/j.cmet.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulikova V, Shabalin K, Nerinovski K, Dolle C, Niere M, Yakimov A et al. (2015) Generation, release, and uptake of the NAD precursor nicotinic acid riboside by human cells. J. Biol. Chem. 290, 27124–27137 10.1074/jbc.M115.664458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdin E (2015) NAD(+) in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 10.1126/science.aac4854 [DOI] [PubMed] [Google Scholar]
- 10.Imai S and Guarente L (2014) NAD+ and sirtuins in aging and disease. Trends Cell Biol. 24, 464–471 10.1016/j.tcb.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolle C, Skoge R, VanLinden M and Ziegler M (2013) NAD biosynthesis in humans-enzymes, metabolites and therapeutic aspects. Curr. Top. Med. Chem. 13, 2907–2917 10.2174/15680266113136660206 [DOI] [PubMed] [Google Scholar]
- 12.Menzies KJ, Zhang H, Katsyuba E and Auwerx J (2016) Protein acetylation in metabolism — metabolites and cofactors. Nat. Rev. Endocrinol. 12, 43–60 10.1038/nrendo.2015.181 [DOI] [PubMed] [Google Scholar]
- 13.Mouchiroud L, Houtkooper RH and Auwerx J (2013) NAD+ metabolism: a therapeutic target for age-related metabolic disease. Crit. Rev. Biochem. Mol. Biol. 48, 397–408 10.3109/10409238.2013.789479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P et al. (2016) NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443 10.1126/science.aaf2693 [DOI] [PubMed] [Google Scholar]
- 15.Mitchell SJ, Bernier M, Aon MA, Cortassa S, Kim EY, Fang EF et al. (2018) Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metab. 27, 667–676.e4 10.1016/j.cmet.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger F, Ramirez-Hernandez MH and Ziegler M (2004) The new life of a centenarian: signalling functions of NAD(P). Trends Biochem. Sci. 29, 111–118 10.1016/j.tibs.2004.01.007 [DOI] [PubMed] [Google Scholar]
- 17.Hill LJ and Williams AC (2017) Meat intake and the dose of vitamin B3 — nicotinamide: cause of the causes of disease transitions, health divides, and health futures? Int. J. Tryptophan. Res. 10, 1178646917704662 10.1177/1178646917704662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savvidou S (2014) Pellagra: a non-eradicated old disease. Ciin. Pract. 4, 637 10.4081/cp.2014.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R, Yu K, Wang Q, Wang L, Mao J and Qian J (2016) Pellagra secondary to medication and alcoholism: a case report and review of the literature. Nut. Ciin. Pract. 31, 785–788 10.1177/0884533616660991 [DOI] [PubMed] [Google Scholar]
- 20.Terada N, Kinoshita K, Taguchi S and Tokuda Y (2015) Wernicke encephalopathy and pellagra in an alcoholic and malnourished patient. BMJ Case Rep. 2015, bcr2015209412 10.1136/bcr-2015-209412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crook MA (2014) The importance of recognizing pellagra (niacin deficiency) as it still occurs. Nutrition 30, 729–730 10.1016/j.nut.2014.03.004 [DOI] [PubMed] [Google Scholar]
- 22.Garrido A and Djouder N (2017) NAD+ deficits in age-related diseases and cancer. Trends Cancer 3, 593–610 10.1016/j.trecan.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 23.Srivastava S (2016) Emerging therapeutic roles for NAD(+) metabolism in mitochondrial and age-related disorders. Clin. Transl. Med. 5, 25 10.1186/s40169-016-0104-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee S, Chellappa K, Moffitt A, Ndungu J, Dellinger RW, Davis JG et al. (2017) Nicotinamide adenine dinucleotide biosynthesis promotes liver regeneration. Hepatology 65, 616–630 10.1002/hep.28912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hershberger KA, Martin AS and Hirschey MD (2017) Role of NAD+ and mitochondrial sirtuins in cardiac and renal diseases. Nat. Rev. Nephrol. 13, 213–225 10.1038/nrneph.2017.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonkowski MS and Sinclair DA (2016) Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 17, 679–690 10.1038/nrm.2016.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belenky P, Bogan KL and Brenner C (2007) NAD+ metabolism in health and disease. Trends Biochem. Sci. 32, 12–19 10.1016/j.tibs.2006.11.006 [DOI] [PubMed] [Google Scholar]
- 28.Katsyuba E and Auwerx J (2017) Modulating NAD+ metabolism, from bench to bedside. EMBO J. 36, 2670–2683 10.15252/embj.201797135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiarugi A, Dolle C, Felici R and Ziegler M (2012) The NAD metabolome-a key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752 10.1038/nrc3340 [DOI] [PubMed] [Google Scholar]
- 30.VanLinden MR, Dolle C, Pettersen IKN, Kulikova VA, Niere M, Agrimi G et al. (2015) Subcellular distribution of NAD+ between cytosol and mitochondria determines the metabolic profile of human cells. J. Biol. Chem. 290, 27644–27659 10.1074/jbc.M115.654129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gazzaniga F, Stebbins R, Chang SZ, McPeek MA and Brenner C (2009) Microbial NAD metabolism: lessons from comparative genomics. Microbiol. Mol. Biol. Rev. 73, 529–541 10.1128/MMBR.00042-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerdes SY, Scholle MD, D’Souza M, Bernal A, Baev MV, Farrell M et al. (2002) From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184, 4555–4572 10.1128/JB.184.16.4555-4572.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bi J, Wang H and Xie J (2011) Comparative genomics of NAD(P) biosynthesis and novel antibiotic drug targets. J. Cell Physiol. 226, 331–340 10.1002/jcp.22419 [DOI] [PubMed] [Google Scholar]
- 34.Liu L, Su X, Quinn WJ, Hui S, Krukenberg K, Frederick DW et al. (2018) Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 27, 1067–1080.e5 10.1016/j.cmet.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori V, Amici A, Mazzola F, Di Stefano M, Conforti L, Magni G et al. (2014) Metabolic profiling of alternative NAD biosynthetic routes in mouse tissues. PLoS ONE 9, e113939 10.1371/journal.pone.0113939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata K, Kobayashi R and Fukuwatari T (2015) Vitamin B1 deficiency inhibits the increased conversion of tryptophan to nicotinamide in severe food-restricted rats. Biosci. Biotechnol. Biochem. 79, 103–108 10.1080/09168451.2014.962473 [DOI] [PubMed] [Google Scholar]
- 37.Majewski M, Kozlowska A, Thoene M, Lepiarczyk E and Grzegorzewski WJ (2016) Overview of the role of vitamins and minerals on the kynurenine pathway in health and disease. J. Physiol. Pharmacol. 67, 3–19 PMID: [PubMed] [Google Scholar]
- 38.Jacobson EL, Kim H, Kim M and Jacobson MK (2012) Niacin: vitamin and antidyslipidemic drug. Subcell. Biochem. 56, 37–47 10.1007/978-94-007-2199-9_3 [DOI] [PubMed] [Google Scholar]
- 39.Tempel W, Rabeh WM, Bogan KL, Belenky P, Wojcik M, Seidle HF et al. (2007) Nicotinamide riboside kinase structures reveal new pathways to NAD+. PLoS Biol. 5, e263 10.1371/journal.pbio.0050263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davila A, Liu L, Chellappa K, Redpath P, Nakamaru-Ogiso E, Paolella LM et al. (2018) Nicotinamide adenine dinucleotide is transported into mammalian mitochondria. eLife 7, e33246 10.7554/eLife.33246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang DX, Zhang J-P, Hu J-Y and Huang Y-S (2016) The potential regulatory roles of NAD(+) and its metabolism in autophagy. Metabolism 65, 454–462 10.1016/jmetabol.2015.11.010 [DOI] [PubMed] [Google Scholar]
- 42.Tong L and Denu JM (2010) Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. Biochim. Biophys. Acta 1804, 1617–1625 10.1016/j.bbapap.2010.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long A, Klimova N and Kristian T (2017) Mitochondrial NUDIX hydrolases: a metabolic link between NAD catabolism, GTP and mitochondrial dynamics. Neurochem. Int. 109, 193–201 10.1016/j.neuint.2017.03.009 [DOI] [PubMed] [Google Scholar]
- 44.Koch-Nolte F, Fischer S, Haag F and Ziegler M (2011) Compartmentation of NAD+-dependent signalling. FEBS Lett. 585, 1651–1656 10.1016/j.febslet.2011.03.045 [DOI] [PubMed] [Google Scholar]
- 45.Camacho-Pereira J, Tarrago MG, Chini CCS, Nin V, Escande C, Warner GM et al. (2016) CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 23, 1127–1139 10.1016/j.cmet.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen MS and Chang P (2018) Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat. Chem. Biol. 14, 236–243 10.1038/nchembio.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aravind L, Zhang D, de Souza RF, Anand S and Iyer LM (2015) The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr. Top. Microbiol. Immunol. 384, 3–32 10.1007/82_2014_414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leslie Pedrioli DM, Leutert M, Bilan V, Nowak K, Gunasekera K, Ferrari E et al. (2018) Comprehensive ADP-ribosylome analysis identifies tyrosine as an ADP-ribose acceptor site. EMBO Rep. 19, e45310 10.15252/embr.201745310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen B, Zang W, Wang J, Huang Y, He Y, Yan L et al. (2015) The chemical biology of sirtuins. Chem. Soc. Rev. 44, 5246–5264 10.1039/C4CS00373J [DOI] [PubMed] [Google Scholar]
- 50.Chini CCS, Tarrago MG and Chini EN (2017) NAD and the aging process: role in life, death and everything in between. Mol. Cell Endocrinol. 455, 62–74 10.1016/j.mce.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calcraft PJ, Ruas M, Pan Z, Cheng X, Arredouani A, Hao X et al. (2009) NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature 459, 596–600 10.1038/nature08030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee HC (2012) Cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate (NAADP) as messengers for calcium mobilization. J. Biol. Chem. 287, 31633–31640 10.1074/jbc.R112.349464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Opitz CA and Heiland I (2015) Dynamics of NAD-metabolism: everything but constant. Biochem. Soc. Trans. 43, 1127–1132 10.1042/BST20150133 [DOI] [PubMed] [Google Scholar]
- 54.Dolle C, Rack JG and Ziegler M (2013) NAD and ADP-ribose metabolism in mitochondria. FEBS J. 280, 3530–3541 10.1111/febs.12304 [DOI] [PubMed] [Google Scholar]
- 55.Kasamatsu A, Nakao M, Smith BC, Comstock LR, Ono T, Kato J et al. (2011) Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3. J. Biol. Chem. 286, 21110–21117 10.1074/jbc.M111.237636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei H and Yu X (2016) Functions of PARylation in DNA damage repair pathways. Genomics Proteomics Bioinformatics 14, 131–139 10.1016/j.gpb.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupte R, Liu Z and Kraus WL (2017) PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 31, 101–126 10.1101/gad.291518.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drenichev MS and Mikhailov SN (2015) Poly(ADP-ribose)—a unique natural polymer structural features, biological role and approaches to the chemical synthesis. Nucleosides Nucleotides Nucleic Acids 34, 258–276 10.1080/15257770.2014.984073 [DOI] [PubMed] [Google Scholar]
- 59.Jones KM (1959) The mechanism of nicotinuric acid synthesis. Biochem. J. 73, 714–719 10.1042/bj0730714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shibata K, Fukuwatari T and Suzuki C (2014) Pharmacological doses of nicotinic acid and nicotinamide are independently metabolized in rats. J. Nutr. Sci. Vitaminol. 60, 86–93 10.3177/jnsv.60.86 [DOI] [PubMed] [Google Scholar]
- 61.Nomura K, Shin M, Sano K, Umezawa C and Shimada T (1983) Nicotinamide N-oxide formation by rat liver microsomes. Biochem. Pharmacol 32 934–936 10.1016/0006-2952(83)90603-2 [DOI] [PubMed] [Google Scholar]
- 62.Kraus D, Yang Q, Kong D, Banks AS, Zhang L, Rodgers JT et al. (2014) Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature 508, 258–262 10.1038/nature13198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong S, Moreno-Navarrete JM, Wei X, Kikukawa Y, Tzameli I, Prasad D et al. (2015) Nicotinamide N-methyltransferase regulates hepatic nutrient metabolism through Sirt1 protein stabilization. Nat. Med. 21, 887–894 10.1038/nm.3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou J, Chan L and Zhou S (2012) Trigonelline: a plant alkaloid with therapeutic potential for diabetes and central nervous system disease. Curr. Med. Chem. 19, 3523–3531 10.2174/092986712801323171 [DOI] [PubMed] [Google Scholar]
- 65.Baspinar B, Eskici G and Ozcelik AO (2017) How coffee affects metabolic syndrome and its components. Food Fund 8, 2089–2101 10.1039/C7FO00388A [DOI] [PubMed] [Google Scholar]
- 66.Stretch C, Eastman T, Mandal R, Eisner R, Wishart DS, Mourtzakis M et al. (2012) Prediction of skeletal muscle and fat mass in patients with advanced cancer using a metabolomic approach. J. Nutr. 142, 14–21 10.3945/jn.111.147751 [DOI] [PubMed] [Google Scholar]
- 67.Lenglet A, Liabeuf S, Bodeau S, Louvet L, Mary A, Boullier A et al. (2016) N-methyl-2-pyridone-5-carboxamide (2PY)-major metabolite of nicotinamide: an update on an old uremic toxin. Toxins 8, E339 10.3390/toxins8110339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kremer JI, Gompel K, Bakuradze T, Eisenbrand G and Richling E (2018) Urinary excretion of niacin metabolites in humans after coffee consumption. Mol. Nutr. Food Res. 62, e1700735 10.1002/mnfr.201700735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shibata K, Morita N, Shibata Y and Fukuwatari T (2013) Enzymes that control the conversion of L-tryptophan-nicotinamide and the urinary excretion ratio (N1)-methyl-2-pyridone-5-carboxamide+N1-methyl-4-pyridone-3-carboxamide)/N1-methylnicotinamide in mice. Biosci. Biotechnol. Biochem. 77, 2105–2111 10.1271/bbb.130467 [DOI] [PubMed] [Google Scholar]
- 70.Shibata K and Matsuo H (1989) Correlation between niacin equivalent intake and urinary excretion of its metabolites, N’-methylnicotinamide, N’-methyl-2-pyridone-5-carboxamide, and N’-methyl-4-pyridone-3-carboxamide, in humans consuming a self-selected food. Am. J. Clin. Nutr. 50, 114–119 10.1093/ajcn/50.H14 [DOI] [PubMed] [Google Scholar]
- 71.Rutkowski B, Slominska E, Szolkiewicz M, Smolenski RT, Striley C, Rutkowski P et al. (2003) N-methyl-2-pyridone-5-carboxamide: a novel uremic toxin? Kidney Int. Suppl. 63, S19–S21 10.1046/j1523-1755.63.s84.36x [DOI] [PubMed] [Google Scholar]
- 72.Schmeisser K, Mansfeld J, Kuhlow D, Weimer S, Priebe S, Heiland I et al. (2013) Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat. Chem. Biol. 9, 693–700 10.1038/nchembio.1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rutkowski P, Slominska EM, Wotyniec W, Smoleński RT, Szolkiewicz M, Świerczyński J et al. (2008) Nicotinamide metabolites accumulate in the tissues of uremic rats. J. Ren. Nutr. 18, 56–59 10.1053/j.jrn.2007.10.012 [DOI] [PubMed] [Google Scholar]
- 74.Slominska EM, Kowalik K, Smolenski RT, Szolkiewicz M, Rutkowski P, Rutkowski B et al. (2006) Accumulation of poly(ADP-ribose) polymerase inhibitors in children with chronic renal failure. Pediatr. Nephrol. 21, 800–806 10.1007/s00467-006-0072-z [DOI] [PubMed] [Google Scholar]
- 75.Slominska EM, Smolenski RT, Osborne F, Swierczynski J and Yacoub MH (2005) The effect of N-methyl-2-pyridone-5-carboxamide—a nicotinamide catabolite on poly ADP-rybosylation and oxidative stress injury in endothelial cells. Nucleosides Nucleotides Nucleic Acids 24, 259–262 10.1081/NCN-59697 [DOI] [PubMed] [Google Scholar]
- 76.Pelikant-Matecka I, Sielicka A, Kaniewska E, Smoleńs ki RT and Stomińs ka EM (2014) 4-Pyridone-3-carboxamide-1 p-D-ribonucieoside metabolism in endothelial cells and its impact on cellular energetic balance. Nucleosides Nucleotides Nucleic Acids 33, 338–341 10.1080/15257770.2014.889303 [DOI] [PubMed] [Google Scholar]
- 77.Pelikant-Matecka I, Sielicka A, Kaniewska E, Smolens ki RT and Stomins ka EM (2016) Influence of 4-pyridone-3-carboxamide-1 β-D-ribonucleoside (4PYR) on activities of extracellular enzymes in endothelial human cells. Nucleosides Nucleotides Nucleic Acids 35, 732–736 10.1080/15257770.2016.1174263 [DOI] [PubMed] [Google Scholar]
- 78.Synesiou E, Fairbanks LD, Simmonds HA, Slominska EM, Smolenski RT and Carrey EA (2011) 4-Pyridone-3-carboxamide-1-beta-D-ribonucleoside triphosphate (4PyTP), a novel NAD metabolite accumulating in erythrocytes of uremic children: a biomarker for a toxic NAD analogue in other tissues? Toxins 3, 520–537 10.3390/toxins3060520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Slominska EM, Carrey EA, Foks H, Orlewska C, Wieczerzak E, Sowinski P et al. (2006) A novel nucleotide found in human erythrocytes, 4-pyridone-3-carboxamide-1-beta-D-ribonucleoside triphosphate. J. Biol. Chem. 281, 32057–32064 10.1074/jbc.M607514200 [DOI] [PubMed] [Google Scholar]
- 80.Slominska EM, Orlewska C, Yuen A, Osman L, Romaszko P, Sokolowska E et al. (2008) Metabolism of 4-pyridone-3-carboxamide-1-beta-D-ribonucleoside triphosphate and its nucleoside precursor in the erythrocytes. Nucleosides Nucleotides Nucleic Acids 27, 830–834 10.1080/15257770802146452 [DOI] [PubMed] [Google Scholar]
- 81.Romaszko P, Slominska EM and Smolenski RT (2014) Effect of 4-pyridone-3-carboxamide ribonucleoside (4PYR)-potential cardiovascular toxin in perfused rat heart. Nucleosides Nucleotides Nucleic Acids 33, 333–337 10.1080/15257770.2013.872793 [DOI] [PubMed] [Google Scholar]
- 82.Romaszko P, Slominska EM, Orlewska C, Lipinski M and Smolenski RT (2011) Metabolism of 4-pyridone-3-carboxamide-1-beta-D-ribonucleoside (4PYR) in rodent tissues and in vivo. Mol. Cell Biochem. 351, 143–148 10.1007/s11010-011-0721-9 [DOI] [PubMed] [Google Scholar]
- 83.Hanukoglu I (2017) Conservation of the enzyme-coenzyme interfaces in FAD and NADP binding adrenodoxin reductase—a ubiquitous enzyme. J. Mol. Evol. 85, 205–218 10.1007/s00239-017-9821-9 [DOI] [PubMed] [Google Scholar]
- 84.de Rosa M, Pennati A, Pandini V, Monzani E, Zanetti G and Aliverti A (2007) Enzymatic oxidation of NADP+ to its 4-oxo derivative is a side-reaction displayed only by the adrenodoxin reductase type of ferredoxin-NADP+ reductases. FEBS J. 274, 3998–4007 10.1111/].1742-4658.2007.05934.x [DOI] [PubMed] [Google Scholar]
- 85.Bossi RT, liverti A, Raimondi D, Fischer F, Zanetti G, Ferrari D et al. (2002) A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase. Biochemistry 41, 8807–8818 10.1021/bi025858a [DOI] [PubMed] [Google Scholar]
- 86.Pelikant-Malecka I, Kaniewska-Bednarczuk E, Szrok S, Sielicka A, Sledzinski M, Orlewska C et al. (2017) Metabolic pathway of 4-pyridone-3-carboxamide-1 β-D-ribonucleoside and its effects on cellular energetics. Int. J. Biochem. Cell Biol. 88, 31–43 10.1016/j.biocel.2017.03.012 [DOI] [PubMed] [Google Scholar]
- 87.Becker-Kettern J, Paczia N, Conrotte J-F, Zhu C, Fiehn O, Jung PP et al. (2018) NAD(p)HX repair deficiency causes central metabolic perturbations in yeast and human cells. FEBS J. 285, 3376–3401 10.1111/febs.14631 [DOI] [PubMed] [Google Scholar]
- 88.Marbaix AY, Tyteca D, Niehaus TD, Hanson AD, Linster CL and Van Schaftingen E (2014) Occurrence and subcellular distribution of the NADPHX repair system in mammals. Biochem. J. 460, 49–58 10.1042/BJ20131482 [DOI] [PubMed] [Google Scholar]
- 89.Hoag MR, Roman J, Beaupre BA, Silvaggi NR and Moran GR (2015) Bacterial renalase: structure and kinetics of an enzyme with 2- and 6-dihydro-beta-NAD(P) oxidase activity from Pseudomonas phaseolicola. Biochemistry 54, 3791–3802 10.1021/acs.biochem.5b00451 [DOI] [PubMed] [Google Scholar]
- 90.Oppenheimer NJ and Kaplan NO (1975) The alpha beta epimerization of reduced nicotinamide adenine dinucleotide. Arch. Biochem. Biophys. 166, 526–535 10.1016/0003-9861(75)90416-6 [DOI] [PubMed] [Google Scholar]
- 91.Klemm A, Steiner T, Cumme GA and Horn A (1993) Determination, purification, and characterization of alpha-NADH. Anal. Biochem. 212, 375–380 10.1006/abio.1993.1343 [DOI] [PubMed] [Google Scholar]
- 92.Beaupre BA, Hoag MR, Carmichael BR and Moran GR (2013) Kinetics and equilibria of the reductive and oxidative half-reactions of human renalase with alpha-NADPH. Biochemistry 52, 8929–8937 10.1021/bi401185m [DOI] [PubMed] [Google Scholar]
- 93.Moran GR and Hoag MR (2017) The enzyme: renalase. Arch. Biochem. Biophys. 632, 66–76 10.1016/jabb.2017.05.015 [DOI] [PubMed] [Google Scholar]
- 94.Trammell SA, Schmidt MS, Weidemann BJ, Redpath P, Jaksch F, Dellinger RW et al. (2016) Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun. 7, 12948 10.1038/ncomms12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dellinger RW, Santos SR, Morris M, Evans M, Alminana D, Guarente L et al. (2017) Repeat dose NRPT (nicotinamide riboside and pterostilbene) increases NAD+ levels in humans safely and sustainably: a randomized, double-blind, placebo-controlled study. NPJ Aging Mech. Dis. 3, 17 10.1038/s41514-017-0016-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martens CR, Denman BA, Mazzo MR, Armstrong ML, Reisdorph N, McQueen MB et al. (2018) Chronic nicotinamide riboside supplementation is well-tolerated and elevates NAD+ in healthy middle-aged and older adults. Nat. Commun. 9, 1286 10.1038/s41467-018-03421-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Okamoto H and Hayaishi O (1967) Flavin adenine dinucleotide requirement for kynurenine hydroxylase of rat liver mitochondria. Biochem. Biophys. Res. Commun. 29, 394–399 10.1016/0006-291X(67)90469-X [DOI] [PubMed] [Google Scholar]
- 98.Dalgliesh CE, Knox WE and Neuberger A (1951) Intermediary metabolism of tryptophan. Nature 168, 20 10.1038/168020a0 [DOI] [PubMed] [Google Scholar]
- 99.Hara N, Yamada K, Shibata T, Osago H, Hashimoto T and Tsuchiya M (2007) Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J. Biol. Chem. 282, 24574–24582 10.1074/jbc.M610357200 [DOI] [PubMed] [Google Scholar]
- 100.Rongvaux A, Shea R, Mulks M, Gigot D, Urbain J, Leo O et al. (2002) Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 32, 3225–3234 [DOI] [PubMed] [Google Scholar]
- 101.Revollo JR, Grimm AA and Imai S (2004) The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 279, 50754–50763 10.1074/jbc.M408388200 [DOI] [PubMed] [Google Scholar]
- 102.van der Veer E, Nong Z, O’Neil C, Urquhart B, Freeman D and Pickering JG (2005) Pre-B-cell colony-enhancing factor regulates NAD+-dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ. Res. 97, 25–34 10.1161/01.RES.0000173298.38808.27 [DOI] [PubMed] [Google Scholar]
- 103.Fletcher RS, Ratajczak J, Doig CL, Oakey LA, Callingham R, Da Silva Xavier G et al. (2017) Nicotinamide riboside kinases display redundancy in mediating nicotinamide mononucleotide and nicotinamide riboside metabolism in skeletal muscle cells. Mol. Metab. 6, 819–832 10.1016/j.molmet.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frederick DW, Loro E, Liu L, Davila A, Chellappa K, Silverman IM et al. (2016) Loss of NAD homeostasis leads to progressive and reversible degeneration of skeletal muscle. Cell Metab. 24, 269–282 10.1016/j.cmet.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mills KF, Yoshida S, Stein LR, Grozio A, Kubota S, Sasaki Y et al. (2016) Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 24, 795–806 10.1016/jcmet.2016.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uddin GM, Youngson NA, Doyle BM, Sinclair DA and Morris MJ (2017) Nicotinamide mononucleotide (NMN) supplementation ameliorates the impact of maternal obesity in mice: comparison with exercise. Sei. Rep. 7, 15063 10.1038/s41598-017-14866-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ratajczak J, Joffraud M, Trammell SAJ, Ras R, Canela N, Boutant M et al. (2016) NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat. Commun. 7, 13103 10.1038/ncomms13103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kornberg A (1948) The participation of inorganic pyrophosphate in the reversible enzymatic synthesis of diphosphopyridine nucleotide. J. Biol. Chem. 176, 1475 PMID: [PubMed] [Google Scholar]
- 109.Berger F, Lau C, Dahlmann M and Ziegler M (2005) Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 280, 36334–36341 10.1074/jbc.M508660200 [DOI] [PubMed] [Google Scholar]
- 110.Zerez CR, Wong MD and Tanaka KR (1990) Partial purification and properties of nicotinamide adenine dinucleotide synthetase from human erythrocytes: evidence that enzyme activity is a sensitive indicator of lead exposure. Blood 75, 1576–1582 PMID: [PubMed] [Google Scholar]
- 111.Pollak N, Niere M and Ziegler M (2007) NAD kinase levels control the NADPH concentration in human cells. J. Biol. Chem. 282, 33562–33571 10.1074/jbc.M704442200 [DOI] [PubMed] [Google Scholar]
- 112.Zhang LQ, Van Haandel L, Xiong M, Huang P, Heruth DP, Bi C et al. (2017) Metabolic and molecular insights into an essential role of nicotinamide phosphoribosyltransferase. Cell Death Dis. 8, e2705 10.1038/cddis.2017.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Trammell SA, Yu L, Redpath P, Migaud ME and Brenner C (2016) Nicotinamide riboside is a major NAD+ precursor vitamin in cow milk. J. Nutr. 146, 957–963 10.3945/jn.116.230078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ummarino S, Mozzon M, Zamporlini F, Amici A, Mazzola F, Orsomando G et al. (2017) Simultaneous quantitation of nicotinamide riboside, nicotinamide mononucleotide and nicotinamide adenine dinucleotide in milk by a novel enzyme-coupled assay. Food Chem. 221, 161–168 10.1016/j.foodchem.2016.10.032 [DOI] [PubMed] [Google Scholar]
- 115.Cooper DL, Murrell DE, Roane DS and Harirforoosh S (2015) Effects of formulation design on niacin therapeutics: mechanism of action, metabolism, and drug delivery. Int. J. Pharm. 490, 55–64 10.10167/j.ijpharm.2015.05.024 [DOI] [PubMed] [Google Scholar]
- 116.Kaplan NO and Stolzenbach FE (1957) [129] Preparation of DPN derivatives and analogs. Methods Enzymol. 3, 899–905 10.1016/S0076-6879(57)03473-4 [DOI] [Google Scholar]
- 117.Saunders PP, Tan MT, Spindler CD and Robins RK (1989) Phosphorylation of 3-deazaguanosine by nicotinamide riboside kinase in Chinese hamster ovary cells. Cancer Res. 49, 6593–6599 [PubMed] [Google Scholar]
- 118.Velasquez JEC, Green PR and Wos JA (2017) Method For Preparing Nicotinamide Riboside, Procter & Gamble Company, Cincinnati, OH, U.S.A. [Google Scholar]
- 119.Lee J, Churchil H, Choi W-B, Lynch JE, Roberts FE, Volante RP et al. (1999) A chemical synthesis of nicotinamide adenine dinucleotide (NAD+). Chem. Commun. 8, 729–730 10.1039/a809930h [DOI] [Google Scholar]
- 120.Jeck R and Woenckhaus C (1980) Simple methods for preparing nicotinamide mononucleotide and related analogs. Methods Enzymol. 66, 62–70 10.1016/0076-6879(80)66439-8 [DOI] [PubMed] [Google Scholar]
- 121.Jeck R, Heik P and Woenckhaus C (1974) Simple methods of preparing nicotinamide mononucleotide. FEBS Lett. 42, 161–164 10.1016/0014-5793(74)80776-3 [DOI] [PubMed] [Google Scholar]
- 122.Martìnez-Moñino A-B, Zapata-Pérez R, Garcia-Saura A-G, Cabanes J and Sánchez-Ferrer Á (2017) A new cross-linked enzyme aggregate biocatalyst for NAD+-booster production. RSCAdv. 7, 14272–14278 10.1039/C7RA00505A [DOI] [Google Scholar]
- 123.Haynes LJ and Todd AR (1950) 66. Codehydrogenases. Part I. The synthesis of dihydronicotinamide-D-ribofuranoside [N-D-ribofuranosidyl-1: 2(or 6)-dihydronicotinamide]. J. Chem. Soc. 303–308 10.1039/jr9500000303 [DOI] [Google Scholar]
- 124.Haynes LJ, Hughes NA, Kenner GW and Todd A (1957) 734. Codehydrogenases. Part II. A synthesis of nicotinamide nucleotide. J. Chem. Soc. 3727–3732 10.1039/JR9570003727 [DOI] [Google Scholar]
- 125.Ness RK, Diehl HW and Fletcher HG (1954) New benzoyl derivatives of D-ribofuranose and aldehydo-D-ribose. The preparation of crystalline 2,3,5-Tri-O-benzoyl-ß-D-ribose from D-Ribose1. J. Am. Chem. Soc. 76, 763–767 10.1021/ja01632a038 [DOI] [Google Scholar]
- 126.Vorbrüggen H and Ruh-Pohlenz C (2004) Synthesis of Nucleosides. Organic Reactions 10.1002/0471264180.or055.01 [DOI] [Google Scholar]
- 127.Franchetti P, Pasqualini M, Petrelli R, Ricciutelli M, Vita P and Cappellacci L (2004) Stereoselective synthesis of nicotinamide β-riboside and nucleoside analogs. Bioorg. Med. Chem. Lett. 14, 4655–4658 10.1016/j.bmcl.2004.06.093 [DOI] [PubMed] [Google Scholar]
- 128.Tanimori S, Ohta T and Kirihata M (2002) An efficient chemical synthesis of nicotinamide riboside (NAR) and analogues. Bioorg. Med. Chem. Lett. 12, 1135–1137 10.1016/S0960-894X(02)00125-7 [DOI] [PubMed] [Google Scholar]
- 129.Yang T, Chan NY and Sauve AA (2007) Syntheses of nicotinamide riboside and derivatives: effective agents for increasing nicotinamide adenine dinucleotide concentrations in mammalian cells. J. Med. Chem. 50, 6458–6461 10.1021/jm701001c [DOI] [PubMed] [Google Scholar]
- 130.Zhang N and Sauve AA (2017) Synthesis of β-nicotinamide riboside using an efficient two-step methodology. Curr. Protoc. Nucleic Acid Chem. 71, 14.14.1–14.14.9 10.1002/cpnc.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Szcepankiewicz BPF, Koppetsch K and Perni RB (2015) Nicotinamide riboside analogs and pharmaceutical compositions and uses thereof. U.S. Pat. WO/2015/186114
- 132.Fouquerel E, Goellner EM, Yu Z, Gagné J-P, Barbi de Moura M, Feinstein T et al. (2014) ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 8, 1819–1831 10.1016/jcelrep.2014.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marie Migaud PR, Crossey K and Doherty M (2016) Methods of preparing nicotinamide riboside and derivatives thereof U.S. Pat. 20160168184
- 134.Yang ASFSMY (2016) Syntheses, activities and methods of use of dihydronicotinamide riboside derivatives in PCT 15/744,602
- 135.Dellinger RM, Marie E, Redpath P, Rhonemus T and Cunningham R (2016) Nicotinic acid riboside or nicotinamide riboside composition, reduced derivatives thereof, and use thereof. U.S. Pat. PCT/US2016/022682
- 136.Normington KDS, David A, Livingston D, McKearin JM, Szczepankiewicz B and Kremsky JN (2017) Nicotinamide mononucleotide derivatives and their uses in WO/2017/024255
- 137.Eisner U and Kuthan J (1972) Chemistry of dihydropyridines. Chem. Rev. 72, 1–42 10.1021/cr60275a001 [DOI] [Google Scholar]
- 138.Stout DM and Meyers AI (1982) Recent advances in the chemistry of dihydropyridines. Chem. Rev. 82, 223–243 10.1021/cr00048a004 [DOI] [Google Scholar]
- 139.Silva EMP, Varandas PAMM and Silva AMS (2013) Developments in the synthesis of 1,2-dihydropyridines. Synthesis 45, 3053–3089 10.1055/s-0033-1338537 [DOI] [Google Scholar]
- 140.Sharma VK and Singh SK (2017) Synthesis, utility and medicinal importance of 1,2- & 1,4-dihydropyridines. RSCAdv. 7, 2682–2732 10.1039/C6RA24823C [DOI] [Google Scholar]
- 141.Dutta SP, Crain PF, McCloskey JA and Chheda GB (1979) Isolation and characterization of 1-β-D-ribofuranosylpyridin-4-one-3-carboxamide from human urine. LifeSci. 24, 1381–1388 10.1016/0024-3205(79)90008-0 [DOI] [PubMed] [Google Scholar]
- 142.Lang R, Wahl A, Skurk T, Yagar EF, Schmiech L, Eggers R et al. (2010) Development of a hydrophilic liquid interaction chromatography-high- performance liquid chromatography-tandem mass spectrometry based stable isotope dilution analysis and pharmacokinetic studies on bioactive pyridines in human plasma and urine after coffee consumption. Anal. Chem. 82,1486–1497 10.1021/ac902616k [DOI] [PubMed] [Google Scholar]
- 143.Lappin G (2015) A historical perspective on radioisotopic tracers in metabolism and biochemistry. Bioanalysis 7, 531–540 10.4155/bio.14.286 [DOI] [PubMed] [Google Scholar]
- 144.Kim I-Y, Suh S-H, Lee I-K and Wolfe RR (2016) Applications of stable, nonradioactive isotope tracers in in vivo human metabolic research. Exp. Mol. Med. 48, e203 10.1038/emm.2015.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jang C, Chen L and Rabinowitz JD (2018) Metabolomics and isotope tracing. Cell 173, 822–837 10.1016/j.cell.2018.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vella A and Rizza RA (2009) Application of isotopic techniques using constant specific activity or enrichment to the study of carbohydrate metabolism. Diabetes 58, 2168–2174 10.2337/db09-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Satyanarayana U and Rao BS (1983) In vivo conversion of tryptophan to nicotinic acid in rats studied by simultaneous incorporation of [3H]-tryptophan and [14C]-nicotinic acid into liver NAD and NADP. Ann. Nutr. Metab. 27, 1–7 10.1159/000176617 [DOI] [PubMed] [Google Scholar]
- 148.Sen A, Stojkovic V and Kohen A (2012) Synthesis of radiolabeled nicotinamide cofactors from labeled pyridines: versatile probes for enzyme kinetics. Anal Biochem. 430, 123–129 10.1016/j.ab.2012.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ueda K, Yamamura H and Nishizuka Y (1971) [106] Preparation of labeled pyridine ribonucleotides and ribonucleosides. Methods Enzymol 18, 55–60 10.1016/S0076-6879(71)18063-9 [DOI] [Google Scholar]
- 150.Friedmann HC (1971) [105] Preparation of DPN+ and NMN labeled with14C in the pyridine moiety. Methods Enzymol 18, 51–55 10.1016/S0076-6879(71)18062-7 [DOI] [Google Scholar]
- 151.Williams TJ, Zens AP, Wisowaty JC, Fisher RR, Dunlap RB, Bryson TA et al. (1976) Nuclear magnetic resonance studies on pyridine dinucleotides. The pH dependence of the carbon-13 nuclear magnetic resonance of NAD+ analogs. Arch. Biochem. Biophys. 172, 490–501 10.1016/0003-9861(76)90102-8 [DOI] [PubMed] [Google Scholar]
- 152.Oppenheimer NJ and Davidson RM (1980) 15N nuclear magnetic resonance studies of [1–15N]nicotinamide adenine dinucleotides. Org. Magn Res. 13, 14–16 10.1002/mrc.1270130104 [DOI] [Google Scholar]
- 153.Ueda K and Yamamura H (1971) [107] Preparation of various labeled NAD’s. Methods Enzymol 18, 60–67 10.1016/S0076-6879(71)18064-0 [DOI] [Google Scholar]
- 154.Colowick SP and Kaplan NO (1957) [34] Preparation and analysis of labeled coenzymes. Methods Enzymol 4, 840–855 10.1016/0076-6879(57)04082-3 [DOI] [Google Scholar]
- 155.Markham KA, Sikorski RS and Kohen A (2004) Synthesis and utility of 14C-labeled nicotinamide cofactors. Anal. Biochem. 325, 62–67 10.1016/j.ab.2003.10.027 [DOI] [PubMed] [Google Scholar]
- 156.Kasăsarov LB and Moat AG (1980) [18] Convenient method for enzymic synthesis of [14C]nicotinamide riboside. Methods Enzymol 66, 120–122 10.1016/0076-6879(80)66448-9 [DOI] [PubMed] [Google Scholar]
- 157.Cen Y and Sauve AA (2010) Transition state of ADP-ribosylation of acetyllysine catalyzed by Archaeoglobus fulgidus Sir2 determined by kinetic isotope effects and computational approaches. J. Am. Chem. Soc. 132, 12286–12298 10.1021/ja910342d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bull HG, Ferraz JP, Cordes EH, Ribbi A and Apitz-Castro R (1978) Concerning the mechanism of the enzymatic and nonenzymatic hydrolysis of nicotinamide nucleotide coenzymes. J. Biol Chem. 253, 5186–5192 PMID: [PubMed] [Google Scholar]
- 159.Rising KA and Schramm VL (1997) Transition state analysis of NAD+ hydrolysis by the cholera toxin catalytic subunit. J. Am. Chem. Soc. 119, 27–37 10.1021/ja9621915 [DOI] [Google Scholar]
- 160.Berti PJ, Blanke SR and Schramm VL (1997) Transition state structure for the hydrolysis of NAD+ catalyzed by diphtheria toxin. J. Am. Chem. Soc. 119, 12079–12088 10.1021/ja971317a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rising KA and Schramm VL (1994) Enzymic synthesis of NAD+ with the specific incorporation of atomic labels. J. Am. Chem. Soc. 116, 6531–6536 10.1021/ja00094a006 [DOI] [Google Scholar]
- 162.Scheuring J, Berti PJ and Schramm VL (1998) Transition-state structure for the ADP-ribosylation of recombinant Giα1 subunits by pertussis toxin. Biochemistry 37, 2748–2758 10.1021/bi972594x [DOI] [PubMed] [Google Scholar]
- 163.Tran A, Yokose R and Cen Y (2018) Chemo-enzymatic synthesis of isotopically labeled nicotinamide riboside. Org. Biomol. Chem. 16, 3662–3671 10.1039/C80B00552D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lu W, Wang L, Chen L, Hui S and Rabinowitz JD (2018) Extraction and quantitation of nicotinamide adenine dinucleotide redox cofactors. Antioxid. Redox Signal. 28, 167–179 10.1089/ars.2017.7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yang T and Sauve AA (2006) NAD metabolism and sirtuins: metabolic regulation of protein deacetylation in stress and toxicity. AAPS J. 8, E632–E643 10.1208/aapsj080472 [DOI] [PMC free article] [PubMed] [Google Scholar]


