Abstract
Inclusion of biospecimens in population-based studies is an integral part of understanding disease etiology, identifying biomarkers and developing prevention and treatment strategies. The Prostate, Lung, Colorectal, and Ovarian (PLCO) cancer screening trial collected, processed and stored biospecimens from participants to create a biorepository of specimens which serves as a useful resource for a broad research community. PLCO collected blood samples from consented screening arm participants at six screening rounds and a buccal sample from consented control arm participants. In addition, formalin-fixed paraffin embedded tumor tissue specimens were collected for participants in both arms for selected cancer sites. Collection of biospecimens at multiple timepoints (i.e. serial samples) and prior to cancer diagnosis, paired with rich epidemiologic and screening data, makes the PLCO collection of biospecimens a uniquely valuable resource. As such, access to the PLCO biorepository is granted to investigators by a rigorous scientific review process and guided by a steering committee which is responsible for developing and implementing the biospecimen use policies. Here, we describe the procedures for biospecimen collection, processing, storage, requisition, and distribution, as well as data management employed in PLCO. We also provide examples of how the biospecimens have been used to advance cancer research and describe relevant lessons learned to help inform cohorts wishing to add or modify biospecimen collection.
Keywords: Biorepository, biospecimen, blood, early marker, etiologic, PLCO, tissue
1. INTRODUCTION
Inclusion of biospecimens in population-based studies is becoming an integral part of creating a research resource for understanding disease etiology, identifying early detection biomarkers and developing prevention and treatment strategies [1]. In 1992, the National Cancer Institute (NCI) approved the initiative to add a biorepository component to the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening trial to help meet the NCI’s research priorities in molecular epidemiology and early cancer detection [2, 3]. Since this component was added to the PLCO trial, the research community (even beyond PLCO investigators) has found the PLCO biorepository to be an extremely valuable resource for investigating cancer etiology and early markers [3, 4].
Study participants were randomized into two arms, with screening-arm participants receiving a series of periodic examinations for prostate, lung, colorectal, and ovarian cancer, while participants in the control arm received usual care from their health providers. All participants completed a baseline questionnaire (BQ) and screening-arm participants also completed a dietary questionnaire (DQX) at baseline. Beginning in 1998, a dietary history questionnaire (DHQ) was administered to all participants, at baseline for the non-screening arm and approximately 3 years after baseline for the screening arm. The active screening component of the trial was concluded in 2006.
At the baseline screening visit (T0) and each annual screening visit thereafter (T1-T5), intervention arm participants (n ~ 77,500) were asked to provide blood samples (Fig. 1) [5]. Approximately 85% of intervention arm participants provided at least one biospecimen to be used for future research purposes. This resulted in a large repository of serial specimens from participants prior to cancer diagnosis. Several years later, a one-time collection of buccal cells from control arm participants (n ~ 77,500) was added to the study. Approximately 91% of control arm participants who consented to provide buccal cells (i.e. 70% of all control arm participants) returned specimens for future research purposes. The PLCO biospecimen repository was further supplemented through an on-going collection effort initiated in 2006 whereby formalin-fixed paraffin embedded (FFPE) pathology tissue samples from PLCO participants who developed a selected cancer were obtained. Finally, in 2007, residual serum samples that remained after testing for CA-125 and prostate specific antigen (PSA) were added to the PLCO biospecimen repository.
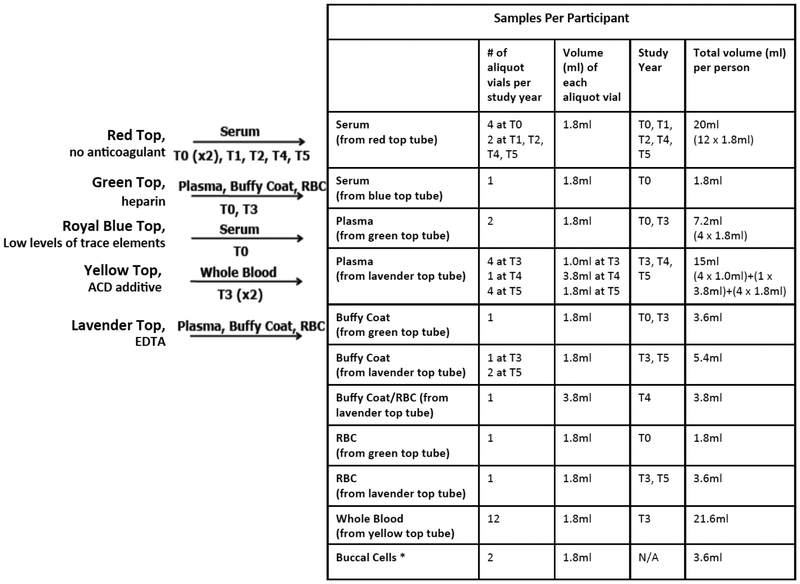
Fig. (1).
Specimens collected from PLCO participants. Listed in the figure and table are the maximum number of samples and volumes per participant. Blood specimens were only collected from screening arm participants. *Buccal cell samples were only collected from control arm participants.
Collection of biospecimens at multiple timepoints (i.e. serial samples) and prior to cancer diagnosis, paired with the rich epidemiologic and screening data, make the PLCO collection of biospecimens a uniquely valuable resource. As such, access to the PLCO biorepository is granted to investigators after undergoing a rigorous scientific review process and guided by a steering committee which developed well-thought-out biospecimen governance policies. Here, we describe the procedures for biospecimen collection, processing, storage, requisition, and distribution, and data management implemented in the PLCO. We also provide examples of how the biospecimens have been used to advance cancer research and describe relevant lessons learned to help inform cohorts wishing to add or modify biospecimen collection.
2. BIOSPECIMEN COLLECTION AND PROCESSING
2.1. Blood Repository: Collection, Processing, and Storage
Serum from one 10ml red top blood tube was collected at each of the 6 screening visits and used for PSA and CA-125II screening tests, for prostate and ovarian cancers, respectively. Additional blood (up to 43ml) was collected at each visit for future research. The additional blood was processed either on site at the screening centers or at a designated processing laboratory, and stored at either −70°C or at −157°C (vapor phase liquid nitrogen); details of the blood processing protocols are available (see reference [5]). As these specimens were collected for broad scientific research purposes and without a specific predefined usage, a range of biospecimen types were collected (Fig. 1) and processed using standardized protocols across the 10 screening sites in the United States (Birmingham AL, Denver CO, Detroit MI, Honolulu HI, Marshfield WI, Minneapolis MN, Pittsburgh PA, Salt Lake City UT, St Louis MO, and Washington DC). Participants were not requested to fast prior to blood draw. While the decision not to require fasting may limit some future uses of the specimens, it was made out of practicality. The trial was approved by the institutional review boards of the National Cancer Institute and the 10 screening centers. All participants signed a consent form to participate in the trial, and only specimens from participants who signed an additional etiologic studies consent form to contribute their biospecimens for research in cancer and/or other diseases are eligible for inclusion in research studies.
Standard operation procedures (SOPs) for biospecimen collection and handling were implemented, quality control (QC) steps were included, and collection and processing details were documented. SOPs were updated as needed and PLCO screening centers, Frederick National Laboratory (central processing laboratory), and biorepository site staff were educated about changes to the SOPs. Common preanalytic variables (see Table 1) were documented and recorded (e.g., time between blood draw and freezer) in the PLCO study management system and then added to the biospecimen inventory system.
Table 1.
Preanalytic variables documented for PLCO specimens.
| Vacutainer tube type (and additive) Glass/plastic tube |
| Volume |
| Freeze thaw cycles |
| Hemolyzed |
| samples Icteric |
| samples Lipemic |
| samples |
| Uncontrolled thaw |
| Deviations from SOPs (e.g. blood drawn in incorrect tube at a specific screening visit) |
| Biospecimen receipt anomalies (e.g. upon receipt at the repository, were the samples thawed when they should have been frozen?) |
After collection, the red top (with only clot activator in the vacutainer tube) and royal blue top (with low levels of trace-elements in the vacutainer tube) serum tubes were allowed to clot at room temperature for one hour prior to centrifugation (see Fig. 2). The green top (with heparin additive in the vacutainer tube) and lavender top (with EDTA additive in the vacutainer tube) tubes were refrigerated until they were centrifuged. The specimens were processed and frozen within two hours of the blood draw. Tubes for all study years, also referred to as T0-T5 (e.g. T0 corresponds to baseline and T5 study year 6), except for the yellow top tubes collected at T3, were centrifuged at either 1200 × g for 15 minutes or 3900 × g for 6 minutes, and then aliquoted into cryovials of serum, plasma, buffy coat, RBC, and RBC and buffy coat. After processing at the screening sites, the frozen cryovials were shipped to the PLCO biorepository located in Frederick, Maryland. The two yellow top tubes (with acid citrate dextrose (ACD) additive in the vacutainer tube) collected at T3 were shipped on the same day at ambient temperature to a central facility for processing into cryopreserved whole blood [6].
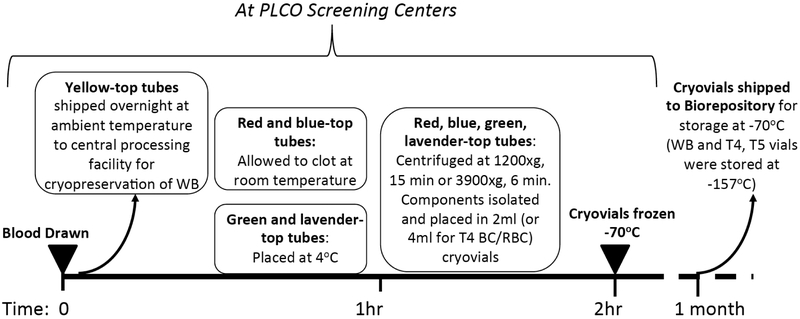
Fig. (2).
Approximate timeline for processing and handling of blood collected from participants in the randomized arm of the PLCO Trial. Blood was drawn according to the protocols at the PLCO screening centers. Yellow top tubes were drawn at the T3 visits only. For the red, blue-top, green, and lavender top tubes, the time between blood draw through to centrifugation, isolation of components, aliquoting into cryovials and placing the cryovials in the freezer was 2 hours. Within a month after freezing the cryovials, they were shipped to the biorepository for long term storage. Cryopreserved whole blood was also shipped to the biorepository for long term storage.
Prior to shipment, the specimens were stored in −70°C freezers at the screening centers. Specimens were shipped at least monthly, or more frequently if necessary depending on the screening center’s volume, to the biorepository. Specimen shipments were made on dry ice in full compliance with all IATA regulations [7]. At the biorepository, specimens from the T0-T3 study years, except for the whole blood specimens collected at T3, were stored in −70°C freezers. The T3 whole blood specimens and specimens collected at T4 and T5 were stored in liquid nitrogen freezers at −157°C. Freezer temperature monitoring and power/liquid nitrogen backups were implemented at the repository.
2.2. Additional Sample Collection: Buccal Cells, Tumor Tissue, and Others
The main PLCO repository, which stores the blood-based specimens from intervention arm participants, has been expanded in three waves: 1) buccal cells were collected from control arm participants; 2) formalin-fixed, paraffin-embedded (FFPE) tissues were collected, from which tissue microarrays (TMAs) were created and other tissue types were retained for selected cancer sites; and 3) residual serums from CA-125II and PSA testing were added to the repository.
The buccal cell collection began in 2000, as a supplement to the main PLCO Screening Trial biorepository. These cells, lining the inner cheek of the human mouth, are easily collected and have proven to be a good source of DNA. All control arm study participants who were willing to sign the etiologic studies consent form were invited to participate, starting with those previously enrolled into PLCO and continuing with new enrollees until enrollment closed in 2001. Consents were obtained either at randomization (n=over 32,000) or by mail (n=over 26,000) for a total of 58,000 (77%) control arm participants.
Eligible buccal cell study participants supplied a single saliva sample at home using a collection kit sent by mail. This kit contained all the materials required to collect and return the sample to the biorepository for processing. Participants collected buccal cells simply by rinsing with the provided mouthwash for 45 seconds and then spitting the mouthwash into the provided sample vial. The filled vial was then returned by mail for processing. More than 51,000 (91%) consented control arm participants (i.e. 70% of all control arm participants) returned their vials for processing. Buccal cell specimens are stored as washed cell suspensions in Tris-EDTA buffer at −70°C.
TMAs were created and additional tissue cores obtained for a number of cancer sites including colorectal, prostate, lung, ovarian, breast, and bladder cancers. In order to create the TMAs, FFPE blocks were obtained from pathology labs and shipped to collaborating pathologists for tissue annotation, and then to tissue processing facilities for microarray construction and additional tissue cores collection.
The laboratory that conducted CA-125II and PSA testing for PLCO was located at the University of California, Los Angeles (UCLA). The laboratory saved residual serum from CA-125II and PSA tests, which were stored in −80°C freezers. Because these samples have undergone one or more freeze- thaw cycles (for CA125II or PSA testing and in some cases, re-testing), they were not initially intended to be part of the PLCO biorepository. However, because of the sheer volume of the samples (~330,000 vials), and also the realization that certain analytes are unaffected by freeze- thaw cycles, in 2007, the NCI made the decision to transfer all residual serum samples to the main repository in Frederick, MD. Ten upright −80°C freezers containing the residual serum samples were transported from UCLA to Frederick, Maryland in a commercial truck operated by Thermo Scientific. The “UCLA” collection provided one complete set of serial serum samples per participant, including the only T3 serum available in the trial.
2.3. Lessons Learned about Collection and Processing of Specimens
The main lesson learned was that the decision to collect serial samples has proven to be extremely useful for the research community [8]. As one of the few sources of serial, pre-diagnostic blood specimens, PLCO biorepository offers a much sought-after resource for studies wishing to assess the stability of analytes over time or the detection of biomarkers in distant (furthest from cancer diagnosis) vs proximate samples (closest to cancer diagnosis) for cancer etiology and early detection [4].
A second lesson learned was that implementing an SOP for each of the operational processes is a critical part of establishing and maintaining a high quality PLCO biorepository. SOPs for the initial sample collection, processing, shipping and storage, as well as downstream sample processing procedures (e.g. DNA extraction, serum or plasma aliquoting) are extremely important. Variability in sample processing and handling procedures could often impact subsequent marker measurements. Having a detailed process-oriented SOP also allows a root cause to be readily identified when problems arise. For example, PLCO shipping SOPs stipulate that shipping schedules be established and communicated with the labs and the repository prior to sample shipments. Complications with shipments tended to arise when such procedures were not followed.
A third lesson learned was that flexibility and planning are required when building a specimen biorepository. One should carefully consider, to the extent possible, what materials would be most useful for future research when designing the biospecimen collection. However, despite careful consideration in the sample collection stage, it is inevitable that advances in research fields and technologies may change the interest and demand for certain types of specimens. Some of the PLCO specimens more commonly used include serum (particularly T0 serum), buffy coat for DNA, and buccal cells for DNA. Less commonly used specimens include plasma (particularly T3 heparin and T3 EDTA) and RBC (red blood cells). Flexibility and foresight are also needed when merging biospecimen collections into the main repository. For example, it is important to consider how the aliquotted (child) samples should be integrated, potential uses of these samples (i.e. what sample volumes will be in potential high demand, how often will they be accessed, etc.) and costs needed for storing these additional samples.
Additional lessons learned involved management of the biorepository. Once the samples were in the repository, periodic quality assessments of the samples took place - for both the parent and child (i.e. aliquot) vials. Additionally, PLCO realized that they needed to carefully consider informed consent for the participants when providing specimens to researchers. For example, in April 2003, during annual IRB review of the PLCO protocols, the Hawaii IRB stipulated that “these samples will not be used for developing commercial products” in its approval letter. In order to ensure that this restriction is upheld, samples from PLCO participants providing consent at Hawaii from April 1, 2003 onward were excluded from biospecimen studies. Only a small number of samples were affected by this new restriction, incurring minimal impact on PLCO sample availability for research.
Another lesson learned was that programmatic and logistic coordination is beneficial when multiple studies addressing related scientific questions are being conducted. For example, studies focusing on the same cancer might benefit from the same subject/specimen sampling plan and a common, core dataset. Use of a common sample and data set facilitates direct comparisons and integration of data across studies. In addition, it is often necessary to coordinate among multiple studies so that the samples can be aliquoted at the same time, thereby minimizing freeze-thaw cycles, loss of sample volumes due to aliquoting, and labor costs.
3. BIOSPECIMEN USE AND MANAGEMENT
3.1. Accessing the PLCO Biospecimens
The PLCO Etiology and Early Marker Study (EEMS) program was created to allow researchers access to specimens and associated data from the trial. Since 2005, the EEMS program has extended its call for proposals to researchers, beyond PLCO investigators, in the general scientific community (2). The PLCO biorepository currently stores approximately 2.9 million biologic specimens collected from PLCO participants. These specimens and their associated data are available to all qualified researchers through a peer-review process. Investigators must submit an online proposal application to EEMS in order to initiate a request to access to the resource [9]. The scientific utilization of the PLCO biorepository is actively managed by intramural and extramural PLCO project leaders and scientific staff with the goal of maximizing the scientific potential of the resource. Information about the review process, review criteria, and application forms can be found at http://www.plcostars.com and https://biometry.nci.nih.gov/cdas/studies/plco.
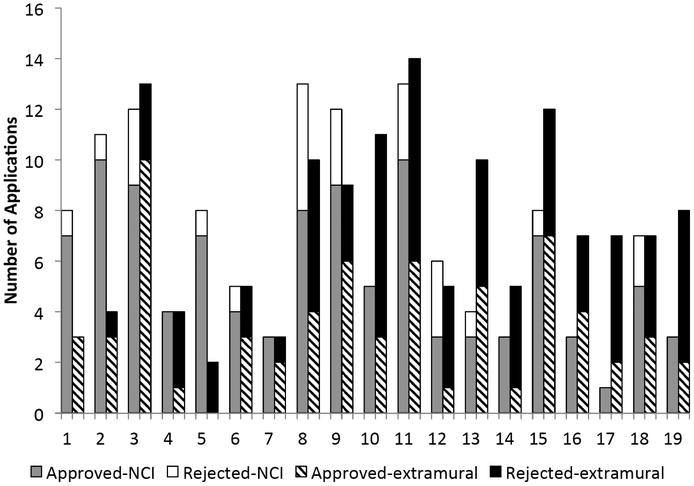
As of the end of 2014, a total of 259 EEMS applications requesting PLCO biospecimens were received. Of these, 127 were from NCI and 132 were from extramural investigators. The overall approval rate is about 63% (Fig. 3). Specific review criteria used in evaluating proposal application are listed in the PLCO EEMS policies & Procedures document, which posted online at www.plcostars.com. Some reasons for denying requests have included: lack of substantial preliminary data, lack of sufficient justification for using PLCO samples, lack of appropriate study design or statistical analysis plan. Approval rate for NCI intramural applications has historically been higher than that of extramural applications, perhaps in part due to institutional knowledge, research experience/coordination in the PLCO, dedicated laboratory facilities, and relatively stable funding [4].
Fig. (3).
EEMS Panel Review Results by Round. Two rounds of EEMS were held each year (one in the winter, and one in the summer). For each round, two bars are shown on the graph. The first bar shows the number of applications from NCI investigators that were ultimately approved (gray) or not approved (white) after EEMS review. The second bar in set shows the number of applications from non-NCI investigators (i.e. extramural) that were ultimately approved (striped) or not approved (black) after EEMS review.
Additionally, an NIH funding program (PAR-13-036) was initiated in December 2012 to facilitate the use of the PLCO biospecimens by extramural investigators. This program allows simultaneous application for both the specimens and funding to conduct studies using the specimens. A total of 7 applications have been funded to date through this mechanism.
3.2. Scientific Use of the PLCO Biospecimens
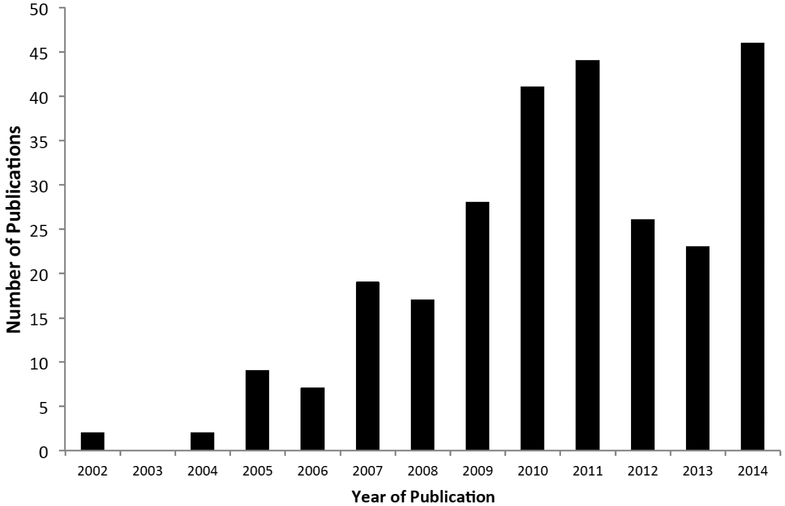
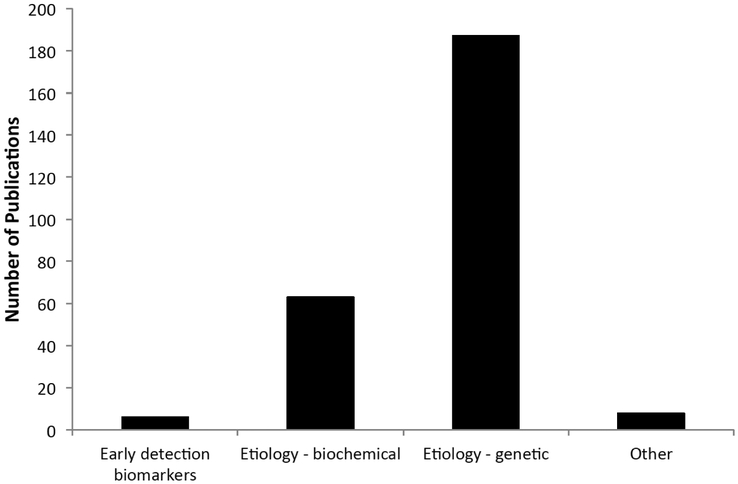
PLCO biospecimens have been used for a wide range of cancer research [4]. We have conducted an updated publication analysis that covers the period from 2002 to 2014. Approximately 270 research publications resulted from research using PLCO biospecimens during the period between 2002 and 2014 (Fig. 4). The majority were genetic (n=192) and serologic (n=63) epidemiology studies (Fig. 5). At the lower end of the spectrum are publications involving PLCO biospecimens for early detection biomarker studies (n=6). The small number of early detection biomarker studies may be a reflection of the higher bar set by EEMS for approving such applications, and/or the scarcity of early detection biomarkers with performance characteristics high enough to justify validation in PLCO (detailed below).
Fig. (4).
PLCO EEMS publications by year of publication. The number of publications reporting results generated from analyses using PLCO specimens is shown here.
Fig. (5).
PLCO EEMS publications by study type. Broad categories of research questions examined using PLCO specimens and reported in publications are shown here.
Genome-wide association studies (GWAS), including meta-analyses and secondary value-added analyses on outcomes different from the original GWAS, make up more than half of all genetic epidemiological studies. Currently, GWAS data have been generated for a large number of cancer sites in the PLCO, including prostate, breast, lung, colorectum, ovary, kidney, bladder, pancreas, endometrium, glioma and non-Hodgkin Lymphoma (chronic lymphocytic leukemia, follicular lymphoma, large B cell lymphoma) [10-20]. Several GWAS follow-up studies, such as fine mapping and resequencing, have been done extensively around several chromosome regions associated with prostate cancer (e.g. 8q24 and 19q13.33) [21, 22]. Secondary GWAS value-added analyses, facilitated by access to the existing GWAS data through the NIH database Genotypes and Phenotypes (dbGaP), proliferated in recent years. These studies often combined data from multiple independent GWAS to increase statistical power in meta-analyses or pooled analyses, leading to the identification of novel genetic markers associated with cancer risk [12, 23, 24] and allowing for assessment of gene-environment interactions in the risk associations [24, 25]. The PLCO GWAS data have also been used to assess polygenic modeling across cancers [26, 27], as well as for estimating cancer heritability [28]. GWAS with PLCO samples have also been conducted for non-cancer outcomes including height and body mass index [29, 30], smoking behavior [31], vitamin D and its binding protein [32, 33], caffeine consumption [34], serum selenium [35] and telomere length [36].
Other genetic epidemiologic studies have included candidate gene studies. Such studies focused on genes involved in DNA repair pathway, immunity, inflammation, metabolism of carcinogens, and nutrient (e.g. Vitamin D) or hormone regulation [37-47]. Epigenetic studies, telomere length and mitochondrial DNA copy number analyses have also been conducted in PLCO for a number of cancers [48-53].
In addition to DNA analyses, serologic markers and viral antibodies also have been associated with cancer risk [54-59]. Further advancement in multiplex detection technologies have made it possible to simultaneously measure a large number of markers with a limited sample volume. Multiplexed inflammation and immunity markers and metabolomics are among the examples [60-63]. Cryopreserved whole blood samples also have been utilized to study early disease markers and transformed to cell lines for studying DNA damage and repair markers [64, 65].
PLCO biospecimens have contributed to a large number of research projects that resulted in a voluminous body of research publications. However, there is also a glaring gap: Only 6 out of 270 research publications using PLCO specimens were on early detection biomarker studies.
Traditionally, biomarker discovery and development follows a phased approach analogous to the drug development process, as outlined in the landmark paper by Pepe et al. [66]. The consensus among NCI PLCO leadership has been that PLCO resource would be most suitable for Phase III validation studies. As such, the low number of early detection biomarker studies likely reflected the lack of biomarkers that merit a Phase III validation study.
The PLCO EEMS steering committee has considered a modified policy to help move the field forward. PLCO has made an explicit policy to support Phase II as well as laboratory discovery studies when the preliminary data and the overall study design are compellingly strong. Additionally, an active sample management strategy has been put in place to conserve the samples that are most valuable for early detection biomarker research. For example, pre-diagnostic serum and plasma samples that are within two years of diagnosis are specifically allocated for early detection biomarker studies, while more distant (to diagnosis) samples are allocated for etiologic studies.
3.3. Sample Depletion
Currently, only a small portion of the PLCO biorepository, about 5% of all samples and 13% of samples from cancer patients, has been depleted. However, sample depletion rates for prostate, lung, ovarian, colorectal and breast cancer patients have been higher than those of other cancer sites, due to higher number of studies focusing on these cancers. Additionally, certain sample types have been depleted more than others. The most depleted sample type is buccal cells; more than 50% of the buccal cell source vials for cancer patients have been accessed (with returned DNA samples stored in the repository). The demand for these samples has been high. Every GWAS involving PLCO samples has used buccal cell DNA for the control arm cases, and in recent years, buccal cells have been used and requested for microbiome studies.
Buffy coat is the next most depleted sample type. 13% of all buffy coat samples and 30% of the buffy coat samples from cancer cases have been accessed (with returned DNA stored in the repository). Requests for buffy coat (or buffy coat DNA) specimens came from the many genetic studies conducted in PLCO, as described in the previous section.
Serum is the third most depleted sample type. More than 8% of all pre-diagnostic serum samples and 30% of the baseline pre-diagnostic serum samples from cancer cases have been accessed, resulting in residual aliquots of these serum samples stored in the repository, reflecting an overall 4% and 16% sample volume depletion, respectively. Requests for serum may in part reflect the fact that assays of circulating biomarkers have traditionally been developed using serum samples.
3.4. Biospecimen Data Management
Rigorous data management procedures were essential to maintaining high quality PLCO biospecimens. PLCO maintained a detailed inventory of the biospecimens beginning at collection, through shipping, processing, storage, and final deposition. This included tracking the number of freeze-thaw cycles for a sample, the SOPs for each processing step, residual volume of specimens, samples from coordinated study sets of subjects (such as for the Cancer Genetic Markers of Susceptibility (CGEMS) Project), and other preanalytic variables (Table 1). As requests for access to the biospecimens increased, detailed inventory updates on a routine basis were vital.
However, real-time (or close to real-time) accounting of PLCO biospecimens was difficult to accomplish in the past. As an alternative to real-time updates to the PLCO inventory, the PLCO coordinating center, analysis group, repository, and laboratory established routine communications. If questions arose about the availability, quality, or quantity of a biospecimen, the appropriate group was contacted.
3.5. Lessons Learned About Providing Biospecimens to Researchers and Managing Biospecimen Data
Over the course of EEMS, lessons have been learned and new policies and procedures have been implemented regarding the provision of PLCO biospecimens to investigators for a wide range of research uses. Here, we describe some of the main lessons that might be of value for future studies.
PLCO learned that it is important for potential investigators to familiarize themselves with the trial design and the various types of biospecimens and data available. Doing so allows investigators to design well-thought out studies that capitalized upon the PLCO resource. Although not a requirement, PLCO recommended that potential investigators contact a PLCO investigator or a relevant PLCO subcommittee to vet the proposed study before submitting a full application. This subcommittee structure has since been discontinued after the main trial outcome data was published. Today, much of the trial-related information as well as trial data have been made available on the above-mentioned PLCO websites, making it possible for anyone to gather enough relevant information to propose a well-designed new study. Biospecimen availability tables and an interactive querying tool are also available online to facilitate the assessment of what PLCO data and/or specimens are potentially available. Additionally, to avoid duplication of effort and repeated use of material resources, current and past research activities in PLCO are viewable on the public websites, and the PLCO management team maintains a searchable database to help inform potential duplication of projects in the proposal approval process.
As the number of studies in PLCO increased, it became apparent that study-generated laboratory data could be reused for other studies. Adding such data to the PLCO databases could facilitate data sharing and preserve PLCO samples from being used unnecessarily. PLCO established post approval processes and policies that require return of laboratory (or biomarker) results to the main PLCO database. Several curated datasets, including serum levels of a set of 28 ovarian cancer biomarkers, are posted on the website (https://biometry.nci.nih.gov/cdas/studies/plco/). However, there have been several challenges in integrating laboratory data into the PLCO database. Primarily, PLCO realized that standard guidelines need to be developed so that when study data are submitted, the data are in the correct format and include enough information to allow accurate interpretation of the data (e.g. the proper units of measurements and the assays used to generate the measurements). Additionally, a system to store the returned data and link them with subjects is also necessary.
Another problem that surfaced over the years is that many of the approved studies were never completed. This situation happened more often with the extramural studies, likely due to lack of funding. Inactive studies could prevent other able investigators from examining the same research question. To prevent lagging studies and maximize the utility of the PLCO resource, the PLCO steering committee instituted sun-setting polices, where projects that have not requisitioned samples within 3 years post approval are considered expired. In addition, PLCO required investigators with an approved study to provide annual progress reports.
Biospecimen Use Guidelines were established to help maintain critical levels of samples for potential future research [67]. Such guidelines helped guide the repository, laboratories, and others responsible for managing the biospecimen resource. Included in the guidelines document are requirements for the minimal amount of each type of biospecimen to remain in the repository after selecting vials for a study. When multiple materials are available for a subject (such as DNA source materials: T0, T3, and T5 buffy coat, T4 RBC/buffy coat, and T3 whole blood), the guidelines provide information on a pre-determined selection order of the vials. PLCO guidelines for the use of serum or plasma indicate that in general, serum or plasma samples that are close to cancer diagnosis (less than one year from the date of diagnosis) should be reserved for early detection biomarker studies, while more distal samples should be reserved for etiologic studies. Rules about using parent vials versus child vials (i.e. a parent vial is originally aliquotted from the blood collection tube; a child vial is an aliquot from that parent tube, and has thus undergone more freeze-thaw cycles) are also stipulated in the guidelines document. If a child vial of serum or plasma from a subject selected for a study exists, that child vial will be preferentially used unless the investigator justifies why a parent vial is required. Additionally, the PLCO guidelines stipulate how special sets of samples (e.g. specimens from participants with rare and aggressive cancers, serial samples, residual serum samples from the “UCLA” collection, etc.) may be used. For early marker validation studies, investigators will be asked to test their markers using pre-diagnostic samples most proximate to diagnosis. Serial samples may be granted if a marker has been validated with justifiable data in the most proximate samples. Additional consideration is given to requests for UCLA serum samples in order to preserve sets of serial samples that included all six time points, since that set of samples is the only source of serum collected at T3.
Samples that can be used for quality control (QC) purposes have proven to be essential for pilot work as well as assay development and refinement. PLCO has dedicated samples that were collected in the same manner, according to the same SOPs as other PLCO specimens, but do not have outcome data for QC and assay development purposes. In addition, PLCO allows the use of samples from the main collection to be used for QC purposes if certain criteria outlined in the Biospecimen Use Guidelines are met (e.g. samples from control subjects who have the largest amount of samples in the repository).
The PLCO EEMS Steering Committee has realized that as technology evolves and laboratory analysis techniques change, so should repository management practices. As such, PLCO Biospecimen Use Guidelines have been updated on a routine basis to reflect changes in preparation of samples for studies, such as serum volumes (an example is illustrated in Table 2). Another change was regarding the storage of extracted DNA. Initially, PLCO stored DNA in multiple aliquots of varying amounts of DNA in varying concentrations. As technologies advanced, smaller amounts of DNA were required for assays, and it became apparent that freeze-thaw cycles do not materially affect most DNA-based assay results. Thus, PLCO started storing extracted DNA in standardized concentrations in “source” tubes, from which future requested amounts of DNA could be aliquoted as needed, thereby reducing storage costs.
Table 2.
PLCO Serum and Plasma Aliquoting Scheme (last updated: March 2013) for 1.8ml Parent Vials Specimens Requested from the Biorepository
| Assay Volume (ml) for Requested Study | Aliquot (Child Vial) Volume (ml) for Storage | ||||
|---|---|---|---|---|---|
| Aliquot 1 | Aliquot 2 * | Aliquot 3 * | Aliquot 4 * | Aliquot 5 * | |
| 0.1 | 0.2 | 0.35 | 0.2 | 0.5 | 0.45 |
| 0.2 | 0.2 | 0.35 | 0.2 | 0.5 | 0.35 |
| 0.3 | 0.2 | 0.35 | 0.2 | 0.5 | 0.25 |
| 0.4 | 0.2 | 0.35 | 0.2 | 0.5 | 0.15 |
| 0.5 | 0.2 | 0.35 | 0.2 | 0.55 | |
| 0.6 | 0.2 | 0.35 | 0.2 | 0.45 | |
| 0.7 | 0.2 | 0.35 | 0.2 | 0.35 | |
| 0.8 | 0.2 | 0.35 | 0.20 | 0.25 | |
| 0.9 | 0.2 | 0.35 | 0.35 | ||
| 1.0 | 0.2 | 0.35 | 0.25 | ||
| 1.1 | 0.2 | 0.5 | |||
| 1.2 | 0.2 | 0.4 | |||
| 1.3 | 0.2 | 0.3 | |||
| 1.4 | 0.2 | 0.2 | |||
| 1.5 | 0.3 | ||||
| 1.6 | 0.2 | ||||
If the remainder volume will be less than 0.15ml, then the aliquot will not be made.
Finally, another lesson learned was that it was critical to request and receive processing data from laboratories in a timely manner. While the main biorepository inventory might not have been updated regarding the individual biospecimen quality and quantity on a daily or weekly basis, the analysis group maintained an ancillary database with such updates. Once specimens were returned to the main repository (after DNA was extracted and aliquotted, serum aliquots prepared, etc.), the main biorepository inventory was then updated. As a reflection of how information technology has advanced, more recently, the NCI central processing lab has integrated their Laboratory Information Management System (LIMS) with the main biorepository inventory to provide nightly real-time updating of the biospecimen status. This has greatly improved real-time updating of the inventory database.
4. CONCLUSION
The PLCO biospecimen repository has substantially added to the conduct of etiologic and early detection studies of cancer. The uniqueness of this resource stems from the pairing of a variety of well-annotated, serial, pre-diagnostic specimens and pathology tissue samples with a rich epidemiologic and clinical database. Well-documented, standardized guidelines and procedures for each step in the complex workflows of biospecimen collection, processing, storage, requisition, and distribution, as well as data management, were essential for ensuring maximum use of this unique resource for a variety of research studies.
ACKNOWLEDGEMENTS
We are grateful for the efforts by many within PLCO that were dedicated to establishing, maintaining, and overseeing the PLCO biorepository and to the PLCO participants for donating their biospecimens.
Biography

Danielle M. Carrick
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare. Funding for this publication was from the National Cancer Institute, National Institutes of Health.
REFERENCES
- [1].Carrick DM, Mette E, Hoyle B, et al. The use of biospecimens in population-based research: a review of the National Cancer Institute's Division of Cancer Control and Population Sciences grant portfolio. Biopreserv Biobank 2014; 12: 240–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hayes RB, Reding D, Kopp W, et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin Trials 2000; 21: 349S–55S. [DOI] [PubMed] [Google Scholar]
- [3].Hayes RB, Sigurdson A, Moore L, et al. Methods for etiologic and early marker investigations in the PLCO trial. Mutat Res 2005; 592: 147–54. [DOI] [PubMed] [Google Scholar]
- [4].Zhu CS, Pinsky PF, Kramer BS, et al. The prostate, lung, colorectal, and ovarian cancer screening trial and its associated research resource. J National Can Inst 2013; 105: 1684–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].PLCO. PLCO Biorepository Blood Specimen Collection Protocols. 2014. Retrieved from: https://www.plcostars.com/Public/Documents/PLCO/BiospecimenCollectProtocol.pdf.
- [6].Hayes RB, Smith CO, Huang WY, Read Y, Kopp WC. Whole blood cryopreservation in epidemiological studies. Cancer epidemiology, biomarkers & prevention: Am Assoc Cancer Res, cosponsored by the Am Soc Prev Oncol 2002; 11: 1496–8. [PubMed] [Google Scholar]
- [7].IATA. IATA Dangerous Good Regulations (DGR). 2015. Retrieved from: http://www.iata.org/publications/dgr/Pages/index.aspx.
- [8].Zhu CS, Pinsky PF, Cramer DW, et al. A framework for evaluating biomarkers for early detection: validation of biomarker panels for ovarian cancer. Cancer Prev Res 2011; 4: 375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].PLCO. PLCO EEMS. 2014. Retrieved from: http://www.plcostars.com.
- [10].Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nature Gen 2007; 39: 870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Landi MT, Chatterjee N, Yu K, et al. A Genome-wide Association Study of Lung Cancer Identifies a Region of Chromosome 5p15 Associated with Risk for Adenocarcinoma. Am J Human Gen 2009; 85: 679–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Peters U, Hutter CM, Hsu L, et al. Meta-analysis of new genomewide association studies of colorectal cancer risk. Human Gen 2012; 131: 217–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wentzensen N, Black A, Jacobs K, et al. Genetic variation on 9p22 is associated with abnormal ovarian ultrasound results in the prostate, lung, colorectal, and ovarian cancer screening trial. PloS one 2011; 6: e21731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Purdue MP, Johansson M, Zelenika D, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nature Gen 2011; 43: 60–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rothman N, Garcia-Closas M, Chatterjee N, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nature Gen 2010; 42: 978–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genomewide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nature Gen 2009; 41: 986–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].De Vivo I, Prescott J, Setiawan VW, et al. Genome-wide association study of endometrial cancer in E2C2. Human Gen 2014; 133: 211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rajaraman P, Melin BS, Wang Z, et al. Genome-wide association study of glioma and meta-analysis. Human Gen 2012; 131: 1877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cerhan JR, Berndt SI, Vijai J, et al. Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nature Gen 2014; 46: 1233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Berndt SI, Skibola CF, Joseph V, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nature Gen 2013; 45: 868–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yeager M, Xiao N, Hayes RB, et al. Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers. Human Gen 2008; 124: 161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Parikh H, Deng Z, Yeager M, et al. A comprehensive resequence analysis of the KLK15-KLK3-KLK2 locus on chromosome 19q13.33. Human Gen 2010; 127: 91–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Al Olama AA, Kote-Jarai Z, Berndt SI, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nature Gen 2014; 46: 1103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Siddiq A, Couch FJ, Chen GK, et al. A meta-analysis of genomewide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet 2012; 21: 5373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Campa D, Kaaks R, Le Marchand L, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J National Cancer Inst; 103: 1252–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Witte JS, Hoffmann TJ. Polygenic modeling of genome-wide association studies: an application to prostate and breast cancer. Omics : J Integ Biol 2011; 15: 393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Machiela MJ, Chen CY, Chen C, et al. Evaluation of polygenic risk scores for predicting breast and prostate cancer risk. Gen Epidemiol 2011; 35: 506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jiao S, Peters U, Berndt S, et al. Estimating the heritability of colorectal cancer. Human Mol Genet 2014; 23: 3898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Gen 2010; 42: 937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Berndt SI, Gustafsson S, Magi R, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nature Gen 2013; 45: 501–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Caporaso N, Gu F, Chatterjee N, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PloS one 2009; 4: e4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ahn J, Yu K, Stolzenberg-Solomon R, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet 2010: ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Moy KA, Mondul AM, Zhang H, et al. Genome-wide association study of circulating vitamin D- binding protein. Am J Clin Nutr 2014; 99: 1424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cornelis MC, Monda KL, Yu K, et al. Genome-wide meta-analysis identifies regions on 7p21 (AHR) and 15q24 (CYP1A2) as determinants of habitual caffeine consumption. PLoS Genet 2011; 7: e1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gong J, Hsu L, Harrison T, et al. Genome-wide association study of serum selenium concentrations. Nutrients 2013; 5: 1706–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Prescott J, Kraft P, Chasman DI, et al. Genome-wide association study of relative telomere length. PloS one 2011; 6: e19635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shiels MS, Engels EA, Shi J, et al. Genetic variation in innate immunity and inflammation pathways associated with lung cancer risk. Cancer 2012; 118: 5630–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ryan BM, Zanetti KA, Robles AI, et al. Germline variation in NCF4, an innate immunity gene, is associated with an increased risk of colorectal cancer. Int J Cancer J Int du Cancer 2014; 134: 1399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huang WY, Berndt SI, Kang D, et al. Nucleotide excision repair gene polymorphisms and risk of advanced colorectal adenoma: XPC polymorphisms modify smoking-related risk. Cancer Epidemiol, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the Amer Soc Prev Oncol 2006; 15: 306–11. [DOI] [PubMed] [Google Scholar]
- [40].Peters U, Hayes RB, Chatterjee N, et al. Circulating vitamin D metabolites, polymorphism in vitamin D receptor, and colorectal adenoma risk. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the Amer Soc Prev Oncol 2004; 13: 546–52. [PubMed] [Google Scholar]
- [41].Canzian F, Cox DG, Setiawan VW, et al. Comprehensive analysis of common genetic variation in 61 genes related to steroid hormone and insulin-like growth factor-I metabolism and breast cancer risk in the NCI breast and prostate cancer cohort consortium. Hum Mol Genet 2010; 19: 3873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Church TR, Haznadar M, Geisser MS, et al. Interaction of CYP1B1, cigarette-smoke carcinogen metabolism, and lung cancer risk. Int J Mol Epidemiol Genet 2010; 1: 295–309. [PMC free article] [PubMed] [Google Scholar]
- [43].Berndt SI, Huang WY, Fallin MD, et al. Genetic variation in base excision repair genes and the prevalence of advanced colorectal adenoma. Cancer Res 2007; 67: 1395–404. [DOI] [PubMed] [Google Scholar]
- [44].Huang WY, Chatterjee N, Chanock S, et al. Microsomal epoxide hydrolase polymorphisms and risk for advanced colorectal adenoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the Amer Soc Prev Oncol 2005; 14: 152–7. [PubMed] [Google Scholar]
- [45].Koutros S, Berndt SI, Sinha R, et al. Xenobiotic metabolizing gene variants, dietary heterocyclic amine intake, and risk of prostate cancer. Cancer Res 2009; 69: 1877–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Michaud DS, Daugherty SE, Berndt SI, et al. Genetic polymorphisms of interleukin-1B (IL-1B), IL-6, IL- 8, and IL-10 and risk of prostate cancer. Cancer Res 2006; 66: 4525–30. [DOI] [PubMed] [Google Scholar]
- [47].Moore LE, Huang WY, Chatterjee N, et al. GSTM1, GSTT1, and GSTP1 polymorphisms and risk of advanced colorectal adenoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the Amer Soc Prev Oncol 2005; 14: 1823–7. [DOI] [PubMed] [Google Scholar]
- [48].Purdue MP, Hofmann JN, Colt JS, et al. A case-control study of peripheral blood mitochondrial DNA copy number and risk of renal cell carcinoma. PloS one 2012; 7: e43149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Andreotti G, Karami S, Pfeiffer RM, et al. LINE1 methylation levels associated with increased bladder cancer risk in pre-diagnostic blood DNA among US (PLCO) and European (ATBC) cohort study participants. Epigenetics : J DNA Methyl Soc 2014; 9: 404–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Huang WY, Su LJ, Hayes RB, et al. Prospective study of genomic hypomethylation of leukocyte DNA and colorectal cancer risk. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the Amer Soc Prev Oncol 2012; 21: 2014–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Seow WJ, Cawthon RM, Purdue MP, et al. Telomere length in white blood cell DNA and lung cancer: a pooled analysis of three prospective cohorts. Cancer Res 2014; 74: 4090–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kim C, Bassig BA, Seow WJ, et al. Pooled analysis of mitochondrial DNA copy number and lung cancer risk in three prospective studies. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the Amer Soc Prev Oncol 2014; 23: 2977–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mirabello L, Huang WY, Wong JY, et al. The association between leukocyte telomere length and cigarette smoking, dietary and physical variables, and risk of prostate cancer. Aging Cell 2009; 8: 405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ahn J, Peters U, Albanes D, et al. Serum vitamin D concentration and prostate cancer risk: a nested case-control study. J Nat Can Ins 2008; 100: 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Breyer BN, Huang WY, Rabkin CS, et al. Sexually transmitted infections, benign prostatic hyperplasia and lower urinary tract symptom-related outcomes: Results from the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. BJU Int 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Brock K, Huang WY, Fraser DR, et al. Low vitamin D status is associated with physical inactivity, obesity and low vitamin D intake in a large US sample of healthy middle-aged men and women. The J Ster Biochem Mol Biol 2010; 121: 462–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Cross AJ, Sinha R, Wood RJ, et al. Iron homeostasis and distal colorectal adenoma risk in the prostate, lung, colorectal, and ovarian cancer screening trial. Cancer Prev Res 2011; 4: 1465–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gunter MJ, Cross AJ, Huang WY, et al. A prospective evaluation of C-reactive protein levels and colorectal adenoma development. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the Amer Soc Prev Oncol 2011; 20: 537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Huang WY, Hayes R, Pfeiffer R, et al. Sexually transmissible infections and prostate cancer risk. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the Am Soc Prev Oncol 2008; 17: 2374–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Purdue MP, Hofmann JN, Kemp TJ, et al. A prospective study of 67 serum immune and inflammation markers and risk of non-Hodgkin lymphoma. Blood 2013; 122: 951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shiels MS, Pfeiffer RM, Hildesheim A, et al. Circulating inflammation markers and prospective risk for lung cancer. J Nat Can Inst 2013; 105: 1871–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Trabert B, Pinto L, Hartge P, et al. Pre-diagnostic serum levels of inflammation markers and risk of ovarian cancer in the prostate, lung, colorectal and ovarian cancer (PLCO) screening trial. Gynecol Oncol 2014; 135: 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Guertin KA, Moore SC, Sampson JN, et al. Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am J Clin Nutr 2014; 100: 208–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Landgren O, Albitar M, Ma W, et al. B-cell clones as early markers for chronic lymphocytic leukemia. New Eng J Med 2009; 360: 659–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sigurdson AJ, Jones IM, Wei Q, et al. Prospective analysis of DNA damage and repair markers of lung cancer risk from the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Carcinogenesis 2011; 32: 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Nat Can Inst 2001; 93: 1054–61. [DOI] [PubMed] [Google Scholar]
- [67].PLCO. Biospecimen Use Guidelines. 2014. Retrieved from: https://www.plcostars.com/Project/Plco/Documents/Biospecimen_UserGuide.pdf.