Abstract
Background:
Organoid is an in vitro three-dimensional organ-bud that shows realistic microanatomy and physiologic relevance. The progress in generating organoids that faithfully recapitulate human in vivo tissue composition has extended organoid applications from being just a basic research tool to a translational platform with a wide range of uses. Study of host-microbial interactions relies on model systems to mimic the in vivo infection. Researchers have developed various experimental models in vitro and in vivo to examine the dynamic host-microbial interactions. For some infectious pathogens, model systems are lacking whereas some of the used systems are far from optimal.
Objective:
In the present work, we will review the brief history and recent findings using organoids for studying host-microbial interactions.
Methods:
A systematic literature search was performed using the PubMed search engine. We also shared our data and research contribution to the field.
Results:
we summarize the brief history of 3D organoids. We discuss the feasibility of using organoids in studying host-microbial interactions, focusing on the development of intestinal organoids and gastric organoids. We highlight the advantage and challenges of the new experimental models. Further, we discuss the future direction in using organoids in studying host-microbial interactions and its potential application in biomedical studies.
Conclusion:
In combination with genetic, transcriptome and proteomic profiling, both murine- and human-derived organoids have revealed crucial aspects of development, homeostasis and diseases. Specifically, human organoids from susceptible host will be used to test their responses to pathogens, probiotics, and drugs. Organoid system is an exciting tool for studying infectious disease, microbiome, and therapy.
Keywords: Bacteria, colonoids, enteroids, gastric organoids, host-microbial interactions, H. pylori, inflammation, intestinal organoids, microbiome, organoids, tight junctions, Salmonella, stem-cell differentiation, ZO-1
1. Introduction
Researchers have employed various in vitro and in vivo experimental models to investigate interactions taking place between microbes (e.g. bacteria, parasites, virus) and their host (e.g. humans, animals). These models consist of cell cultures deriving from human or animal cells (Dingli and Nowak 2006), animals that can be inoculated with pathogens orally or parenterally (Fang, Kapikian et al. 2013), and organoids modeling host-microbial interactions (Fatehullah, Tan et al. 2016). The ultimate aim of these models is to create an environment in vitro that can imitate the real circumstances of the human to elucidate physiological mechanisms of host responses in health and diseases.
In this review, we will summarize the brief history of 3D organoids, focusing on the development of intestinal organoids and gastric organoids. We will discuss the feasibility of using organoids in studying the effects and mechanisms of host-microbial interactions. Also, we will highlight the advantage and challenges of the newly developed experimental models. Further, we will discuss the future direction in using organoids as an in vitro model to study host-microbial interactions and its potential application in biomedical studies.
2. History of organoids
Cell culture was first invented at the beginning of the twentieth century, and was used to study frog embryo nerve fiber outgrowth (Harrison 1907). Using of dispersed cell cultures rapidly increased in the second half of the twentieth century (Hilleman 1990). Organ culture is a development from cell culture methods. It retains histological structure and the architecture characteristic of the tissue. However, compared with other types of tissue culture, organ culture has several shortcomings, including difficulty in quantification of tissues or cells, limitation in the amounts of cultured samples, the requirement of skillful manipulation of the samples, and the challenge of reproducibility.
The 3D culture system has made it possible to recapitulate partially the complexity of mammalian organogenesis in vitro. The newly developed organoid system acts as a bridge between in vivo and in vitro systems. They can be derived from pluripotent stem cells (PSCs), both from embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs), as well as from adult stem cells (AdSCs) (Kristin, Weitz et al. 2016).
ESCs are derived from the inner cell mass (ICM) of the pre-implantation blastocyst. Under appropriate in vitro culture conditions, ESCs proliferate indefinitely without differentiation, a property hereinafter referred to as “self-renewal”, and at the same time retain the developing potential to generate cells of all three primary germ layers (Huang, Ye et al. 2015).
Over the past three decades, researchers have developed methods to derive ESCs from the epiblast and expand them continuously in vitro. More recently, induced pluripotent stem cells (iPSCs) can be generated from almost any mature cell type in our bodies. These breakthroughs have allowed the differentiation of various PSC populations into somatic cell derivatives (Huch and Koo 2015). Culturing human derivatives (hESCs/hiPSCs/hAdSCs) in three-dimension (3D) has opened up new horizons for the exploration of development and regenerative medicine approaches.
In the late 1980s, organoids derived from adult stem cells experiments performed on skin proved that epidermal stem cells were expandable and could generate vast amounts of epithelium in vitro, supported by lethally irradiated 3T3 cells(Barrandon and Green 1987). Until recently, the AdSC maintenance and tissue repair have been instrumental for the development of primary AdSCs cultures. AdSCs have gained much attention also for their intrinsic abilities to self-renew and differentiate into the cell types present in adult tissues while retaining genomic stability. Thus, organoids have all been derived in vitro from AdSCs, including mammary gland, bone, stomach, small intestine, colon and liver. A recent review from Nature Cell Biology has excellently summarized the organoids from different tissues (Fatehullah, Tan et al. 2016). Here, we will focus on the intestinal organoids derived from adult stem/progenitor cells. We follow the definition from (Fatehullah, Tan et al. 2016) an organoid is an in vitro 3D cellular cluster derived exclusively from primary tissue, ESCs or iPSCs, capable of self-renewal and self-organization, and exhibiting similar organ functionality as the tissue of origin. Most of the documented organoid cultures contain functional tissue units that lack the mesenchymal, stromal, immune and neural cells that intersperse the tissue in vivo. These organoids rely on artificial extracellular matrices to facilitate self-organization into structures that resemble native tissue architecture.
In 2009, the development of intestinal organoid culture was an outstanding technological advance for the stem cell field (Ootani, Li et al. 2009; Sato, Vries et al. 2009). Establishing intestinal organoids is challenging because it requires tissue-specific modifications that reflect the individual niche and lineage commitment factors for the resident stem cell populations and their progeny. Ootani et al. has reported a complex culture system from minced whole intestinal tissue embedded in a 3D collagen structure with support of stromal cells12. Sato et al has established a relatively simple organoid culture system, using Matrigel as an extracellular matrix (ECM) substitute (Sato, Vries et al. 2009), supplemented with growth factors constituting key endogenous niche signals. The system was used to create 3D structures with distinct crypt-like and villus-like domains bordering a central lumen containing dead cells extruded from the constantly renewing epithelial layer (Sato, Vries et al. 2009). Furthermore, these organoids from small intestine could be expanded for over a year in vitro (Huch and Koo 2015). In the organoids, the domains bordering a central lumen containing dead cells extruded from the constantly renewing epithelial layer. This new method takes advantage of our knowledge of endogenous intestinal stem cell niche components to deliver a well-defined, stable culture system capable of sustaining the long-term growth of near-physiological epithelia from purified Lgr5+ stem cells or isolated crypts.
Two years later, mouse- and human-derived colonic stem cells could also be expanded in culture with a minor modification to the culture medium composition (Jung, Sato et al. 2011; Sato, Stange et al. 2011). Human colonic organoids could be expanded for at least 1 month. From 1 month onward, the colonic organoids changed their morphology from budding structures into cystic structures (Sato, Stange et al. 2011). Coinciding with the morphologic conversion, proliferation progressively decreased. Occasionally, cystic organoids regained their proliferative potential. However, all organoids eventually arrested growth within 3 months. Microarray analysis revealed that the human small intestinal and colonic organoids possess comparable molecular signatures of intestinal crypts, including the expression of intestinal stem cell genes. Alk receptor and p38 signaling negatively regulate long-term maintenance of human intestinal epithelial cells (Sato, Stange et al. 2011).
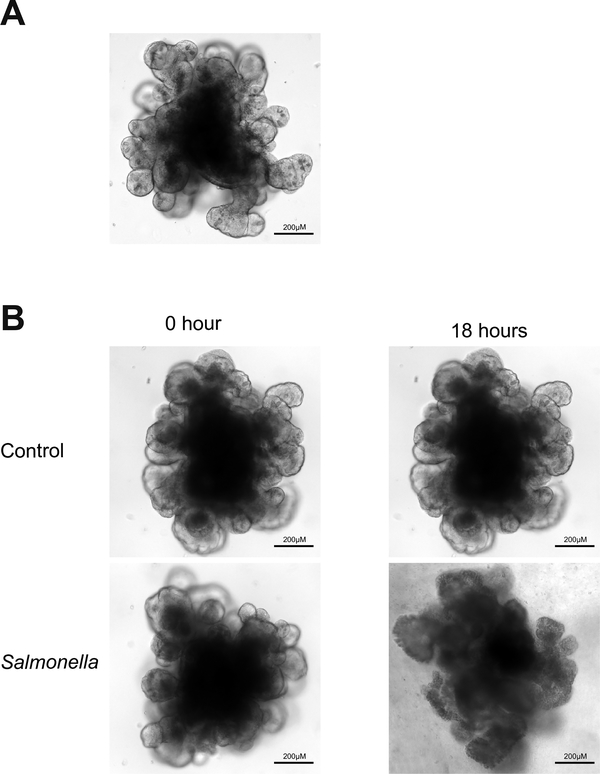
Miyoshi and Stappenbeck in 2013 published a very nice paper on how to perform expansion and genetic modification of organoids (Miyoshi and Stappenbeck 2013). Based on their protocol, isolation of epithelial cell units from mice takes up to 2 hours and stem cell-enriched gastrointestinal organoids are obtained within 3 days. Genetically modified organoids with lentiviruses can be obtained in 2 weeks. As shown in Fig. 1, the organoids derived from mouse small intestinal stem cells show organ-buds 7 days after isolation from crypts and recapitulated the in vivo tissue architecture.
Figure 1.
Normal and Salmonella-infected crypt-derived intestinal organoids. A. Normal crypt-derived mouse intestinal organoids in day 7 after isolation. B. Salmonella-infected crypt-derived intestinal organoids.
Despite the similarities with the murine orgaoind system, the human organoids require specific molecules to enhance and sustain their growth over time. Compared to the culture of mouse organoids, the other challenge in human organoid culture is to obtain fresh samples and establish feasible protocol. 2015, Mahe et al reported the detailed protocol and procedure to establish human epithelial enteroids and colonoids using whole tissue and biopsy(Mahe, Sundaram et al. 2015). In this methodological paper, the authors emphasize the crypt collection from whole tissue and biopsies. They recapitulate the culture modalities that are critical for the successful growth and maintenance of human epithelial organoids (enteroids)(Mahe, Aihara et al. 2013) and colonoids. Commonly, termed “enteroids” when derived from small intestine and “colonoids” when derived from colon (Mahe, Sundaram et al. 2015).
The intestinal epithelium of mammals consists of absorptive enterocytes and of three secretory cell types, paneth, goblet, and enteroendocrine cells, which are continuously replenished from stem cells that reside in niches in the lower parts of the crypts (Crosnier, Stamataki et al. 2006). Organoids contain the full complement of stem, progenitor and differentiated cell types (Fatehullah, Tan et al. 2016), (Sato, Vries et al. 2009). Marker-specific antibodies for mucin 2 (goblet cells), lysozyme (Paneth cells), and chromogranin A (enteroendocrine cells) are used to validate the presence of intestinal secretory cell lineages in orgaoinds. The enteroids/colonoids continuously produce all cell types found normally within the intestinal epithelium. Researchers who focus on specialized cell types, such as goblet cells or Paneth cells, find the organoid as a very useful in vitro model for insights into GI development, tissue homeostasis, and diseases (Dedhia, Bertaux-Skeirik et al. 2016). In human intestinal organoids treated with Notch inhibition dibenzazepine (DBZ, 10 mol/ L), the intestinal organoids ceased their proliferation and most cells converted into goblet cells within 3 days (Sato, Stange et al. 2011). It is reported that Mitogen-activated Protein Kinase (MAPK) signaling controls goblet/Paneth cell fate decisions in the intestine (Heuberger, Kosel et al. 2014). Ablation of the tyrosine phosphatase Shp2 in the intestinal epithelium reduced MAPK signaling and led to a reduction of goblet cells while promoting Paneth cell development. Inhibition of MAPK signaling in mouse intestinal organoids changed the relative abundance of T-cell factor 4 isoforms, which promoted Wnt/β-catenin activity. The data show that Shp2-mediated MAPK signaling controls the choice between goblet and Paneth cell fates by regulating Wnt/β-catenin activity.
Goblet and Paneth cells represent two secretory cell types in the intestinal epithelium It is believed that the intestinal organoids derived from human IPSCs are more small intestine like in structure than mouse organoids because of the presence of Paneth-like cells and the villus-like protrusions (Forbester, Goulding et al. 2015). The organoids provide a new tool to study the development and functions of these cell types that are critical for the host-microbe interactions in health and diseases.
The organoids subsequently adapted for generating human organoids and also from animal models with different genetic modification have been used for various basic and clinic research, including host-microbial interactions.
3. Feasibility and application of organoids in studying host-microbial interactions
Study of host-microbial interactions relies on model systems to mimic the in vivo infection. For some infectious pathogens, model systems are lacking whereas some of the used systems are far from optimal. Recent reports have shown the feasibility to use organoids as an in vitro model to study host-pathogen interactions (Zhang, Wu et al. 2014) (Bartfeld and Clevers 2015). These studies have used organoids to model infections with bacteria, such as Salmonella (Forbester, Goulding et al. 2014; Zhang, Wu et al. 2014; Forbester, Goulding et al. 2015; Wilson, Tocchi et al. 2015), Helicobacter pylori (McCracken, Cata et al. 2014; Bartfeld, Bayram et al. 2015; Huang, Sweeney et al. 2015; Schumacher, Feng et al. 2015; Sigal, Rothenberg et al. 2015; Schlaermann, Toelle et al. 2016), Bacterioides thetaiotaomicron (Engevik, Aihara et al. 2013), Clostridium difficile (Leslie, Huang et al. 2015)), viruses (e.g. rotavirus (Finkbeiner, Zeng et al. 2012; Finkbeiner, Zeng et al. 2012; Yin, Bijvelds et al. 2015), Cytomegalovirus (D’Aiuto, Di Maio et al. 2012; Penkert and Kalejta 2013), Zika virus (Garcez, Loiola et al. 2016) or Hepatitis C Virus (Yoshida, Takayama et al. 2011; Roelandt, Obeid et al. 2012; Schwartz, Trehan et al. 2012; Wu, Robotham et al. 2012; Shlomai, Schwartz et al. 2014)) and parasites, such as Plasmodium falciparum (Ng, Schwartz et al. 2015) or Toxoplasma gondii (Klotz, Aebischer et al. 2012).
3.1. Intestinal organoids in host-pathogenic bacterial interactions
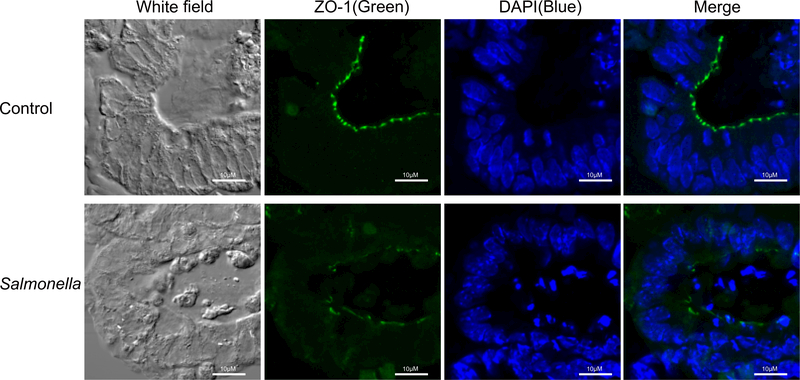
Salmonella Typhimurium is a primary enteric pathogen infecting both humans and animals. Infection begins with the ingestion of contaminated food or water so that Salmonella reach the intestinal epithelium and trigger gastrointestinal disease. In 2014, we reported using an intestinal organoid culture system to study pathophysiology of bacterial-epithelial interactions post S. Typhimurium infection (Zhang, Wu et al. 2014). Using crypt-derived mouse intestinal organoids, we were able to visualize the invasiveness of S. Typhimurium and the morphologic changes of the organoids (Fig. 1B). S. Typhimurium entered the epithelial cells of the organoids and this resulted in disruption of the tight junctions (Fig. 2). We showed that distribution of ZO-1, a tight junction protein, was weak and disconnected in the organoids infected with Salmonella. Using the organoids, we also established methods for western blot, PCR, and immunofluorescence to demonstrate the changes of stem cell markers (Lgr5 and Bmi1). We found that Lgr5 and Bmi1 were significantly decreased by Salmonella infection. We also cultured GFP-labeled Lgr5 organoids to study the pathogen regulation of stem cells (Zhang, Wu et al. 2014).
Figure 2.
Distribution of tight junction protein ZO-1 in organoids. Salmonella-induced disruption of tight junctions in the mouse intestinal organoids. Please note the disorganized structure of ZO-1 (green staining) in organoids infected with Salmonella. Scale bars: 10 lm. Images for ZO-1 protein represent three separate experiments.
The NF-κB pathway in intestine is activated by Salmonella infection in vitro and in vivo. We examined the changes of NF-κB pathway in the organoids infected with Salmonella. Salmonella-infected organoids had a significantly decreased total IkBα and increased phospho-IκBα. The phospho-NF-κB p65 was also increased in the Salmonella-infected organoids. By confocal microscopy, we found that NF-κB p65 was translocated into the nucleus in organoids-infected with Salmonella. As the downstream targets of NF-κB activation, inflammatory cytokines (e.g.IL-2, IL-4, IL-6, and TNF-a) were significantly increased in the infected organoids compared to the organoids without any infection. Moreover, we found that the ELISA was sensitive enough to detect IL-6 protein in the culture medium 1 hour post Salmonella infection. IL-6 protein was significantly enhanced in the culture medium post 1 hour, 2-, and 4- hours post infection (Zhang, Wu et al. 2014). For the first time, we have created an in vitro model system that recapitulated a number of observations from in vivo studies of the Salmonella-infected intestine: bacterial invasion, altered tight junctions, inflammatory responses, and decreased stem cells during host-bacterial interactions. We have demonstrated that the Salmonella-infected organoid culture system is a new and feasible experimental tool for studying host–bacterial interactions (Zhang, Wu et al. 2014). Our study has demonstrated the complexity of the host response to bacterial infection, even in the absence of immune cells.
Using intestinal organoids (iHOs) derived from human induced pluripotent stem cells (hIPSCs), Forbester et al. (Forbester, Goulding et al. 2014; Forbester, Goulding et al. 2015) established microinjection of S. Typhimurium into the lumen of iHOs. They reported that 1,448 genes significantly upregulated in iHOs infected with S. Typhimurium and 577 genes significantly downregulated compared to controls, by RNA sequencing. Genes encoding proinflammatory cytokines, including CCL20, IL1B, and IL23A, were significantly upregulated. Utilizing a S. Typhimurium mutant strains that lacked the invA component of the SPI-1 type III secretion system, they have demonstrated that this system could be utilized to functionally assess the pathogenesis of defined mutants (Forbester, Goulding et al. 2014; Forbester, Goulding et al. 2015). It is believed that the intestinal organoids derived from human IPSCs are more small intestine like in structure because of the presence of Paneth-like cells and the villus-like protrusions (Forbester, Goulding et al. 2015).
Microinjection of organoids with bacteria can mimic bacterial infection in a relatively well-controlled environment, allowing for direct examination of pathogen interactions with epithelial cells in the absence of confounding variables introduced by immune cells or the commensal microbiota. Wilson et al. reported that Paneth cells in organoids from both wild-type mice and Mmp7−/− mice produced granules containing pro-α-defensins. Organoids form a sealed lumen that contains concentrations of α-defensins capable of restricting growth of S. Typhimurium for at least 20 h postinfection (Wilson, Tocchi et al. 2015). In human intestinal organoids (Leslie, Huang et al. 2015), toxin production by clostridium difficile result in disruption of epithelial paracellular barrier function.
Organoids also provide a novel and powerful ex vivo model for studying commensal bacteria, probiotics, and microbiome studies. For example, inoculation of Bacterioides thetaiotaomicron, a Bacteroidetes member, in wild-type and NHE3−/− terminal ileum organoids displayed increased fut2 and fucosylation. These data suggest that B. thetaiotaomicron alone is sufficient for the increased fucosylation seen in vivo. (Engevik, Aihara et al. 2013).
Probiotic Lactobacillus rhamnosus GG (LGG) has been reported to be therapeutically effective against acute secretory diarrhea by rotavirus infection; however, the underlying mechanisms remain to be completely elucidated. Intestinal organoids derived from small intestinal crypts treated with LGG showed increased Toll-like receptor 3 (TLR3) mRNA levels, by quantitative real-time polymerase chain reaction (Aoki-Yoshida, Saito et al. 2016).
A recent study has examined the transcriptional response of organoids upon exposure to short-chain fatty acids (SCFAs) and products generated by two abundant microbiota constituents, Akkermansia muciniphila and Faecalibacterium prausnitzii (Arnold, Roach et al. 2016). A. muciniphila metabolites affect various transcription factors and genes involved in cellular lipid metabolism and growth, supporting previous in vivo findings. Contrastingly, F. prausnitzii products exerted only weak effects on host transcription. In addition, A. muciniphila and its metabolite propionate modulated expression of Fiaf, Gpr43, histone deacetylases, and peroxisome proliferator-activated receptor gamma, important regulators of transcription factor regulation, cell cycle control, lipolysis, and satiety. A. muciniphila induces stronger effects on the host than F. prausnitzii. This study thus illustrates that specific bacteria and their metabolites differentially modulate epithelial transcription in mouse organoids.
VanDussen et al reported a culture system for human gastrointestinal epithelial cells from multiple regions of the gastrointestinal tract (VanDussen, Marinshaw et al. 2015). Key advantages of this system include use of endoscopic biopsy tissue as starting material and the rapid expansion of the spheroids, which allows for line establishment from an individual patient within a time frame that is commensurate with patient care (~2–3 weeks). The mucus layer is a critical component of the physical barrier separating the host from the luminal environment, thus providing protection from pathogens. The spheroid-derived intestinal epithelial cells produced a mucus layer that could be mechanically dissociated. The adherence phenotypes of diarrheagenic E. coli was also test in this culture system. It has great potential for use in patient-specific assays (VanDussen, Marinshaw et al. 2015).
3.2. Gastric organoids/enteroids in host-bacterial interactions
Gastric organoids have evolved to a new state-of-the-art in vitro tool for Helicobacter pylori (H. pylori) research. H. pylori is a gastric pathogen that colonizes approximately 50% of the world’s population. Infection with H. pylori causes chronic inflammation, peptic ulcers and ultimately leads to gastric cancer (Wroblewski, Peek et al. 2010; Salama, Hartung et al. 2013). Bartfeld et al. generated human gastric organoids (hGO) from surgical samples of gastric corpus. hGO maintained many characteristics of their respective tissues based on their histology, expression of markers, and euploidy. They performed microinjection of GFP-expressing H. pylori into the lumen of gastric organoids. Plating of bacteria from organoids 2 hours after injection verified that the bacteria are alive inside the organoids. Electron microscopy showed that bacteria were engaged in very intimate contact with the epithelial cells. The primary response of the hGO to H. pylori showed robust NF-κB pathway, like IL-8, by microarray (Bartfeld, Bayram et al. 2015).
McCracken et al reported that bacteria were tightly associated with the hGO epithelium. The major physiological changes include an increase in proliferation due to oncogenic H.pylori protein CagA and increased β-catenin signaling (McCracken, Cata et al. 2014). Schumacher and colleagues has examined the epithelial response to infection with H. pylori, using both mouse and human gastric enteroids. Enteroids are epithelial organoids. H.pylori infection of gastric organoids induced Shh expression, triggering Shh expression via CagA dependent activation of NF-κB (Schumacher, Feng et al. 2015). Sigal et al. described a direct colonization of Lgr5+ stem cells by H. pylori (Sigal, Rothenberg et al. 2015). Gland-associated H pylori induce increased Lgr5-lineage tracing at the level of individual glands. Antral glands from infected mice formed organoids with a significantly higher capacity and larger sizes (Sigal, Rothenberg et al. 2015). These data indicate bacterial ability to alter the stem cells and important implications for gastrointestinal stem cell biology and H pylori–induced gastric pathology.
The studies in gastric enteroids allow researcher to identify new mechanisms which was not found in the animal models. Using spent media from both gastric enteroids and polarized cell lines, Huang et al. demonstrated that H. pylori used the chemoreceptor TlpB to sense urea emanating. It suggests that H. pylori concurrently senses and modulates its environment while colonizing its gastric niche (Huang, Sweeney et al. 2015). Surprisingly, the authors noted that TlpB is sensitive to low levels of urea and that physiological levels of urea in the stomach would inactivate the receptor. Ultimately, they found that H. pylori’s ability to break down urea via its urease facilitates its ability to sense host urea. Gastric enteroids were particularly useful as the absence of immune cells in this model enabled the authors to determine that NF-kB activation resulted directly from H. pylori infection rather than recruitment of other cell types, a finding that would be more difficult to make in an animal model. This group has continued to use enteroids to demonstrate that the host receptor CD44 played a functional role in the epithelial cell proliferation triggered by the H. pylori Cag pathogenicity island (Bertaux-Skeirik, Feng et al. 2015)
3.3. Organoids/enteroids for host-virus interactions
Organoids have been used for host-virus interactions, including rotavirus (Finkbeiner, Zeng et al. 2012; Yin, Bijvelds et al. 2015), Cytomegalovirus (D’Aiuto, Di Maio et al. 2012; Penkert and Kalejta 2013), Zika virus (Garcez, Loiola et al. 2016) or Hepatitis C Virus (Yoshida, Takayama et al. 2011; Roelandt, Obeid et al. 2012; Schwartz, Trehan et al. 2012; Wu, Robotham et al. 2012; Shlomai, Schwartz et al. 2014). Organoids/enteroids potentially can be used to determine ways to correct the diarrhea-induced ion transport abnormalities via drug therapy (VanDussen, Marinshaw et al. 2015).
Spence et al. has reported directed differentiation of stem cell lines into intestine-like tissue called “induced human intestinal organoids” (iHIOs)(Spence, Mayhew et al. 2011). iHIOs is used as a new model to cultivate and study enteric viruses (Finkbeiner, Zeng et al. 2012). iHIOs support replication of rotavirus, on the basis of detection of nonstructural viral proteins by immunofluorescence, increased levels of viral RNA, and production of infectious progeny virus. iHIOs also support replication of 12/13 clinical rotavirus isolates directly from stool samples. Interestingly, rotavirus infection is not only detected in the epithelial cells, but also in the mesenchymal cell population of the iHIOs. Thus, iHIOs offer a new model to study rotaviruses and other gastrointestinal viruses (Finkbeiner, Zeng et al. 2012).
Garcez et al. used human iPS-derived human neural stem cells (NSCs) infection with Zika virus. They demonstrated that Zika virus was detected in NSCs after 24 hours. Zika Virus targets human brain cells by reducing their viability and growth and causing cell death in neurospheres and brain organoids (Garcez, Loiola et al. 2016).
Researchers have made advancement by using human enteroids for rotavirus and norovirus research (Foulke-Abel, In et al. 2014). (Saxena, Blutt et al. 2015; Ettayebi, Crawford et al. 2016; In, Foulke-Abel et al. 2016). Particularly, norovirus culture is an important advancement made by using enterioids (Ettayebi et al 2016). Bile is required for strain-dependent human norovirus replication. Lack of appropriate histoblood group antigen expression in intestinal cells restricts virus replication, and infectivity is abrogated by inactivation (e.g., irradiation, heating) and serum neutralization. Multiple human norovirus strains are cultured in enterocytes in stem cell–derived, nontransformed human intestinal enteroid monolayer cultures. This culture system permits human host-pathogen studies of previously noncultivatable pathogens, and allows the assessment of strategies to prevent and treat norovirus infections.
3.4. Organoids for studying host-parasite interaction
Studying human protozoan parasites and their interaction with the host remain severely limited, because of non-existent or inappropriate animal models and challenge to culture parasites in vitro due to strict human-host specificity or physiology. Using organoids is a strategy to address many of these experimental bottlenecks(Klotz, Aebischer et al. 2012). Studies on Plasmodium falciparum(Ng, Schwartz et al. 2015) and Toxoplasma gondii (Klotz, Aebischer et al. 2012) in organoids allow us to address questions of cell and developmental biology, immunology, and pharmacology in unprecedented ways.
4. Advantage and challenges of using organoid system in studying host-microbial interactions.
The first advantages of using organoid systems in studying host-microbial interactions is that adult stem cells from many murine and human tissues can be grown in vitro and self-organize into organoids that resemble the in vivo counterpart. Using the traditional in vitro models to investigate interactions between microbes and intestinal epithelial cells, many studies fail to recreate the differentiated tissue components and structure observed in the normal intestine. One approach to creating differentiated cells is through a suspension culture technology using a rotating wall vessel bioreactor that allows cells to remain in suspension with bubble free aeration (Unsworth and Lelkes 1998). However, this system may lack normal stem cell niches, which are responsible for the renewal of normal intestinal tissues.
Stem cells of the gastrointestinal tract, pancreas, liver and other columnar epithelia are known to resist cloning in their elemental states. A recent study has reported cloning and propagation of highly clonogenic, ‘ground state’ stem cells of the human intestine and colon (Wang, Yamamoto et al. 2015). Interestedly, derived stem-cell pedigrees sustain limited copy number and sequence variation despite extensive serial passaging and display exquisitely precise, cell-autonomous commitment to epithelial differentiation consistent with their origins along the intestinal tract. Using clonally derived colonic epithelium, toxins A or B of the Clostridium difficile recapitulate the salient features of pseudomembranous colitis (Wang, Yamamoto et al. 2015). These stem cells may have certain advantages for use in host-microbial interactions.
The second advantage of organoids is that researcher can work on the tissue-specific or site-specific host interaction with a particular pathogen. Organoids can be generated from different organs or specific sites of the gut, including the small intestine and colon (Sato, Stange et al. 2011; Sato, van Es et al. 2011; Yui, Nakamura et al. 2012). Organoids represent more closely the intestinal epithelium than often-used colon cancer cell lines (e.g. CaCo2 or HCT116). Thirdly, studies in organoids allow researchers to have findings that would be more difficult to make in an animal model. Small biopsy specimens taken from adult donors can be expanded without any apparent limit or genetic harm, the technology may serve to generate transplantable epithelium for regenerative purposes (Sato, van Es et al. 2011). Schwank and colleagues have demonstrated the possibility to use the CRISPR/Cas9 system to edit the genome with and correct the mutation on the CFTR gene causing a cystic fibrosis (Schwank, Koo et al. 2013). Overall, intestinal organoids in host-microbe interactions allow us to address questions of cell and developmental biology, microbiology, immunology, and pharmacology in unprecedented ways.
Ideally, the cell-cell interactions are needed in host-microbial interactions. However, the intestinal organoids still have their disadvantages/limits. First, organoids lack several components of the intestine in vivo, such as the enteric nervous system and the vascular, lymphatic and immune systems. Organoids co-cultured with immune cells are needed for more comprehensive studies. Second, studying tissue patterning and organ morphogenesis has still been hindered by the lack of optimal culture condition for lab usage. Last, organoids relied on animal-derived matrices, which can be highly variable and are poorly defined, a problem that also makes them unsuitable for clinical application. A recent study has reported now designed modular synthetic hydrogen networks to support the formation of intestinal organoids from mouse and human intestinal stem cells (Gjorevski, Sachs et al. 2016). A highly accurate, reproducible culture model could help to overcome current limitations that hinder the technology’s transition from bench to bedside.
5. Conclusion and future direction
Organoids are one of the most accessible and physiologically relevant models to study the dynamics of host-microbial interaction in a controlled environment. The progress in generating organoids that recapitulate the human in vivo tissue composition has extended organoid applications from being just a basic research tool to a translational platform with a wide range of uses. In combination with genetic, transcriptome and proteomic profiling, both murine- and human-derived organoids have revealed crucial aspects of development, homeostasis and diseases. The commercial development of more standardized, validated organoid culture media, and affordable materials will be valuable in ensuring that the organoid system becomes accessible to a wide range of academic and clinical researchers, to further maximize its potential. Specifically, human organoids from susceptible host will be used to test their responses to pathogens, probiotics, and drugs. The physiological relevance of the system makes organoids one of the most exciting and promising technologies for studying human development, infectious disease, microbiome, and therapy.
Acknowledgements
This work was supported by the NIDDK 1R01DK105118–01 and the UIC Cancer Center to Jun Sun
Footnotes
Declaration of financial interests
No financial interests
Reference:
- Aoki-Yoshida A, Saito S, et al. (2016). “Lactobacillus rhamnosus GG increases Toll-like receptor 3 gene expression in murine small intestine ex vivo and in vivo.” Benef Microbes 7(3): 421–429. [DOI] [PubMed] [Google Scholar]
- Arnold JW, Roach J, et al. (2016). “Emerging Technologies for Gut Microbiome Research.” Trends Microbiol 24(11): 887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon Y and Green H (1987). “Three clonal types of keratinocyte with different capacities for multiplication.” Proceedings of the National Academy of Sciences of the United States of America 84(8): 2302–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S, Bayram T, et al. (2015). “In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection.” Gastroenterology 148(1): 126–U554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S and Clevers H (2015). “Organoids as Model for Infectious Diseases: Culture of Human and Murine Stomach Organoids and Microinjection of Helicobacter Pylori.” Jove-Journal of Visualized Experiments(105). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaux-Skeirik N, Feng R, et al. (2015). “CD44 plays a functional role in Helicobacter pylori-induced epithelial cell proliferation.” Plos Pathogens 11(2): e1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Stamataki D, et al. (2006). “Organizing cell renewal in the intestine: stem cells, signals and combinatorial control.” Nature reviews. Genetics 7(5): 349–359. [DOI] [PubMed] [Google Scholar]
- D’Aiuto L, Di Maio R, et al. (2012). “Human Induced Pluripotent Stem Cell-Derived Models to Investigate Human Cytomegalovirus Infection in Neural Cells.” PLoS One 7(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedhia PH, Bertaux-Skeirik N, et al. (2016). “Organoid Models of Human Gastrointestinal Development and Disease.” Gastroenterology 150(5): 1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingli D and Nowak MA (2006). “Cancer biology: infectious tumour cells.” Nature 443(7107): 35–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engevik MA, Aihara E, et al. (2013). “Loss of NHE3 alters gut microbiota composition and influences Bacteroides thetaiotaomicron growth.” American Journal of Physiology-Gastrointestinal and Liver Physiology 305(10): G697–G711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettayebi K, Crawford SE, et al. (2016). “Replication of human noroviruses in stem cell-derived human enteroids.” Science 353(6306): 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang SB, Kapikian AZ, et al. (2013). “Human Intestinal In Vitro Organ Culture as a Model for Investigation of Bacteriae-Host interactions.” Journal of Experimental and Clinical Medicine 5(2): 43–50. [Google Scholar]
- Fatehullah A, Tan SH, et al. (2016). “Organoids as an in vitro model of human development and disease.” Nature Cell Biology 18(3): 246–254. [DOI] [PubMed] [Google Scholar]
- Finkbeiner SR, Zeng XL, et al. (2012). “Stem cell-derived human intestinal organoids as an infection model for rotaviruses.” MBio 3(4): e00159–00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner SR, Zeng XL, et al. (2012). “Stem Cell-Derived Human Intestinal Organoids as an Infection Model for Rotaviruses.” MBio 3(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbester JL, Goulding D, et al. (2014). “Intestinal organoids are a novel system to study Salmonella enterica Serovar Typhimurium interaction with the intestinal epithelial barrier.” Immunology 143: 111–112. [Google Scholar]
- Forbester JL, Goulding D, et al. (2015). “Interaction of Salmonella enterica Serovar Typhimurium with Intestinal Organoids Derived from Human Induced Pluripotent Stem Cells.” Infect Immun 83(7): 2926–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulke-Abel J, In J, et al. (2014). “Human enteroids as an ex-vivo model of host-pathogen interactions in the gastrointestinal tract.” Exp Biol Med (Maywood) 239(9): 1124–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, et al. (2016). “Zika virus impairs growth in human neurospheres and brain organoids.” Science 352(6287): 816–818. [DOI] [PubMed] [Google Scholar]
- Gjorevski N, Sachs N, et al. (2016). “Designer matrices for intestinal stem cell and organoid culture.” Nature 539(7630): 560–564. [DOI] [PubMed] [Google Scholar]
- Harrison RG (1907). “Observations on the living developing fiber.” Proc Soc Exp Biol Med 4: 140–143. [Google Scholar]
- Heuberger J, Kosel F, et al. (2014). “Shp2/MAPK signaling controls goblet/paneth cell fate decisions in the intestine.” Proceedings of the National Academy of Sciences of the United States of America 111(9): 3472–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleman MR (1990). “History, precedent, and progress in the development of mammalian cell culture systems for preparing vaccines: safety considerations revisited.” J Med Virol 31(1): 5–12. [DOI] [PubMed] [Google Scholar]
- Huang G, Ye S, et al. (2015). “Molecular basis of embryonic stem cell self-renewal: from signaling pathways to pluripotency network.” Cell Mol Life Sci 72(9): 1741–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Sweeney EG, et al. (2015). “Chemodetection and Destruction of Host Urea Allows Helicobacter pylori to Locate the Epithelium.” Cell Host Microbe 18(2): 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M and Koo BK (2015). “Modeling mouse and human development using organoid cultures.” Development 142(18): 3113–3125. [DOI] [PubMed] [Google Scholar]
- In JG, Foulke-Abel J, et al. (2016). “Human mini-guts: new insights into intestinal physiology and host-pathogen interactions.” Nat Rev Gastroenterol Hepatol 13(11): 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P, Sato T, et al. (2011). “Isolation and in vitro expansion of human colonic stem cells.” Nat Med 17(10): 1225–1227. [DOI] [PubMed] [Google Scholar]
- Klotz C, Aebischer T, et al. (2012). “Stem cell-derived cell cultures and organoids for protozoan parasite propagation and studying host-parasite interaction.” International Journal of Medical Microbiology 302(4–5): 203–209. [DOI] [PubMed] [Google Scholar]
- Kristin W, Weitz J, et al. (2016). “Organoids as model systems for gastrointestinal diseases: tissue engineering meets.” Curr. Pathobiol. Rep 4: 1–9. [Google Scholar]
- Leslie JL, Huang S, et al. (2015). “Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function.” Infect Immun 83(1): 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahe MM, Aihara E, et al. (2013). “Establishment of Gastrointestinal Epithelial Organoids.” Current protocols in mouse biology 3(4): 217–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahe MM, Sundaram N, et al. (2015). “Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy.” Journal of visualized experiments : JoVE(97). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Cata EM, et al. (2014). “Modelling human development and disease in pluripotent stem-cell-derived gastric organoids.” Nature 516(7531): 400–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H and Stappenbeck TS (2013). “In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture.” Nat Protoc 8(12): 2471–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Schwartz RE, et al. (2015). “Human iPSC-Derived Hepatocyte-like Cells Support Plasmodium Liver-Stage Infection In Vitro.” Stem Cell Reports 4(3): 348–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ootani A, Li X, et al. (2009). “Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche.” Nat Med 15(6): 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkert RR and Kalejta RF (2013). “Human Embryonic Stem Cell Lines Model Experimental Human Cytomegalovirus Latency.” MBio 4(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelandt P, Obeid S, et al. (2012). “Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus.” Journal of Hepatology 57(2): 246–251. [DOI] [PubMed] [Google Scholar]
- Salama NR, Hartung ML, et al. (2013). “Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori.” Nature Reviews Microbiology 11(6): 385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange DE, et al. (2011). “Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium.” Gastroenterology 141(5): 1762–1772. [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, et al. (2011). “Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts.” Nature 469(7330): 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, et al. (2009). “Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche.” Nature 459(7244): 262–265. [DOI] [PubMed] [Google Scholar]
- Saxena K, Blutt SE, et al. (2015). “Human Intestinal Enteroids: a New Model To Study Human Rotavirus Infection, Host Restriction, and Pathophysiology.” J Virol 90(1): 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaermann P, Toelle B, et al. (2016). “A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro.” Gut 65(2): 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Feng R, et al. (2015). “Helicobacter pylori-induced Sonic Hedgehog Expression is Regulated by NF kappa B Pathway Activation: The Use of a Novel In vitro Model to Study Epithelial Response to Infection.” Helicobacter 20(1): 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G, Koo BK, et al. (2013). “Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients.” Cell stem cell 13(6): 653–658. [DOI] [PubMed] [Google Scholar]
- Schwartz RE, Trehan K, et al. (2012). “Modeling hepatitis C virus infection using human induced pluripotent stem cells.” Proceedings of the National Academy of Sciences of the United States of America 109(7): 2544–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai A, Schwartz RE, et al. (2014). “Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems.” Proceedings of the National Academy of Sciences of the United States of America 111(33): 12193–12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal M, Rothenberg ME, et al. (2015). “Helicobacter pylori Activates and Expands Lgr5(+) Stem Cells Through Direct Colonization of the Gastric Glands.” Gastroenterology 148(7): 1392–1404. [DOI] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, et al. (2011). “Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro.” Nature 470(7332): 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth BR and Lelkes PI (1998). “Growing tissues in microgravity.” Nature medicine 4(8): 901–907. [DOI] [PubMed] [Google Scholar]
- VanDussen KL, Marinshaw JM, et al. (2015). “Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays.” Gut 64(6): 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yamamoto Y, et al. (2015). “Cloning and variation of ground state intestinal stem cells.” Nature 522(7555): 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SS, Tocchi A, et al. (2015). “A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions.” Mucosal Immunology 8(2): 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski LE, Peek RM, et al. (2010). “Helicobacter pylori and Gastric Cancer: Factors That Modulate Disease Risk.” Clin Microbiol Rev 23(4): 713–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XF, Robotham JM, et al. (2012). “Productive Hepatitis C Virus Infection of Stem Cell-Derived Hepatocytes Reveals a Critical Transition to Viral Permissiveness during Differentiation.” Plos Pathogens 8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YB, Bijvelds M, et al. (2015). “Modeling rotavirus infection and antiviral therapy using primary intestinal organoids.” Antiviral Research 123: 120–131. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Takayama K, et al. (2011). “Use of human hepatocyte-like cells derived from induced pluripotent stem cells as a model for hepatocytes in hepatitis C virus infection.” Biochemical and Biophysical Research Communications 416(1–2): 119–124. [DOI] [PubMed] [Google Scholar]
- Yui S, Nakamura T, et al. (2012). “Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell.” Nat Med 18(4): 618–623. [DOI] [PubMed] [Google Scholar]
- Zhang YG, Wu S, et al. (2014). “Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions.” Physiol Rep 2(9). [DOI] [PMC free article] [PubMed] [Google Scholar]