Abstract
Purpose
Pet animals have been considered a potential carrier of clinically important multi-drug-resistant Escherichia coli. However, little is known about the role of pets as reservoirs of extended-spectrum β-lactamase (ESBL) producing E. coli in Pakistan. This study was designed to determine the prevalence and genetic relatedness of ESBL-producing multidrug-resistant E. coli in pets, their owners, and veterinary professionals.
Methods
A total of 105 fecal samples were collected from dogs, cats, their owners, and veterinary professionals from veterinary clinics. Isolates of ESBL-producing E. coli were subjected to antimicrobial susceptibility testing. The presence of blaCTX-M genes and CTX-M groups I and II in multidrug-resistant E. coli was detected using PCR. Clonal diversity was checked using BOX-PCR.
Results
Of the 105 fecal samples screened, 73 (69.5%) were found to contain ESBL-producing E. coli. The percentage of ESBL-producing E. coli isolates in dogs and dog owners was found to be 81.8% (18/22) and 59% (13/22), respectively. In cats, this percentage was 73.9% (17/23) and in cat owners, 56.5% (13/23). Furthermore, 80% (12/15) of E. coli isolates in veterinary professionals were ESBL producers. Of these 73 ESBL-producing E. coli isolates, 23 isolates exhibited a multidrug-resistant phenotype. The most prevalent multidrug-resistant pattern (17.4%) identified was resistant to ampicillin, cefotaxime, ciprofloxacin, and nitrofurantoin. In the multidrug-resistant E. coli, blaCTX-M was identified as the most common ESBL-producing genotype (19/23), with blaCTX-M-1 dominating in all 19 isolates. Furthermore, BOX-PCR analysis exhibited genetically diverse clonal groups among isolates of the CTX-M-1 group.
Conclusion
Our results provide important baseline information on the potential burden of multidrug-resistant E. coli among companion animals in Pakistan. Further studies are needed to understand the drivers of antimicrobial resistance at human–animal–environmental intersections.
Keywords: antimicrobial resistance, ESBL-producing E. coli, pets, zoonosis, multidrug resistance, one-health
Introduction
Antimicrobial resistance has become a global challenge for public health. The extended-spectrum cephalosporins are considered critically important antibiotics in both human and veterinary medicine. However, resistance to extended-spectrum cephalosporins mediated by extended-spectrum β-lactamase (ESBL) enzymes has become a well-recognized problem among Gram-negative bacteria. The main ESBL enzymes include CTX-M, TEM, and SHV classes.1 However, the dominance of CTX-M type ESBLs in human hospitals and community settings, and its occurrence in different animal hosts, suggests a one-health approach to determine the genetic relationship between Escherichia coli isolates from different hosts.2
Household pets could be potential causes of the spread of antimicrobial resistance, owing to the extensive use of antimicrobial agents in these animals and their close contact with human beings. The relationship between pet animals and human beings has drastically changed, with dogs in increasingly close contact with their owners. Nevertheless, the use of antimicrobial agents in companion animals implies selection as well as the potential to harbor and spread antimicrobial drug resistance, which constitutes a potential risk to public health.3 E. coli is a Gram-negative bacterium commonly present in the gastrointestinal tracts of human beings and animal species. The emergence of multidrug resistance in E. coli isolates of human and animal origin has raised serious concerns over the increasing risk of therapeutic failure.4,5 ESBL-producing E. coli has been considered an indicator for the transmission of antimicrobial resistance at the human–animal interface.6 Previous reports have suggested pets as reservoirs of clinically relevant ESBL-producing E. coli.7
High rates of ESBL-producing human clinical E. coli isolates have been reported in numerous studies from Pakistan in the last decade,8,9 but there is a scarcity of data on antimicrobial resistance among pet animals and their putative zoonotic connections. The objective of this study was to determine the occurrence and genetic relationship between ESBL-producing E. coli from pets, their owners, and veterinary professionals.
Materials and methods
Sample collection
A total of 105 fecal sample pairs from pets including dogs and cats (n=23 and n=22, respectively) and their owners (n=45) and veterinary professionals (n=15) were collected from July to December 2016 from the Veterinary Teaching Hospital, University of Agriculture, Faisalabad, and two private veterinary clinics in the Faisalabad district of Pakistan. These samples were transferred to the Zoonotic and Food Pathogen Research Laboratory, Institute of Microbiology, University of Agriculture, Faisalabad, under refrigerated conditions for further processing.
Isolation of ESBL-producing E. coli
All 105 rectal swabs were streaked directly onto Chrom agar ESBL (CHROMagar, Paris, France) for presumptive detection of ESBL-producing E. coli. Biochemical identification of E. coli was achieved using the RapID kit identification system (Remel, Kent, UK) according to the manufacturer’s instructions.
Antimicrobial susceptibility testing
Antimicrobial susceptibility of presumptive ESBL-producing E. coli isolates was determined on Muller Hinton agar (Oxoid, Basingstoke, UK) by the disk diffusion method, following the Clinical and Laboratory Standards Institute criteria with 30 µg each of ceftazidime, cefotaxime (CTX), and ceftriaxone. Phenotypic detection of ESBL production was achieved using the double-disk synergy test according to Clinical and Laboratory Standards Institute guidelines.10 Additionally, antimicrobial susceptibility to the following antimicrobials was tested: ampicillin (10 µg), CTX (30 µg), ertapenem (10 µg), ciprofloxacin (5 µg), tobramycin (30 µg), tigecycline (15 µg), nitrofurantoin (100 µg), and chloramphenicol (30 µg). The reference strain E. coli ATCC 25922 was used for quality control in antimicrobial susceptibility testing. The results were compared with Clinical and Laboratory Standards Institute standards; this strain was found to be susceptible to the given classes of antibiotics. Multidrug resistance was defined as phenotypic resistance to at least three antimicrobial drugs belonging to three different classes.11
Detection of blaCTX-M gene and blaCTX-M groups using PCR
DNA extraction was done manually by phenol-chloroform method.12 ESBL-producing E. coli isolates with multidrug-resistant phenotypes were investigated for the blaCTX-M gene with the primers and thermal cycler conditions reported previously.13 blaCTX-M-positive E. coli were further tested for the detection of major blaCTX-M groups, ie, CTX-M-1, CTX-M-2, and CTX-M-9, using a multiplex PCR approach.14 The primer sequence, gene target, amplified product size, and annealing conditions are listed in Table S1.
Plasmid extraction and replicon typing
Plasmid was extracted using Plasmid DNA kit (GeneJET plasmid miniprep kit; Fermentas, Glen Burnie, MD, USA), and PCR-based replicon typing was performed to determine plasmid incompatibility (Inc) groups.15 In addition, we further detected ESBL blaCTX-M gene on the extracted plasmids.
Genetic relationship between ESBL-producing E. coli isolates
To determine the clonal relationship between ESBL-producing E. coli with multidrug resistance, BOX-PCR was performed, as described earlier.16 The BOX-AIR primer 5′-CTA CGG CAA GGC GAC GCT GAC G-3′ was used with amplification conditions as follows: 1 cycle of 95°C for 4 minutes, 30 cycles of 94°C for 30 seconds, 92°C for 30 seconds, 50°C for 1 minute, 65°C for 3 minutes, and 1 cycle of 65°C for 8 minutes. The PCR products were run on 1% agarose gel at 90 V for ~1.5 hours. Gels were made and run in premade 1X TBE buffer. After gel electrophoresis, amplicons were visualized under an ultraviolet transilluminator (Dolphin-Doc, Wealtec, NV, USA). The BOX-PCR fingerprints were analyzed using Bionumerics software (Applied Maths, Kortrijk, Belgium). A dendrogram was constructed according to the unweighted pair group method, based on Dice coefficients. Isolates with a Dice similarity index ≥80% were considered the same clone.
Results
Prevalence of ESBL-producing E. coli
A total of 73/105 (69.5%) ESBL-producing E. coli isolates were detected in this study. The maximum percentage of ESBL-producing E. coli isolates was observed in dogs (18/22, 81.8%), followed by cats (17/23, 73.9%), whereas the detection rate in dog and cat owners was 13/22 (59.1%) and 13/23 (56.5%), respectively. Furthermore, 12/15 (80%) veterinary professionals carried ESBL E. coli.
Multidrug resistance pattern
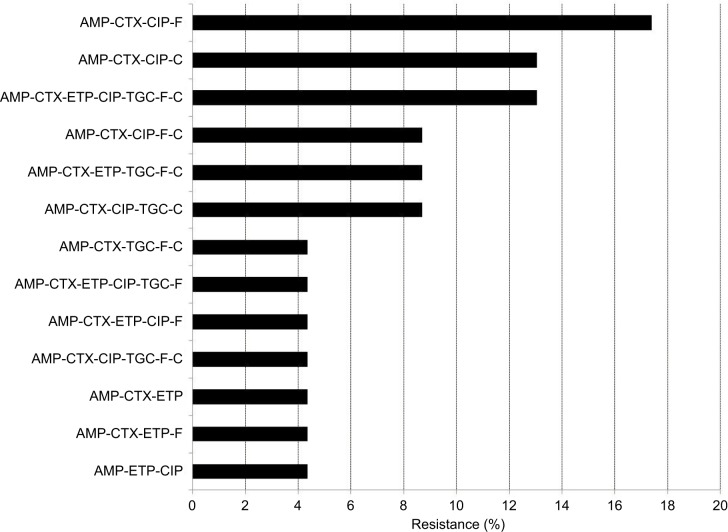
The multidrug resistance phenotype, defined as resistance to three or more antimicrobial classes, was detected in 23/73 (31.5%) isolates. The most common multidrug-resistant phenotype was resistance to ampicillin, CTX, ciprofloxacin, and nitrofurantoin (Figure 1).
Figure 1.
Multidrug resistance phenotypes.
Abbreviations: AMP, ampicillin; C, chloramphenicol; CIP, ciprofloxacin; CTX, cefotaxime; ETP, ertapenem; F, nitrofurantoin; TGC, tigecycline.
Frequency of blaCTX-M gene and CTX-M group
The overall prevalence of the blaCTX-M gene in ESBL-producing multidrug-resistant E.coli was 19/23 (82.6%). Of these, results showed that seven out of eight isolates of dog origin and three out of four isolates from dog owners carried the blaCTX-M gene. In cats 4/5 of isolates and in cat owners 3/4 of isolates harbored the blaCTX-M gene. All of the isolates (2/2) from veterinary professionals were found to carry the blaCTX-M gene. All CTX-M-positive multidrug-resistant ESBL-producing E. coli belonged to the CTX-M-1 group; both CTX-M-2 and CTX-M-9 groups were absent (Table 1).
Table 1.
Distribution of CTX-M gene and groups and plasmid replicon types
| Sample ID | Origin | CTX-M | CTX-M-1 | CTX-M-2 | CTX-M-9 | Replicon type |
|---|---|---|---|---|---|---|
|
| ||||||
| D115 | Dog | + | + | − | − | IncFIA |
| D302 | Dog | + | + | − | − | IncFII |
| D306 | Dog | − | − | − | − | IncFII |
| D108 | Dog | + | + | − | − | IncFII |
| D103 | Dog | + | + | − | − | IncFIA |
| D104 | Dog | + | + | − | − | IncFIA |
| D105 | Dog | + | + | − | − | IncFII |
| D101 | Dog | + | + | − | − | IncFIA |
| O105 | Dog owner | + | + | − | − | IncFIA |
| O109 | Dog owner | + | + | − | − | IncFIA |
| O114 | Dog owner | + | + | − | − | IncFIA |
| O301 | Dog owner | − | − | − | − | IncFII |
| C203 | Cat | + | + | − | − | IncFII |
| C207 | Cat | + | + | − | − | IncFII |
| C211 | Cat | − | − | − | − | IncFII |
| C312 | Cat | + | + | − | − | IncFII |
| C317 | Cat | + | + | − | − | IncFII |
| O202 | Cat owner | + | + | − | − | IncFII |
| O204 | Cat owner | + | + | − | − | IncFIA |
| O315 | Cat owner | + | + | − | − | IncB/O |
| O320 | Cat owner | − | − | − | − | IncFIA |
| Vp106 | Veterinary professional | + | + | − | − | IncFII |
| Vp108 | Veterinary professional | + | + | − | − | IncFIA |
Notes: In the first column, the letter represents the sources of the isolate and the numbers represent the isolate numbers.
Plasmid replicon types
Overall, replicon typing of the 23 ESBL-producing E. coli plasmids showed the predominant presence of IncFII (n=14) and IncFIA (n=8). One isolate carried IncB/O (Table 1). The ESBL encoding gene blaCTX-M was detectable from all the extracted plasmids.
Genetic diversity
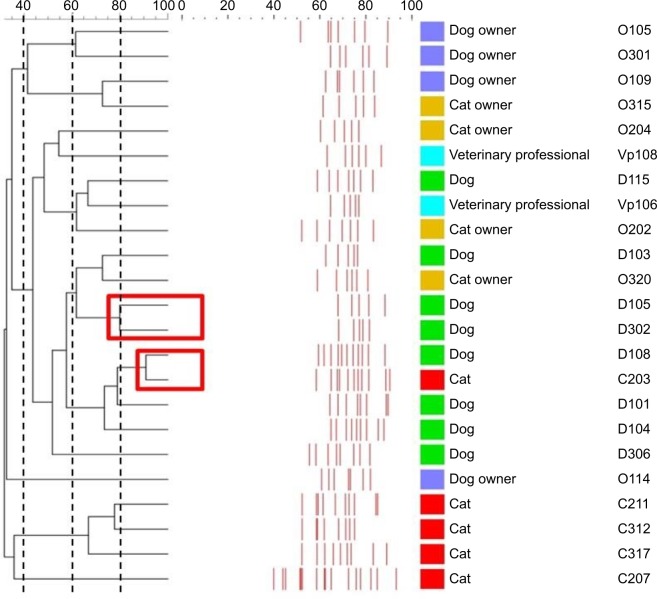
A large genetic diversity was found among ESBL-producing E. coli isolates in this study. Of 23 multidrug-resistant ESBL-producing E. coli isolates, 21 different clonal groups were identified using BOX-PCR analysis. The Dice’s coefficient among isolates varied from 0.3 (high genomic dissimilarity) to 0.8 (full genomic similarity), demonstrating the high polymorphism of the analyzed genotypes. Two clonal groups were found to have high genetic relatedness. The first clonal group consisted of two isolates of dog origin and the second comprised one isolate from a cat and one from a dog (Figure 2).
Figure 2.
Genetic relationship between multidrug-resistant Escherichia. coli isolates.
Notes: Dendrogram constructed using Bionumerics v 6.0 (Applied Maths) using Dice similarity coefficient. Isolates with a Dice similarity index ≥80% were considered to belong to the same clone.
Discussion
Antimicrobial resistance is an ongoing public health problem of global dimensions. In this study, we detected multidrug-resistant ESBL-producing E. coli from pets, their owners, and veterinary professionals, and determined their genetic relationships for the first time, to our knowledge, in Pakistan.
Our results demonstrated high rates of CTX-M-producing E. coli with multidrug resistance phenotypes in pets and their owners in Pakistan. Previously, several studies have reported the presence of ESBL-producing Enterobacteriaceae in clinical samples of human origin in Pakistan. One of these studies, probing ESBL-producing isolates among tertiary care hospital laboratories in Pakistan, detected 41% of ESBL-producing E.coli among 2,840 samples.17 Another study conducted on urinary tract pathogens showed high rates of ESBL-producing E. coli (53.4%). Most of these isolates were of multidrug-resistant pathogens.18 However, there is a lack of data on ESBL-producing E. coli among companion animals in Pakistan.
Globally, various studies have been conducted on the zoonotic impact of multidrug-resistant E. coli in pets and owners and their associated environments. We found 23/73 (31.5%) isolates of multidrug-resistant ESBL-producing E. coli from different sources, with the highest prevalence in dogs (8/18, 44.4%), followed by dog owners (4/13, 30.77%), cat owners 4/13 (30.8%), cats (5/17, 29.4%), and veterinary professionals (2/12, 16.7%). In a previous study conducted on dogs and dog owners, 31.3% of the total number of isolates collected contained multidrug-resistant E. coli.19 In another study, the prevalence of multidrug-resistant E. coli was reported as 8% in dogs and cats, which is quite low compared with our findings.20 This low occurrence of multidrug-resistant E. coli can be attributed to the antibiotic treatment of animals prior to the study.21 The persistence of multidrug resistance in dogs or dog owners has been variably reported in different studies. A study concluded that dogs and their owners harbor less percentage of multidrug-resistant isolates than the control groups.22 The current situation of resistance to antibiotics has reached a serious point in urinary tract infections. Presently, multidrug-resistant bacteria, including ESBL-producing bacteria, can be readily encountered in clinics,21 and animals presented for medical attention in hospitals are at greater risk of acquiring the antimicrobial-resistant bacteria.23
In our study, the highest frequency of resistance was found against ampicillin (80.8%) followed by CTX (39.7%) and nitrofurantoin (34.2%), while the least resistance was observed against ertapenem (12.3%) and tigecycline (19.1%) among E. coli isolates, while none of the isolates was resistant to tobramycin. Many studies reported resistance phenotype patterns similar to our results and described ampicillin and CTX as the most resistant phenotypes detected in pets.4,17 Almost similar but more drastic patterns of resistance have been reported in the clinical isolates of humans and animals by Tuerena et al.24 The use of antimicrobials in companion animals not only implies selection but also the potential to carry and spread antimicrobial drug resistance, which is a probable risk to public health.
We detected the globally prevalent type of multidrug-resistant gene, blaCTX-M, in our study. We found 19/23 (82.6%) of the multidrug-resistant isolates carrying the blaCTX-M gene in dogs, cats, pet owners, and veterinary professionals. Cluster analysis of the isolates revealed that all the CTX-M-producing isolates had the CTX-M-1 genotype. Previous reports also suggest that CTX-M-1 is the most prevalent genotype to be isolated from multidrug-resistant and ESBL-producing isolates.25 It has been reported that CTX-M was the most common (73.5%) genotype to be isolated from dogs and cats in the USA,26 which was comparable to our study. Another study illustrated the presence of CTX-M-1 in pets,27 although the prevalence was quite low when compared with our findings; similarly, veterinary professionals also carried the CTX-M-1 genotype. The results of our study suggest and favor the idea that CTX-M-1 is not only limited to human beings or animals in Pakistan, but is more prevalent than any other type of multidrug-resistant genotype in human beings and animals globally. In this study, plasmid replicon typing showed two common plasmids, FII and FIB. Similarly, in other Asian countries, IncFII plasmids associated with CTX-M genotypes have been reported from pets.28,29
Clonal diversity of the multidrug-resistant isolates was checked by BOX-PCR to determine any link between pets and pet owner’s isolates. Repetitive element sequence-based PCR fingerprinting (eg, BOX-PCR) has been shown to be a valuable tool for classifying and typing a variety of Gram-negative and several Gram-positive genera.30 The results of that study suggest that multidrug-resistant isolates of pets were genetically more similar to each other than those of pet owners or veterinary professionals. Interestingly, most of the multidrug-resistant isolates showed almost similar phenotypic profiles and resistant genotypes, while they were different genetically. This type of pattern has already been observed in studies of pets and pet owners in Brazil;19 it was elucidated that only the isolates from pets were genetically similar, rather than owners, but another study reported the genetical similarity of pets’ and pet owners’ isolates, based on pulsed-field gel electrophoresis.22 This inconsistency in genetic linkage between isolates from pets and pet owners may be because different E. coli strains acquire the antibiotic-resistant phenotypes and genotypes from the same source or because the same strain carrying multidrug-resistant phenotypes and genotypes was transmitted to both pets and pet owners.31 Transmission dynamics of antimicrobial resistance is complex and need more in-depth studies including multilocus sequence typing or whole genome sequencing to establish epidemiological evidence of transmission of ESBL-producing E. coli between human and animals.
Conclusion
Although the sample size of this study was limited, our results provide important baseline information on the potential burden of multidrug-resistant E. coli among the companion animals in Pakistan. Further studies are needed to understand the epidemiological and molecular characteristics of CTX-M type ESBL-producing E. coli in the companion animals for infection control and prevention in veterinary as well as in human medicine.
Ethics approval
The Institutional Bioethics and Biosafety Committee of the University of Agriculture, Faisalabad, has approved this study (D. No. 43/ORIC). This study was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all human participants.
Supplementary Materials
Table S1.
Primers used in study with target gene and amplicon size
| Target gene | Primer name | Sequence | Product size | Reference |
|---|---|---|---|---|
| blaCTX-M | CTX-M-F CTX-M-R |
5′SCS ATG TGC AGY ACC AGT3′ 5′ACC AGA AYV AGC GGB GC3′ |
585 bp | 13, 14 |
| blaCTX-M-1 | CTX-M-1-F CTX-M-1-R |
5′GCG TGA TAC CAC TTC ACC TC3′ 5′TGA AGT AAG TGA CCA GAA TC3′ |
260 bp | 13, 14 |
| blaCTX-M-2 | CTX-M-2-F CTX-M-2-R |
5′TGA TAC CAC CAC GCC GCT C3′ 5′TAT TGC ATC AGA AAC CGT GGG3′ |
341 bp | 13, 14 |
| blaCTX-M-9 | CTX-M-9-F CTX-M-9-R |
5′ATC AAG CCT GCC GAT CTG GTT A3′ 5′GTA AGC TGA CGC AAC GTC TGC3′ |
293 bp | 13, 14 |
Acknowledgments
We thank all pet owners and veterinary professionals involved in this study for their cooperation and help in sampling. This work was supported by the Federal German Foreign Office (grant reference number AA-OR 12–370.43), BIOS FLI PAK, and the Higher Education Commission, Pakistan (NRPU Grant number 4681).
Footnotes
Author contributions
Dr Mashkoor Mohsin is responsible for the experimental plan and is the corresponding author. All authors contributed to data analysis, preparing, and drafting the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Cantón R, González-Alba JM, Galán JC. CTX-M enzymes: origin and diffusion. Front Microbiol. 2012;3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chainier D, Barraud O, Masson G, et al. Integron digestive carriage in human and cattle: A “one health” cultivation-independent approach. Front Microbiol. 2017;1891:8. doi: 10.3389/fmicb.2017.01891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pomba C, Rantala M, Greko C, et al. Public health risk of antimicrobial resistance transfer from companion animals. J Antimicrob Chemother. 2016;72(4):957–968. doi: 10.1093/jac/dkw481. [DOI] [PubMed] [Google Scholar]
- 4.da Costa PM, Loureiro L, Matos AJF. Transfer of multidrug-resistant bacteria between intermingled ecological niches: the interface between humans, animals and the environment. Int J Environ Res Public Health. 2013;10(1):278–294. doi: 10.3390/ijerph10010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guenther S, Ewers C, Wieler LH. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol. 2011;2:246. doi: 10.3389/fmicb.2011.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohsin M, Raza S, Schaufler K, et al. High Prevalence of CTX-M-15-Type ESBL-Producing E. coli from Migratory Avian Species in Pakistan. Front Microbiol. 2017;8:2476. doi: 10.3389/fmicb.2017.02476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ljungquist O, Ljungquist D, Myrenås M, et al. Evidence of household transfer of ESBL-/pAmpC-producing Enterobacteriaceae between humans and dogs – a pilot study. Infect Ecol Epidemiol. 2016;6(1):31514. doi: 10.3402/iee.v6.31514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Habeeb MA, Haque A, Iversen A, Giske CG. Occurrence of virulence genes, 16S rRNA methylases, and plasmid-mediated quinolone resistance genes in CTX-M-producing Escherichia coli from Pakistan. Eur J Clin Microbiol Infect Dis. 2014;33(3):399–409. doi: 10.1007/s10096-013-1970-1. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad M, Hassan M, Khalid A, et al. Prevalence of Extended Spectrum β -Lactamase and Antimicrobial Susceptibility Pattern of Clinical Isolates of Pseudomonas from Patients of Khyber Pakhtunkhwa, Pakistan. BioMed Research International. 2016;2016(2):1–10. doi: 10.1155/2016/6068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2016. pp. M100–S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magiorakos A-P, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 13.Tofteland S, Haldorsen B, Dahl KH, et al. Effects of phenotype and genotype on methods for detection of extended-spectrum-beta-lactamase-producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in Norway. J Clin Microbiol. 2007;45(1):199–205. doi: 10.1128/JCM.01319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Ensor V, Gossain S, Nye K, Hawkey P. Rapid and simple detection of blaCTX-M genes by multiplex PCR assay. J Med Microbiol. 2005;54(Pt 12):1183–1187. doi: 10.1099/jmm.0.46160-0. [DOI] [PubMed] [Google Scholar]
- 15.Carattoli A, Bertini A, Villa L, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Versalovic J, Schneider M, de Bruin FJ, Lupski Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5(1):25–40. [Google Scholar]
- 17.Jabeen K, Zafar A, Hasan R. Frequency and sensitivity pattern of extended spectrum beta lactamase producing isolates in a tertiary care hospital laboratory of Pakistan. J Pak Med Assoc. 2005;55(10):436–439. [PubMed] [Google Scholar]
- 18.Ahmed I, Sajed M, Sultan A, et al. The erratic antibiotic susceptibility patterns of bacterial pathogens causing urinary tract infections. EXCLI J. 2015;14:916–925. doi: 10.17179/excli2015-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho AC, Barbosa AV, Arais LR, et al. Resistance patterns, ESBL genes, and genetic relatedness of Escherichia coli from dogs and owners. Braz J Microbiol. 2016;47(1):150–158. doi: 10.1016/j.bjm.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton E, Kruger JM, Schall W, et al. Acquisition and persistence of antimicrobial-resistant bacteria isolated from dogs and cats admitted to a veterinary teaching hospital. J Am Vet Med Assoc. 2013;243(7):990–1000. doi: 10.2460/javma.243.7.990. [DOI] [PubMed] [Google Scholar]
- 21.Lalak A, Wasyl D, Zając M, et al. Mechanisms of cephalosporin resistance in indicator Escherichia coli isolated from food animals. Vet Microbiol. 2016;194:69–73. doi: 10.1016/j.vetmic.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 22.Harada K, Okada E, Shimizu T, et al. Antimicrobial resistance, virulence profiles, and phylogenetic groups of fecal Escherichia coli isolates: a comparative analysis between dogs and their owners in Japan. Comp Immunol Microbiol Infect Dis. 2012;35(2):139–144. doi: 10.1016/j.cimid.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Lee DS, Lee CB, Lee S-J. Prevalence and risk factors for extended spectrum beta-lactamase-producing uropathogens in patients with urinary tract infection. Korean J Urol. 2010;51(7):492–497. doi: 10.4111/kju.2010.51.7.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuerena I, Williams NJ, Nuttall T, Pinchbeck G. Antimicrobial-resistant Escherichia coli in hospitalised companion animals and their hospital environment. J Small Anim Pract. 2016;57(7):339–347. doi: 10.1111/jsap.12525. [DOI] [PubMed] [Google Scholar]
- 25.Schaufler K, Bethe A, Lübke-Becker A, et al. Putative connection between zoonotic multiresistant extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in dog feces from a veterinary campus and clinical isolates from dogs. Infect Ecol Epidemiol. 2015;5(1):25334. doi: 10.3402/iee.v5.25334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Thungrat K, Boothe DM. Occurrence of OXA-48 carbapenemase and other β-lactamase genes in ESBL-producing multidrug resistant Escherichia coli from dogs and cats in the United States, 2009-2013. Front Microbiol. 2016;7:1057. doi: 10.3389/fmicb.2016.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randall LP, Clouting C, Horton RA, et al. Prevalence of Escherichia coli carrying extended-spectrum β-lactamases (CTX-M and TEM-52) from broiler chickens and Turkeys in Great Britain between 2006 and 2009. J Antimicrob Chemother. 2011;66(1):86–95. doi: 10.1093/jac/dkq396. [DOI] [PubMed] [Google Scholar]
- 28.So JH, Kim J, Bae IK, et al. Dissemination of multidrug-resistant Escherichia coli in Korean veterinary hospitals. Diagn Microbiol Infect Dis. 2012;73(2):195–199. doi: 10.1016/j.diagmicrobio.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Tamang MD, Nam H-M, Jang G-C, et al. Molecular characterization of extended-spectrum-β-lactamase-producing and plasmid-mediated AmpC β-lactamase-producing Escherichia coli isolated from stray dogs in South Korea. Antimicrob Agents Chemother. 2012;56(5):2705–2712. doi: 10.1128/AAC.05598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Versalovic J, Schneider M, de Bruijn FJ, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 31.Leite-Martins L, Meireles D, Beça N, et al. Spread of multidrug-resistant Escherichia coli within domestic aggregates (humans, pets, and household environment) J Vet Behav. 2015;10(6):549–555. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Primers used in study with target gene and amplicon size
| Target gene | Primer name | Sequence | Product size | Reference |
|---|---|---|---|---|
| blaCTX-M | CTX-M-F CTX-M-R |
5′SCS ATG TGC AGY ACC AGT3′ 5′ACC AGA AYV AGC GGB GC3′ |
585 bp | 13, 14 |
| blaCTX-M-1 | CTX-M-1-F CTX-M-1-R |
5′GCG TGA TAC CAC TTC ACC TC3′ 5′TGA AGT AAG TGA CCA GAA TC3′ |
260 bp | 13, 14 |
| blaCTX-M-2 | CTX-M-2-F CTX-M-2-R |
5′TGA TAC CAC CAC GCC GCT C3′ 5′TAT TGC ATC AGA AAC CGT GGG3′ |
341 bp | 13, 14 |
| blaCTX-M-9 | CTX-M-9-F CTX-M-9-R |
5′ATC AAG CCT GCC GAT CTG GTT A3′ 5′GTA AGC TGA CGC AAC GTC TGC3′ |
293 bp | 13, 14 |