Abstract
Background
Prostate cancer (PCa) is the second leading cause of cancer death in men. Several articles have reported that microRNA-21 (miR-21) and microRNA-30c (miR-30c) have diagnostic values for PCa, but the results are inconclusive. In order to precisely assess the diagnostic values of miR-21 and miR-30c for PCa, this meta-analysis is performed.
Methods
Articles were searched in the databases of PubMed, Embase, and Web of Knowledge (search date: September 6, 2018). Studies were included if they were designed to evaluate the diagnostic performance of miR-21 or miR-30c for PCa. Using Stata 12.0 and Meta-Disc 1.4, the pooled sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under curve (AUC) of the summary receiver-operating characteristic (SROC) curve with the corresponding 95% CI were calculated.
Results
Overall, ten studies (six studies for miR-21 and four for miR-30c) involving1,371 participants were included in this meta-analysis. For miR-21, the pooled SEN and SPE were, respectively, 0.91 (95% CI: 0.87–0.94) and 0.88 (95% CI: 0.82–0.93), the pooled PLR and NLR were, respectively, 7.74 (95% CI: 4.81–12.47), 0.1 (95% CI: 0.06–0.15), the DOR was 77.64 (95% CI: 34.64–174.02), AUC of SROC was 0.95 (95% CI: 0.93–0.97). For miR-30c, the pooled SEN and SPE were, respectively, 0.74 (95% CI: 0.65–0.81) and 0.78 (95% CI: 0.72–0.83), the pooled PLR and NLR were, respectively, 3.39 (95% CI: 2.69–4.26), and 0.34 (95% CI: 0.26–0.44), the DOR was 10.06 (95% CI: 6.96–14.55), and AUC of SROC was 0.83 (95% CI: 0.79–0.86).
Conclusion
For PCa, miR-21 is a good diagnostic biomarker and miR-30c is a moderate diagnostic biomarker.
Keywords: microRNA-21, microRNA-30c, prostate cancer, meta-analysis
Introduction
As a complex disorder resulting from the combined effects of multiple environmental and genetic factors, prostate cancer (PCa) is the second leading cause of cancer death in men.1 In Europe, it accounts for >92,300 deaths annually.2 As a routine assay test in clinic, prostate-specific antigen (PSA) in serum is not specific for PCa, some evaluated PSA levels may induce false-positive due to infection or hyperplasia. Therefore, it is necessary to find novel effective biomarkers to diagnose PCa.
MicroRNAs, a family of small noncoding RNAs (19±22 nucleotides), are stable and easy to accurately measure. MicroRNAs are differentially expressed in normal tissues and cancers, contributing to cancer development and progression.3 MicroRNA-21(miR-21), an anti-apoptotic agent functioning via p53 network, can target and inhibit tumor suppressor gene PTEN expression to promote PCa cell proliferation and invasion.4,5 MicroRNA-30c (miR-30c) has been acknowledged as a tumor suppressor in various human cancers, such as ovarian cancer, gastric cancer, and PCa.6
With respect to diagnostic abilities of miR-21 and miR-30c for PCa, several studies have been conducted, but their results are inconclusive.7–14 For example, Porzycki et al found specificity (SPE) of miR-21 to diagnose PCa was 0.75, while Yang et al found it could reach 0.93.8,9 What’s more, Huang found sensitivity (SEN) of miR-30c was 0.82, while Chen et al found it was only 0.6.11,14 In order to precisely assess the diagnostic values of miR-21 and miR-30c for PCa, this meta-analysis is performed.
Methods
This meta-analysis was conducted according to the guidelines of the PRISMA statement (Table S1).15
Eligibility criteria
This meta-analysis selected the patients diagnosed as PCa and the control consisted of healthy people or patients with benign prostate disease. The index tests, which included miR-21 and miR-30c from serum, plasma, mononuclear cell, and tissue, were assessed by fluorescence quantitative PCR. Reference standard was histopathological test, PCa was diagnosed based on histopathological confirmation.
Studies were included if they met following inclusion criteria: 1) studies were designed to evaluate the diagnostic performance of miR-21 or miR-30c for PCa; 2) PCa was diagnosed based on histopathological confirmation and the control group consisted of healthy people or patients with benign prostate disease; 3) 2×2 contingency tables could be directly extracted or calculated; and 4) articles were written in English.
Search strategy
Articles published prior to September 6, 2018, were systematically searched in the databases of PubMed, Embase, and Web of Knowledge. Search terms were as follows: (“microRNA” or “miRNA” or “miR”) and (“prostate cancer” or “prostate carcinoma”) and (“diagnosis” or “sensitivity” or “specificity”). Search strategy is shown in Table S2. In addition, cited references from relevant articles were examined to select other relevant articles. Two reviewers independently searched the articles. Any disagreements between the two reviewers were resolved by discussion.
Study selection
The titles and abstracts of the identified articles were examined by two reviewers. Those articles falling under the exclusion criteria (reviews, case reports, letters, comments, conference abstracts, or meta-analyses; animal studies; duplicate studies) were not considered. The full texts were then rescreened and evaluated more thoroughly for eligibility using the same exclusion criteria. Disagreement between the two reviewers was discussed until a consensus was reached.
Data extraction
The following data were extracted or calculated from retrieved articles by the two reviewers: first author, year of publication, country of participants, specimen type, source of control group, sample size, microRNAs as control for normalization, microRNAs as biomarker, relative expressions in case group compared with control group, diagnostic SEN and SPE values, values of true positive (TP), false positive (FP), false negative (FN) and true negative (TN).
Methodological quality assessment
Two reviewers assessed the methodological quality of included studies according to the quality assessment of diagnostic accuracy studies-2 (QUADAS-2) tool.16 Patient selection, index test, reference standard, flow, and timing were used to evaluate the risk of bias and the first three domains were applied to assess applicability concerns.
Statistical analysis
Statistical analyses were undertaken utilizing Stata 12.0 (StataCorp, College Station, TX, USA) and Meta-Disc 1.4 (Cochrane Colloquium, Barcelona, Spain). Diagnostic threshold effects were inspected with the Spearman rank correlation analysis. Heterogeneity was measured using Cochrane-Q test and I2 index. P<0.05 for Q test or I2 >50% designated heterogeneity. The pooled SEN and SPE, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic score (DS), diagnostic odds ratio (DOR), and area under curve (AUC) of the summary receiver operating characteristic (SROC) curve with the corresponding 95% CI were calculated using a bivariate binomial mixed model. Deeks’ funnel plot asymmetry analysis was performed to identify publication bias. Sensitivity analysis was conducted to assess the stability.
Results
Literature search and study characteristics
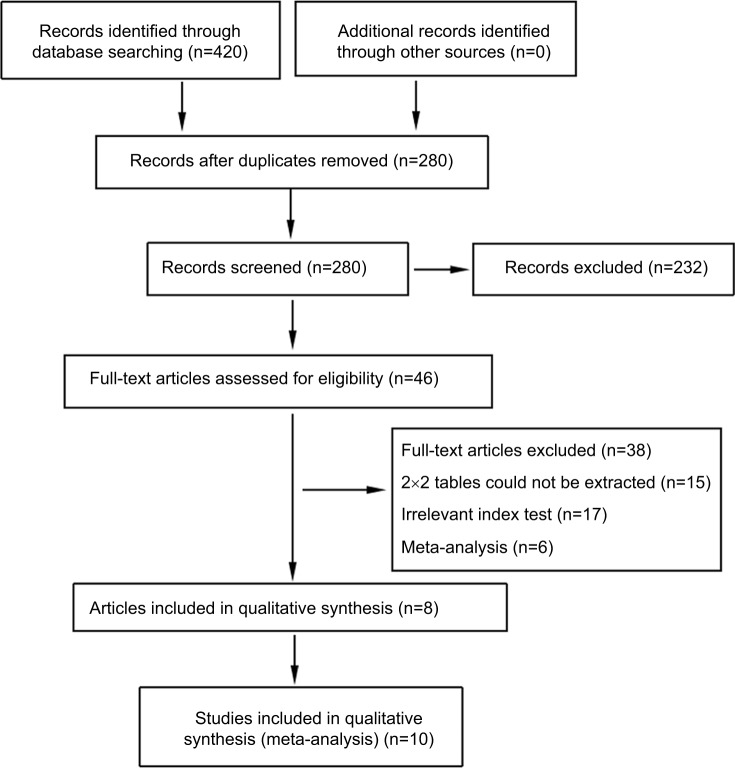
A total of 420 records were identified through database searching. After removing duplicates, the titles and abstracts for 280 records were screened for eligibility. Of these, 46 records were identified and full-text articles were retrieved. Thirty-eight manuscripts were excluded through assessment of the full-text articles and eight remaining articles encompassing ten studies were included in the meta-analysis (Figure 1).
Figure 1.
Flow diagram of study selection.
The ten included studies owned 1,371 participants (737 PCa patients and 634 controls). Their characteristics and the numbers of TP, FP. FN, and TN with the corresponding SEN, SPE are listed in Table 1. The number of included studies for miR-21 and miR-30c were, respectively, six and four. Countries of participants for miR-21 involved Poland, China, and Egypt, while miR-30c only involved China. Relative expressions of miR-21 in case group were higher than that in control group. On the contrary, relative expressions of miR-30c were lower in case group. Two studies assessed miR-30c from PCa cancer tissue and PCa adjacent normal tissue,7, 11 and other two studies assessed miR-30c from plasma of PCa patients and control group.14 Studies assessed miR-21 from serum, plasma, mononuclear cell of PCa patients, and control group.
Table 1.
Characteristics of included studies
| Author | Year | Country | Specimen | Control group | MiR as control | Case size | Control size | MiR as biomarker | Relative expressiona | SEN | SPE | TP | FP | FN | TN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Zhu et al7 | 2018 | China | Tissueb | U6 | 187 | 187 | miR-30c | Low | 0.72 | 0.80 | 135 | 37 | 52 | 150 | |
| Porzycki et al8 | 2018 | Poland | Serum | HP | U6 | 20 | 10 | miR-21 | High | 0.90 | 0.75 | 18 | 2 | 2 | 8 |
| Yang et al9 | 2016 | China | Mononuclear cell | PBPD | miR-16 | 92 | 85 | miR-21 | High | 0.94 | 0.93 | 86 | 6 | 6 | 79 |
| Yang et al9 | 2016 | China | Mononuclear cell | HP | miR-16 | 92 | 97 | miR-21 | High | 0.95 | 0.93 | 87 | 7 | 5 | 90 |
| Gao et al10 | 2016 | China | Plasma | PBPD | U6 | 57 | 28 | miR-21 | High | 0.88 | 0.75 | 50 | 7 | 7 | 21 |
| Huang et al11 | 2016 | China | Tissueb | 44 | 44 | miR-30c | Low | 0.82 | 0.80 | 36 | 9 | 8 | 35 | ||
| Huang et al12 | 2015 | China | Mononuclear cell | HP | U6 | 75 | 75 | miR-21 | High | 0.88 | 0.86 | 66 | 11 | 9 | 64 |
| Kotb et al13 | 2014 | Egypt | Serum | PBPD | SNORD47 | 10 | 10 | miR-21 | High | 0.90 | 0.90 | 9 | 1 | 1 | 9 |
| Chen et al14 | 2012 | China | Plasma | HP | U6 | 80 | 54 | miR-30c | Low | 0.79 | 0.69 | 64 | 17 | 16 | 37 |
| Chen et al14 | 2012 | China | Plasma | PBPD | U6 | 80 | 44 | miR-30c | Low | 0.61 | 0.83 | 49 | 8 | 31 | 36 |
Notes:
Relative expression of miR-21 or miR-30c as biomarker in case group compared with control group.
Include PCa cancer tissue and PCa adjacent normal tissue, cancer tissue was regarded as case group and adjacent normal tissue as control group.
Abbreviations: PBPD, patients with benign prostate disease; HP, healthy people; SEN, sensitivity; SPE, specificity; TP, true positive; FP, false positive; FN, false negative; TN, true negative.
Methodological quality assessment
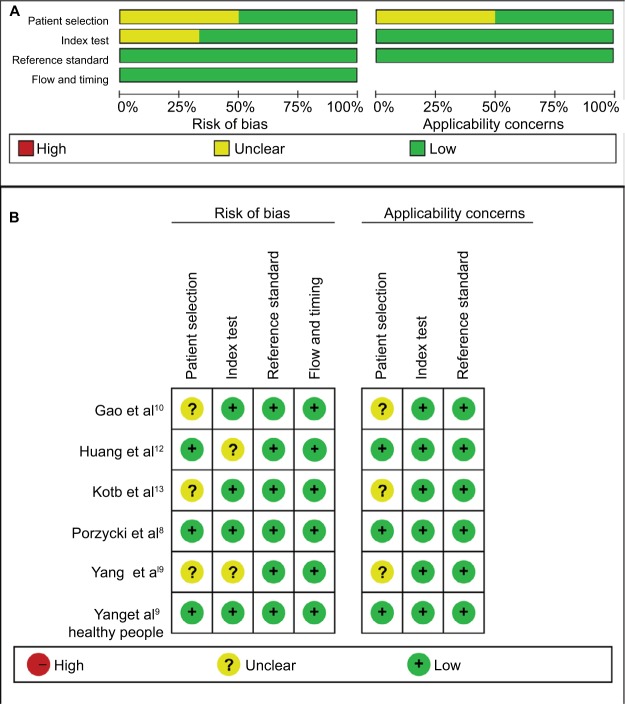
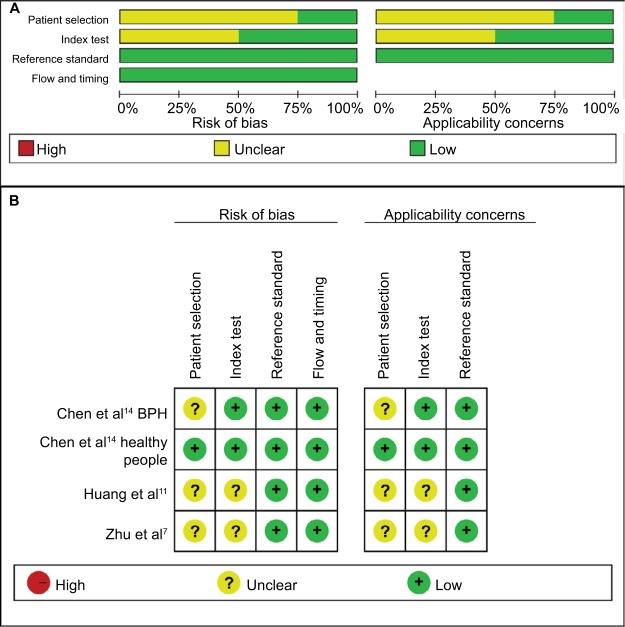
Quality assessment of included studies was conducted using the QUADAS-2 tool (Figures 2 and 3). The majority of included studies satisfied most domains of QUADAS-2. The unclear situation existed in the domains of patient selection and index test because there were variations in control group, specimen, and microRNAs as control for normalization.
Figure 2.
Quality assessment of miR-21 according to QUADAS-2 guidelines.
Note: (A) Risk of bias, (B) applicability concerns.
Figure 3.
Quality assessment of miR-30c according to QUADAS-2 guidelines.
Note: (A) Risk of bias, (B) applicability concerns.
Diagnostic values
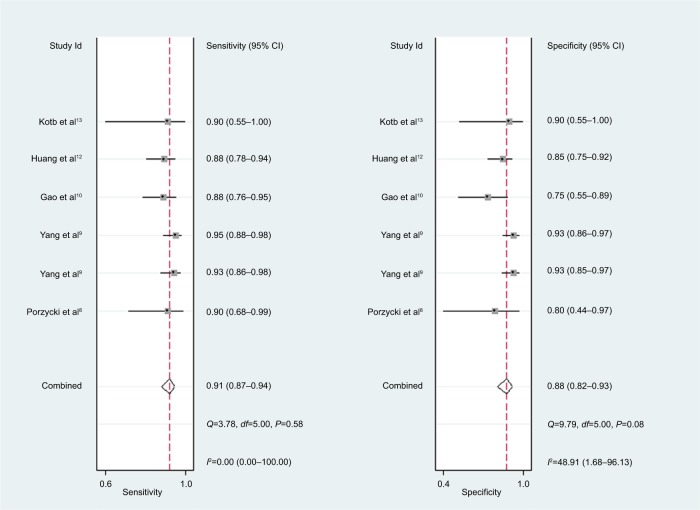
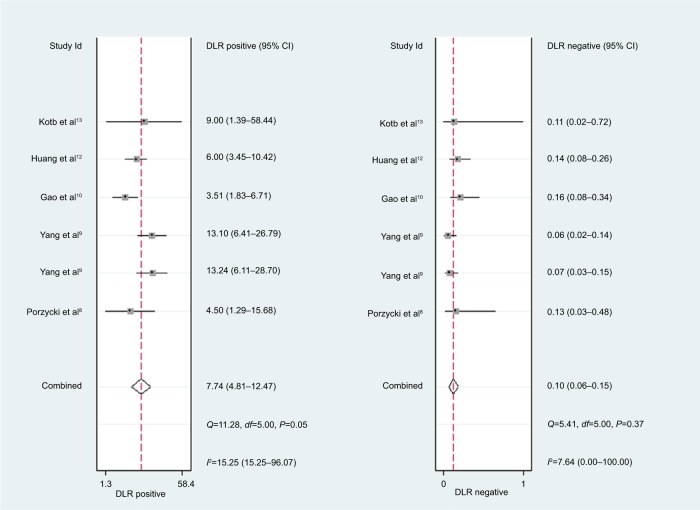
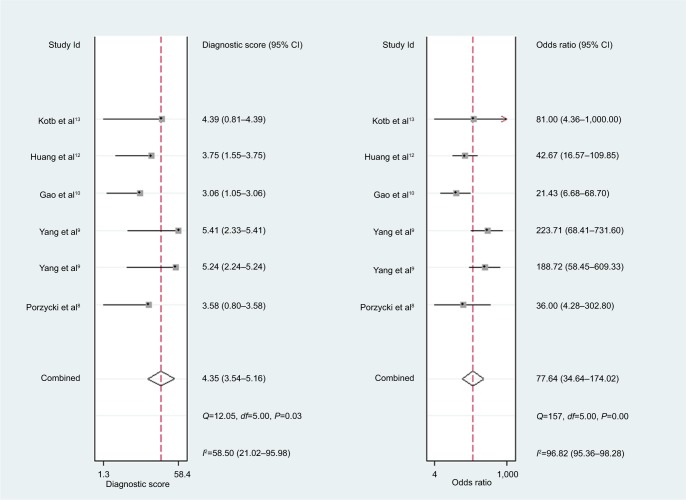
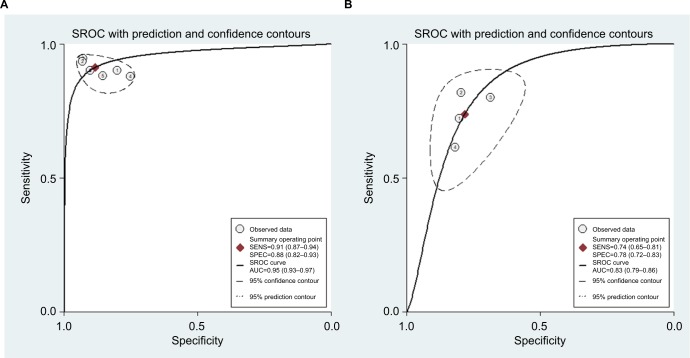
For miR-21, the Spearman rank correlation coefficient was –0.841 (P=0.036), suggesting that there were no diagnostic threshold effects. The pooled SEN and SPE were, respectively, 0.91 (95% CI: 0.87–0.94) and 0.88 (95% CI: 0.82–0.93) (Figure 4). I2 for SEN and SPE were, respectively, 0% (P=0.58) and 48.9% (P=0.08), showing no significant heterogeneity among studies. The pooled PLR and NLR were, respectively, 7.74 (95% CI: 4.81–12.47) and 0.1 (95% CI: 0.06–0.15) (Figure 5). The pooled DS and DOR were, respectively, 4.35 (95% CI: 3.54–5.16) and 77.64 (95% CI: 34.64–174.02) (Figure 6). AUC of SROC curve was 0.95 (95% CI: 0.93–0.97) (Figure 7).
Figure 4.
Forest plots of sensitivity and specificity for miR-21.
Figure 5.
Forest plots of positive likelihood ratio and negative likelihood ratio for miR-21.
Abbreviation: DLR, diagnostic likelihood ratio.
Figure 6.
Forest plots of diagnostic score and diagnostic odds ratio for miR-21.
Figure 7.
Summary receiver operating characteristic curves.
Note: (A) miR-21, (B) miR-30c.
Abbreviations: AUC, area under curve; SROC, summary receiver- operating characteristic; SENS, sensitivity; SPEC, specificity.
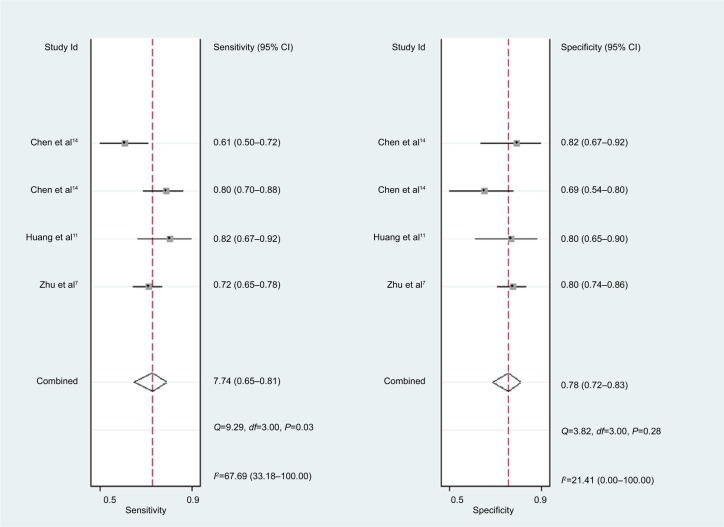
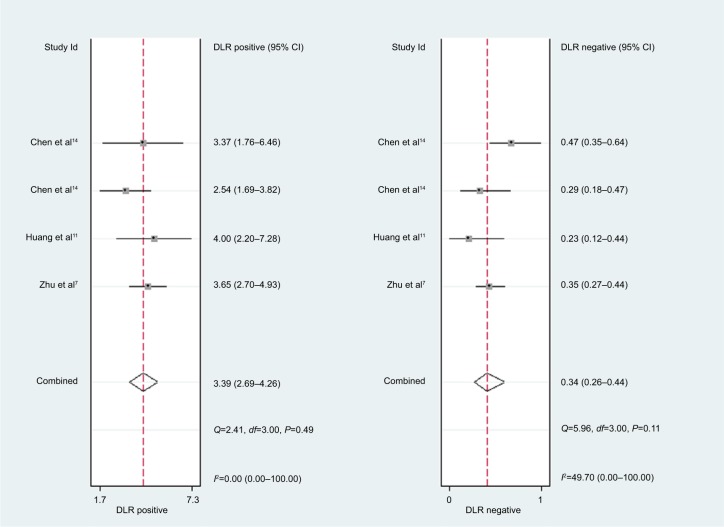
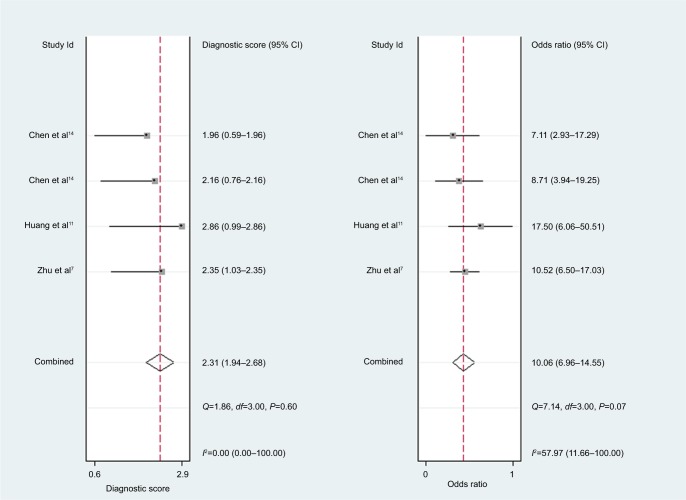
For miR-30c, the Spearman rank correlation coefficient was 0.80 (P=0.2), suggesting no existence of diagnostic threshold effects. The pooled SEN and SPE were, respectively, 0.74 (95% CI: 0.65–0.81) and 0.78 (95% CI: 0.72–0.83) (Figure 8). I2 for SEN and SPE were, respectively, 67.69% (P=0.03) and 21.41% (P=0.28), revealing that significant heterogeneities existed among studies. The pooled PLR and NLR were, respectively, 3.39 (95% CI: 2.69–4.26) and 0.34 (95% CI: 0.26–0.44) (Figure 9). The pooled DS and DOR were, respectively, 2.31 (95% CI: 1.94–2.68) and 10.06 (95% CI: 6.96–14.55) (Figure 10). AUC of SROC curve was 0.83 (95% CI: 0.79–0.86) (Figure 7).
Figure 8.
Forest plots of sensitivity and specificity for miR-30c.
Figure 9.
Forest plots of positive likelihood ratio and negative likelihood ratio for miR-30c.
Abbreviation: DLR, diagnostic likelihood ratio.
Figure 10.
Forest plots of diagnostic score and diagnostic odds ratio for miR-30c.
Publication bias
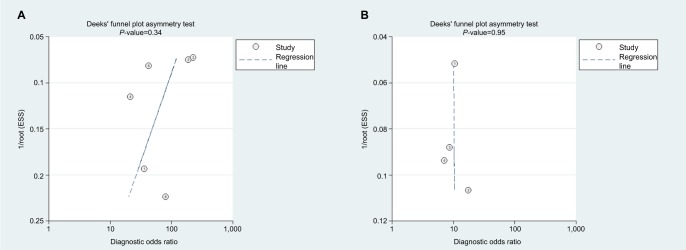
As shown in Deeks’ funnel plots (Figure 11), publication biases were likely absent with symmetrical funnel shapes. The P-values for the funnel plot asymmetry tests (P=0.43 for miR-21 and P=0.95 for miR-30c) confirmed lack of publication biases.
Figure 11.
Deeks’ funnel plots.
Note: (A) miR-21, (B) miR-30c.
Sensitivity analysis
Sensitivity analysis was performed by eliminating studies one by one. As shown in Table S3, the pooled SEN and corresponding heterogeneities of miR-21 and miR-30c were not dominantly influenced by removing any study.
Discussion
The pooled SEN and SPE of miR-21 were, respectively, 0.91 and 0.88, and the AUC of SROC was 0.95, presenting a good diagnostic ability of miR-21. The pooled PLR of 7.74 indicated that PCa probability increased by 7.74-fold with positive miR-21 result. Conversely, the pooled NLR of 0.1 showed the probability could only be 10% when miR-21 result was negative. As a combination of PLR and NLR, DOR is a meaningful diagnostic index. With a DOR value of 77.64, it was confirmed that miR-21 could be a good diagnostic biomarker for PCa. Sensitivity analysis showed that the conclusion of this meta-analysis was robust. There is a research which suggests that miR-21 and androgen receptor axis can exert oncogenic effects in prostate tumors by downregulating transforming growth factor beta receptor II.17
For miR-30c, this meta-analysis showed that pooled SEN and SPE were, respectively, 0.74 and 0.78; furthermore AUC of SROC was 0.83, presenting a moderate diagnostic ability of miR-30c. Significant heterogeneities were found among included studies. Although sensitivity analysis was carried out, it was still hard to identify which study was the source of heterogeneities.
Three previous meta-analyses (by Yin et al, Ouyang et al, and Yang et al) also reported the diagnostic value of microRNAs for PCa,18–20 we read them carefully with great interest. The number for miR-21 of enrolled studies by a previous meta-analysis (by Yin)18 was less than our meta-analysis. Other two meta-analyses (by Ouyang et al and Yang et al)19,20 only focused on overall microRNAs but not each single microRNA, inevitably brought about significant heterogeneities, lower quality assessments, and meaninglessness in application. Another previous meta-analysis by Song et al provided mean ± SD or fold change of microRNAs expression levels in PCa patients vs controls,21 but did not provide diagnostic information, such as pooled SEN and SPE.
Limitations
There were several limitations in our meta-analysis. First, the situation of unclear risk in the domains of patient selection and index could lower methodological qualities. Second, significant heterogeneities existed among included studies of miR-30c. Despite sensitive analysis being performed, potential sources of heterogeneities were still hard to been found. Third, comparison with blood-based specimen, tissue-based specimen is difficult to obtain in real life. Fourth, the number of included studies were few. It is necessary to adopt standardization of control group, specimen, and index test and to conducted more related studies.
Conclusion
Our meta-analysis showed that, for PCa, miR-21 is a good diagnostic biomarker and miR-30c is a moderate diagnostic biomarker.
Supplementary material
Table S1.
PRISMA Checklist
| Section/topic | # | Checklist item | Reported on page # |
|---|---|---|---|
| TITLE | |||
| Title | 1 | Identify the report as a systematic review, meta-analysis, or both. | 1 |
| ABSTRACT | |||
| Structured summary | 2 | Provide a structured summary including, as applicable: background; objectives; data sources; study eligibility criteria, participants, and interventions; study appraisal and synthesis methods; results; limitations; conclusions and implications of key findings; systematic review registration number. | 2 |
| INTRODUCTION | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known. | 3 |
| Objectives | 4 | Provide an explicit statement of questions being addressed with reference to participants, interventions, comparisons, outcomes, and study design (PICOS). | 3 |
| METHODS | |||
| Protocol and registration | 5 | Indicate if a review protocol exists, if and where it can be accessed (e.g., Web address), and, if available, provide registration information including registration number. | Not applicable |
| Eligibility criteria | 6 | Specify study characteristics (e.g., PICOS, length of follow-up) and report characteristics (e.g., years considered, language, publication status) used as criteria for eligibility, giving rationale. | 4 |
| Information sources | 7 | Describe all information sources (e.g., databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched. | 4 |
| Search | 8 | Present full electronic search strategy for at least one database, including any limits used, such that it could be repeated. | 4 (Table S2) |
| Study selection | 9 | State the process for selecting studies (i.e., screening, eligibility, included in systematic review, and, if applicable, included in the meta-analysis). | 4 |
| Data collection process | 10 | Describe method of data extraction from reports (e.g., piloted forms, independently, in duplicate) and any processes for obtaining and confirming data from investigators. | 4 |
| Data items | 11 | List and define all variables for which data were sought (e.g., PICOS, funding sources) and any assumptions and simplifications made. | 5 |
| Risk of bias in individual studies | 12 | Describe methods used for assessing risk of bias of individual studies (including specification of whether this was done at the study or outcome level), and how this information is to be used in any data synthesis. | 5 |
| Summary measures | 13 | State the principal summary measures (e.g., risk ratio, difference in means). | 5 |
| Synthesis of results | 14 | Describe the methods of handling data and combining results of studies, if done, including measures of consistency (e.g., I2) for each meta-analysis. | 5–6 |
| Section/topic | # | Checklist item | Reported on page # |
| Risk of bias across studies | 15 | Specify any assessment of risk of bias that may affect the cumulative evidence (e.g., publication bias, selective reporting within studies). | 6 |
| Additional analyses | 16 | Describe methods of additional analyses (e.g., sensitivity or subgroup analyses, meta-regression), if done, indicating which were pre-specified. | 6 |
| RESULTS | |||
| Study selection | 17 | Give numbers of studies screened, assessed for eligibility, and included in the review, with reasons for exclusions at each stage, ideally with a flow diagram. | 6 |
| Study characteristics | 18 | For each study, present characteristics for which data were extracted (e.g., study size, PICOS, follow-up period) and provide the citations. | 6–7 |
| Risk of bias within studies | 19 | Present data on risk of bias of each study and, if available, any outcome level assessment (see item 12). | 7 |
| Results of individual studies | 20 | For all outcomes considered (benefits or harms), present, for each study: (a) simple summary data for each intervention group (b) effect estimates and confidence intervals, ideally with a forest plot. | 7–8 |
| Synthesis of results | 21 | Present results of each meta-analysis done, including confidence intervals and measures of consistency. | 7–8 |
| Risk of bias across studies | 22 | Present results of any assessment of risk of bias across studies (see Item 15). | 7 |
| Additional analysis | 23 | Give results of additional analyses, if done (e.g., sensitivity or subgroup analyses, meta-regression [see Item 16]). | 8 |
| DISCUSSION | |||
| Summary of evidence | 24 | Summarize the main findings including the strength of evidence for each main outcome; consider their relevance to key groups (e.g., healthcare providers, users, and policy makers). | 8–9 |
| Limitations | 25 | Discuss limitations at study and outcome level (e.g., risk of bias), and at review-level (e.g., incomplete retrieval of identified research, reporting bias). | 9 |
| Conclusions | 26 | Provide a general interpretation of the results in the context of other evidence, and implications for future research. | 10 |
| FUNDING | |||
| Funding | 27 | Describe sources of funding for the systematic review and other support (e.g., supply of data); role of funders for the systematic review. | 13 |
Table S2.
Search strategy
| Search strategy | |
|---|---|
|
| |
| #1 | “microRNA” OR “miRNA” OR “miR” AND “prostate cancer” AND “diagnosis” |
| #2 | “microRNA” OR “miRNA” OR “miR” AND “prostate cancer” AND “sensitivity” |
| #3 | “microRNA” OR “miRNA” OR “miR” AND “prostate cancer” AND “specificity” |
| #4 | “microRNA” OR “miRNA” OR “miR” AND “prostate carcinoma” AND “diagnosis” |
| #5 | “microRNA” OR “miRNA” OR “miR” AND “prostate carcinoma” AND “sensitivity” |
| #6 | “microRNA” OR “miRNA” OR “miR” AND “prostate carcinoma” AND “specificity” |
Table S3.
Sensitivity analysis for pooled sensitivities and corresponding heterogeneities
| A | |||
|---|---|---|---|
|
| |||
| Studya | SEN (95% CI) | I2 index (%) | Pb |
|
| |||
| Porzycki et al1 | 0.91(0.87–0.94) | 0 | 0.43 |
| Yang et al2,c | 0.90 (0.85–0.94) | 0 | 0.58 |
| Yang et al2,d | 0.90 (0.85–0.93) | 0 | 0.75 |
| Gao et al3 | 0.92 (0.88–0.95) | 0 | 0.58 |
| Huang et al4 | 0.92 (0.87–0.95) | 0 | 0.61 |
| Kotb et al5 | 0.91(0.88–0.94) | 0 | 0.434 |
|
| |||
| B | |||
|
| |||
| Studya | SEN (95% CI) | I2 index (%) | Pb |
|
| |||
| Zhu et al6 | 0.73(0.66–0.79) | 78.3 | 0.01 |
| Huang et al7 | 0.72 (0.66–0.76) | 71.3 | 0.03 |
| Chen et al8,c | 0.71 (0.65–0.76) | 68.5 | 0.04 |
| Chen et al8,d | 0.76 (0.70–0.80) | 33.4 | 0.22 |
Notes: (A) miR-21, (B) miR-30c.
Given named study is omitted.
P-value of the Cochrane-Q test.
Control group consisted of patients with benign prostate disease.
Control group consisted of healthy people.
References
- 1.Porzycki P, Ciszkowicz E, Semik M, Tyrka M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int Urol Nephrol. 2018;50(9):1619–1626. doi: 10.1007/s11255-018-1938-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang B, Liu Z, Ning H, et al. MicroRNA-21 in peripheral blood mono-nuclear cells as a novel biomarker in the diagnosis and prognosis of prostate cancer. Cancer Biomark. 2016;17(2):223–230. doi: 10.3233/CBM-160634. [DOI] [PubMed] [Google Scholar]
- 3.Gao Y, Guo Y, Wang Z, et al. Analysis of circulating miRNAs 21 and 375 as potential biomarkers for early diagnosis of prostate cancer. Neoplasma. 2016;63(4):623–628. doi: 10.4149/neo_2016_417. [DOI] [PubMed] [Google Scholar]
- 4.Huang W, Kang XL, Cen S, Wang Y, Chen X. High-level expression of microRNA-21 in peripheral blood mononuclear cells is a diagnostic and prognostic marker in prostate cancer. Genet Test Mol Biomarkers. 2015;19(9):469–475. doi: 10.1089/gtmb.2015.0088. [DOI] [PubMed] [Google Scholar]
- 5.Kotb S, Mosharafa A, Essawi M, Hassan H, Meshref A, Morsy A. Circulating miRNAs 21 and 221 as biomarkers for early diagnosis of prostate cancer. Tumour Biol. 2014;35(12):12613–12617. doi: 10.1007/s13277-014-2584-7. [DOI] [PubMed] [Google Scholar]
- 6.Zhu C, Hou X, Zhu J, Jiang C, Wei W. Expression of miR-30c and miR-29b in prostate cancer and its diagnostic significance. Oncol Lett. 2018;16(3):3140–3144. doi: 10.3892/ol.2018.9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Z, Zhang L, Yi X, Yu X. Diagnostic and prognostic values of tissue hsa-miR-30c and hsa-miR-203 in prostate carcinoma. Tumour Biol. 2016;37(4):4359–4365. doi: 10.1007/s13277-015-4262-9. [DOI] [PubMed] [Google Scholar]
- 8.Chen ZH, Zhang GL, Li HR, et al. A panel of five circulating microRNAs as potential biomarkers for prostate cancer. Prostate. 2012;72(13):1443–1452. doi: 10.1002/pros.22495. [DOI] [PubMed] [Google Scholar]
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Collak FK, Demir U, Ozkanli S, Kurum E, Zerk PE. Increased expression of YAP1 in prostate cancer correlates with extraprostatic extension. Cancer Biol Med. 2017;14(4):405–413. doi: 10.20892/j.issn.2095-3941.2017.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson HC, Shah SI, Abel PD, et al. Contemporary hormone therapy with LHRH agonists for prostate cancer: avoiding osteoporosis and fracture. Cent European J Urol. 2015;68(2):165–168. doi: 10.5173/ceju.2015.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W, Zhang E, Lin C. MicroRNAs in tumor angiogenesis. Life Sci. 2015;136:28–35. doi: 10.1016/j.lfs.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 4.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68(19):8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y, Guo JX, Shao ZQ. miR-21 targets and inhibits tumor suppressor gene PTEN to promote prostate cancer cell proliferation and invasion: an experimental study. Asian Pac J Trop Med. 2017;10(1):87–91. doi: 10.1016/j.apjtm.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Li X, Zhuang S. MiR-30C impedes glioblastoma cell proliferation and migration by targeting SOX9. Oncol Res. 2019;27(2):165–171. doi: 10.3727/096504018X15193506006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu C, Hou X, Zhu J, Jiang C, Wei W. Expression of miR-30C and miR-29b in prostate cancer and its diagnostic significance. Oncol Lett. 2018;16(3):3140–3144. doi: 10.3892/ol.2018.9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porzycki P, Ciszkowicz E, Semik M, Tyrka M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int Urol Nephrol. 2018;50(9):1619–1626. doi: 10.1007/s11255-018-1938-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang B, Liu Z, Ning H, et al. MicroRNA-21 in peripheral blood mono-nuclear cells as a novel biomarker in the diagnosis and prognosis of prostate cancer. CBM. 2016;17(2):223–230. doi: 10.3233/CBM-160634. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y, Guo Y, Wang Z, et al. Analysis of circulating miRNAs 21 and 375 as potential biomarkers for early diagnosis of prostate cancer. Neoplasma. 2016;63(4):623–628. doi: 10.4149/neo_2016_417. [DOI] [PubMed] [Google Scholar]
- 11.Huang Z, Zhang L, Yi X, Yu X. Diagnostic and prognostic values of tissue hsa-miR-30c and hsa-miR-203 in prostate carcinoma. Tumor Biol. 2016;37(4):4359–4365. doi: 10.1007/s13277-015-4262-9. [DOI] [PubMed] [Google Scholar]
- 12.Huang W, Kang XL, Cen S, Wang Y, Chen X. High-level expression of microRNA-21 in peripheral blood mononuclear cells is a diagnostic and prognostic marker in prostate cancer. Genet Test Mol Biomarkers. 2015;19(9):469–475. doi: 10.1089/gtmb.2015.0088. [DOI] [PubMed] [Google Scholar]
- 13.Kotb S, Mosharafa A, Essawi M, Hassan H, Meshref A, Morsy A. Circulating miRNAs 21 and 221 as biomarkers for early diagnosis of prostate cancer. Tumor Biol. 2014;35(12):12613–12617. doi: 10.1007/s13277-014-2584-7. [DOI] [PubMed] [Google Scholar]
- 14.Chen ZH, Zhang GL, Li HR, et al. A panel of five circulating microRNAs as potential biomarkers for prostate cancer. Prostate. 2012;72(13):1443–1452. doi: 10.1002/pros.22495. [DOI] [PubMed] [Google Scholar]
- 15.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8(5):336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 17.Mishra S, Deng JJ, Gowda PS, et al. Androgen receptor and microRNA-21 axis downregulates transforming growth factor beta receptor II (Tgfbr2) expression in prostate cancer. Oncogene. 2014;33(31):4097–4106. doi: 10.1038/onc.2013.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin C, Fang C, Weng H, Yuan C, Wang F. Circulating microRNAs as novel biomarkers in the diagnosis of prostate cancer: a systematic review and meta-analysis. Int Urol Nephrol. 2016;48(7):1087–1095. doi: 10.1007/s11255-016-1281-4. [DOI] [PubMed] [Google Scholar]
- 19.Ouyang H, Zhou Y, Zhang L, Shen G. Diagnostic value of microRNAs for urologic cancers: a systematic review and meta-analysis. Medicine. 2015;94(37):e1272. doi: 10.1097/MD.0000000000001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Q, Zheng Y, Zhu D. Diagnostic performance of microRNAs expression in prostate cancer. Tumor Biol. 2014;35(10):10529–10538. doi: 10.1007/s13277-014-2351-9. [DOI] [PubMed] [Google Scholar]
- 21.Song CJ, Chen H, Chen LZ, Ru GM, Guo JJ, Ding QN. The potential of microRNAs as human prostate cancer biomarkers: a meta-analysis of related studies. J Cell Biochem. 2018;119(3):2763–2786. doi: 10.1002/jcb.26445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
PRISMA Checklist
| Section/topic | # | Checklist item | Reported on page # |
|---|---|---|---|
| TITLE | |||
| Title | 1 | Identify the report as a systematic review, meta-analysis, or both. | 1 |
| ABSTRACT | |||
| Structured summary | 2 | Provide a structured summary including, as applicable: background; objectives; data sources; study eligibility criteria, participants, and interventions; study appraisal and synthesis methods; results; limitations; conclusions and implications of key findings; systematic review registration number. | 2 |
| INTRODUCTION | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known. | 3 |
| Objectives | 4 | Provide an explicit statement of questions being addressed with reference to participants, interventions, comparisons, outcomes, and study design (PICOS). | 3 |
| METHODS | |||
| Protocol and registration | 5 | Indicate if a review protocol exists, if and where it can be accessed (e.g., Web address), and, if available, provide registration information including registration number. | Not applicable |
| Eligibility criteria | 6 | Specify study characteristics (e.g., PICOS, length of follow-up) and report characteristics (e.g., years considered, language, publication status) used as criteria for eligibility, giving rationale. | 4 |
| Information sources | 7 | Describe all information sources (e.g., databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched. | 4 |
| Search | 8 | Present full electronic search strategy for at least one database, including any limits used, such that it could be repeated. | 4 (Table S2) |
| Study selection | 9 | State the process for selecting studies (i.e., screening, eligibility, included in systematic review, and, if applicable, included in the meta-analysis). | 4 |
| Data collection process | 10 | Describe method of data extraction from reports (e.g., piloted forms, independently, in duplicate) and any processes for obtaining and confirming data from investigators. | 4 |
| Data items | 11 | List and define all variables for which data were sought (e.g., PICOS, funding sources) and any assumptions and simplifications made. | 5 |
| Risk of bias in individual studies | 12 | Describe methods used for assessing risk of bias of individual studies (including specification of whether this was done at the study or outcome level), and how this information is to be used in any data synthesis. | 5 |
| Summary measures | 13 | State the principal summary measures (e.g., risk ratio, difference in means). | 5 |
| Synthesis of results | 14 | Describe the methods of handling data and combining results of studies, if done, including measures of consistency (e.g., I2) for each meta-analysis. | 5–6 |
| Section/topic | # | Checklist item | Reported on page # |
| Risk of bias across studies | 15 | Specify any assessment of risk of bias that may affect the cumulative evidence (e.g., publication bias, selective reporting within studies). | 6 |
| Additional analyses | 16 | Describe methods of additional analyses (e.g., sensitivity or subgroup analyses, meta-regression), if done, indicating which were pre-specified. | 6 |
| RESULTS | |||
| Study selection | 17 | Give numbers of studies screened, assessed for eligibility, and included in the review, with reasons for exclusions at each stage, ideally with a flow diagram. | 6 |
| Study characteristics | 18 | For each study, present characteristics for which data were extracted (e.g., study size, PICOS, follow-up period) and provide the citations. | 6–7 |
| Risk of bias within studies | 19 | Present data on risk of bias of each study and, if available, any outcome level assessment (see item 12). | 7 |
| Results of individual studies | 20 | For all outcomes considered (benefits or harms), present, for each study: (a) simple summary data for each intervention group (b) effect estimates and confidence intervals, ideally with a forest plot. | 7–8 |
| Synthesis of results | 21 | Present results of each meta-analysis done, including confidence intervals and measures of consistency. | 7–8 |
| Risk of bias across studies | 22 | Present results of any assessment of risk of bias across studies (see Item 15). | 7 |
| Additional analysis | 23 | Give results of additional analyses, if done (e.g., sensitivity or subgroup analyses, meta-regression [see Item 16]). | 8 |
| DISCUSSION | |||
| Summary of evidence | 24 | Summarize the main findings including the strength of evidence for each main outcome; consider their relevance to key groups (e.g., healthcare providers, users, and policy makers). | 8–9 |
| Limitations | 25 | Discuss limitations at study and outcome level (e.g., risk of bias), and at review-level (e.g., incomplete retrieval of identified research, reporting bias). | 9 |
| Conclusions | 26 | Provide a general interpretation of the results in the context of other evidence, and implications for future research. | 10 |
| FUNDING | |||
| Funding | 27 | Describe sources of funding for the systematic review and other support (e.g., supply of data); role of funders for the systematic review. | 13 |
Table S2.
Search strategy
| Search strategy | |
|---|---|
|
| |
| #1 | “microRNA” OR “miRNA” OR “miR” AND “prostate cancer” AND “diagnosis” |
| #2 | “microRNA” OR “miRNA” OR “miR” AND “prostate cancer” AND “sensitivity” |
| #3 | “microRNA” OR “miRNA” OR “miR” AND “prostate cancer” AND “specificity” |
| #4 | “microRNA” OR “miRNA” OR “miR” AND “prostate carcinoma” AND “diagnosis” |
| #5 | “microRNA” OR “miRNA” OR “miR” AND “prostate carcinoma” AND “sensitivity” |
| #6 | “microRNA” OR “miRNA” OR “miR” AND “prostate carcinoma” AND “specificity” |
Table S3.
Sensitivity analysis for pooled sensitivities and corresponding heterogeneities
| A | |||
|---|---|---|---|
|
| |||
| Studya | SEN (95% CI) | I2 index (%) | Pb |
|
| |||
| Porzycki et al1 | 0.91(0.87–0.94) | 0 | 0.43 |
| Yang et al2,c | 0.90 (0.85–0.94) | 0 | 0.58 |
| Yang et al2,d | 0.90 (0.85–0.93) | 0 | 0.75 |
| Gao et al3 | 0.92 (0.88–0.95) | 0 | 0.58 |
| Huang et al4 | 0.92 (0.87–0.95) | 0 | 0.61 |
| Kotb et al5 | 0.91(0.88–0.94) | 0 | 0.434 |
|
| |||
| B | |||
|
| |||
| Studya | SEN (95% CI) | I2 index (%) | Pb |
|
| |||
| Zhu et al6 | 0.73(0.66–0.79) | 78.3 | 0.01 |
| Huang et al7 | 0.72 (0.66–0.76) | 71.3 | 0.03 |
| Chen et al8,c | 0.71 (0.65–0.76) | 68.5 | 0.04 |
| Chen et al8,d | 0.76 (0.70–0.80) | 33.4 | 0.22 |
Notes: (A) miR-21, (B) miR-30c.
Given named study is omitted.
P-value of the Cochrane-Q test.
Control group consisted of patients with benign prostate disease.
Control group consisted of healthy people.