Abstract
Emerging studies have reported that coatomer protein complex subunit β2 (COPB2) is overexpressed in several types of malignant tumor; however, to the best of our knowledge, no studies regarding COPB2 in gastric cancer have been published thus far. Therefore, the present study aimed to determine the significance and function of COPB2 in gastric cancer. COPB2 expression in gastric cancer cell lines was measured using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. In addition, lentivirus-short hairpin RNA (shRNA) COPB2 (Lv-shCOPB2) was generated and used to infect BGC-823 cells to analyze the effects of COPB2 on the cancerous phenotype. The effects of shRNA-mediated COPB2 knockdown on cell proliferation were detected using MTT, 5-bromo-2-deoxyuridine and colony formation assays. In addition, the effects of COPB2 knockdown on apoptosis were analyzed by flow cytometry. Nude mice and fluorescence imaging were used to characterize the regulation of tumor growth in vivo, and qPCR and immunohistochemistry were subsequently conducted to analyze COPB2 expression in xenograft tumor tissues. Furthermore, a receptor tyrosine kinase (RTK) signaling pathway antibody array was used to explore the relevant molecular mechanisms underlying the effects of COPB2 knockdown. The results revealed that COPB2 mRNA was abundantly overexpressed in gastric cancer cell lines, whereas knockdown of COPB2 significantly inhibited cell growth and colony formation ability, and led to increased cell apoptosis in vitro. The tumorigenicity assay revealed that knockdown of COPB2 reduced tumor growth in nude mice, and fluorescence imaging indicated that the total radiant efficiency of mice in the Lv-shCOPB2-infected group was markedly reduced compared with the mice in the Lv-shRNA control-infected group in vivo. The antibody array assay revealed that the levels of phosphorylation in 23 target RTKs were significantly reduced: In conclusion, COPB2 was highly expressed in gastric cancer cell lines, and knockdown suppressed colony formation and promoted cell apoptosis via inhibiting the RTK signaling and its downstream signaling cascade molecules. Therefore, COPB2 may present a valuable target for gene silencing strategy in gastric cancer.
Keywords: coatomer protein complex subunit β2, gastric cancer, apoptosis, receptor tyrosine kinase signaling pathway
Introduction
Gastric cancer is one of the most common types of cancer, with high incidence rates in the Asia-Pacific region. In addition, it is the third most common cause of cancer-associated mortality in this region (1); notably, high incidence rates of gastric cancer have been reported in China, Korea and Japan in the 20th century, and gastric cancer remains a key cause of morbidity and mortality worldwide (2,3). Gastric cancer has been ranked second in the top 10 most common cancer types, and third in the primary causes of cancer-associated mortality in China (2). Therefore, there is an urgent need to identify and study novel targets for gene therapy, and to determine their clinical significance in early diagnosis and prognostic estimates. Coatomer protein complex subunit β2 (COPB2) is a subunit of a typical cytoplasmic protein complex, which binds dilysine motifs and associates with Golgi-derived nonclathrin-coated vesicles. COPB2 functions as a mediator to transport proteins from the endoplasmic reticulum to the Golgi apparatus in the process of protein biosynthesis (4). It has previously been reported that COPB2 may be a target gene in prostate cancer cell lines (5), whereby silencing COPB2 inhibits cell proliferation, arrests the cell cycle at G1 and G2 phases, and induces apoptosis. RNA interference (RNAi)-mediated knockdown of COPB2 has also been observed to significantly inhibit the growth and invasion of pulmonary cancer A549 cells (6). Furthermore, knockdown of COPB2 inhibits the growth and colony formation ability of human colon cancer cell lines, including RKO and HCT116, and leads to cell cycle arrest at G0/G1 or S phases via regulating cell cycle-associated proteins (7). These previous findings suggested that COPB2 may be considered a promising target for cancer gene therapy. However, to the best of our knowledge, the significance and function of COPB2 in gastric cancer remains unknown. Therefore, there is an urgent requirement to investigate the effects of silencing COPB2 on the cancerous phenotype of gastric cancer cells to elucidate its role in gastric cancer tumorigenesis, and to explore the possibility of uncovering a novel potential biomarker and target gene for therapy.
In the present study, lentivirus-short hairpin RNA (shRNA) COPB2 (Lv-shCOPB2) was designed and constructed, and transduced into gastric cancer BGC-823 cells, in order to analyze the effects of COPB2 knockdown on the cancerous phenotype. Briefly, MTT, 5-bromo-2-deoxyuridine (BrdU) and colony formation assays were used to detect cell proliferation, and flow cytometry was conducted to analyze apoptosis. In addition, to characterize the regulatory role of COPB2 in tumor growth in vivo, a nude mouse model was constructed and fluorescence imaging was conducted, and COPB2 expression was detected in xenograft tumor tissues using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and immunohistochemistry. In addition, a receptor tyrosine kinase (RTK) signaling pathway antibody array was used to determine the molecular mechanisms underlying the effects of COPB2 knockdown.
Materials and methods
Cell lines and culture conditions
The gastric cancer cell lines, BGC-823, SGC-7901, MGC-803 and MKN45, were purchased from the Type Culture Collection of Cancer Institute and Hospital, Chinese Academy of Medical Sciences (Beijing, China). The normal gastric mucous membrane epithelial cell line, GES-1, was purchased from OBiO Technology (Shanghai) Corp., Ltd. (Shanghai, China). All cell lines were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 100 IU/ml penicillin, 100 µg/ml streptomycin and 10% heat-inactivated fetal bovine serum (Zhejiang Tianhang Biotechnology Co. Ltd., Hangzhou, China). The cells were maintained in a humidified incubator at 37°C in an atmosphere containing 5% CO2. Gastric cancer cells in the exponential growth phase were used for subsequent experiments.
Lentiviral transduction of BGC-823 cells
Human gastric cancer BGC-823 cells were seeded in 6-well plates at 5×104 cells/well and incubated at 37°C until 30% confluence was reached. Cells were then divided into the lentivirus-short hairpin RNA (shRNA) control group (Lv-shCtrl), where cells were infected with empty green fluorescent protein (GFP) lentiviruses (sequence: 5′-CCGGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTG-3′), and the Lv-shCOPB2 (sequence: 5′-CCGGGCAGATTAGAGTGTTCAATTACTCGAGTAATTGAACACTCTAATCTG CTTTTT-3′) group, where cells were infected with GFP-tagged Lv-shCOPB2 lentiviruses. Lentviruses were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China). A suitable amount of lentivirus (multiplicity of infection, 10) was added to the culture medium of BGC-823 cells for transduction, according to the multiplicity of infection, and the cells were incubated for a further 8 h. The medium containing lentiviruses was then removed and the cells were cultured in normal culture medium for 12 h. GFP expression was observed under a fluorescence microscope 3 days following infection, and gastric cancer cells with an infection efficiency of >80% were selected for subsequent analyses. Cells were harvested 48 h post-transduction for further analysis.
Expression of COPB2 mRNA and detection of transduction efficiency by RT-qPCR analysis
To determine the expression of COPB2 in gastric cancer cells and to confirm the silencing efficiency of COPB2 knockdown in BGC-823 cells, RT-qPCR analysis was conducted. Briefly, gastric cancer cells in the exponential growth phase, and BGC-823 cells infected with Lv-shCOPB2 or Lv-shCtrl, were collected and lysed for total RNA extraction using the RNAiso Plus kit (Takara Biotechnology Co., Ltd., Dalian, China). RNA purity and concentration were determined using the NanoDrop-2000 spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Total RNA was reverse transcribed using the Prime Script™ RT Reagent kit (Takara Biotechnology Co., Ltd.), according to manufacturer's protocol at 37°C for 15 min and 85°C for 20 sec. Subsequently, PCR amplification was performed using the SYBR® Premix Ex Taq™ Master Mix (Takara Biotechnology Co., Ltd.) and the Bio-Rad CFX96 Real-time PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The primers used were as follows: COPB2, forward 5′-GTGGGGACAAGCCATACCTC-3′, reverse 5′-GTGCTCTCAAGCCGGTAGG-3′; and GAPDH, forward 5′-TGACTTCAACAGCGACACCCA-3′ and reverse 5′-CACCCTGTTGCTGTAGCCAAA-3′. PCR was performed on a final volume of 10 µl, comprising 2 µl cDNA, 0.5 mM of each primer and 1X SYBR® Premix Ex Taq™ Master Mix. The amplification program was as follows: Initial denaturation step at 95°C for 10 min, followed by 40 cycles at 95°C for 10 sec, 60°C for 30 sec and 72°C for 30 sec, where fluorescence signals were acquired. Amplification was followed by a melting curve analysis, which was used to determine the dissociation characteristics of the PCR products. Each sample was run in triplicate and the mRNA expression levels of COPB2 were calculated relative to the internal reference gene, GAPDH, using the 2−ΔΔCq method (8).
Cell proliferation analysis
Following transduction, cells from both groups were trypsinized and counted. Cells (3×103 cells/well) were plated in each well of a 96-well plate (five replicate wells for each group) and incubated at 37°C for 24, 48, 72, 96 and 108 h. The cell counts were monitored over 5 consecutive days. Briefly, 20 µl MTT (5 µmol/l; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) dissolved in PBS was added to each well, and the cells were incubated at 37°C for 4 h. Cells were subsequently centrifuged at 1,000 × g for 2 min before the medium was removed and 150 µl dimethyl sulfoxide was added to each well. After incubation at 37°C for 5 min, the absorbance was measured using a Benchmark microtiter plate reader (Bio-Rad Laboratories, Inc.) at a wavelength of 490 nm.
BrdU assay
BGC-823 gastric cancer cells in the Lv-shCtrl and Lv-shCOPB2 groups were seeded into triplicate wells of a 96-well plate at 2×104 cells/well with 100 µl complete medium. The following day, 20 µmol/ml BrdU (Roche Diagnostics, Shanghai, China) was added to the cultured cells before they were incubated with complete medium for a sufficient culture time (≤24 h). The culture medium was discarded, 200 µl FixDenat (Roche Diagnostics) was added to each well and cells were fixed for 30 min in the dark at room temperature. Following removal of the fixation solution, 10% bovine serum albumin (BSA: Zhejiang Tianhang Biotechnology Co. Ltd.) was added to block the cells for 30 min at room temperature in the dark. An anti-BrdU monoclonal antibody (dilution, 1:100; cat. no. 11669915001; Roche Diagnostics) with peroxidase activity was then added to the cells, which were incubated for 90 min at room temperature in the dark. The cells were subsequently washed three times with washing buffer, and the substrate solution (Component A containing luminol and 4-iodophenol + Component B containing a stabilized form of H2O2) was added (100 µl/well) for ~30 min at room temperature in the dark until the solution became blue in color. H2SO4 (10%; 50 µl) was added to each well for colorimetry. The optical density was measured using a spectrophotometer microplate reader (Tecan Group, Ltd., Mannedorf, Switzerland) at 450/550 nm. Each data point was calculated from the average of six replicates and each experiment was repeated in triplicate.
Colony formation assay
BGC-823 gastric cancer cells in the Lv-shCtrl and Lv-shCOPB2 groups were digested in 0.25% trypsin and diluted to a concentration of 5×104 cells/ml. A hemocytometer was used to count the cells, and cell suspensions were seeded into 6-well plates at 400 cells/well. The medium was refreshed every 3 days, and cell growth was observed as normal. A total of 14 days after the cells were seeded, or at the point where the number of cells in each colony reached >50, the colonies were visualized under a fluorescence microscope. Cells were washed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. The fixed cells were stained with 500 µl Giemsa (Beijing Dingguo Changsheng Biotechnology Co., Ltd., Beijing, China) for 20 min, washed with ddH2O and air-dried at room temperature. The total number of colonies with >50 cells were counted, and images were captured using light and fluorescence microscopes. The assay was repeated three times.
Quantification of apoptosis by flow cytometry
Cells were harvested using 0.25% trypsin and washed once with ice-cold (4°C) D-Hanks solution (pH 7.2-7.4). Cells were centrifuged at 300 × g for 5 min and washed with 1X binding buffer (eBioscience™ Annexin V Apoptosis Detection kit APC; cat, no. 88-8007; eBioscience; Thermo Fisher Scientific, Inc.), prior to further centrifugation at 300 × g for 3 min. The cells were then resuspended in 200 µl binding buffer to a final concentration of 106 cells/ml for subsequent analysis. The cell suspension (100 µl) was mixed with 10 µl APC Annexin V (Apoptosis Detection kit; eBioscience; Thermo Fisher Scientific, Inc.) and incubated in the dark for 15 min at room temperature. If necessary, 400-800 µl 1X binding buffer was added to the stained cells, according to the quantity of cells. The percentage of apoptotic cells was analyzed by flow cytometry (Guava® easyCyte 6HT; EMD Millipore, Billerica, MA, USA). This assay was repeated in triplicate.
Tumorigenesis in nude mice and in vivo imaging
Male BALB/c nude mice (n=20; weight, 15-19 g; age, 4 weeks; Shanghai Lingchang Biotechnology Co., Ltd., Shanghai, China) were maintained under the following conditions: Temperature, 22-24°C, humidity, 40-70%; ad libitum food/water access; artificial feeding for 2-3 days; 12-h light/dark cycle). Eligible nude mice were inoculated with Lv-shCOPB2-infected and Lv-shCtrl-infected BGC-823 cells. Briefly, a total of 20 mice were divided into two equal groups at random. BGC-823 cells from both groups were resuspended in physiological saline solution at a density of 5×107 cells/ml before a 0.2 ml cell suspension was injected subcutaneously into the mice using a 6-gauge, 1 ml syringe. The mice were maintained until the tumors were visible, and tumor diameter and size were measured 8, 11, 14, 16 and 18 days following inoculation. Tumor volume was monitored frequently and was recorded on days 8, 11, 14, 16 and 18; volume was calculated using the following formula for hemi-ellipsoids: Volume = length (cm) x width (cm) x height (cm) x 3.14/6. At 28 days following inoculation of the gastric cancer cells, the mice were injected intraperitoneally with 10 µl/g D-Luciferin (Shanghai Yeasen Biotechnology Co., Ltd., Shanghai, China). After 15 min, 70 mg/kg pentobarbital sodium was injected intraperitoneally to anesthetize the mice. A few minutes later, the anesthetized mice were placed under a small animal live imaging system (LT Lumina; PerkinElmer, Inc., Waltham, MA, USA) to observe the fluorescence results. The mice were then sacrificed, and the tumors were dissected and photographed. All animal experiments were performed in strict accordance with international ethical guidelines and the National Institutes of Health Guide for the Care and Use of Laboratory Animals (9). The experiments were authorized and approved by the Institutional Animal Care and Use Committee of Gansu University of Chinese Medicine (Lanzhou, China).
Detection of COPB2 mRNA and protein expression in xenograft tumor tissues
The mRNA and protein expression levels of COPB2 were detected in xenograft tumor tissues to validate knockdown of COPB2 in gastric cancer cells. After the mice were sacrificed, the xenograft tumor tissues from both groups were dissected and collected for further detection of COPB2 mRNA and protein expression. Fresh tumor tissues were stored at −80°C. COPB2 mRNA expression was detected using RT-qPCR analysis, as aforementioned. In addition, different tumor tissue sections were immediately fixed in 4% paraformaldehyde for 12 h at room temperature and embedded in paraffin for subsequent histological and immunohistochemical analysis of COPB2. To perform immunohistochemical analysis, tissues from both groups were deparaffinized and cut into thin sections (5 µm). All sections were rehydrated and heated for antigen retrieval with 0.3% H2O2 for 30 min. Prior to staining, non-specific binding was blocked by incubation with 10% BSA in PBS at 37°C for 1 h. The section slides were then incubated with an anti-COPB2 primary antibody (dilution, 1:200; cat. no. HPA036867, Sigma-Aldrich; Merck KGaA) at 4°C overnight. The following day, slides were incubated with a biotin-conjugated secondary antibody (dilution, 1:100; cat. no. TA130016; OriGene Technologies, Inc., Rockville, MD, USA) for ~2 h at room temperature. Subsequently, horseradish enzyme (HRP) labeled streptavidin was added for 15 min to form a complex and 3,3′-diaminobenzidine solution was added to detect HRP activity. The tissue sections were also stained with hematoxylin and eosin (H&E) for 10 min at 37°C to analyze the growth of xenograft tumor tissues with a microscope (Leica DM2700M; Leica Microsystems GmbH, Wetzlar, Germany). The staining percentage was graded as follows: 0 (0-5%), 1 (6-20%), 2 (21-60%) and 3 (61-100%), and the staining intensity was graded as follows: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). The final sum of the staining percentage and intensity scores was considered the staining score (0-6). Tumors with final staining scores of 0, 1, 2-4 and 5 or 6 were considered negative (−), slightly positive (+), moderately positive (++) and strongly positive (+++), respectively; the method was described in previous reports (10,11).
PathScan® RTK signaling antibody array assay
To investigate the activation of intracellular signaling pathways associated with tumor growth, the PathScan® RTK Signaling Antibody array (Cell Signaling Technology, Inc., Danvers, MA, USA) was used, according to the manufacturer's protocol. This antibody array is a slide-based antibody array that uses the sandwich immunoassay principle for the detection of signaling nodes and downstream target nodes that have undergone phosphorylation of tyrosine or other residues. Briefly, a total of 5 days post-lentiviral infection, BGC-823 cells were collected and lysed. Detection of COPB2-silenced and negative control cells was performed in triplicate. Images were captured by briefly exposing the slide to obtain the chemiluminescent film signal at all sites of the array slide, and aberrantly expressed protein targets were compared and calculated between two groups.
Statistical analysis
All experimental data are expressed as the means ± standard deviation from at least three separate experiments. Statistical analyses were performed using a Student's two-tailed t-test or one-way analysis of variance, and Dunnett method was used to test multiple comparisons. P<0.05 was considered to indicate a statistically significant difference, and all statistical analyses were performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA).
Results
COPB2 is upregulated in gastric cancer cell lines
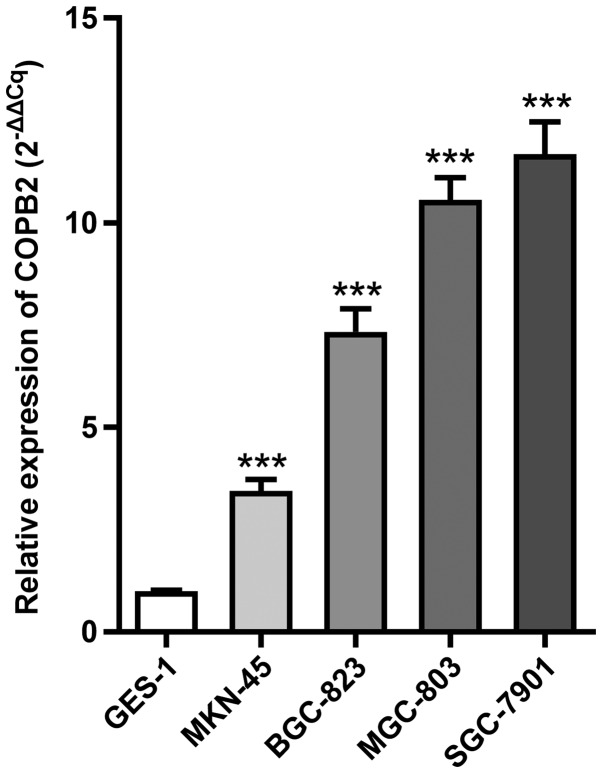
To investigate COPB2 expression in gastric cancer cell lines, RT-qPCR analysis was employed to analyze its expression in four gastric cancer cell lines (BGC-823, SGC-7901, MGC-803 and MKN45) and the normal gastric mucous membrane epithelial cell line, GES-1. The results demonstrated that COPB2 mRNA expression was increased in all four gastric cancer cell lines compared with in the GES-1 cell line (Fig. 1).
Figure 1.
mRNA expression levels of COPB2 in gastric cancer cell lines. Reverse transcription-quantitative polymerase chain reaction analysis results demonstrated that COPB2 was expressed at high levels in MKN-45, MGC-803, BGC-823 and SGC-7901 gastric cancer cell lines relative to the internal reference gene GAPDH. ***P<0.01, compared with GES-1. COPB2, coatomer protein complex subunit β2.
Efficiency of shRNA-mediated COPB2 knockdown in gastric cancer cells
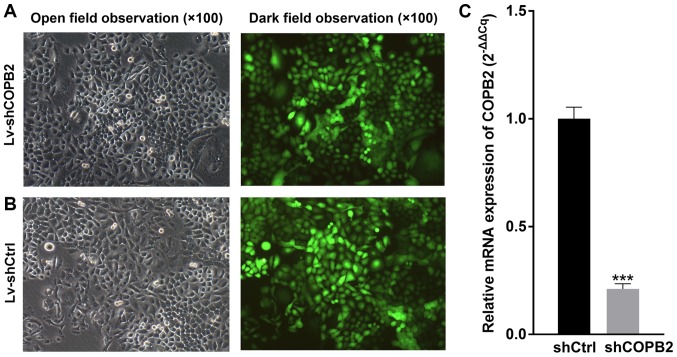
BGC-823 cells infected with Lv-shCtrl or Lv-shCOPB2 were observed under a fluorescence microscope to determine infection efficiency, and the infection rate was measured by monitoring GFP fluorescence emitted by cells. The results indicated that the infection efficiency was >80% (Fig. 2A and B). As determined by RT-qPCR analysis, the mRNA expression levels of COPB2 were significantly decreased in the Lv-shCOPB2 group, with a knockdown efficiency of 78.9% (P<0.01; Fig. 2C). These findings indicated that lentivirus-mediated targeting of COPB2 effectively silenced COPB2 expression in the BGC-823 gastric cancer cell line.
Figure 2.
COPB2 infection efficiency and gene knockdown efficiency in BGC-823 cells. (A) Lv-shCOPB2-infected BGC-823 cells under open field observation and dark field observation. Magnification, ×100. (B) Lv-shCtrl-infected BGC-823 cells under open field observation and dark field observation. The results demonstrated that Lv-shCOPB2 and Lv-shCtrl effectively infected BGC-823 cells. Infection rate of Lv-shCOPB2 and Lv-shCtrl was >80%. (C) mRNA expression levels of COPB2 were significantly decreased in the Lv-shCOPB2 group, with a significant knockdown efficiency of 78.9% (P<0.01). ***P<0.01, compared with the Lv-shCtrl group. Ctrl, control; COPB2, coatomer protein complex subunit β2; Lv, lentivirus; sh, short hairpin RNA.
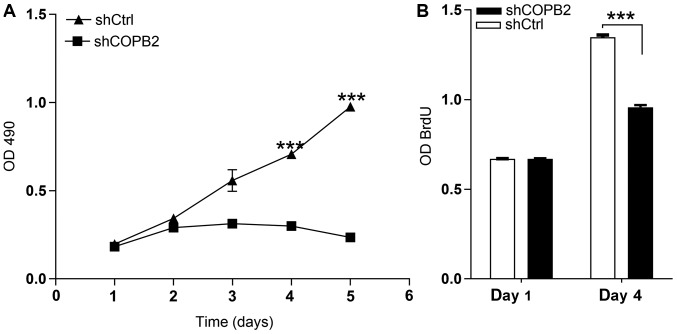
COPB2 silencing inhibits gastric cancer cell proliferation
The MTT assay results demonstrated that the number and fold-change in proliferation of cells in the Lv-shCOPB2 group was markedly reduced when compared with the Lv-shCtrl group at 4 and 5 days following transduction of BGC-823 cells (P<0.05; Fig. 3A). These findings suggested that knockdown of COPB2 may be associated with a reduction in cell proliferation. The BrdU thymidine analog naturally incorporates into the DNA of proliferating cells during cell division. In the present study, the effects of COPB2 knockdown on BrdU incorporation were measured. Cell proliferation in the Lv-shCOPB2 group was significantly reduced compared with the in Lv-shCtrl group at 4 days following transduction (P<0.05; Fig. 3B). Therefore, the results of the BrdU incorporation assay indicated that inhibition of COPB2 expression may significantly suppress the proliferation of BGC-823 cells.
Figure 3.
Effects of COPB2 gene knockdown on cell proliferation, as determined by MTT and BrdU assays. (A) MTT assay was conducted and absorbance at 490 nm was compared, indicating the number of active BGC-823 cells in the Lv-shCOPB2 and Lv-shCtrl groups. The proliferation rate of BGC-823 cells was significantly inhibited in the Lv-shCOPB2 infection group at 4 and 5 days post-infection (P<0.05), indicating that the COPB2 gene may be significantly associated with the proliferation ability of BGC-823 cells. (B) BrdU incorporation assay was conducted and OD was detected. Cell proliferation in the Lv-shCOPB2 group was significantly reduced compared with in the Lv-shCtrl group 4 days post-transduction. ***P<0.01, compared with the Lv-shCtrl group. BrdU, 5-bromo-2-deoxyuridine; COPB2, coatomer protein complex subunit β2; Lv, lentivirus; OD, optical density; sh, short hairpin RNA.
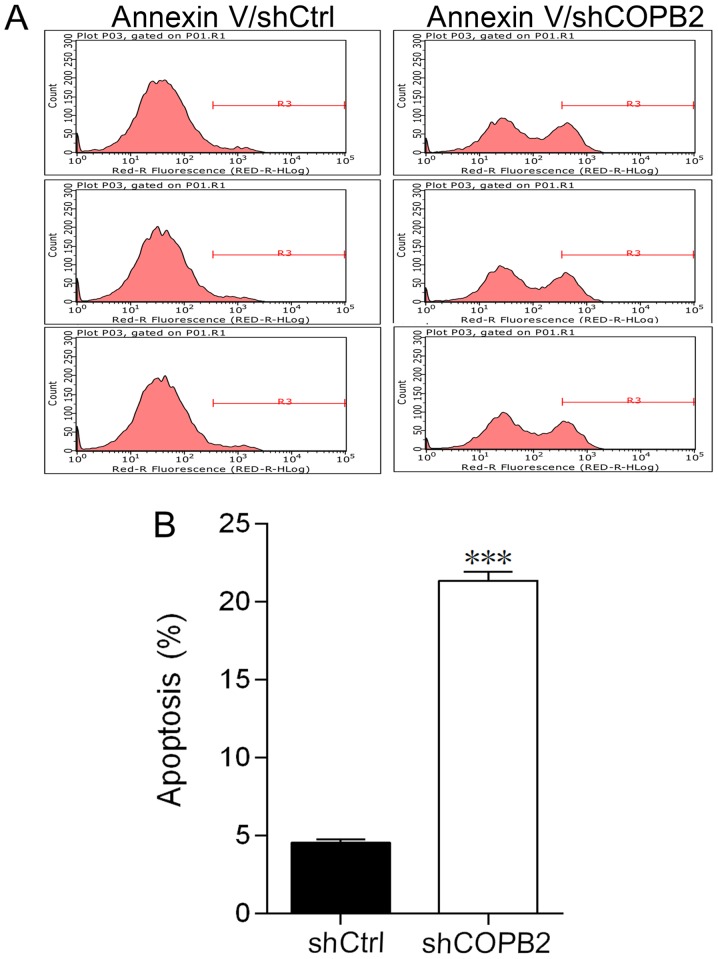
COPB2 silencing induces cell apoptosis
In order to investigate the effects of COPB2 knockdown on apoptosis of BGC-823 cells, the levels of apoptosis were compared in Lv-shCOPB2 and Lv-shCtrl groups. The apoptotic rate was measured and evaluated by flow cytometry. The percentage of apoptotic BGC-823 cells in the Lv-shCOPB2 group was significantly higher compared with in the Lv-shCtrl group (P<0.001; Fig. 4A and B). These results suggested that knockdown of COPB2 may affect cell survival and induce apoptosis.
Figure 4.
Effects of COPB2 gene knockdown on apoptosis, as determined by fluorescence-activated cell sorting detection. (A) Images of Lv-shCOPB2 and Lv-shCtrl groups demonstrating the level of apoptosis in BGC-823 cells following infection. (B) A total of 5 days post-infection, the percentage of apoptosis in BGC-823 cells from the Lv-shCOPB2 group was significantly increased compared with in the Lv-shCtrl group (P<0.001), thus suggesting that the COPB2 gene may be associated with apoptosis of BGC-823 cells. ***P<0.01, compared with the Lv-shCtrl group. COPB2, coatomer protein complex subunit β2; Lv, lentivirus; sh, short hairpin RNA.
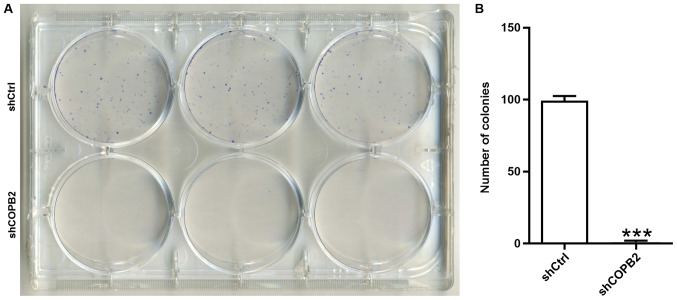
COPB2 silencing reduces gastric cancer cell colony formation
A colony formation assay is used to assess the proliferative potential of cells. In the present study, the colony formation assay results demonstrated that the Lv-shCOPB2 group formed significantly fewer colonies in soft agar when compared with the Lv-shCtrl group (P<0.001; Fig. 5A and B). These results indicated that silencing COPB2 may reduce the anchorage-independent proliferative potential of BGC-823 gastric cancer cells.
Figure 5.
Effects of COPB2 gene knockdown on cell colony formation ability, as determined using light microscopy. (A) Images of Lv-shCOPB2 and Lv-shCtrl groups indicating the BGC-823 cell colony formation ability post-infection. (B) A total of 11 days following infection with shRNA lentiviruses, the number of colonies in each well was observed. The number of BGC-823 cell colonies from the Lv-shCOPB2 infection group was decreased compared with from the Lv-shCtrl group (P<0.001). These findings suggested that the COPB2 gene may be associated with the colony formation ability of BGC-823 cells. ***P<0.01, compared with the Lv-shCtrl group. COPB2, coatomer protein complex subunit β2; Lv, lentivirus; sh, short hairpin RNA.
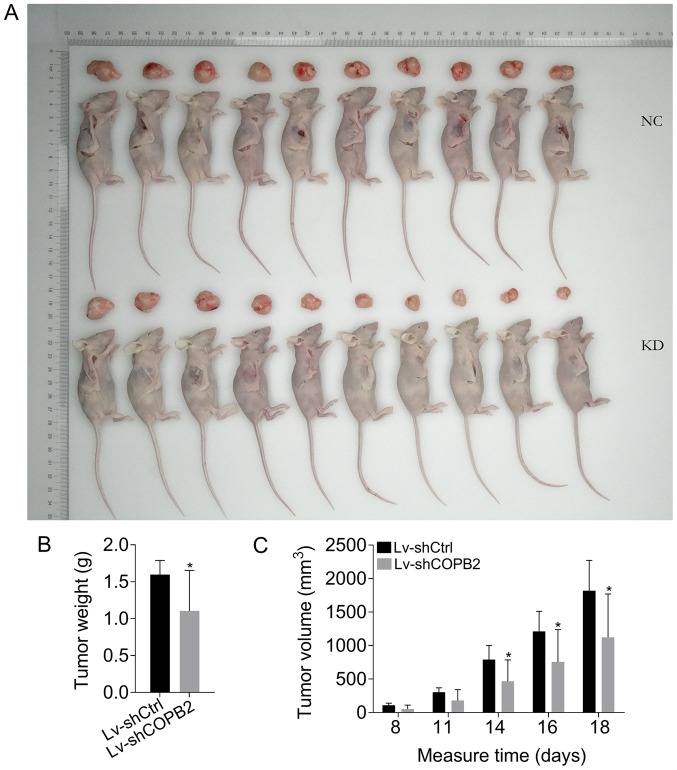
COPB2 silencing inhibits tumor growth in vivo
To confirm the results of COPB2 knockdown in vitro, an in vivo mouse tumorigenesis model, where mice were injected with BGC-823 cells from the Lv-shCtrl or Lv-shCOPB2 groups, was generated. Over the course of 18 days, the rate of tumor growth and the tumor volume were significantly reduced at 14, 16 and 18 days following injection with BGC-823 cells in the Lv-shCOPB2 group compared with in the Lv-shCtrl group (P<0.05). The results of tumor weight analysis revealed that COPB2-silenced BGC-823 cells generated smaller subcutaneous xenograft tumors in nude mice compared with in the Lv-shCtrl group (P<0.05; Fig. 6A-C). The results demonstrated that silencing COPB2 using the Lv-shCOPB2 vector may significantly inhibit the tumorigenicity of BGC-823 cells in a xenograft nude mouse model.
Figure 6.
Effects of COPB2 gene knockdown on tumorigenesis in nude mice in vivo. (A) Images of nude mice in the Lv-shCOPB2 and Lv-shCtrl groups. (B) Weight of subcutaneous xenograft tumors from the Lv-shCOPB2 group was lower than the weight of those from the Lv-shCtrl group (P<0.05). (C) Volume of subcutaneous xenograft tumors from the Lv-shCOPB2 group was lower than the volume of those from the Lv-shCtrl group at 14, 16 and 18 days (P<0.05). *P<0.05, compared with the Lv-shCtrl group at the same day. COPB2, coatomer protein complex subunit β2; KD, knockdown; Lv, lentivirus; sh, short hairpin RNA.
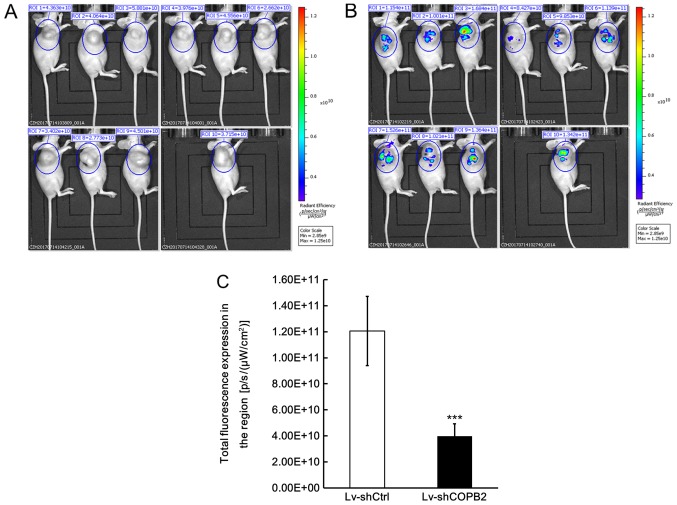
In order to confirm that knockdown of COPB2 was directly associated with the observed effects on tumor growth, a fluorescence imaging test was also conducted using a small animal live imaging system, which monitors the fluorescence signals emitted from cells and tissues. The Lv-shCOPB2-infected and Lv-shCtrl-infected BGC-823 cells were also transduced with GFP; therefore, tumor xenografts in both groups emit fluorescence signals when triggered by specific fluorescence in the live imaging system. The recorded fluorescence signal was used to calculate the total radiant efficiency, which reflects the number of xenograft tumor cells. The fluorescence imaging results demonstrated that the total radiant efficiency of mice in the Lv-shCOPB2-infected group was markedly reduced compared with in the Lv-shCtrl-infected group (P<0.05; Fig. 7A and B). These results confirmed successful infection of BGC-823 cells with the lentiviral vectors and verified the effects of COPB2 on cell proliferation in vivo.
Figure 7.
Effects of COPB2 gene knockdown on tumorigenesis in nude mice using an imaging assay in vivo. The total radiant efficiency of the ROI in xenografts from mice injected with (A) Lv-shCOPB2-infected BGC-823 cells and (B) Lv-shCtrl-infected BGC-823 cells. (C) Fluorescence imaging assay indicated that the total radiant efficiency of the ROI in xenograft tumors from the Lv-shCOPB2-infected group was lower when compared with the Lv-shCtrl-infected group (P<0.05). ***P<0.001, compared with the Lv-shCtrl group. COPB2, coatomer protein complex subunit β2; Lv, lentivirus; ROI, region of interest; sh, short hairpin RNA.
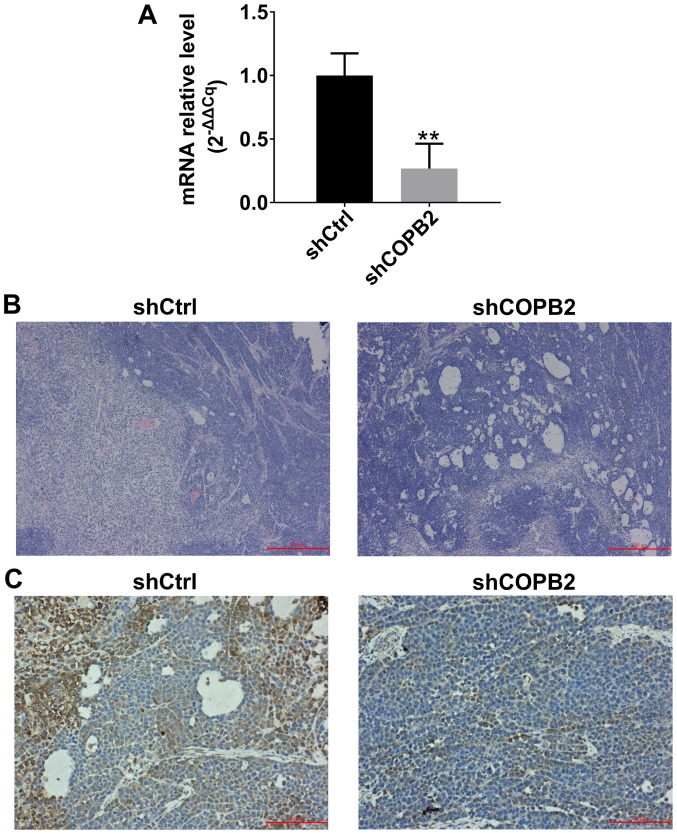
Validation of COPB2 silencing effects in xenograft tumor tissues
In order to confirm that COPB2 was successfully silenced in xenograft tumor tissues at the mRNA and protein levels, RT-qPCR, histological and immunohistochemical methods were employed to analyze COPB2 expression. The results indicated that the mRNA expression levels of COPB2 were significantly lower in xenograft tumor tissues from mice in the Lv-shCOPB2 group compared with in the Lv-shCtrl group (Fig. 8A). H&E analysis of tissues from the Lv-shCtrl group indicated that the tumor cells were scattered and varied in size, the nuclei were enlarged, and exhibited intense staining. A high degree of pathological mitosis in this group was observed, as characterized by an abundance of cell nuclei, and a marked number of large tumor cells was detected. In addition, necrotic areas were observed in tumor tissues. H&E staining analysis of tissues from the Lv-shCOPB2 group also indicated that the tumor cells were scattered and varied in size, the nuclei were enlarged, and exhibited intense staining. A high degree of pathological mitosis was also observed in this group; however, only a small number of multinucleated giant cells were identified in the tumor tissues. In addition, a number of small necrotic areas were observed scattered throughout the tumor tissues (Fig. 8B). Immunohistochemical staining analysis of Lv-shCtrl group tissues indicated that COPB2 expression could be observed in the cytoplasm of tumor cells and was markedly increased in the cytoplasm of tumor cells surrounding the necrotic regions. Immunohistochemical analysis of Lv-shCOPB2 group tissues demonstrated that the expression of COPB2 was increased in the cytoplasm of tumor cells and in the central region of the tumor cell mass. In conclusion, COPB2 expression in the Lv-shCOPB2 group tissues was markedly lower than in the Lv-shCtrl group tissues (Fig. 8C).
Figure 8.
Detection of COPB2 mRNA and protein expression in xenograft tumor tissues. (A) mRNA expression levels of COPB2 in xenograft tumor tissues from the Lv-shCOPB2 group were significantly lower than in the Lv-shCtrl group (P<0.01). (B) H&E analysis of Lv-shCtrl group tissues indicated that the tumor cells were scattered and varied in size, the nuclei were enlarged, and the staining was intense. A high degree of pathological mitosis was observed in this group, characterized by an abundance of cell nuclei, and a marked number of large tumor cells were detected. In addition, necrotic areas were observed in tumor tissues. H&E staining analysis of tissues from the Lv-shCOPB2 group indicated that the tumor cells were scattered and varied in size. The nuclei were enlarged, and the staining was intense. A high degree of pathological mitosis was also observed in this group; however, only a small number of multinucleated giant cells was identified in the tumor tissues. In addition, a number of small necrotic areas were observed scattered throughout the tumor tissues. Scale bar, 200 µm. (C) Immunohistochemical staining analysis of Lv-shCtrl group tissues demonstrated that COPB2 expression was observed in the cytoplasm of tumor cells and was markedly increased in the cytoplasm of tumor cells surrounding the necrotic regions. Immunohistochemical staining analysis of Lv-shCOPB2 group tissues indicated that COPB2 expression of was increased in the cytoplasm of tumor cells and in the central region of the tumor cell mass. The semi-quantitative score of COPB2 expression in Lv-shCtrl group tissues was moderately positive (++), whereas the semi-quantitative score of COPB2 expression in Lv-shCOPB2 group tissues was slightly positive(+). In conclusion, COPB2 expression in the Lv-shCOPB2 group was markedly reduced compared with in the Lv-shCtrl group. Scale bar, 50 µm. **P<0.01, compared with the Lv-shCtrl group. COPB2, coatomer protein complex subunit β2; H&E, hema-toxylin and eosin; Lv, lentivirus; sh, short hairpin RNA.
COPB2 silencing induces alterations in the RTK signaling pathway
Tyrosine kinases serve important roles in the modulation of growth factor signaling pathways; therefore, the PathScan® RTK Signaling Pathway Antibody array was employed to investigate the regulatory mechanisms underlying the effects of COPB2 silencing on the tumorigenesis of BGC-823 gastric cancer cells. The results demonstrated that knockdown of COPB2 significantly downregulated the expression (to varying degrees) of phosphorylated target factors from the RTK signaling pathway (n=23 in total), including epidermal growth factor receptor (EGFR)/ErbB1, human epidermal growth factor receptor (HER)2/ErbB2, HER3/ErbB3, fibroblast growth factor receptor (FGFR)4, insulin receptor (InsR), tropomyosin-related kinase (Trk)A/neurotrophic receptor tyrosine kinase (NTRK)1, TrkB/NTRK2, recepteur d'origine nantais (Ron)/macrophage stimulating 1 receptor (MST1R), Ret, c-Kit/stem cell growth factor receptor (SCFR), FMS-like receptor tyrosine kinase 3 (FLT3)/Flk2, EPH receptor (Eph)A3, EphB1, EphB4, TYRO3 protein tyrosine kinase (TYRO3)/Dtk, vascular endothelial growth factor receptor (VEGFR)2/kinase insert domain receptor (KDR), Akt/PKB/Rac (Thr308), Akt/PKB/Rac (Ser473), ribosomal S6 kinase (RSK), c-Abl, Src, Lck and signal transducer and activator of transcription 3 (Stat3; P<0.05 or P<0.01; Fig. 9 and Table I). These results indicated that COPB2 affects the proliferation of BGC-823 cells potentially via the phosphorylation-activated RTK signaling pathway. In addition, knockdown of COPB2 decreased the expression levels of downstream targets of the RTK signaling pathway in gastric cancer cells, indicating that RTKs and downstream targets may serve important roles in the apoptosis of COPB2-silenced BGC-823 cells. Further studies are required to clarify the function and regulatory mechanisms of COPB2 in gastric cancer development.
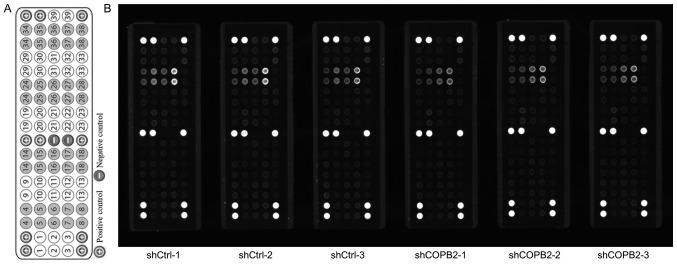
Figure 9.
Effects of COPB2 gene knockdown on specific RTK signaling pathway genes in BGC-823 cells. (A) Target map of the PathScan® RTK Signaling Antibody Array kit (Fluorescent Readout). This map image was taken from the Cell Signaling Technology website (https://www.cst-c.com.cn/product/product-Detail.jsp?productId=7949). Each site represents a target, which was indicated numerically and corresponds to the numbers presented in Table I. (B) Effects of COPB2 gene knockdown on relevant genes from the RTK signaling pathway in BGC-823 cells. This slide-based antibody array image indicated the reaction results for Lv-shCOPB2-infected and Lv-shCtrl-infected BGC-823 cells. The results demonstrated that knockdown of COPB2 significantly reduced the phos-phorylation of 23 RTK signaling pathway targets, including epidermal growth factor receptor/ErbB1, HER2/ErbB2, HER3/ErbB3, fibroblast growth factor 4, insulin receptor, tropomyosin-related kinase A/NTRK1, TrkB/NTRK2, recepteur d'origine nantais/macrophage stimulating 1 receptor, Ret, c-Kit/stem cell growth factor receptor, FMS-like receptor tyrosine kinase 3/Flk2, EphA3, EphB1, EphB4, TYRO3 protein tyrosine kinase/Dtk, vascular endothelial growth factor receptor 2/kinase insert domain receptor, Akt/PKB/Rac (Thr308), Akt/PKB/Rac (Ser473), ribosomal S6 kinase, c-Abl, Src, Lck and signal transducer and activator of transcription 3 at different levels (P<0.05 or P<0.01). COPB2, coatomer protein complex subunit β2; Eph, EPH receptor; HER, human epidermal growth factor receptor; Lv, lentivirus; NTRK, neurotrophic receptor tyrosine kinase 1; RTK, receptor tyrosine kinase; sh, short hairpin RNA.
Table I.
Detected proteins screened and validated by PathScan® RTK Signaling Antibody array between shCtrl-infected cells and shCOPB2-infected cells.
| Site | Target | Phosphorylation site | Family | Average gray value
|
P-value | Up/down | |
|---|---|---|---|---|---|---|---|
| shCtrl | shCOPB2 | ||||||
| 1 | EGFR/ErbB1 | pan-Tyr | EGFR | 20.95±1.10 | 16.63±1.56 | 0.0002 | −20.60% |
| 2 | HER2/ErbB2 | pan-Tyr | EGFR | 15.43±0.82 | 13.03±0.81 | 0.0005 | −15.55% |
| 3 | HER3/ErbB3 | pan-Tyr | EGFR | 22.87±0.99 | 19.73±2.17 | 0.0092 | −13.70% |
| 4 | FGFR1 | pan-Tyr | EGFR | 16.25±3.39 | 15.07±3.55 | 0.5682 | −7.28% |
| 5 | FGFR3 | pan-Tyr | EGFR | 12.25±0.59 | 11.25±1.26 | 0.1090 | −8.16% |
| 6 | FGFR4 | pan-Tyr | EGFR | 13.72±0.70 | 12.13±0.90 | 0.0068 | −11.54% |
| 7 | InsR | pan-Tyr | Insulin R | 14.52±0.60 | 12.05±0.43 | <0.00001 | −16.99% |
| 8 | IGF-IR | pan-Tyr | Insulin R | 14.75±2.82 | 13.5±4.19 | 0.5582 | −8.47% |
| 9 | TrkA/NTRK1 | pan-Tyr | NGFR | 11.73±0.20 | 10.07±0.65 | 0.0010 | −14.20% |
| 10 | TrkB/NTRK2 | pan-Tyr | NGFR | 11.72±0.78 | 10.05±0.44 | 0.0010 | −14.22% |
| 11 | Met/HGFR | pan-Tyr | HGFR | 11.83±0.74 | 11.2±0.26 | 0.1007 | −5.21% |
| 12 | Ron/MST1R | pan-Tyr | HGFR | 12.88±0.98 | 10.7±0.52 | 0.0007 | −16.95% |
| 13 | Ret | pan-Tyr | Ret | 11.27±0.68 | 8.93±0.41 | <0.00001 - | 20.71% |
| 14 | ALK | pan-Tyr | LTK | 14.73±4.62 | 12.37±4.24 | 0.3769 | −16.06% |
| 15 | PDGFR | pan-Tyr | PDGFR | 15.65±4.59 | 13.20±4.33 | 0.3639 | −15.65% |
| 16 | c-Kit/SCFR | pan-Tyr | PDGFR | 13.03±0.66 | 10.58±0.29 | <0.00001 - | 18.80% |
| 17 | FLT3/Flk2 | pan-Tyr | PDGFR | 11.77±0.60 | 9.43±0.29 | <0.00001 - | 19.83% |
| 18 | M-CSFR/CSF-1R | pan-Tyr | PDGFR | 15.05±4.40 | 12.57±3.97 | 0.3257 | −16.50% |
| 19 | EphA1 | pan-Tyr | EphR | 14.52±3.25 | 13.97±3. 40 | 0.7802 | −3.79% |
| 20 | EphA2 | pan-Tyr | EphR | 17.55±4.40 | 17.07±3.43 | 0.8361 | −2.75% |
| 21 | EphA3 | pan-Tyr | EphR | 13.93±1.26 | 11.72±1.12 | 0.0092 | −15.91% |
| 22 | EphB1 | pan-Tyr | EphR | 12.07±0.89 | 10.28±1.06 | 0.0101 | −14.78% |
| 23 | EphB3 | pan-Tyr | EphR | 12.53±2.60 | 12.25±3.98 | 0.8868 | −2.26% |
| 24 | EphB4 | pan-Tyr | EphR | 13.27±1.76 | 11.08±0.97 | 0.0237 | −16.46% |
| 25 | TYRO3/Dtk | pan-Tyr | Axl | 12.9±1.80 | 10.63±0.95 | 0.0213 | −17.57% |
| 26 | Axl | pan-Tyr | Axl | 13.1±1.70 | 11.5±1.95 | 0.1613 - | 12.21% |
| 27 | Tie2/TEK | pan-Tyr | Tie | 16.43±3.71 | 13.55±2.99 | 0.1694 | −17.55% |
| 28 | VEGFR2/KDR | pan-Tyr | VEGFR | 12.4±0.92 | 10.17±0.63 | 0.0006 | −18.01% |
| 29 | Akt/PKB/Rac | Thr308 | Akt | 22.42±3.13 | 16.65±1.45 | 0.0022 | −25.72% |
| 30 | Akt/PKB/Rac | Ser473 | Akt | 31.33±4.25 | 21.23±1.54 | 0.0013 | −32.23% |
| 31 | p44/42 MAPK (ERK1/2) | Thr202/Tyr204 | MAPK | 31.12±3.54 | 30.1±3.11 | 0.6087 | −3.27% |
| 32 | S6 Ribosomal Protein | Ser235/236 | RSK | 42.5±4.69 | 32.63±3.55 | 0.0021 | −23.22% |
| 33 | c-Abl | pan-Tyr | Abl | 16.58±1.81 | 12.9±1.22 | 0.0020 | −22.21% |
| 34 | IRS-1 | pan-Tyr | IRS | 18.93±4.60 | 16.1±3.69 | 0.2666 | −14.96% |
| 35 | Zap-70 | pan-Tyr | Zap-70 | 19.25±3.82 | 16.2±3.81 | 0.1957 | −15.84% |
| 36 | Src | pan-Tyr | Src | 15.65±0.91 | 13.32±0.90 | 0.0012 | −14.91% |
| 37 | Lck | pan-Tyr | Src | 15.7±2.57 | 12.15±1.48 | 0.0148 | −22.61% |
| 38 | Stat1 | Tyr701 | Stat | 19.8±4.13 | 16.287±3.74 | 0.1532 | −17.76% |
| 39 | Stat3 | Tyr705 | Stat | 22.38±1.37 | 20.067±1.89 | 0.0354 | −10.35% |
The results indicated that knockdown of COPB2 could significantly induce downregulation of phosphorylation of 23 targets in the RTK signaling pathway, including EGFR/ErbB1, HER2/ErbB2, HER3/ErbB3, FGFR4, InsR, TrkA/NTRK1, TrkB/NTRK2, Ron/MST1R, Ret, c-Kit/SCFR, FLT3/Flk2, EphA3, EphB1, EphB4, TYRO3/Dtk, VEGFR2/KDR, Akt/PKB/Rac (Thr308), Akt/PKB/Rac (Ser473), Ribosomal S6 kinase, c-Abl, Src, Lck and Stat3 at different levels.
Discussion
In the present study, the mRNA expression levels of COPB2 were abundant in gastric cancer cell lines, including MKN-45, MGC-803, BGC-823 and SGC-7901 cells. In addition, silencing of COPB2 expression inhibited gastric cancer cell proliferation in vitro and in vivo, and reduced cell colony formation and induced apoptosis of BGC-823 cells. These data indicated that COPB2 upregulation may serve an essential role in mediating the tumorigenicity of human gastric cancer cells. In addition, these observations provided strong evidence to suggest that COPB2 may contribute to the pathogenesis of gastric cancer, as indicated by the anti-proliferative and apoptosis-inducing effects of COPB2 knockdown. Therefore, COPB2 may represent a novel promising target for gene therapy of gastric cancer. The results of the current study were consistent with the results of COPB2 silencing in prostate cancer cells (5), lung adenocarcinoma cells (6) and colon cancer cells (7), whereby cell growth was inhibited and apoptosis was increased. Taken together, these data suggested that COPB2 may serve an essential role in gastric cancer cell growth. In addition, the size of gastric cancer tumors was significantly decreased when COPB2 expression was silenced, indicating that downregulation of COPB2 may inhibit the progression of gastric cancer cells. Furthermore, silencing of COPB2 using lentivirus-mediated shRNA effectively downregulated gastric cancer progression in BGC-823 cells potentially via the RTK signaling pathway.
The PathScan® RTK Signaling Antibody Array kit (Fluorescent Readout) is a slide-based antibody array product based on the sandwich immunoassay principle. This array kit is capable of simultaneously detecting the levels of 28 RTKs and 11 important signaling nodes that have been phosphorylated at tyrosine or other residues. RTKs, such as cell surface receptors, emit signals primarily via tyrosine phosphorylation reactions, which alter the function of downstream growth factors, including EGF, nerve growth factor, platelet-derived growth factor (PDGF), VEGF, FGF, insulin-like growth factor (IGF), ephrins and angiopoietins. RTKs activate a wide range of downstream signaling cascades, including the phosphatidylinositol 3-kinase (PI3K)/Akt, mitogen-activated protein kinase (MAPK) and Janus kinase (Jak)/Stat signaling pathways (12). It is known that these pathways modulate fundamental cellular functions, including cell division, growth, metabolism, differentiation, migration and survival.
In addition to these normal cellular functions, it has been reported that RTKs participate in the development and progression of human cancer (13); therefore, aberrantly expressed RTK signaling nodes are regarded as novel therapeutic targets for pharmaceutical intervention (14). Following activation, indispensable tyrosine kinases may stimulate numerous signaling pathways that serve important roles in DNA repair, apoptosis and cell proliferation. It has been suggested that tyrosine kinases may be attractive targets for cancer therapy, and tyrosine kinase inhibitors have been effective in the treatment of various tumor types, including head and neck, gastric, prostate and breast cancer, and leukemia (15). In the present study, the phosphorylated forms of a total of 23 tyrosine kinases were downregulated following knockdown of COPB2 in BGC-823 cells. Among these 23 targets, the EGFR family consists of four members that belong to the ErbB lineage of proteins (ErbB1-4), which, as a ligand of RTKs, may stimulate and modulate cell function. Therefore, the EGFR family may be considered therapeutic targets; EGFR inhibitors have been used for the treatment of various types of cancer, including lung, breast, pancreas, bladder, and head and neck cancers (16-19). Additional tyrosine kinases in the RTK signaling pathway identified in the present study were also confirmed to serve important roles in modulating cancer behavior, and are regarded as therapeutic targets in cancer, particularly in gastric cancer.
It has been demonstrated that silencing FGFR4 may disrupt the biological features of gastric cancer cells, and it is therefore considered a novel target molecule for therapy (20). The EphA2 RTK was also observed to be a promising target for cancer therapy (21). The Raf protein kinases are key intermediates in cellular signal transduction pathways, and Raf inhibitors, such as vemurafenib and dabrafenib, are considered to present a novel strategy for antitumor therapy (22). Targeting Stat3 signaling is also a molecular strategy for therapeutic intervention (23,24). The InsR and the IGF receptor 1 (IGF1R) exert oncogenic functions (25), and participate in cancer development and progression via the IGF network (26). RTKs and their associated growth factors, including IGF1R, InsR, MET and HER3, have been observed to be enriched in HER2-positive gastric cancer tissue samples from patients, and have been identified as an important cause of lapatinib resistance in HER2-positive gastric cancer cells (27).
Trk receptors have been observed to be significantly associated with tumor progression and survival (28). TrkC activates Akt and suppresses transforming growth factor-β signaling, and is considered a potential therapeutic target for colorectal cancer (29). Activation of Trks stimulates tumor cell proliferation, aggressiveness and metastasis (30).
Ron, also known as MST1R, is an RTK from the Met proto-oncogene family; its dimerization and subsequent phosphorylation activates classical downstream signaling pathways, including MAPK, Jak/Stat3 and PI3K/Akt (31). Previous studies have demonstrated that knockdown of Ron by small interfering (si)RNA could suppress tumor cell migration and invasion, induce apoptosis, and arrest the cell cycle at S phase in AGS cells and G2/M phase in MKN28 cells (31,32). In addition, Ron expression has been significantly associated with tumor size, depth of invasion, lymph node metastasis, tumor stage and poor survival (32).
The Ret proto-oncogene RTK may serve an important role in cell growth, differentiation and survival. Following ligand binding, it activates numerous downstream signaling pathways, such as MAPK/extracellular signal-regulated kinase (ERK) and PI3k/Akt (33). The tyrosine kinase domain of Ret is considered to be a crucial therapeutic target (34).
The c-Kit proto-oncogene RTK belongs to the type III receptor family. As c-Kit signaling serves a role in tumorigenesis, imatinib has been used to treat tumors by inhibiting c-Kit signaling (35). c-Kit activation is pivotal in the majority of gastrointestinal stromal tumors, and is likely an initiating tumorigenic event; therefore, c-Kit is almost a universal therapeutic target (36).
FLT3, together with KIT, FMS and PDGF receptor (PDGFR), belongs to the class III RTKs (37). Signal transduction pathways activated by FLT3 include several conserved pathways, such as RAS/MAPK and PI3K/Akt (38). Combined inhibition of PI3Kδ and FLT3 exerts synergistic antitumor activities in FLT3-activated acute myeloid leukemia (39).
The Eph RTK has been demonstrated to exert a complex role in tumor formation, progression and metastasis. EphA2, EphA3 and EphB4 are some of the most widely overexpressed Eph RTKs in cancer, which are associated with tumor aggressiveness. EphB4 signaling may activate downstream signaling factors, including VEGF expression (40). EphB1-targeting small interfering (si)RNA may reduce cell viability and growth, alter cell cycle progression, decrease the expression of important cell cycle regulators, and increase the percentage of cells in G1 phase of the cell cycle. Knockdown of EphB1 in DAOY cells results in a significant reduction in the migration of medulloblastoma cells (41). Silencing EphA2 may also suppress the growth and haptotaxis of malignant mesothelioma cells (42). In addition, knockdown of EphA2 expression has been observed to inhibit gastric cancer cell proliferation and invasion in vitro and in vivo (43). An additional study (44) demonstrated that EphB4-targeting siRNA decreases non-small cell lung cancer cell viability and the volume of established tumors in vivo.
FGF signaling regulates cell fate, angiogenesis, immunity and metabolism via its receptors, FGFR1, FGFR2, FGFR3 and FGFR4. Aberrantly expressed FGF signaling may lead to the development of breast, gastric and lung cancer. Therefore, anti-FGFR therapy, in the form of anti-FGF/FGFR monoclonal antibodies and small-molecule FGFR inhibitors, is considered an effective means of cancer treatment (45).
TYRO3 is involved in the process of controlling cell survival and proliferation, and upregulation of TYRO3 may enhance cell motility, invasion, anchorage-independent growth and metastatic ability. Conversely, knockdown of TYRO3 may reverse these biological behaviors (46,47). A previous study demonstrated that overexpression of TYRO3 is associated with a hepatocellular carcinoma serum biomarker, α-fetoprotein (AFP), and alanine aminotransferase expression and tumor diameter. Knockdown of TYRO3 in the Hep3B hepatocellular carcinoma cell line reduces cell proliferation, ERK phosphorylation, cyclin D1 expression and AFP levels (48). RNAi-targeting of TYRO3 inhibits the proliferation of luminal-type cells in estradiol-rich and estradiol-null conditions, and is associated with G0-G1/S cell cycle arrest (49).
VEGFR2, also known as KDR, functions as the main mediator of VEGF-induced endothelial proliferation, survival and migration in response to numerous factors. VEGFR2 is a molecular target for the treatment of gastric cancer. Ramucirumab is a recombinant monoclonal antibody that targets VEGFR2, which has been used to treat specific cancer types.
Once downstream signals of PI3K have been activated by various growth factors, including PDGF, EGF and IGF-I, Akt is activated by phosphorylation on Thr-308 and Tyr-474 residues. Activation of the PI3K/AKT signaling pathway is frequently associated with human malignancies and serves a key role in cancer progression (50,51), including gastric cancer (52). Therefore, targeting the PI3K-AKT-mTOR pathway may be considered an anticancer therapeutic strategy. RSK contains two non-identical catalytic kinase domains, which phosphorylate various substrates, including members of the MAPK signaling pathway, and control cell growth and differentiation. For example, RSK regulates cell migration and invasion of metastatic breast cancer cells via stimulating the phosphorylation of EphA2, thus activating the RSK-EphA2 signaling pathway (53). In response to the RSK inhibitor, or via RNAi-targeting of RSK2, EGF-induced cell proliferation is suppressed (54), and knockdown of RSK2, but not RSK1, in L3.6pl pancreatic cancer cells significantly inhibits the macrophage-stimulating protein-induced epithelial-mesenchymal-transition (EMT)-like phenotype and cell migration. RSK2 activation serves a critical determinant role in linking Ron signaling to the cellular EMT program. Consequently, inhibition of RSK2 activity may provide a therapeutic opportunity for inhibiting Ron-mediated cancer cell migration and invasion (55).
The c-Abl proto-oncogene is not an RTK, but participates in numerous cellular processes, such as cell division, adhesion, differentiation and stress responses. c-Abl kinases may promote PDGFR-mediated proliferation and migration (56).
The non-RTK Src proto-oncogene has been observed to be involved in numerous signaling pathways associated with cell proliferation, migration, tumor adhesion and angiogenesis, and may also mediate signaling pathways from many types of receptors, including RTKs (57). Treatment with the novel Src/Abl inhibitor, bosutinib, alone or in combination with additional chemotherapeutic agents, may be a valuable therapeutic strategy for neuroblastoma treatment (58). An inhibitor of Src kinase has been observed to inhibit the growth of cervical cancer cells in vitro and in vivo. Downregulation of phosphorylated-Src (Y416) inhibits cell proliferation and cell cycle arrest in HeLa and SiHa cells by regulating cyclin-dependent kinases and cyclin X. Results from a nude mouse xenograft model indicated that PP2, an inhibitor of Src kinase, may significantly inhibit subcutaneous tumor growth of cervical cancer cells (59).
The Lck proto-oncogene, a Src family member of protein tyrosine kinases, may protect cells from glucocorticoid-induced apoptosis. Small-molecule inhibitors of Lck, such as dasatinib, reverse glucocorticoid resistance in some lymphoid malignancies (60). Inhibition of Lck by treatment with PP2, an Src kinase family inhibitor, or via targeting Lck with siRNA, suppresses sphingosine-induced conformational activation and oligomerization of B-cell lymphoma 2 homologous antagonist/killer, and leads to mitochondrial membrane potential loss and apoptotic cell death (61).
Stat3 is constitutively activated in various tumor types, and serves an important role in tumor survival, metastasis, chemoresistance and escape from immune responses (62,63). Stat3 is activated upon tyrosine phosphorylation, which is mediated by upstream cytokines, including, JAK, Src, Abl or RTKs. Conversely, Stat3 is inhibited by Debio 0617B, a novel RTK inhibitor that inhibits Stat3 in Stat3-activated carcinoma cell lines and causes a dose-dependent decrease in cell proliferation (63). Inhibiting Stat3 reduces cell growth and induces apoptosis in head and neck cancer (64), glioma (65), prostate cancer (66) and pancreatic cancer (67) cells. Specifically, silencing of Stat3 significantly inhibits the growth of gastric cancer cells in vitro and in vivo via cell apoptosis and cell cycle shift (68).
In conclusion, the present study demonstrated that COPB2 was abundantly expressed in human gastric cancer cell lines. Knockdown of COPB2 in BGC-823 cells inhibited cell growth and colony formation abilities, and promoted cell apoptosis, potentially via modulating RTK signaling and its downstream signaling cascades. Factors, including EGFR/ErbB1, HER2/ErbB2, HER3/ErbB3, FGFR4, InsR, TrkA/NTRK1, TrkB/NTRK2, Ron/MST1R, Ret, c-Kit/SCFR, FLT3/Flk2, EphA3, EphB1, EphB4, TYRO3/Dtk, VEGFR2/KDR, Akt/PKB/Rac (Thr308), Akt/PKB/Rac (Ser473), S6 ribosomal protein, c-Abl, Src, Lck and Stat3, may be involved in the effects of COPB2 knockdown. Therefore, COPB2 may be considered a valuable gene therapy target for the treatment of gastric cancer.
Acknowledgments
Not applicable.
Funding
This study was supported by the Lanzhou Science and Technology Planning Project (grant no. 2016-3-113), the 60th Project of China Postdoctoral Foundation (grant no. 2016M602888), the China's National Science and Technology Program for Public Wellbeing (grant no. 2012GS620101) and the National Key Research and Development Plan (grant no. 2017YFC0908302).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
CA, HL and YZ were involved in conception and design. XZ, JW, YQ, XY, QL and QG were involved in the collection and assembly of data. YZ provided study materials and patients. All authors contributed to data analysis and interpretation, and wrote and gave final approved for the manuscript.
Ethics approval and consent to participate
The experiments were authorized and approved by the Institutional Animal Care and Use Committee of Gansu University of Chinese Medicine (Lanzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Yang BH, Parkin DM, Cai L, Zhang ZF. Cancer burden and trends in the Asian Pacific Rim region. Asian Pac J Cancer Prev. 2004;5:96–117. [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng R, Zeng H, Zhang S, Chen W. Estimates of cancer incidence and mortality in China, 2013. Chin J Cancer. 2017;36:66. doi: 10.1186/s40880-017-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jim MA, Pinheiro PS, Carreira H, Espey DK, Wiggins CL, Weir HK. Stomach cancer survival in the United States by race and stage (2001–2009): Findings from the CONCORD-2 study. Cancer. 2017;123(Suppl 24):4994–5013. doi: 10.1002/cncr.30881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck R, Rawet M, Wieland FT, Cassel D. The COPI system: Molecular mechanisms and function. FEBS Lett. 2009;583:2701–2709. doi: 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 5.Mi Y, Yu M, Zhang L, Sun C, Wei B, Ding W, Zhu Y, Tang J, Xia G, Zhu L. COPB2 is upregulated in prostate cancer and regulates PC-3 cell proliferation, cell cycle, and apoptosis. Arch Med Res. 2016;47:411–418. doi: 10.1016/j.arcmed.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Erdogan E, Klee EW, Thompson EA, Fields AP. Meta-analysis of oncogenic protein kinase Ciota signaling in lung adenocarcinoma. Clin Cancer Res. 2009;15:1527–1533. doi: 10.1158/1078-0432.CCR-08-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Chai Z, Wang M, Jin Y, Yang A, Li M. COPB2 suppresses cell proliferation and induces cell cycle arrest in human colon cancer by regulating cell cycle-related proteins. Exp Ther Med. 2018;15:777–784. doi: 10.3892/etm.2017.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 9.Committee for the Update of the Guide for the Care and Use of Laboratory Animals, Institute for Laboratory Animal Research, Division on Earth and Life Studies, National Research Council of The National Academies . Guide For The Care And Use Of Laboratory Animals. 8th edition. The National Academies Press; Washington, DC: 2011. [Google Scholar]
- 10.Schuster C, Malinowsky K, Liebmann S, Berg D, Wolff C, Tran K, Schott C, Reu S, Neumann J, Faber C, et al. Antibody validation by combining immunohistochemistry and protein extraction from formalin-fixed paraffin-embedded tissues. Histopathology. 2012;60(6B):E37–E50. doi: 10.1111/j.1365-2559.2012.04184.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Yu S, Cui L, Wang W, Li J, Wang K, Lao X. Role of SMC1A overexpression as a predictor of poor prognosis in late stage colorectal cancer. BMC Cancer. 2015;15:90. doi: 10.1186/s12885-015-1085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song S, Rosen KM, Corfas G. Biological function of nuclear receptor tyrosine kinase action. Cold Spring Harb Perspect Biol. 2013;5:5. doi: 10.1101/cshperspect.a009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennasroune A, Gardin A, Aunis D, Crémel G, Hubert P. Tyrosine kinase receptors as attractive targets of cancer therapy. Crit Rev Oncol Hematol. 2004;50:23–38. doi: 10.1016/j.critrevonc.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Choura M, Rebaï A. Receptor tyrosine kinases: From biology to pathology. J Recept Signal Transduct Res. 2011;31:387–394. doi: 10.3109/10799893.2011.625425. [DOI] [PubMed] [Google Scholar]
- 15.Pytel D, Sliwinski T, Poplawski T, Ferriola D, Majsterek I. Tyrosine kinase blockers: New hope for successful cancer therapy. Anticancer Agents Med Chem. 2009;9:66–76. doi: 10.2174/187152009787047752. [DOI] [PubMed] [Google Scholar]
- 16.Cripps C, Winquist E, Devries MC, Stys-Norman D, Gilbert R, Head and Neck Cancer Disease Site Group Epidermal growth factor receptor targeted therapy in stages III and IV head and neck cancer. Curr Oncol. 2010;17:37–48. doi: 10.3747/co.v17i3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qing L, Qing W. Development of epidermal growth factor receptor targeted therapy in pancreatic cancer. Minerva Chir. 2018;73:488–496. doi: 10.23736/S0026-4733.18.07512-0. [DOI] [PubMed] [Google Scholar]
- 18.Tiseo M, Loprevite M, Ardizzoni A. Epidermal growth factor receptor inhibitors: A new prospective in the treatment of lung cancer. Curr Med Chem Anticancer Agents. 2004;4:139–148. doi: 10.2174/1568011043482106. [DOI] [PubMed] [Google Scholar]
- 19.Bellmunt J, Hussain M, Dinney CP. Novel approaches with targeted therapies in bladder cancer. Therapy of bladder cancer by blockade of the epidermal growth factor receptor family. Crit Rev Oncol Hematol. 2003;46(Suppl):S85–S104. doi: 10.1016/S1040-8428(03)00067-2. [DOI] [PubMed] [Google Scholar]
- 20.Ye Y, Jiang D, Li J, Wang M, Han C, Zhang X, Zhao C, Wen J, Kan Q. Silencing of FGFR4 could influence the biological features of gastric cancer cells and its therapeutic value in gastric cancer. Tumour Biol. 2016;37:3185–3195. doi: 10.1007/s13277-015-4100-0. [DOI] [PubMed] [Google Scholar]
- 21.Ireton RC, Chen J. EphA2 receptor tyrosine kinase as a promising target for cancer therapeutics. Curr Cancer Drug Targets. 2005;5:149–157. doi: 10.2174/1568009053765780. [DOI] [PubMed] [Google Scholar]
- 22.Durrant DE, Morrison DK. Targeting the Raf kinases in human cancer: The Raf dimer dilemma. Br J Cancer. 2018;118:3–8. doi: 10.1038/bjc.2017.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikitakis NG, Siavash H, Sauk JJ. Targeting the STAT pathway in head and neck cancer: Recent advances and future prospects. Curr Cancer Drug Targets. 2004;4:637–651. doi: 10.2174/1568009043332736. [DOI] [PubMed] [Google Scholar]
- 24.Turkson J, Jove R. STAT proteins: Novel molecular targets for cancer drug discovery. Oncogene. 2000;19:6613–6626. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- 25.Heidegger I, Kern J, Ofer P, Klocker H, Massoner P. Oncogenic functions of IGF1R and INSR in prostate cancer include enhanced tumor growth, cell migration and angiogenesis. Oncotarget. 2014;5:2723–2735. doi: 10.18632/oncotarget.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ofer P, Heidegger I, Eder IE, Schöpf B, Neuwirt H, Geley S, Klocker H, Massoner P. Both IGF1R and INSR knockdown exert antitumorigenic effects in prostate cancer in vitro and in vivo. Mol Endocrinol. 2015;29:1694–1707. doi: 10.1210/me.2015-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Wang J, Ji D, Wang C, Liu R, Wu Z, Liu L, Zhu D, Chang J, Geng R, et al. Functional genetic approach identifies MET, HER3, IGF1R, INSR pathways as determinants of lapatinib unresponsiveness in HER2-positive gastric cancer. Clin Cancer Res. 2014;20:4559–4573. doi: 10.1158/1078-0432.CCR-13-3396. [DOI] [PubMed] [Google Scholar]
- 28.Kamiya A, Inokuchi M, Otsuki S, Sugita H, Kato K, Uetake H, Sugihara K, Takagi Y, Kojima K. Prognostic value of tropomyosin-related kinases A, B, and C in gastric cancer. Clin Transl Oncol. 2016;18:599–607. doi: 10.1007/s12094-015-1407-7. [DOI] [PubMed] [Google Scholar]
- 29.Kim MS, Suh KW, Hong S, Jin W. TrkC promotes colorectal cancer growth and metastasis. Oncotarget. 2017;8:41319–41333. doi: 10.18632/oncotarget.17289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meldolesi J. Neurotrophin Trk receptors: New targets for cancer therapy. Rev Physiol Biochem Pharmacol. 2018;174:67–79. doi: 10.1007/112_2017_6. [DOI] [PubMed] [Google Scholar]
- 31.Yang SY, Nguyen TT, Ung TT, Jung YD. Role of recepteur d'origine nantais on gastric cancer development and progression. Chonnam Med J. 2017;53:178–186. doi: 10.4068/cmj.2017.53.3.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song YA, Park YL, Kim KY, Myung E, Chung CY, Cho SB, Lee WS, Jung YD, Kweon SS, Joo YE. RON is associated with tumor progression via the inhibition of apoptosis and cell cycle arrest in human gastric cancer. Pathol Int. 2012;62:127–136. doi: 10.1111/j.1440-1827.2011.02765.x. [DOI] [PubMed] [Google Scholar]
- 33.Richardson DS, Lai AZ, Mulligan LM. RET ligand-induced internalization and its consequences for downstream signaling. Oncogene. 2006;25:3206–3211. doi: 10.1038/sj.onc.1209349. [DOI] [PubMed] [Google Scholar]
- 34.Plaza-Menacho I, Mologni L, McDonald NQ. Mechanisms of RET signaling in cancer: Current and future implications for targeted therapy. Cell Signal. 2014;26:1743–1752. doi: 10.1016/j.cellsig.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 35.Ali S, Ali S. Role of c-kit/SCF in cause and treatment of gastrointestinal stromal tumors (GIST) Gene. 2007;401:38–45. doi: 10.1016/j.gene.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 36.Fletcher JA. KIT oncogenic mutations: Biologic insights, therapeutic advances, and future directions. Cancer Res. 2016;76:6140–6142. doi: 10.1158/0008-5472.CAN-16-2646. [DOI] [PubMed] [Google Scholar]
- 37.Kiyoi H, Naoe T. Biology, clinical relevance, and molecularly targeted therapy in acute leukemia with FLT3 mutation. Int J Hematol. 2006;83:301–308. doi: 10.1532/IJH97.06071. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt-Arras D, Schwäble J, Böhmer FD, Serve H. Flt3 receptor tyrosine kinase as a drug target in leukemia. Curr Pharm Des. 2004;10:1867–1883. doi: 10.2174/1381612043384394. [DOI] [PubMed] [Google Scholar]
- 39.He Y, Sun L, Xu Y, Fu L, Li Y, Bao X, Fu H, Xie C, Lou L. Combined inhibition of PI3Kδ and FLT3 signaling exerts synergistic antitumor activity and overcomes acquired drug resistance in FLT3-activated acute myeloid leukemia. Cancer Lett. 2018;420:49–59. doi: 10.1016/j.canlet.2018.01.071. [DOI] [PubMed] [Google Scholar]
- 40.Charmsaz S, Boyd AW. Eph receptors as oncotargets. Oncotarget. 2017;8:81727–81728. doi: 10.18632/oncotarget.21045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhatia S, Baig NA, Timofeeva O, Pasquale EB, Hirsch K, MacDonald TJ, Dritschilo A, Lee YC, Henkemeyer M, Rood B, et al. Knockdown of EphB1 receptor decreases medul-loblastoma cell growth and migration and increases cellular radiosensitization. Oncotarget. 2015;6:8929–8946. doi: 10.18632/oncotarget.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nasreen N, Mohammed KA, Antony VB. Silencing the receptor EphA2 suppresses the growth and haptotaxis of malignant mesothelioma cells. Cancer. 2006;107:2425–2435. doi: 10.1002/cncr.22254. [DOI] [PubMed] [Google Scholar]
- 43.Yuan W, Chen Z, Chen Z, Wu S, Guo J, Ge J, Yang P, Huang J. Silencing of EphA2 inhibits invasion of human gastric cancer SGC-7901 cells in vitro and in vivo. Neoplasma. 2012;59:105–113. doi: 10.4149/neo_2012_014. [DOI] [PubMed] [Google Scholar]
- 44.Becerikli M, Merwart B, Lam MC, Suppelna P, Rittig A, Mirmohammedsadegh A, Stricker I, Theiss C, Singer BB, Jacobsen F, et al. EPHB4 tyrosine-kinase receptor expression and biological significance in soft tissue sarcoma. Int J Cancer. 2015;136:1781–1791. doi: 10.1002/ijc.29244. [DOI] [PubMed] [Google Scholar]
- 45.Katoh M. Therapeutics targeting FGF signaling network in human diseases. Trends Pharmacol Sci. 2016;37:1081–1096. doi: 10.1016/j.tips.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Chien CW, Hou PC, Wu HC, Chang YL, Lin SC, Lin SC, Lin BW, Lee JC, Chang YJ, Sun HS, et al. Targeting TYRO3 inhibits epithelial-mesenchymal transition and increases drug sensitivity in colon cancer. Oncogene. 2016;35:5872–5881. doi: 10.1038/onc.2016.120. [DOI] [PubMed] [Google Scholar]
- 47.Schmitz R, Valls AF, Yerbes R, von Richter S, Kahlert C, Loges S, Weitz J, Schneider M, Ruiz de Almodovar C, Ulrich A, et al. TAM receptors Tyro3 and Mer as novel targets in colorectal cancer. Oncotarget. 2016;7:56355–56370. doi: 10.18632/oncotarget.10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duan Y, Wong W, Chua SC, Wee HL, Lim SG, Chua BT, Ho HK. Overexpression of Tyro3 and its implications on hepato-cellular carcinoma progression. Int J Oncol. 2016;48:358–366. doi: 10.3892/ijo.2015.3244. [DOI] [PubMed] [Google Scholar]
- 49.Ekyalongo RC, Mukohara T, Funakoshi Y, Tomioka H, Kataoka Y, Shimono Y, Chayahara N, Toyoda M, Kiyota N, Minami H. TYRO3 as a potential therapeutic target in breast cancer. Anticancer Res. 2014;34:3337–3345. [PubMed] [Google Scholar]
- 50.Koundouros N, Poulogiannis G. Phosphoinositide 3-kinase/Akt signaling and redox metabolism in cancer. Front Oncol. 2018;8:160. doi: 10.3389/fonc.2018.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faes S, Dormond O. PI3K and AKT: Unfaithful partners in cancer. Int J Mol Sci. 2015;16:21138–21152. doi: 10.3390/ijms160921138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasaki T, Kuniyasu H, Luo Y, Kitayoshi M, Tanabe E, Kato D, Shinya S, Fujii K, Ohmori H, Yamashita Y. AKT activation and telomerase reverse transcriptase expression are concurrently associated with prognosis of gastric cancer. Pathobiology. 2014;81:36–41. doi: 10.1159/000351721. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Y, Yamada N, Tanaka T, Hori T, Yokoyama S, Hayakawa Y, Yano S, Fukuoka J, Koizumi K, Saiki I, et al. Crucial roles of RSK in cell motility by catalysing serine phosphorylation of EphA2. Nat Commun. 2015;6:7679. doi: 10.1038/ncomms8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamaoka Y, Negishi M, Katoh H. EphA2 is a key effector of the MEK/ERK/RSK pathway regulating glioblastoma cell proliferation. Cell Signal. 2016;28:937–945. doi: 10.1016/j.cellsig.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Ma Q, Guin S, Padhye SS, Zhou YQ, Zhang RW, Wang MH. Ribosomal protein S6 kinase (RSK)-2 as a central effector molecule in RON receptor tyrosine kinase mediated epithelial to mesenchymal transition induced by macrophage-stimulating protein. Mol Cancer. 2011;10:66. doi: 10.1186/1476-4598-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srinivasan D, Kaetzel DM, Plattner R. Reciprocal regulation of Abl and receptor tyrosine kinases. Cell Signal. 2009;21:1143–1150. doi: 10.1016/j.cellsig.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yezhelyev MV, Koehl G, Guba M, Brabletz T, Jauch KW, Ryan A, Barge A, Green T, Fennell M, Bruns CJ. Inhibition of SRC tyrosine kinase as treatment for human pancreatic cancer growing orthotopically in nude mice. Clin Cancer Res. 2004;10:8028–8036. doi: 10.1158/1078-0432.CCR-04-0621. [DOI] [PubMed] [Google Scholar]
- 58.Bieerkehazhi S, Chen Z, Zhao Y, Yu Y, Zhang H, Vasudevan SA, Woodfield SE, Tao L, Yi JS, Muscal JA, et al. Novel Src/Abl tyrosine kinase inhibitor bosutinib suppresses neuroblastoma growth via inhibiting Src/Abl signaling. Oncotarget. 2017;8:1469–1480. doi: 10.18632/oncotarget.13643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong L, Deng Z, Zhao Y, Wang Y, Sarkar FH, Zhang Y. Down-regulation of phospho-non-receptor Src tyrosine kinases contributes to growth inhibition of cervical cancer cells. Med Oncol. 2011;28:1495–1506. doi: 10.1007/s12032-010-9583-3. [DOI] [PubMed] [Google Scholar]
- 60.Harr MW, Caimi PF, McColl KS, Zhong F, Patel SN, Barr PM, Distelhorst CW. Inhibition of Lck enhances glucocorticoid sensitivity and apoptosis in lymphoid cell lines and in chronic lymphocytic leukemia. Cell Death Differ. 2010;17:1381–1391. doi: 10.1038/cdd.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim MJ, Park MT, Yoon CH, Byun JY, Lee SJ. Activation of Lck is critically required for sphingosine-induced conformational activation of Bak and mitochondrial cell death. Biochem Biophys Res Commun. 2008;370:353–358. doi: 10.1016/j.bbrc.2008.03.084. [DOI] [PubMed] [Google Scholar]
- 62.Kanda N, Seno H, Konda Y, Marusawa H, Kanai M, Nakajima T, Kawashima T, Nanakin A, Sawabu T, Uenoyama Y, et al. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene. 2004;23:4921–4929. doi: 10.1038/sj.onc.1207606. [DOI] [PubMed] [Google Scholar]
- 63.Murone M, Vaslin Chessex A, Attinger A, Ramachandra R, Shetty SJ, Daginakatte G, Sengupta S, Marappan S, Dhodheri S, Rigotti S, et al. Debio 0617B inhibits growth of STAT3-driven solid tumors through combined inhibition of JAK, SRC, and class III/V receptor tyrosine kinases. Mol Cancer Ther. 2016;15:2334–2343. doi: 10.1158/1535-7163.MCT-15-0974. [DOI] [PubMed] [Google Scholar]
- 64.Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, Robbins PD, Gadiparthi S, Burke NA, Watkins SF, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci USA. 2003;100:4138–4143. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu J, Li G, Sun T, Su Y, Zhang X, Shen J, Tian Z, Zhang J. Blockage of the STAT3 signaling pathway with a decoy oligo-nucleotide suppresses growth of human malignant glioma cells. J Neurooncol. 2008;89:9–17. doi: 10.1007/s11060-008-9590-9. [DOI] [PubMed] [Google Scholar]
- 66.Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, Bowman T, Falcone R, Fairclough R, Cantor A, et al. Constitutive activation of Stat3 in human prostate tumors and cell lines: Direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62:6659–6666. [PubMed] [Google Scholar]
- 67.Kim C, Kim JH, Oh EY, Nam D, Lee SG, Lee J, Kim SH, Shim BS, Ahn KS. Blockage of STAT3 signaling pathway by morusin induces apoptosis and inhibits invasion in human pancreatic tumor cells. Pancreas. 2016;45:409–419. doi: 10.1097/MPA.0000000000000496. [DOI] [PubMed] [Google Scholar]
- 68.Sun Y, Guo BF, Xu LB, Zhong JT, Liu ZW, Liang H, Wen NY, Yun WJ, Zhang L, Zhao XJ. Stat3-siRNA inhibits the growth of gastric cancer in vitro and in vivo. Cell Biochem Funct. 2015;33:495–502. doi: 10.1002/cbf.3148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.