Abstract
Clear cell renal cell carcinoma (ccRCC) has been associated with one of the highest mortality rates among all cancers. Fatty acid binding proteins (FABPs) are 14-15 kDa proteins that are highly abundant in the cytosol of most tissues. FABP5, a member of the FABP family, has been observed to promote tumor cell growth in numerous cancer types. In order to investigate the function of FABP5 in ccRCC cells in the present study, RNA sequencing data from The Cancer Genome Atlas were analyzed to determine the expression levels of FABP5 in ccRCC patient samples. Survival and Cox regression analyses were performed to measure the association between FABP5 expression and clinicopathological features of patients with ccRCC. Subsequent in vitro experiments downregulated or overexpressed FABP5 in Caki-1 and 786O ccRCC cells using lentiviral vectors to evaluate cell proliferation ability, and a xenograft transplantation model was established to examine the effect of FABP5 on tumorigenesis in vivo. The results demonstrated that FABP5 expression was significantly upregulated in samples from patients with ccRCC when compared with normal tissue samples. High FABP5 expression was also significantly correlated with tumor and metastasis classifications and predicted poor survival in patients with ccRCC. In ccRCC cells, silencing of FABP5 significantly inhibited cell proliferation, while overexpression of FABP5 promoted cell proliferation when compared to the respective controls. In addition, treatment with the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT inhibitor, LY294002, attenuated the pro-proliferative effects of exogenous FABP5 expression in Caki-1 and 786O cells. This indicated that the PI3K/AKT signaling pathway may be partially involved in the FABP5-mediated increase in ccRCC cell proliferation. Furthermore, FABP5 was observed to regulate tumor growth in nude mice in vivo. In conclusion, the results of the present study suggest that FABP5 may exert a pro-proliferative role in ccRCC and may be associated with malignant progression and tumorigenesis.
Keywords: clear cell renal cell carcinoma; fatty acid binding protein 5; proliferation; phosphatidylinositol-4,5-bisphosphate 3-kinase/AKT signaling pathway
Introduction
The kidney is an important organ that is responsible for maintaining homeostasis in the human body. The urine-producing functional unit of the kidney is the nephron, which consists of a renal corpuscle and tubules. Kidney cancer, also known as renal cancer, originates from cells in the kidney. The two most common types of renal cancer are renal cell carcinoma (RCC) and urothelial cell carcinoma. RCC is a heterogeneous disease that arises from the proximal convoluted tubular epithelium, and is the most common type of adult renal cancer (1, 2). Clear cell (cc) RCC accounts for ~80% of all RCC cases (3). Surgical excision is effective for the majority of patients with ccRCC; however, disease recurrence or distant metastasis occurs in ~30% of patients (4,5).
The phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT signaling pathway is involved in numerous diverse cellular functions, including cell proliferation (6), differentiation (7), angiogenesis (8) and autophagy (9). The PI3K/AKT signaling pathway also affects epithelial-mesenchymal transition (EMT) via multiple regulatory mechanisms, which influences tumor aggressiveness (10). In addition, the PI3K/AKT signaling pathway has been demonstrated to regulate cell proliferation and invasion in ccRCC cell lines (11,12).
Fatty acid binding proteins (FABPs) are 14-15 kDa proteins that are highly abundant in the cytosol of most tissues. Nine mammalian FABPs have thus far been identified, and they contain highly conserved structures (13,14). FABPs display a high affinity for long-chain fatty acids (FAs) and regulate lipid metabolism in different tissues (15), including the brain (16), intestine and liver (13). FABPs also mediate the biological properties of tumor cells. For instance, FABP7, an FABP expressed in the mammalian central nervous system, has been demonstrated to regulate glioblastoma cell proliferation (17).
Recently, FABP5, which promotes tumor cell growth in cervical cancer, was identified as a potential biomarker for lymph node metastasis (18,19). FABP5 gene silencing inhibited the proliferation and invasion of human SGC-7901 gastric cancer cells in vitro (20), and FABP5 stimulated hepatocellular carcinoma progression and metastasis via EMT (21). Considering the pivotal functions of the PI3K/AKT signaling pathway in tumor cells, particularly ccRCC cells, we hypothesized that FABP5 may affect ccRCC cell function via the PI3K/AKT signaling pathway.
In the present study, the function of FABP5 in ccRCC cell lines was investigated and the results suggest that FABP5 may present a putative prognostic biomarker for patients with ccRCC and provide a novel perspective for the role of FABPs in tumor biology.
Materials and methods
Bioinformatics prediction using the The Cancer Genome Atlas (TCGA) database
RNA sequencing data from TCGA (https://cancergenome.nih.gov/) was used to assess the correlation between FABP5 mRNA expression levels and clinicopathological features of patients with ccRCC. The expression of FABP5 in all samples was sorted from low to high, and the median expression was selected as the cutoff value to distinguish patients with low and high expression. The median number was 75.32635. Overall survival and disease-free survival analysis were performed according to a previously described method (22). A total of 246 patient samples with associated clinical parameters were selected for further analysis.
Cell culture and transfection
Caki-1 (cat. no. GCC-KI0004RT) and 786O (cat. no. GCC-KI0003RT) ccRCC cell lines were purchased from Shanghai GeneChem, Co., Ltd. (Shanghai, China). All cells were cultivated in complete medium consisting of Dulbecco’s modified Eagle’s medium/F12 (Corning Inc., Corning, NY, USA) and 10% fetal bovine serum (Clark Bioscience, Richmond, VA, USA). The GV112 RNA interference (RNAi) system (Shanghai GeneChem Co., Ltd.) was used to generate lentiviruses expressing short interfering RNA sequences targeting FABP5 (LV-FABP5-RNAi). This system contains a U6 promoter-driven multiple cloning site (MCS) and a cytomegalovirus promoter-driven puromycin gene. The target sequence of FABP5 was 5′-TGGGAAGGAAAGCACAATA-3′ (20). Lentiviral vectors overexpressing FABP5 (LV-FABP5) were purchased from Shanghai GeneChem Co., Ltd. directly and were generated using the GV492 system (Shanghai GeneChem Co., Ltd.). Briefly, expression from an MCS combined with a 3xFLAG tag is driven by the ubiquitin promoter, and green fluorescent protein (GFP) and puromycin expression are driven by the cellobiohydrolase promoter. The negative control lentiviruses, LV-NC-RNAi and LV-NC, were also purchased from Shanghai GeneChem Co., Ltd. The scrambled sequence used for the LV-NC-RNAi was as follows: 5′-TTCTCCGAACGTGTCACGT-3′. An empty lentiviral vector was used to transfect cells in the LV-NC group.
Prior to transfection, cells were seeded in six-well plates at a density of 1×105 cells/well in complete medium and incubated overnight. Lentiviruses (multiplicity of infection=10) together with 5 µg/ml polybrene, was then added to the cells. At 24 h following infection, the culture medium was replaced with fresh medium containing 2 µg/ml puromycin (Sangon Biotech Co., Ltd., Shanghai, China), followed by positive selection for 3 days. The cells were then maintained in complete medium containing 1 µg/ml puromycin. The PI3K/AKT signaling pathway inhibitor, LY294002, was purchased from MedChem Express (Monmouth Junction, NJ, USA) and added to the culture medium at a concentration of 20 µM for 24 h to inactivate the PI3K/AKT signaling pathway in vitro.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using a UNIQ-10 Spin Column RNA Purification kit (Sangon Biotech Co., Ltd.). A total of 1 µg total RNA was prepared and first-strand cDNA was synthesized using the RevertAid First Strand cDNA Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc., Waltham, MA, USA). cDNA was subsequently analyzed using a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific, Inc.) using AceQ qPCR SYBR Green Master Mix (Vazyme, Piscataway, NJ, USA). The following primers were used: FABP5, forward, 5′-GCATTGGTTCAGCATCAG-3′, and reverse, 5′-ATCCGAGTACAGGTGACA-3′; β-actin (control reference gene), forward, 5′-TAGTTGCGTTACACC CTTTCTTG-3′, and reverse, 5′-CACCTTCACCGTTCCAGT TTT-3′. The thermal cycling parameters were as follows: 95°C for 5 min, followed by 40 cycles at 95°C for 10 sec and 60°C for 30 sec. The expression of FABP5 was normalized to β-actin and the expression level was calculated using the 2−∆∆Cq method (23).
Western blotting
Western blotting was performed according to previously reported methods (24). Briefly, following culture for 24 h, a Tissue or Cell Total Protein Extraction kit (Sangon Biotech Co., Ltd.) was used to extract total protein from cells. Protein concentrations were determined using the Enhanced BCA Protein assay kit (Beyotime Institute of Biotechnology, Haimen, China) and 30 µg total protein was loaded and separated by 10% SDS-PAGE. Proteins were transferred onto polyvinylidene fluoride (PVDF) membranes following electrophoresis. All membranes were blocked with 5% non-fat milk (cat. no. A600669; Sangon Biotech Co., Ltd.) for 1 h at room temperature and then incubated with primary antibodies at 4°C overnight. Following washing with 1X Tris-buffered saline and Tween-20 (TBST) solution (0.1% Tween; Sangon Biotech Co., Ltd.) three times (for 5 min each time), the membranes were then incubated with secondary antibodies at room temperature for 4 h. The PVDF membranes were incubated with the following primary antibodies: Mouse anti-β-actin (dilution, 1:1,000; cat. no. 3700; Cell Signaling Technology, Inc., Danvers, MA, USA); rabbit anti-FABP5 (dilution, 1:1,000; cat. no. 33191; GeneTex, Inc., Irvine, CA, USA), mouse anti-FLAG (dilution, 1:1,000; cat. no. AF519; Beyotime Institute of Biotechnology), rabbit anti-AKT (dilution, 1:1,000; cat. no. 4685; Cell Signaling Technology, Inc.), rabbit anti-phosphorylated (p)-AKT (Ser 473; dilution, 1:1,000; cat. no. 4060; Cell Signaling Technology, Inc.) and rabbit anti-p-AKT (Thr 308; dilution, 1:1,000; cat. no. 13038; Cell Signaling Technology, Inc.). The secondary antibodies used were as follows: Horseradish peroxidase (HRP)-conjugated goat anti-mouse (dilution, 1:3,000; cat. no. D110103; Sangon Biotech Co., Ltd.) and HRP-conjugated goat anti-rabbit (dilution, 1:3,000; cat. no. D110058; Sangon Biotech Co., Ltd.). The membranes were washed with TBST 3 times (for 5 min each time) and detected using enhanced chemiluminescence reagent (cat. no. E411; Vazyme). The immunoreactive membranes were scanned using the Chemidoc XRS system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Relative protein expression levels were calculated using Image Lab software (Bio-Rad Laboratories, Inc.).
Cell viability assay
Cells were first seeded at a density of 2×103 cells/well in 96-well plates for 24, 48 and 72 h. A total of 10 µl Cell Counting kit-8 (CCK-8) reagent (Beyotime Institute of Biotechnology) was then added to each well and cells were incubated for 1 h. The optical density (OD) at 450 nm was then measured using a Synergy 2 Enzyme Mark instrument (BioTek Instruments, Inc., Winooski, VT, USA). Cell viability was expressed as a percentage and was calculated using the following formula: (ODdrug-treated group / ODcontrol group) × 100 (25).
5-ethynyl-2′-deoxyuridine (EdU) assay
Cells were seeded at a density of 2×105 cells/well in 6-well plates and incubated for 24 h. The cells were then treated with 50 µM EdU reagent (Guangzhou RiboBio Co., Ltd., Guangzhou, China) followed by Apollo-567 reaction cocktail (Guangzhou RiboBio Co., Ltd.) for 30 min. Following three washes with phosphate-buffered saline, cells were counterstained with Hoechst (dilution 1:1,000; Guangzhou RiboBio Co., Ltd.) for 10 min at room temperature for nuclear staining. All cells were observed using an EVOS FL Imaging System (Thermo Fisher Scientific, Inc.).
Cell migration assay
A wound-healing assay was performed using a 35 mm μ-Dish with a culture insert (ibidi GmbH, Planegg, Germany). Briefly, 2×104 cells/well were seeded onto the inserts and cultured in 70 µl complete medium. To estimate cell migration ability, the culture inserts were gently removed to create a space ~500 µm in diameter. Each well was then filled with 1 ml fresh medium. Photographs of the wound areas were taken using the EVOS FL Imaging System (Thermo Fisher Scientific, Inc.) immediately (0-h time point), and at 6, 12 and 24 h following generation of the wound. The migration index was subsequently calculated, which was considered as the distance migrated by the treated group relative to the distance migrated in the control group (26).
Cell invasion assay
Transwell chambers with 8-µm pores (EMD Millipore, Billerica, MA, USA) were used to measure the invasion ability of cells. Briefly, the upper chambers were pre-coated with Matrigel (BD Biosciences; Becton, Dickinson and Company, Franklin, Lakes, NJ, USA) for 1 h, and 2×104 cells in 300 µl serum-free medium were seeded into the upper chamber. A total of 1 ml complete medium was added into the lower chamber. Following incubation at 37°C for 24 h, the cells on the upper surface of the chamber were removed, fixed in 4% paraformaldehyde for 10 min at room temperature, and stained with 0.1% crystal violet (Sangon Biotech, Co., Ltd.) for 1 h at room temperature. Stained cells were counted using the EVOS FL Imaging System (Thermo Fisher Scientific, Inc.).
Xenograft model and immunohistochemistry analysis
A total of 24 BALB/c male mice (age, 4-6 weeks) were purchased from the Animal Experimental Center of Nantong University (Nantong, China). All mice were treated under pathogen-free conditions in cages according to protocols approved by Nantong University, and the present study was approved by the Ethics Committee of Nantong University. Briefly, 5×106 Caki-1 cells were diluted in 100 µl serum-free medium and inoculated subcutaneously into the left flank of each mouse. At 60 days following inoculation, all mice were euthanized by CO2, where the flow rate displace no more than 30% of the chamber volume/minute according to the American Veterinary Medical Association guidelines (27). The tumor tissues were then excised, and the tumor volumes (V) were calculated using the following formula: V = ½ (length × width2). Tumor tissues were fixed in 4% paraformaldehyde for 2 h at room temperature, and subsequently placed in a 20% sucrose solution for 24 h. All tissues were then frozen at -20°C and cut into 10-µm sections for immunohistochemistry analysis. Tissue sections were blocked using 5% bovine serum albumin (cat. no. A500023; Sangon Biotech, Co., Ltd.) for 2 h at room temperature. Then, tissue sections were incubated with rabbit anti-Ki67 antibodies (dilution, 1:100; cat. no. PA5-19462; Thermo Fisher Scientific, Inc.) overnight at 4°C, followed by incubation with Alexa Fluor 594-labeled anti-rabbit IgG (dilution, 1:1,000; cat. no. 8889S; Cell Signaling Technology, Inc.) at room temperature for 2 h. Cells were then stained with Hoechst solution (dilution 1:1,000; Guangzhou RiboBio Co., Ltd.) for 10 min at room temperature. Cells were examined using an Olympus laser confocal microscope (Olympus Corporation, Tokyo, Japan).
Statistical analysis
Data from at least three independent experiments were collected and expressed as the mean ± standard error of the mean. A Student’s t-test or one-way analysis of variance followed by Student-Newman-Keuls or least significant difference post hoc tests was used to analyze the results. The SPSS 23.0 (IBM Corp., Armonk, NY, USA) software package was employed for statistical analyses. Survival analysis was performed using the Kaplan-Meier method and groups were compared using the log-rank test, according to a previous report (22). A χ2 test was performed on selected clinical samples to determine the association between FABP5 expression and clinicopathological features of patients with ccRCC. Univariate and multivariate Cox regression analysis was used to evaluate the prognostic significance of specific features. P<0.05 was considered to indicate a statistically significant difference.
Results
FABP5 is upregulated and correlates with poor survival in patients with ccRCC
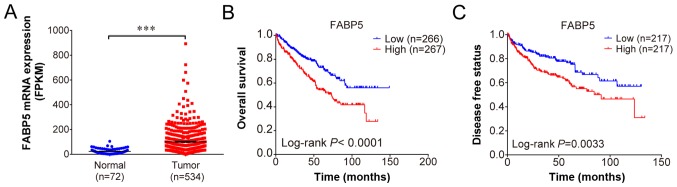
Using the profiles published in TCGA, FABP5 expression was observed to be upregulated in ccRCC samples when compared with adjacent normal samples (P<0.001; Fig. 1A). The overall survival (P<0.001; Fig. 1B) and disease-free survival curves demonstrated that patients with higher FABP5 expression exhibited significantly shorter survival rates when compared with patients exhibiting lower FABP5 expression levels (P<0.01; Fig. 1C). These results indicate that higher FABP5 expression in patients with ccRCC may be associated with poor survival.
Figure 1.
(A) Increased FABP5 expression in clear cell renal cell carcinoma tissues when compared with adjacent normal tissues. (B) Overall and (C) disease free survival curves demonstrating significantly lower survival rates of patients with high expression of FABP5 when compared with patients with low FABP5 expression. ***P<0.001, as indicated. FABP5, fatty acid binding protein 5.
The TNM classification system is a recognized standard for grading the extent of disease in patients with cancer (28). ‘T’ represents the size of the primary tumor, ‘N’ reflects the extent of metastasis to regional lymph nodes, and ‘M’ refers to the distant metastasis of tumor cells. In addition, the grade (G) classification is used to describe the degree of differentiation of cancer cells. As shown in Table I, further analysis revealed that FABP5 was expressed at lower levels in early-stage tumors (Stages I + II) and at higher levels in more advanced stages (III + IV). In addition, an association between FABP5 expression and T and M classification was observed (P<0.05); however, FABP5 expression did not correlate with N or G classification, age or gender (P>0.05; Table I). Furthermore, univariate Cox regression analysis demonstrated that high FABP5 expression was an unfavorable prognostic parameter in patients with ccRCC (P<0.01; Table II). Multivariate Cox regression analysis revealed that age (P<0.05) and M stage (P<0.01) were also unfavorable prognostic indicators.
Table I.
Association between FABP5 expression and the clini-copathological characteristics of patients with clear cell renal cell carcinoma.
| Characteristic | FABP5 gene expression (patients, no.)
|
Total | P-value | χ2 | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age (years) | 0.369 | 1.052 | |||
| ≤60 | 59 | 51 | 110 | ||
| >60 | 64 | 72 | 136 | ||
| Sex | 0.794 | 0.153 | |||
| Female | 47 | 50 | 97 | ||
| Male | 76 | 73 | 149 | ||
| T classification | 0.002 | 10.498 | |||
| T1+T2 | 85 | 60 | 145 | ||
| T3+T4 | 38 | 63 | 101 | ||
| N classification | 0.784 | 0.303 | |||
| N0 | 117 | 115 | 232 | ||
| N1+N2 | 6 | 8 | 14 | ||
| M classification | 0.039 | 4.946 | |||
| M0 | 109 | 96 | 205 | ||
| M1 | 14 | 27 | 41 | ||
| G classification | 0.072 | 3.707 | |||
| G1+G2 | 62 | 47 | 109 | ||
| G3+G4 | 61 | 76 | 137 | ||
| Stage classification | 0.005 | 8.659 | |||
| Stage I+II | 78 | 55 | 133 | ||
| Stage III+IV | 45 | 68 | 113 | ||
| Survival status | 0.002 | 10.805 | |||
| Live | 89 | 64 | 153 | ||
| Dead | 34 | 59 | 93 | ||
FABP5, fatty acid binding protein 5. Bold print denotes a statistically significant difference.
Table II.
Univariate and multivariate Cox regression analysis of FABP5 expression with the clinicopathological characteristics of patients with clear cell renal cell carcinoma.
| Characteristics | Univariate Cox
|
Multivariate Cox
|
||||||
|---|---|---|---|---|---|---|---|---|
| P-Value | HR | 95% CI
|
P-Value | HR | 95% CI
|
|||
| Lower | Upper | Lower | Upper | |||||
| Sex | 0.789 | 1.059 | 0.697 | 1.608 | ||||
| Age | 0.042 | 1.548 | 1.015 | 2.362 | 0.019 | 1.670 | 1.090 | 2.561 |
| T stage | <0.001 | 2.947 | 1.945 | 4.466 | 0.489 | 1.349 | 0.577 | 3.151 |
| N stage | 0.001 | 3.114 | 1.609 | 6.029 | 0.136 | 1.695 | 0.847 | 3.394 |
| M stage | <0.001 | 4.042 | 2.618 | 6.242 | 0.001 | 2.540 | 1.489 | 4.335 |
| G stage | <0.001 | 2.487 | 1.580 | 3.915 | 0.058 | 1.609 | 0.983 | 2.633 |
| Stage | <0.001 | 3.329 | 2.159 | 5.132 | 0.534 | 1.348 | 0.526 | 3.458 |
| FABP5 gene expression | 0.001 | 2.032 | 1.332 | 3.1 | 0.107 | 1.45 | 0.923 | 2.276 |
FABP5, fatty acid binding protein 5; HR, hazard ratio; CI, confidence interval. Bold print denotes a statistically significant difference.
FABP5 knockdown inhibits ccRCC cell growth and decreases p-AKT expression
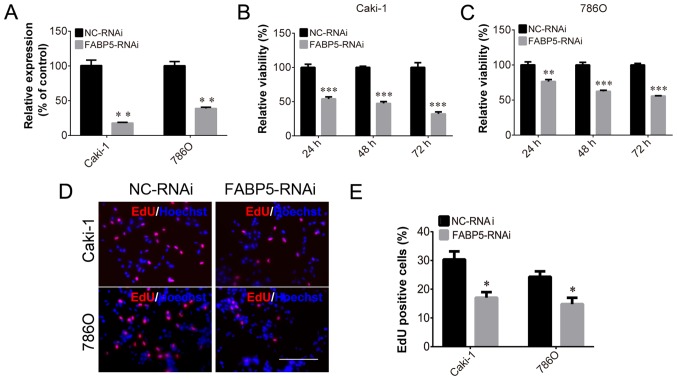
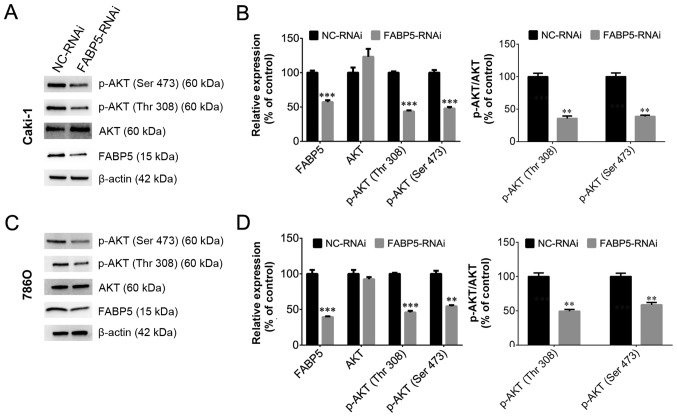
To investigate the function of FABP5 in ccRCC cells, Caki-1 and 786O cell lines transfected with FABP5-RNAi were generated and FABP5 expression was first determined. These cells were observed to be stably transfected with FABP5-RNAi, as indicated by the significantly lower levels of FABP5 mRNA in the FABP5-RNAi group compared with their respective negative controls (P<0.01; Fig. 2A). As demonstrated in Fig. 2B and C, the viability of these cell lines was significantly reduced in the FABP5-RNAi group when compared with the NC-RNAi group at 24, 48 and 72 h following transfection (all P<0.001 vs. NC-RNAi group apart from 786O cells at 24 h, P<0.01). An EdU assay was then used to measure the proliferative ability of the FABP5-RNAi cell lines. The results demonstrated that the proportion of EdU-positive Caki-1 and 786O cells was decreased in the FABP5-RNAi group compared with the respective control cells (P<0.05; Fig. 2D and E). Considering the pivotal functions of the PI3K/AKT signaling pathway in tumor cells, particularly ccRCC cells (11,12), the level of p-AKT in FABP5-RNAi cells was examined. As shown in Fig. 3A-D, FABP5 and p-AKT levels were decreased in Caki-1 and 786O cells [all P<0.001 vs. NC-RNAi group apart from p-AKT (Ser 473), P<0.01] suggesting that reduced FABP5 expression may decrease cell proliferation by inhibiting the PI3K/AKT signaling pathway.
Figure 2.
(A) Decreased FABP5 mRNA levels in Caki-1 and 786O cells in the FABP5-RNAi group. Decreased viability of (B) Caki-1 and (C) 786O cells in the FABP5-RNAi group at all time points. (D) Fewer EdU-positive Caki-1 and 786O cells were observed in the FABP5-RNAi group when compared with the NC-RNAi control group (scale bar, 200 µm). (E) Quantification of the EdU staining results. *P<0.05, **P<0.01 and ***P<0.001 vs. NC-RNAi group. FABP5, fatty acid binding protein 5; RNAi, RNA interference; NC, negative control; EdU, 5-ethynyl-2′-deoxyuridine.
Figure 3.
Western blotting results demonstrating that FABP5 and p-AKT protein levels were decreased in (A and B) Caki-1 and (C and D) 786O cells in the FABP5-RNAi group when compared with the respective NC-RNAi groups. **P<0.01 and ***P<0.001 vs. NC-RNAi. FABP5, fatty acid binding protein 5; p-, phosphorylated; RNAi, RNA interference; NC, negative control.
Overexpression of FABP5 promotes ccRCC cell growth
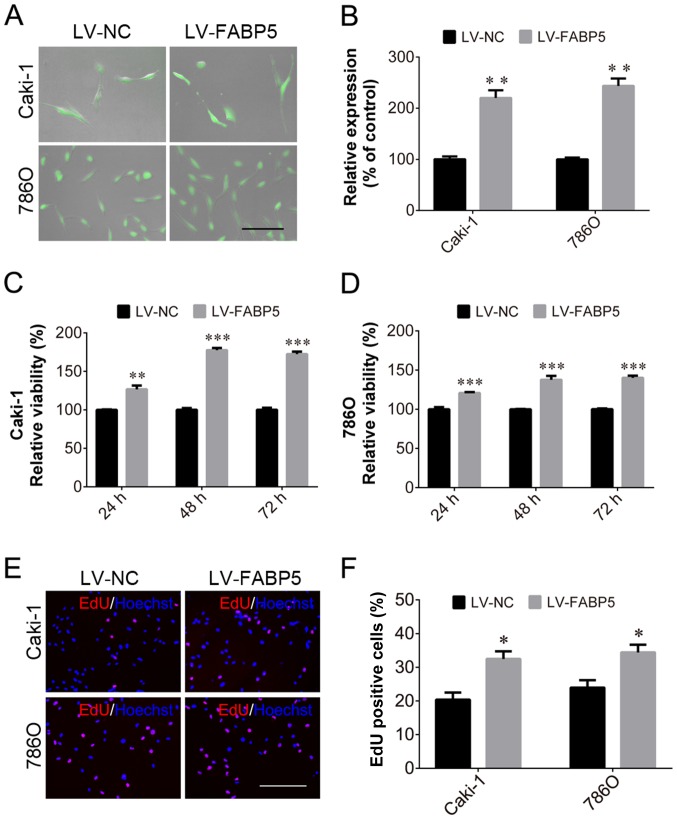
In order to investigate the role of FABP5 in ccRCC further, the effect of exogenous FABP5 expression on ccRCC cells was examined. Transfected Caki-1 and 786O cells expressed GFP (Fig. 4A) and cells in the LV-FABP5 group exhibited significantly higher FABP5 mRNA levels when compared with controls (P<0.01; Fig. 4B). CCK-8 assay analysis indicated that the proliferative ability of FABP5-overexpressing cells was significantly increased in Caki-1 and 786O cells (all P<0.001 vs. LV-NC group apart from Caki-1 cells at 24 h, P<0.01; Fig. 4C and D). Consistent with these results, the EdU assay also demonstrated an increased proportion of proliferating Caki-1 and 786O cells in the FABP5-overexpression group when compared with the negative controls (P<0.05; Fig. 4E and F). Considering that FABP5 knockdown inhibited ccRCC cell growth and decreased p-AKT expression, the authors of the current study hypothesized that exogenous FABP5 may promote the proliferation of ccRCC cells via activating the PI3K/AKT signaling pathway.
Figure 4.
Following transfection of Caki-1 and 786O cells with LV-NC or LV-FABP5, (A) cells were observed to express green fluorescent protein (scale bar, 200 µm). (B) FABP5 mRNA levels were increased in LV-FABP5-transfected Caki-1 and 786O cells. The viability of (C) Caki-1 and (D) 786O cells was increased in the LV-FABP5 group when compared with the LV-NC group at all time points. (E) The number of EdU-positive Caki-1 and 786O cells in the LV-FABP5 group was higher than the LV-NC group (scale bar, 200 µm). (F) Quantification of the EdU staining results. *P<0.05, **P<0.01 and ***P<0.001 vs. LV-NC. LV, lentivirus; NC, negative control; FABP5, fatty acid binding protein 5; EdU, 5-ethynyl-2′-deoxyuridine.
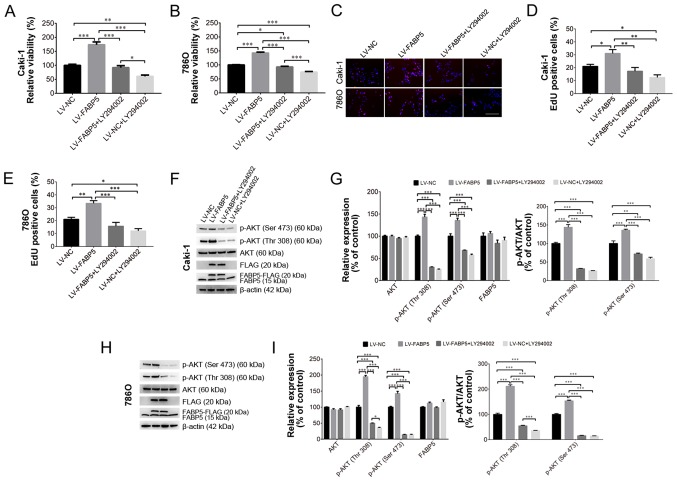
Inhibition of PI3K/AKT signaling alleviates the pro-proliferative effects of exogenous FABP5 expression
To investigate the role of FABP5 in regulating the PI3K/AKT signaling pathway in ccRCC cells further, 20 µM LY294002 was used to inhibit the PI3K/AKT signaling pathway in Caki-1 and 786O cells in vitro. As shown in Fig. 5A and B, LY294002 treatment significantly reduced the viability of FABP5-overexpressing cells, as demonstrated by the CCK-8 assay results in Caki-1 (all P<0.001 vs. LV-FABP5 group; LV-NC group vs. LV-NC+LY294002 group, P<0.01; LV-FABP5+LY294002 group vs. LV-NC+LY294002 group, P<0.05; Fig. 5A) and in 786O (all P<0.001 apart from LV-NC group vs. LV-FABP5+LY294002 group, P<0.05; Fig. 5B) cells. Consistent with these observations, the results of the EdU assay (Fig. 5C-E) also indicated that the number of EdU-positive FABP5-overexpressing Caki-1 (all P<0.01 vs. LV-FABP5 group apart from LV-NC group, P<0.05; LV-NC vs. LV-NC+LY294002, P<0.05; Fig. 5D) and 786O (all P<0.001 vs. LV-FABP5 group apart from LV-NC group, P<0.01; LV-NC group vs. LV-NC+LY294002 group, P<0.05; Fig. 5E) cells were significantly decreased following treatment with LY294002.
Figure 5.
Exogenous FABP5 expression increased the viability of (A) Caki-1 and (B) 786O cells, whereas LY294002 treatment decreased the viability of FABP5-overexpressing cells as determined using the CCK-8 assay. (C) An EdU assay demonstrated that the proportion of EdU-positive Caki-1 and 786O cells in the LV-FABP5 group were decreased following LY294002 treatment (scale bar, 200 µm). Quantification of the EdU staining results in (D) Caki-1 and (E) 786O cells. (F) Western blotting results demonstrating exogenous FABP5 expression in the LV-FABP5 group (indicated as FABP5-FLAG and FLAG) and the upregulation of p-AKT in Caki-1 cells from the LV-FABP5 group. LY294002 treatment decreased the level of p-AKT in FABP5-overexpressing Caki-1 cells. (G) Quantification of the western blotting results in Caki-1 cells. (H) Western blotting results demonstrating exogenous FABP5 expression in the LV-FABP5 group (indicated as FABP5-FLAG and FLAG) and the upregulation of p-AKT in 786O cells from the LV-FABP5 group. LY294002 treatment decreased the level of p-AKT in FABP5-overexpressing 786O cells. (I) Quantification of the western blotting results in 786O cells. *P<0.05, **P<0.01 and ***P<0.001, as indicated. FABP5, fatty acid binding protein 5; CCK-8, Cell Counting kit-8; EdU, 5-ethynyl-2′-deoxyuridine; LV, lentivirus; p-, phosphorylated; NC, negative control.
Western blotting analysis verified that exogenous FABP5 expression (indicated as FABP5-FLAG or FLAG; Fig. 5F and H) could be detected in FABP5-overexpressing cells. FABP5-FLAG or FLAG expression was detected in the LV-FABP5 group, indicating the exogenous FABP5 was successfully expressed in these cells. By contrast, FABP5-FLAG or FLAG was not detected in the LV-NC group, which confirmed that there was no exogenous FABP5 expression in the LV-NC group. These results demonstrated that exogenous FABP5 was successfully expressed in the LV-FABP5 group of cells. As shown in Fig. 5F-I, the level of p-AKT in Caki-1 and 786O cells from the LV-FABP5 group was significantly increased when normalized to β-actin and compared with controls. Accordingly, treatment with LY294002 significantly decreased p-AKT levels in FABP5-overexpressing Caki-1 (P<0.001; Fig. 5G) and 786O cells (all P<0.001 apart from p-AKT (Thr308) in LV-FABP5+LY294002 group vs. LV-NC+LY294002 group, P<0.05; Fig. 5I). However, LY294002 treatment did not affect the expression of endogenous FABP5 (indicated as FABP5 only; Fig. 5F-H). Taken together, these results suggest that the PI3K/AKT signaling pathway may participate in FABP5-induced proliferation of ccRCC cells, and that inhibiting PI3K/AKT signaling may suppress the pro-proliferative effects of FABP5 in ccRCC cells.
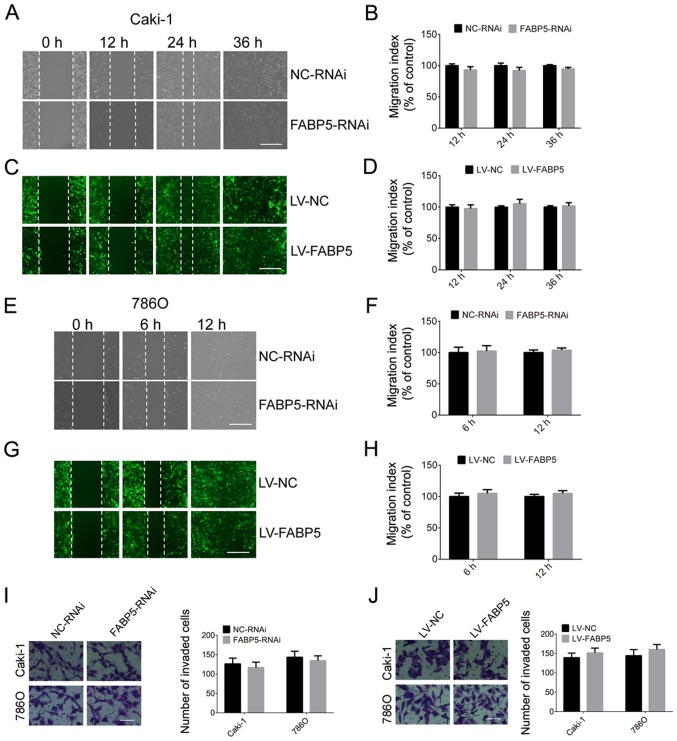
The migration and invasion abilities of Caki-1 and 786O cells in the FABP5-RNAi and NC-RNAi groups were then investigated in the present study. As indicated in Fig. 6, silencing of FABP5 did not affect the migration and invasion abilities of ccRCC cells at all time points. Similarly, overexpression of FABP5 was not associated with a significant effect on the migration or invasion of Caki-1 and 786O cells when compared with controls (Fig. 6).
Figure 6.
Effect of FABP5 (A) knockdown on Caki-1 cell migration (scale bar, 200 µm) and (B) quantification of the results. Effect of FABP5 (C) overexpres-sion on Caki-1 cell migration (scale bar, 200 µm) and (D) quantification of the results. Effect of FABP5 (E) knockdown on 786O cell migration (scale bar, 200 µm) and (F) quantification of the results. Effect of FABP5 (G) overexpression on 786O cell migration (scale bar, 200 µm) and (H) quantification of the results. Effect of FABP5 (I) knockdown and (J) overexpression on the invasion of Caki-1 and 786O cells (scale bar, 100 μm). FABP5, fatty acid binding protein 5; LV, lentivirus; NC, negative control; RNAi, RNA interference.
FABP5 affects tumorigenesis in nude mice
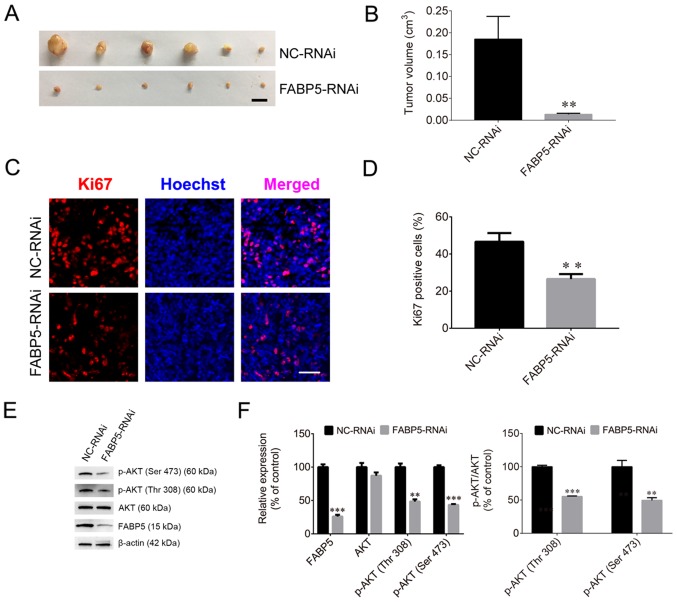
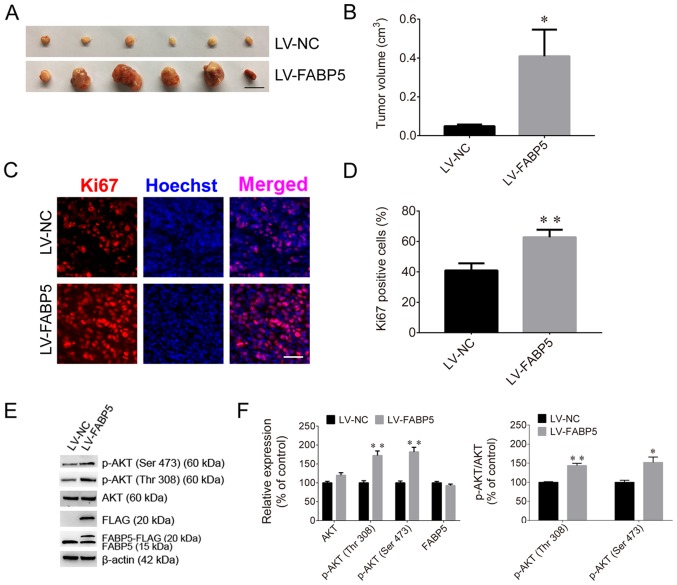
To evaluate the effect of FABP5 on tumorigenesis, Caki-1 cells were injected into nude mice. The tumor volumes in the FABP5-RNAi group of mice were significantly smaller than those in the NC-RNAi groups (P<0.01; Fig. 7A and B), and the maximum tumor diameter was 1.01 cm. The proportion of Ki67-positive cells in the FABP5-RNAi group was also significantly lower than that in the control group (P<0.01; Fig. 7C and D). Furthermore, the protein expression were normalized to β-actin, the FABP5 and p-AKT were decreased in the FABP5-RNAi group (all P<0.001 vs. NC-RNAi group apart from p-AKT (Thr308), P<0.01; Fig. 7E and F). However, following inoculation of mice with FABP5-overexpressing Caki-1 cells, the average volume of tumors in these mice (LV-FABP5 group) was significantly larger than those in the LV-NC group (P<0.05; Fig. 8A and B), and the maximum tumor diameter was 1.41 cm. In addition, the proportion of Ki67-positive cells was increased in LV-FABP5 group (P<0.01; Fig. 8C and D), and the expression of p-AKT in the LV-FABP5 group were significantly higher than that in the LV-NC group when normalized to β-actin (P<0.01; Fig. 8E and F). The primary FABP5 antibody is able to detect both endogenous FABP5 and exogenous FABP5-FLAG expression. Exogenous expression of FABP5 (indicated as FABP5-FLAG or FLAG) was also detected in FABP5-overexpressing cells, indicating that exogenous FABP5 was successfully expressed in cells of LV-FABP5 group Using the GV492 vector, the FABP5 sequence was directly combined with the FLAG tag. Following transfection of cells with LV-FABP5, exogenous FABP5-FLAG could be detected, however this did not affect endogenous FABP5 expression (Fig. 8E and F). In addition, the FLAG tag is ~3 kDa and FABP5 is ~15 kDa; therefore, exogenous FABP5-FLAG is ~18 kDa in size. As a result, the use of the FABP5 or FLAG antibodies to detect exogenous FABP5-FLAG expression identified the ~18 kDa protein band in cells transfected with LV-FABP5, but not in the LV-NC group. This confirmed that there was no exogenous FABP5 expression in the LV-NC group. These results support the notion that FABP5 exerts a pro-proliferative role and contributes to tumorigenesis in ccRCC in vivo.
Figure 7.
(A) Images of xenograft tumors and (B) tumor volumes in the FABP5-RNAi and NC-RNAi groups (scale bar, 1 cm). (C) Fluorescence images and (D) quantified fluorescence levels demonstrating that the proportion of Ki67-positive cells in the FABP5-RNAi group was reduced when compared with the NC-RNAi group (scale bar, 50 µm). (E) Western blotting images and (F) quantified protein expression levels demonstrating that FABP5 and p-AKT were decreased in the FABP5-RNAi group when compared with the NC-RNAi group. **P<0.01 and ***P<0.001 vs. NC-RNAi group. FABP5, fatty acid binding protein 5; RNAi, RNA interference; NC, negative control; p-, phosphorylated.
Figure 8.
(A) Images of xenograft tumors and (B) tumor volumes in the LV-FABP5 and LV-NC groups (scale bar, 1 cm). (C) Fluorescence images and (D) quantified fluorescence levels demonstrating that the proportion of Ki67-positive cells in the LV-FABP5 group was higher than in the LV-NC group (scale bar, 50 µm). (E) Western blotting images and (F) quantified protein expression levels demonstrating that exogenous expression of FABP5 (indicated as FABP5-FLAG and FLAG) was detected, and that p-AKT expression was increased in the LV-FABP5 group when compared with the LV-NC group. *P<0.05 and **P<0.01 vs. LV-NC. LV, lentiviral; FABP5, fatty acid binding protein 5; NC, negative control; p-, phosphorylated.
Discussion
FABPs are a group of small, highly conserved proteins that bind long-chain FAs (29). As FA transportation-associated proteins, FABPs are divided into two groups according to their cellular location; plasma membrane-associated FABPs and cytoplasmic FABPs (30,31). Cytoplasmic FABPs are a group of lipid-binding proteins with low molecular masses of ~14-15 kDa. The primary function of cytoplasmic FABPs is their involvement in lipid metabolism. Various FABPs exhibit unique patterns of expression in tissues; however, FABPs are not exclusively expressed in certain cell types (32). For instance, FABP5 and FABP7 were identified in adult rat hippocampal neural progenitor cells and in newborn rat neurons (33).
FAs function as signaling compounds to regulate metabolic networks, survival pathways and inflammatory responses in various cancers. As transporters of FAs, FABPs also modulate tumor cell growth, metabolism, migration, differentiation, and development (34). FABPs have been identified as target genes of peroxisome proliferator-activated receptors (PPARs) (35,36), and PPARs may also be stimulated by FABPs to enhance their transcriptional activities in cancer cells (37,38). In addition, numerous signaling pathways and molecules may facilitate FABP-induced tumor progression, such as the mitogen-activated protein kinase (MAPK) (39), the Src/focal adhesion kinase/cell division cycle 42 (40), reactive oxygen species (41) and epidermal growth factor receptor (EGFR) (42) signaling pathways.
FABP5, also known as epidermal FABP, is upregulated in many types of tumors, and previous studies have also demonstrated that FABP5 is overexpressed in several types of human cancer cells (13,18,20,21,34). In prostate cancer cells, FABP5 contains a typical CpG island around its promoter region, and the overexpression of FABP5 is attributed to hypomethylation of this CpG island (43). Consequently, silencing of FABP5 led to a significant decrease in cell proliferation. FABP5 facilitates the malignant progression and tumorigenicity of prostate cancer cells via the FABP5-PPARγ-vascular endothelial growth factor signal transduction axis (44). FABP5 is also expressed at higher levels in breast cancer tissues; the FABP5/PPARδ axis induces breast cancer cell proliferation, migration and invasion by activating EGFR (45,46), and suppression of FABP5 may present a strategy to overcome retinoic acid (RA)-resistant breast cancer (36). In addition, FABP5 promotes tumorigenesis in numerous additional types of cancer, such colorectal cancer (47), cervical cancer (18), hepatocellular carcinoma (21), and gastric cancer (20). In the present study, FABP5 expression was observed to be significantly higher in samples from patients with ccRCC when compared with normal control specimens, and FABP5 expression was significantly correlated with tumor stage. These results suggest that FABP5 may be associated with the malignant progression of ccRCC cells and may serve an important role in tumorigenesis. In addition, the overall survival and disease-free status curves indicated that survival rates were significantly decreased in patients with higher FABP5 expression. Univariate Cox regression analysis further revealed that high FABP5 expression was an unfavorable prognostic parameter. Taken together, these results suggest that FABP5 expression significantly correlates with the clinical characteristics and survival of patients with ccRCC.
The PI3K/AKT signaling pathway is normally activated by extracellular signals in cells and functions as a crucial intracellular signaling pathway in tumorigenesis. This pathway regulates a number of functions in cancer cells, such as cellular metabolism, tumor development, growth, proliferation and metastasis (48,49). Progress in uncovering PI3K/AKT alterations and their roles in tumorigenesis has enabled the development of novel targeted molecules for anticancer treatment (48-51). ccRCC is the most aggressive subtype of RCC and accounts for the vast majority of kidney cancer-associated deaths. This subtype is notoriously resistant to traditional chemotherapy and radiotherapy (52). The primary signaling pathways underlying ccRCC pathogenesis include the PI3K/AKT and MAPK signaling pathways (53). Elevated Notch1 signaling exerts its tumor growth-promoting effects via the PI3K/AKT signaling pathway (54,55) and the oncogenic effects of specific microRNAs and the long non-coding RNA, promoter of CDKN1A antisense DNA damage activated RNA, also involve the activation of this pathway (56,57).
In the present study, FABP5 was observed to promote the proliferation of Caki-1 and 786O ccRCC cells. Silencing of FABP5 significantly inhibited ccRCC cell proliferation, while overexpressing FABP5 promoted cell proliferation. The results also revealed that silencing of FABP5 significantly decreased the p-AKT expression levels, while overexpression of FABP5 upregulated p-AKT levels. The authors of the current study hypothesized that FABP5 may regulate ccRCC cell proliferation via the PI3K/AKT signaling pathway. To test this hypothesis, the AKT inhibitor, LY294002, was used to inactivate the PI3K/AKT signaling pathway. The results demonstrated that LY294002 treatment attenuated the pro-proliferative effects of FABP5 in Caki-1 and 786O cells, suggesting that the PI3K/AKT signaling pathway, at least in part, may be involved in FABP5-induced cell proliferation.
It was previously reported that FABP5 silencing inhibited the invasion and migration of gastric cancer cells and breast cancer cells (18-20). By contrast, the present study observed that FABP5 silencing did not affect the migration or invasion of ccRCC cells, indicating that the intrinsic properties of ccRCC cells may differ from gastric and breast cancer cells, and FABP5 might exert different functions in different cancer cells, which required further investigation. To better understand the role of FABP5 in ccRCC, its effects on cell cycle progression, cell proliferation and colony formation require further investigation in future studies. In addition, future experiments investigating the association between FABP5, AKT phosphorylation and cancer metastasis will be performed.
In conclusion, the present study revealed that FABP5 expression was upregulated in ccRCC samples when compared with normal tissues. High FABP5 expression was significantly correlated with tumor stage and predicted poor patient survival. In addition, the results indicated that FABP5 exerted a pro-proliferative role in Caki-1 and 786O ccRCC cells in vitro. Furthermore, the results indicated that the PI3K/AKT signaling pathway may be involved in mediating FABP5-induced ccRCC cell proliferation.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Health and Family Planning Commission of Shanghai (grant no. 201740114), and the Cultivation Project of Tongji Hospital in the National Natural Science Foundation.
Availability of data and materials
All data used or analyzed in this study are included in this published article or are available from the corresponding author on reasonable request.
Authors’ contributions
QL, GW, YZ performed the majority of this experiments. QL, GW and HL wrote the manuscript. XH, HL performed the animal studies. WL, MZ acquired experimental materials. QD, CM and PW contributed to the conception and design of this study. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Nantong University (Nantong, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
References
- 1.Volpe A, Patard JJ. Prognostic factors in renal cell carcinoma. World J Urol. 2010;28:319–327. doi: 10.1007/s00345-010-0540-8. [DOI] [PubMed] [Google Scholar]
- 2.Wood LS. Renal cell carcinoma: Screening, diagnosis, and prognosis. Clin J Oncol Nurs. 2009;13:3–7. doi: 10.1188/09.CJON.S2.3-7. [DOI] [PubMed] [Google Scholar]
- 3.Cheville JC, Lohse CM, Zincke H, Weaver AL, Blute ML. Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am J Surg Pathol. 2003;27:612–624. doi: 10.1097/00000478-200305000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Levy DA, Slaton JW, Swanson DA, Dinney CP. Stage specific guidelines for surveillance after radical nephrectomy for local renal cell carcinoma. J Urol. 1998;159:1163–1167. doi: 10.1016/S0022-5347(01)63541-9. [DOI] [PubMed] [Google Scholar]
- 5.Figlin RA. Renal cell carcinoma: Management of advanced disease. J Urol. 161:381–386. doi: 10.1016/s0022-5347(01)61897-4. discussion 386–387 1999. [DOI] [PubMed] [Google Scholar]
- 6.Zhong C, Chen Y, Tao B, Peng L, Peng T, Yang X, Xia X, Chen L. LIM and SH3 protein 1 regulates cell growth and chemosensitivity of human glioblastoma via the PI3K/AKT pathway. BMC Cancer. 2018;18:722. doi: 10.1186/s12885-018-4649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen GY, Ren H, Huang JJ, Zhang ZD, Zhao WH, Yu X, Shang Q, Qiu T, Zhang YZ, Tang JJ, et al. Plastrum Testudinis Extracts Promote BMSC Proliferation and Osteogenic Differentiation by Regulating Let-7f–5p and the TNFR2/PI3K/AKT Signaling Pathway. Cell Physiol Biochem. 2018;47:2307–2318. doi: 10.1159/000491541. [DOI] [PubMed] [Google Scholar]
- 8.Hou T, Zhou L, Wang L, Kazobinka G, Chen Y, Zhang X, Chen Z. Leupaxin Promotes Bladder Cancer Proliferation, Metastasis, and Angiogenesis Through the PI3K/AKT Pathway. Cell Physiol Biochem. 2018;47:2250–2260. doi: 10.1159/000491536. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Wang W, Li D, Li M, Wang P, Wen J, Liang M, Su B, Yin Y. IGF-1 alleviates NMDA-induced excitotoxicity in cultured hippocampal neurons against autophagy via the NR2B/PI3K-AKT-mTOR pathway. J Cell Physiol. 2014;229:1618–1629. doi: 10.1002/jcp.24607. [DOI] [PubMed] [Google Scholar]
- 10.Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adhes Migr. 2015;9:317–324. doi: 10.1080/19336918.2015.1016686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun P, Wang L, Lu Y, Liu Y, Li L, Yin L, Zhang C, Zhao W, Shen B, Xu W. MicroRNA-195 targets VEGFR2 and has a tumor suppressive role in ACHN cells via PI3K/Akt and Raf/MEK/ERK signaling pathways. Int J Oncol. 2016;49:1155–1163. doi: 10.3892/ijo.2016.3608. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Zhu G, Huang J, Li L, Du Y, Gao Y, Wu D, Wang X, Hsieh JT, He D, et al. Non-canonical GLI1/2 activation by PI3K/AKT signaling in renal cell carcinoma: A novel potential therapeutic target. Cancer Lett. 2016;370:313–323. doi: 10.1016/j.canlet.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Gajda AM, Storch J. Enterocyte fatty acid-binding proteins (FABPs): Different functions of liver and intestinal FABPs in the intestine. Prostaglandins Leukot Essent Fatty Acids. 2015;93:9–16. doi: 10.1016/j.plefa.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- 15.Richieri GV, Ogata RT, Zimmerman AW, Veerkamp JH, Kleinfeld AM. Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry. 2000;39:7197–7204. doi: 10.1021/bi000314z. [DOI] [PubMed] [Google Scholar]
- 16.Maximin E, Langelier B, Aïoun J, Al-Gubory KH, Bordat C, Lavialle M, Heberden C. Fatty acid binding protein 7 and n-3 p oly unsaturated fatty acid supply in early rat brain development. Dev Neurobiol. 2016;76:287–297. doi: 10.1002/dneu.22314. [DOI] [PubMed] [Google Scholar]
- 17.Lin H, Patel S, Affleck VS, Wilson I, Turnbull DM, Joshi AR, Maxwell R, Stoll EA. Fatty acid oxidation is required for the respiration and proliferation of malignant glioma cells. Neuro-oncol. 2017;19:43–54. doi: 10.1093/neuonc/now128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Chu HJ, Liang YC, Huang JM, Shang CL, Tan H, Liu D, Zhao YH, Liu TY, Yao SZ. FABP5 correlates with poor prognosis and promotes tumor cell growth and metastasis in cervical cancer. Tumour Biol. 2016;37:14873–14883. doi: 10.1007/s13277-016-5350-1. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Jia HL, Huang JM, Liang YC, Tan H, Geng HZ, Guo LY, Yao SZ. Identification of biomarkers for lymph node metastasis in early-stage cervical cancer by tissue-based proteomics. Br J Cancer. 2014;110:1748–1758. doi: 10.1038/bjc.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao G, Wu M, Wang X, Du Z, Zhang G. Effect of FABP5 gene silencing on the proliferation, apoptosis and invasion of human gastric SGC-7901 cancer cells. Oncol Lett. 2017;14:4772–4778. doi: 10.3892/ol.2017.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohata T, Yokoo H, Kamiyama T, Fukai M, Aiyama T, Hatanaka Y, Hatanaka K, Wakayama K, Orimo T, Kakisaka T, et al. Fatty acid-binding protein 5 function in hepatocellular carcinoma through induction of epithelial-mesenchymal transition. Cancer Med. 2017;6:1049–1061. doi: 10.1002/cam4.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Zhang M, Peng Y, He J. Ubiquitin Associated Protein 2-Like (UBAP2L) Overexpression in Patients with Hepatocellular Carcinoma and its Clinical Significance. Med Sci Monit. 2017;23:4779–4788. doi: 10.12659/MSM.907071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) μethod. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Han X, Li H, Zhang Y, Qin J, Yang Q, Wang L, Yuan M, Xia C. Brain lipid-binding protein promotes proliferation and modulates cell cycle in C6 rat glioma cells. Int J Oncol. 2017;51:1439–1448. doi: 10.3892/ijo.2017.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang N, Meng C, Han X, Guo J, Li H, Yu Z. Low-dose cisplatin causes growth inhibition and loss of autophagy of rat astrocytes in vitro. Neurosci Lett. 2018;682:112–117. doi: 10.1016/j.neulet.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 26.Shi H, Xu J, Zhao R, Wu H, Gu L, Chen Y. FGF2 regulates proliferation, migration, and invasion of ECA109 cells through PI3K/Akt signalling pathway in vitro. Cell Biol Int. 2016;40:524–533. doi: 10.1002/cbin.10588. [DOI] [PubMed] [Google Scholar]
- 27.Leary S, Underwood W, Anthony R, Cartner S, Corey D, Grandin T, Greenacre CB, Gwaltney-Bran S, McCrackin MA, Meyer R. AVMA Guidelines for the Euthanasia of Animals. 2013 Edition. University of Alaska Anchorage; Schaumburg: 2013. [Google Scholar]
- 28.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Chen R, Yang T, Xu N, Chen J, Gao Y, Stetler RA. Fatty acid transporting proteins: Roles in brain development, aging, and stroke. Prostaglandins Leukot Essent Fatty Acids. 2018;136:35–45. doi: 10.1016/j.plefa.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berk PD, Wada H, Horio Y, Potter BJ, Sorrentino D, Zhou SL, Isola LM, Stump D, Kiang CL, Thung S. Plasma membrane fatty acid-binding protein and mitochondrial glutamic-oxalo-acetic transaminase of rat liver are related. Proc Natl Acad Sci USA. 1990;87:3484–3488. doi: 10.1073/pnas.87.9.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stremmel W, Kochwa S, Berk PD. Studies of oleate binding to rat liver plasma membranes. Biochem Biophys Res Commun. 1983;112:88–95. doi: 10.1016/0006-291X(83)91801-6. [DOI] [PubMed] [Google Scholar]
- 32.Storch J, Thumser AE. The fatty acid transport function of fatty acid-binding proteins. Biochim Biophys Acta. 2000;1486:28–44. doi: 10.1016/S1388-1981(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 33.Matsumata M, Sakayori N, Maekawa M, Owada Y, Yoshikawa T, Osumi N. The effects of Fabp7 and Fabp5 on postnatal hippocampal neurogenesis in the mouse. Stem Cells. 2012;30:1532–1543. doi: 10.1002/stem.1124. [DOI] [PubMed] [Google Scholar]
- 34.Amiri M, Yousefnia S, Seyed Forootan F, Peymani M, Ghaedi K, Nasr Esfahani MH. Diverse roles of fatty acid binding proteins (FABPs) in development and pathogenesis of cancers. Gene. 2018;676:171–183. doi: 10.1016/j.gene.2018.07.035. [DOI] [PubMed] [Google Scholar]
- 35.Boiteux G, Lascombe I, Roche E, Plissonnier ML, Clairotte A, Bittard H, Fauconnet S. A-FABP, a candidate progression marker of human transitional cell carcinoma of the bladder, is differentially regulated by PPAR in urothelial cancer cells. Int J Cancer. 2009;124:1820–1828. doi: 10.1002/ijc.24112. [DOI] [PubMed] [Google Scholar]
- 36.Schug TT, Berry DC, Toshkov IA, Cheng L, Nikitin AY, Noy N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc Natl Acad Sci USA. 2008;105:7546–7551. doi: 10.1073/pnas.0709981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan E, Kannan-Thulasiraman P, Noy N. Involvement of Fatty Acid Binding Protein 5 and PPARβ/δ in Prostate Cancer Cell Growth. PPAR Res. 20102010 doi: 10.1155/2010/234629. pii: 234629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Rosa A, Pellegatta S, Rossi M, Tunici P, Magnoni L, Speranza MC, Malusa F, Miragliotta V, Mori E, Finocchiaro G, et al. A radial glia gene marker, fatty acid binding protein 7 (FABP7), is involved in proliferation and invasion of glioblastoma cells. PLoS One. 2012;7:e52113. doi: 10.1371/journal.pone.0052113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee D, Wada K, Taniguchi Y, Al-Shareef H, Masuda T, Usami Y, Aikawa T, Okura M, Kamisaki Y, Kogo M. Expression of fatty acid binding protein 4 is involved in the cell growth of oral squamous cell carcinoma. Oncol Rep. 2014;31:1116–1120. doi: 10.3892/or.2014.2975. [DOI] [PubMed] [Google Scholar]
- 40.Ku CY, Liu YH, Lin HY, Lu SC, Lin JY. Liver fatty acid-binding protein (L-FABP) promotes cellular angiogenesis and migration in hepatocellular carcinoma. Oncotarget. 2016;7:18229–18246. doi: 10.18632/oncotarget.7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song GX, Shen YH, Liu YQ, Sun W, Miao LP, Zhou LJ, Liu HL, Yang R, Kong XQ, Cao KJ, et al. Overexpression of FABP3 promotes apoptosis through inducing mitochondrial impairment in embryonic cancer cells. J Cell Biochem. 2012;113:3701–3708. doi: 10.1002/jcb.24243. [DOI] [PubMed] [Google Scholar]
- 42.Liang Y, Bollen AW, Aldape KD, Gupta N. Nuclear FABP7 immunoreactivity is preferentially expressed in infiltrative glioma and is associated with poor prognosis in EGFR-overexpressing glioblastoma. BMC Cancer. 2006;6:97. doi: 10.1186/1471-2407-6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawaguchi K, Kinameri A, Suzuki S, Senga S, Ke Y, Fujii H. The cancer-promoting gene fatty acid-binding protein 5 (FABP5) is epigenetically regulated during human prostate carcinogenesis. Biochem J. 2016;473:449–461. doi: 10.1042/BJ20150926. [DOI] [PubMed] [Google Scholar]
- 44.Forootan FS, Forootan SS, Gou X, Yang J, Liu B, Chen D, Al Fayi MS, Al-Jameel W, Rudland PS, Hussain SA, et al. Fatty acid activated PPARγ promotes tumorigenicity of prostate cancer cells by up regulating VEGF via PPAR responsive elements of the promoter. Oncotarget. 2016;7:9322–9339. doi: 10.18632/oncotarget.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levi L, Lobo G, Doud MK, von Lintig J, Seachrist D, Tochtrop GP, Noy N. Genetic ablation of the fatty acid-binding protein FABP5 suppresses HER2-induced mammary tumorigenesis. Cancer Res. 2013;73:4770–4780. doi: 10.1158/0008-5472.CAN-13-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powell CA, Nasser MW, Zhao H, Wochna JC, Zhang X, Shapiro C, Shilo K, Ganju RK. Fatty acid binding protein 5 promotes metastatic potential of triple negative breast cancer cells through enhancing epidermal growth factor receptor stability. Oncotarget. 2015;6:6373–6385. doi: 10.18632/oncotarget.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawaguchi K, Senga S, Kubota C, Kawamura Y, Ke Y, Fujii H. High expression of Fatty Acid-Binding Protein 5 promotes cell growth and metastatic potential of colorectal cancer cells. FEBS Open Bio. 2016;6:190–199. doi: 10.1002/2211-5463.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ciruelos Gil EM. Targeting the PI3K/AKT/mTOR pathway in estrogen receptor-positive breast cancer. Cancer Treat Rev. 2014;40:862–871. doi: 10.1016/j.ctrv.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 49.Singh SS, Yap WN, Arfuso F, Kar S, Wang C, Cai W, Dharmarajan AM, Sethi G, Kumar AP. Targeting the PI3K/Akt signaling pathway in gastric carcinoma: A reality for personalized medicine? World J Gastroenterol. 2015;21:12261–12273. doi: 10.3748/wjg.v21.i43.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polivka J, Jr, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther. 2014;142:164–175. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Fumarola C, Bonelli MA, Petronini PG, Alfieri RR. Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochem Pharmacol. 2014;90:197–207. doi: 10.1016/j.bcp.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 52.Guo H, German P, Bai S, Barnes S, Guo W, Qi X, Lou H, Liang J, Jonasch E, Mills GB, et al. The PI3K/AKT Pathway and Renal Cell Carcinoma. J Genet Genomics. 2015;42:343–353. doi: 10.1016/j.jgg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Apanovich NV, Peters MV, Apanovich PV, Kamolov BS, Matveev VB, Ginter EK, Karpukhin AV. Expression Profiles of Genes-Potential Therapy Targets-and Their Relationship to Survival in Renal Cell Carcinoma. Dokl Biochem Biophys. 2018;478:14–17. doi: 10.1134/S1607672918010040. [DOI] [PubMed] [Google Scholar]
- 54.Xu L, Zhu Y, Xu J, Wu K, Li J, Xu W, Liu H, Wang S, Yin H, Chen L, et al. Notch1 activation promotes renal cell carcinoma growth via PI3K/Akt signaling. Cancer Sci. 2012;103:1253–1258. doi: 10.1111/j.1349-7006.2012.02291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S, Ma X, Ai Q, Huang Q, Shi T, Zhu M, Wang B, Zhang X. NOTCH1 functions as an oncogene by regulating the PTEN/PI3K/AKT pathway in clear cell renal cell carcinoma. Urol Oncol. 2013;31:938–948. doi: 10.1016/j.urolonc.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 56.Nogueira I, Dias F, Teixeira AL, Medeiros R. miRNAs as potential regulators of mTOR pathway in renal cell carcinoma. Pharmacogenomics. 2018;19:249–261. doi: 10.2217/pgs-2017-0160. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y, Tong Y, Zhu J, Lei Z, Wan L, Zhu X, Ye F, Xie L. An increase in long non-coding RNA PANDAR is associated with poor prognosis in clear cell renal cell carcinoma. BMC Cancer. 2017;17:373. doi: 10.1186/s12885-017-3339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used or analyzed in this study are included in this published article or are available from the corresponding author on reasonable request.