Abstract
Accumulating evidence indicates that microRNAs (miRNAs) have a critical role in cell proliferation and metastasis in hepatocellular carcinoma (HCC). However, the effect of miR-300 on the development and progression of HCC remains unclear. In the present study, it was observed that miRNA (miR)-300 expression was significantly decreased in HCC cell lines compared with normal liver cells. Furthermore, we detected the effects of miR-300 on cell proliferation and apoptosis, cell cycle, migration and invasion by using MTT, colony formation assay, wound healing, Transwell assay and flow cytometry methods, respectively. The results demonstrated that miR-300 overexpression inhibited proliferation, induced apoptosis and G1/S cell cycle arrest, and suppressed migration and invasion in Huh-7 cells, whereas miR-300 silencing promoted the proliferation, migration and invasion of Hep3B cells. Mechanistically, the transcription factor lymphoid enhancer-binding factor 1 (LEF-1), which was verified as a direct target gene of miR-300, promoted cell proliferation, migration and invasion and mediates the effects of miR-300 on HCC cells. In addition, low expression of miR-300 and high expression of LEF-1 in HCC tissues were found to be associated with poor prognosis of patients with HCC. These findings indicate that miR-300 may be a potential prognostic predictor and therapeutic target for patients with HCC.
Keywords: hepatocellular carcinoma, microRNA-300, lymphoid enhancer-binding factor 1, proliferation, apoptosis, invasion
Introduction
Liver cancer is estimated to be the sixth most common cancer and the fourth leading cause of cancer-associated morality worldwide in 2018 (1). Hepatocellular carcinoma (HCC) is the major histological subtype of primary liver cancer. Certain factors have been reported to contribute to HCC, including aflatoxin, chronic hepatitis B virus infection, obesity, type 2 diabetes, heavy alcohol consumption and smoking, among others (2). Despite the advances in current therapeutic approaches, the prognosis of HCC is extremely poor and the recurrence rate is high. Consequently, it is crucial to elucidate the molecular mechanisms underlying metastasis and progression and identify novel therapeutic targets for HCC.
MicroRNAs (miRNAs/miRs) are a class of highly conserved, small, non-coding RNAs, ~22 nucleotides in length, which negatively regulate gene expression by targeting the 3′untranslated region (3′UTR) of mRNAs (3-5). A number of studies have confirmed that miRNAs are closely associated with the HCC cell proliferation, metastasis and cell cycle distribution, and the prognosis of HCC patients (6-9). A recent study reported that the expression of miR-124 was significantly decreased in HCC tissues and cell lines, whereas miR-124 overexpression inhibited cell proliferation and migration by targeting baculoviral IAP repeat containing 3 and regulating the nuclear factor-κB signaling pathway (10). Another study confirmed that increased miR-142-3p expression suppressed the proliferation, migration and invasion and reversed epithelial-to-mesenchymal transition (EMT) of HCC cells by targeting zinc finger E-box binding homeobox 1 (11). Recently, attention has focused on miR-300, a novel microRNA that has been reported to be involved in cell proliferation, migration and invasion in certain types of cancer, including colorectal (12), lung (13) and pancreatic (14) cancer. Although there is only one study reporting that miR-300 may regulate the migration and invasion of HCC cells by targeting the focal adhesion kinase/phosphoinositide-3-kinase/protein kinase B signaling pathway and is associated with poor prognosis (15), the role and mechanism of action of miR-300 in the development of HCC has not been fully elucidated.
In the present study, the function of miR-300 in HCC was found to be associated with lymphoid enhancer-binding factor 1 (LEF-1), a member of the LEF/T-cell-specific factor family, which has been shown to be associated with the development of tumors and serves as a prognostic marker for certain types of cancer. Shang et al (16) demonstrated that the expression of LEF-1 was increased in stage III/IV and grade 3 human renal cell carcinoma (RCC) compared with that in early-stage, low-grade RCC and normal kidney tissues, and further demonstrated that LEF-1 overexpression increased cell proliferation by reversing G2/M arrest in HCC cells. In addition, Xu et al (17) reported that increased levels of LEF-1 were correlated with poor prognosis of BRAF and NRAS mutation-negative acral melanoma. A recent study confirmed that LEF-1 overexpression promoted cell proliferation and metastasis through the miR-371a-5p/SRC kinase signalling inhibitor 1 (SRCIN1)/pleiotrophin/Slug pathway in HCC cells (18); however, to the best of our knowledge, whether miR-300 is involved in the regulation of cell proliferation and metastasis induced LEF-1 in HCC has not been reported to date.
The aim of the present study was to measure miR-300 expression in HCC and determine whether it is involved in the proliferation, migration and invasion of HCC cells. It was also aimed to investigate whether the effects of miR-300 on HCC cells are mediated through regulation of LEF-1, and their association with the prognosis of patients with HCC.
Materials and methods
Patient tissue
A total of 86 samples, including 62 HCC tissues (male 41 and female 21; age range 26-74 years old; mean 52.3±9.8) and 24 non-tumor liver tissues (male 15 and female 9; age range 26-68 years old; mean 52.0±10.9), were collected from patients with HCC that underwent surgery at the First Affiliated Hospital of Bengbu Medical College (Bengbu, China) between September 2011 and December 2015. The specimens were stored at −80°C immediately after harvesting for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. None of the patients received any preoperative chemotherapy or radiotherapy prior to surgery. Informed consent was obtained from each patient, and all the protocols of this study were approved by the Ethics Committee of Bengbu Medical College.
Cell culture
Human HCC cell lines (Huh-7, Li-7, Hep3B and SNU-449) and the normal hepatocyte cell line L02 were purchased from Cellcook Cell Biotechnology Co., Ltd. (Guangzhou, China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1% penicillin/streptomycin (Beyotime Institute of Biotechnology, Haimen, China). All cell lines were cultured at 37°C in 5% CO2.
RT-qPCR analysis
Total RNA was purified using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer’s instructions. RT was performed with 2 µg total RNA using the Revert Aid First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). The incubation conditions were 25°C for 5 min, 42°C for 60 min, 72°C for 5 min and 4°C until the end of the reaction. Subsequently, PCR was performed using SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd., Dalian, China) with primers against target genes on the ABI 7500 Real-Time PCR System (Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Holding stage, 50°C for 2 min, 95°C for 30 sec; PCR stage (40 cycles), 95°C for 15 sec, 53.6°C for 30 sec, 72°C for 34 sec. For miRNA, RT and PCR were performed using TransScript Green miRNA Two-Step RT-qPCR SuperMix (Beijing Transgen Biotech Co., Ltd., Beijing, China) with miR-specific primers on the ABI 7500 Real-Time PCR System. The incubation conditions for RT were 37°C for 60 min and 85°C for 5 sec. The thermocycling conditions for PCR were: Holding stage, 50°C for 2 min and 94°C for 30 sec; PCR stage (40 cycles) 94°C for 5 sec, 55°C for 34 sec and 72°C for 34 sec. All primers were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). GAPDH and U6 were used as endogenous control for normalizing mRNA and miRNA, respectively. Quantification was conducted to analyze mRNA or microRNA expression relative to the endogenous control using the 2−ΔΔCq method (19). ΔΔCq = [(Cq gene of interest - Cq internal control) sample A - (Cq gene of interest - Cq internal control) sample B]; sample A is the treated sample and sample B, the untreated control. Each sample was run in triplicate. The primers used in the present study were as follows: LEF-1, forward 5′-GCAGCACTTAGAAGGGGCTT-3′ and reverse 5′-GCAGCACTTAGAAGGGGCTT-3′; E-cadherin, forward 5′-GAAGTGTCCGAGGACTTTGG-3′ and reverse 5′-CAGTGTCTCTCCAAA TCCGATA-3′; vimentin, forward 5′-TGTCCAAATCGATGT GGATGTTTC-3′ and reverse 5′-TTGTACCATTCTTCTGCC TCCTG-3′; miR-300, forward, 5′-CGTATACAAGGGCAGACTCTCTCT-3′; GAPDH, forward 5′-CAGCCTCAAGATCATCAGCA-3′ and reverse 5′-TGTGGTCATGAGTCCTTCCA-3′; and U6, forward, 5′-AGAGAAGATTAGCATGGCCCCT-3′.
Vectors and cell transfections
LEF-1 overexpression and knockdown lentiviral vectors, and their negative controls, were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China). Huh-7 and Hep3B cells were infected with lentivirus-transducing units plus 10 µg/ml polybrene (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). When infected, the cell paving density remained around 30-40%. Huh-7 and Hep3B cells were infected with recombinant lentiviruses at a multiplicity of infection of 10. LEF-1 overexpression vector and short hairpin RNA (shRNA) were designed and co-transfected with miR-300 mimic and inhibitor. LEF-1 coding sequence was inserted into pcDNA 3.1 vector. Huh-7 and Hep3B cells transfected by LEF-1 overexpression vector were set as LEF-1 group. Furthermore, pcDNA 3.1 empty vector was used to transfect Huh-7 and Hep3B cells, and these cells were named as control group. miRNAs and their corresponding negative controls were synthesized by Shanghai GeneChem Co., Ltd. The sequences were as follows: LEF-1 shRNA1, 5′-GCAGCTATCAACCAGATTCTT-3′; LEF-1 shRNA2, 5′-CCATCAGATGTCAACTCCAAA-3′; LEF-1 shRNA3, 5′- GCACGGAAAGAAAGACAGCTA-3′; sh R NA non-targeting sequences for negative control, 5′-TTCTCCGAA CGTGTCACGT-3′; miR-300 mimic, sense 5′-UAUACAAGGGCAGACUCUCUCU-3′ and anti-sense 5′-AGAGAGAGUCUGCCCUUGUAUA-3′; miR negative control, sense 5′-UUCUCCGAACGUGUCACGUTT-3′ and anti-sense 5′-ACGUGACAC GUUCGGAGAATT-3′; miR-300 inhibitor, sense 5′-GAGAGA GUCUGCCCUUGUAU-3′; miR-300-inhibitor NC, sense 5′-CAGUACUUUUGUGUAGUACAA-3′. Target cells were transiently transfected with miR-300 mimic and miR-300 inhibitor using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. A total of 3×105 cells per well were seeded on 6-well-plates and transfected with 100 pmol miR-300 mimic/inhibitor or 4 µg LEF-1 overexpression vector. Puromycin (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) was used to select lentivirus stably transfected clones in accordance with the manufacturer’s protocol, and validation was performed by western blot and RT-PCR assays. At 24 h after transfection, total RNA from cells was isolated. At 48 h after transfection, HCC cells were collected for western blot, cell proliferation and apoptosis, cell cycle, migration and invasion, colony formation assay, wound healing, Transwell assay and flow cytometry. Wild-type or mutant 3′UTR sequences of LEF-1 were inserted into the pGL3-promoter vector (Promega Corporation, Madison, WI, USA) to generate LEF-1 expression vectors. The luciferase-mutant vector, in which the nucleotides complementary to the miR-300 seed-region were mutated by site-directed mutagenesis, was constructed as a mutant control. Luciferase and control signals were measured at 48 h after transfection.
Western blot analysis
HCC cells were lysed in radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology) supplemented with protease and phosphatase inhibitor cocktail (Beyotime Institute of Biotechnology) and placed on ice for 40 min. The cell lysates were centrifuged at 15,294 × g at 4°C for 20 min. Protein concentration was quantified using the Bicinchoninic Acid Protein Assay kit (Beyotime Institute of Biotechnology). The cell extracts were boiled with loading dye for 10 min. The protein samples (40 µg) were separated by SDS-PAGE on 10% gels and then transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked in Western Blocking Buffer at room temperature (Beyotime Institute of Biotechnology) for 2 h, followed by incubation with primary antibodies against LEF-1 (cat. no. 76010S; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), E-cadherin (cat. no. ab1416; 1:1,000; Abcam, Cambridge, MA, USA), vimentin (cat. no. ab20346; 1:1,000; Abcam) and GAPDH (cat. no. AC002; 1:1,000; ABclonal Biotech Co., Ltd., Woburn, MA, USA) according to the manufacturer’s recommendations overnight at 4°C. Following incubation with goat-anti-rabbit IgG (cat. no. BA1054) and goat-anti-mouse IgG (cat. no. BA1050) antibody (1:5,000; horseradish peroxidase conjugates; Beyotime Institute of Biotechnology) at 37°C for 2 h, chemiluminescence was detected with enhanced chemiluminescence (EMD Millipore) to analyze the protein levels. GAPDH (cat. no. AC002; 1:1,000; ABclonal Biotech Co., Ltd.) was used as the loading control. The western blot experiment was analyzed three times, and representative images are shown in the figures.
miRNA target prediction and luciferase reporter assay
miR-300 target prediction was performed using the miRanda and miRDB (mirdb.org/) algorithms. Cells (5×104) were seeded in 24-well plates and transiently transfected with 0.2 µg reporter plasmid and 20 pmol miRNA using Lipofectamine® 2000. After 48 h, the cells were harvested and lysed, and luciferase activity was measured using a GloMax 20/20 Luminometer (Promega Corporation). PmirGLO vector Renilla luciferase (hRluc-neo) was used for normalization.
Cell proliferation and colony formation assays
Cell proliferation was measured using MTT and colony formation assays. To evaluate cell viability, 3×103 cells were plated in 96-well plates and incubated for 24 h. Subsequently, 20 µl MTT dye solution (5 mg/ml; Beijing Solarbio Science & Technology Co., Ltd.) was added to each well and incubation was continued for 3-4 h. The supernatant was then removed, 200 µl DMSO was added to stop the reaction and the mixture was oscillated for 20 min. Finally, the optical density was determined using an Epoch microplate spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA) at a wavelength of 490 nm. A cell viability curve was generated based on absorbance and time. For the colony formation assay, 3,000 cells were seeded into each well of a 6-well plate and cultured for 7-14 days. The colonies (>50 cells) were fixed with 20% methanol for 15 min at room temperature, stained with 0.1% crystal violet solution for 20 min at room temperature. Visible cell colonies were counted.
Wound healing assay
Huh-7 and Hep3B cells were seeded in 6-well plates (4-6×105 cells per well). Longitudinal scratch wounds were created with a sterile 10 µl pipette tip 48 h after transfection and floating cell debris was removed by washing three times with PBS. Subsequently, the cells were cultured in serum-free medium. Typical wound healing images were observed and captured at 0 and 24 h under a light microscope.
Transwell assay
Cell migration and invasion ability was assessed by 24-well Transwell chambers (Costar; Corning Inc., Corning, NY, USA). At 48 h after transfection, the cells were trypsinized and counted. Approximately 2-3×104 cells resuspended in 100 µl serum-free DMEM were seeded into the upper chambers with 1:10 diluted Matrigel-coated (BD Biosciences; Becton, Dickinson and Company, Franklin, Lakes, NJ, USA) for the invasion assay; uncoated inserts were used for migration assay, whereas the bottom chamber was filled with 800 µl 10% FBS-supplemented medium. After 24 h, non-migrating/non-invading cells were wiped off with a cotton bud, and migrating/invading cells underneath the chamber were fixed with 4% polyoxymethylene for 5 min at room temperature and stained with 0.1% crystal violet solution for 10 min at room temperature. The cells were counted in five randomly selected fields under a light microscope.
Apoptosis detection and cell cycle distribution
For cell apoptosis detection, 5 µl Annexin V-phycoerythrin and 5 µl 7-aminoactinomycin D (BD Biosciences; Becton, Dickinson and Company) were added to the cells, followed by incubation for 20 min at room temperature. Cell apoptosis was evaluated by flow cytometer (BD Biosciences; Becton, Dickinson and Company) and apoptosis rates were analyzed with FlowJo software (FlowJo LLC, Ashland, OR, USA).
To synchronize cell cultures, cells were cultured in medium supplemented with 10% FBS overnight, rinsed with PBS, and then transferred to serum-free medium for 24 h. To test the cell cycle distribution, transfected cells were digested using trypsin, centrifuged at 425 × g (cell density, 1×106 cells/ml), washed twice with PBS and fixed in −20°C ethanol for 2 h. For cell cycle distribution detection, 1 µl DAPI was added per 100 µl cell suspension, and the cells were incubated for 40 min in the dark. All samples were evaluated by flow cytometry (BD Biosciences; Becton, Dickinson and Company).
Statistical analysis
All data were analyzed using GraphPad Prism 6.0 (GraphPad Software, Inc., San Diego, CA, USA). The results are presented as mean ± standard deviation of at least three independent experiments. The data were statistically analyzed by Student’s t-test for comparing two groups and one-way analysis of variance for comparing multiple groups. Dunnett and Tukey methods were used as post-hoc tests. Survival curves were plotted by the Kaplan-Meier method and compared using the log-rank test. Correlations between two indices were analyzed using Spearman’s rank correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-300 suppresses cell proliferation and induces apoptosis and cell cycle arrest in HCC cells
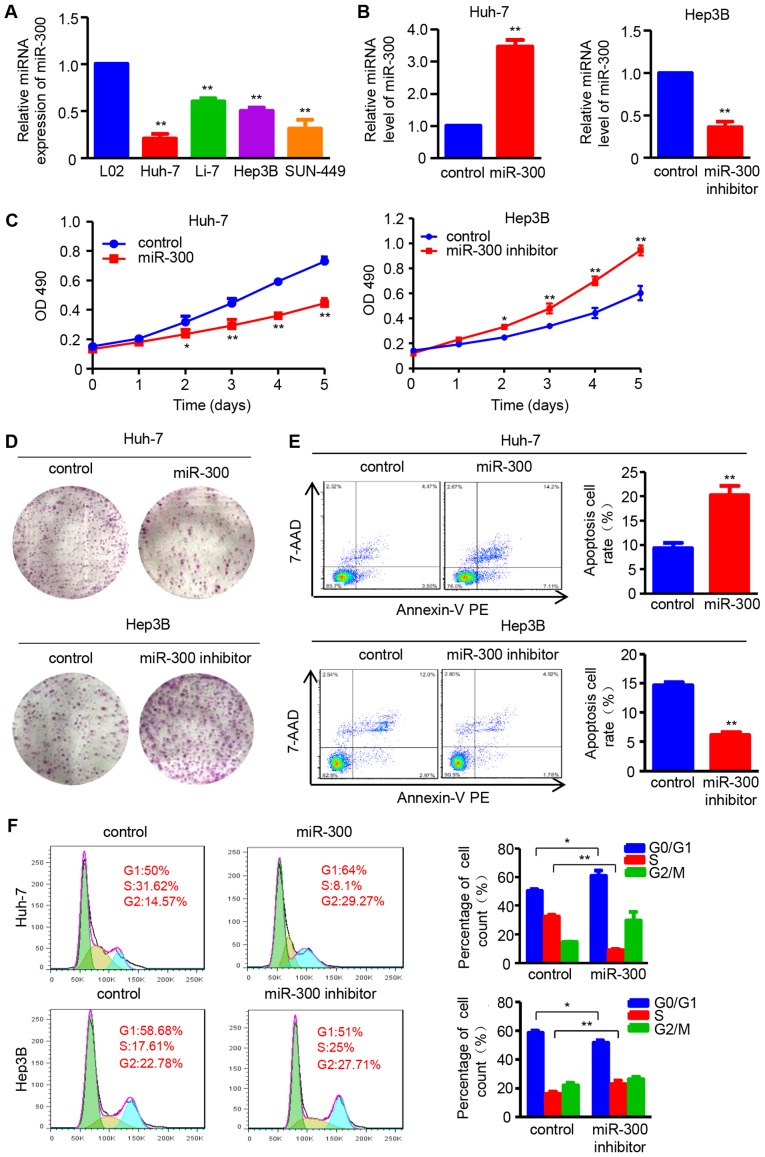
The expression of miR-300 was examined in the normal hepatic cell line L02 and the HCC cell lines, Huh-7, Li-7, Hep3B and SNU-449, by RT-qPCR. The results demonstrated that miR-300 expression was decreased in all four HCC cell lines compared with in L02 cells (Fig. 1A). Based on these results, the effects of miR-300 on the proliferation and apoptosis of HCC cells were investigated by up- and downregulating miR-300 expression using miR-300 mimic and inhibitor, respectively (Fig. 1B). The MTT and colony formation assays were performed to analyze the effect of miR-300 on cell proliferation. The results demonstrated that cell viability was suppressed in miR-300 mimic-transfected Huh-7 cells and enhanced in miR-300 inhibitor-transfected Hep3B cells compared with that in negative control cells on days 2, 3, 4 and 5 (Fig. 1C). Furthermore, the colony formation was significantly decreased in Huh-7 cells transfected with miR-300 mimic, whereas it was increased in Hep3B cells transfected with miR-300 inhibitor compared with the respective controls (Fig. 1D).
Figure 1.
miR-300 suppresses cell proliferation, and induces apoptosis and cell cycle arrest in HCC cells. (A) RT-qPCR analysis was used to detect the miR-300 mRNA expression in the HCC cell lines Huh-7, Li-7, Hep3B and SNU-449, and the normal hepatic cell line L02 (n=3; **P<0.01 vs. L02). (B) Expression of miR-300 mRNA was analyzed by RT-qPCR after transfection with miR-300 mimic (miR-300) or miR-300 inhibitor in Huh-7 or Hep3B cells, respectively (n=3; **P<0.01 vs. control). (C) MTT assay was performed to detect the proliferation of Huh-7 and Hep3B cells transfected with miR-300 mimic (miR-300) or miR-300 inhibitor, respectively (n=3; *P<0.05, **P<0.01 vs. control). (D) Colony formation ability was assessed using colony formation assay; representative images are shown. (E) Apoptosis rate of Huh-7 and Hep3B cell lines following transfection with miR-300 mimic (miR-300) or miR-300 inhibitor was examined by flow cytometric analysis (n=3; **P<0.01 vs. control). (F) DNA content was analyzed in Huh-7 and Hep3B cells transfected with miR-300 mimic (miR-300) or miR-300 inhibitor using flow cytometry, and the percentage of cells in the G0/G1, S and G2/M phases of the cell cycle was calculated (n=3; **P<0.01 vs. control). HCC, hepatocellular carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, microRNA; OD, optical density; 7-AAD, 7-aminoactinomycin D; PE, phycoerythrin.
Furthermore, the effect of miR-300 on HCC cell apoptosis was investigated. The number of apoptotic cells was increased in miR-300 mimic-transfected Huh-7 cells, whereas it was decreased in miR-300 inhibitor-transfected Hep3B cells (Fig. 1E). In addition, flow cytometry was applied to evaluate cell cycle distribution. The results demonstrated that miR-300 overexpression induced G1/S cell cycle arrest, as reflected by an increased percentage of cells in the G0/G1 phase and a decreased percentage of cells in the S phase in miR-300-overexpressing Huh-7 cells. By contrast, number of cells in the S phase was increased by miR-300 downregulation in Hep3B cells transfected with miR-300 inhibitor (Fig. 1F). These data indicate that miR-300 suppresses cell proliferation, and promotes apoptosis and G1/S cell cycle arrest in HCC cells.
miR-300 suppresses migration, invasion and EMT in HCC cells
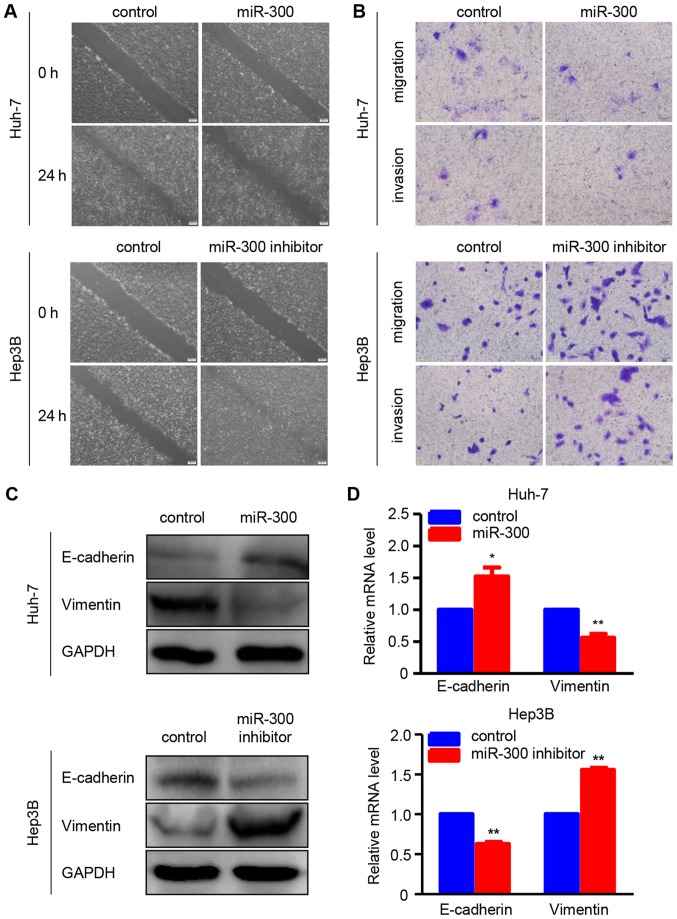
To determine whether miR-300 is associated with HCC metastasis, wound healing and Transwell assays were performed to examine the effects of miR-300 on the migration and invasion abilities of HCC cells. As shown in the wound healing assay, the migration ability of Huh-7 cells was suppressed by miR-300 overexpression, whereas miR-300 downregulation increased the migratory ability of Hep3B cells (Fig. 2A). In addition, the results of the Transwell migration assay were consistent with those of the wound healing assay in Huh-7 and Hep3B cells. Furthermore, the Transwell Matrigel invasion assay demonstrated that the invasion ability was suppressed in miR-300-overexpressing Huh-7 cells and enhanced in miR-300-silenced Hep3B cells compared wit the respective controls (Fig. 2B). Additionally, the protein and mRNA expressions of EMT-associated markers, including E-cadherin and vimentin, were investigated using western blot and RT-qPCR analyses, respectively. The data revealed that miR-300 overexpression markedly increased the protein and mRNA levels of the epithelial marker E-cadherin, whereas the protein and mRNA levels of the mesenchymal marker vimentin were significantly reduced in Huh-7 cells. As expected, the protein and mRNA expressions of E-cadherin and vimentin exhibited the opposite trend in Hep3B cells transfected with miR-300 inhibitor (Fig. 2C and D). All these data confirmed that miR-300 inhibits the migration, invasion and EMT of HCC cells.
Figure 2.
miR-300 inhibits migration, invasion and EMT in hepatocellular carcinoma cells. (A) Wound healing assay was performed to observe the cell migration capacity at 24 h after transfection with miR-300 mimic (miR-300) or miR-300 inhibitor in Huh-7 and Hep3B cells. Representative images are shown. Scale bars, 200 µm. (B) Transwell migration assay and Transwell Matrigel invasion assay were performed to examine the migration and invasion ability of Huh-7 and Hep3B cells transfected with miR-300 mimic (miR-300) or miR-300 inhibitor. Representative images are shown. Scale bars, 50 µm. (C) The protein expression of the EMT markers E-cadherin and vimentin was detected in Huh-7 and Hep3B cells transfected with miR-300 mimic (miR-300) or miR-300 inhibitor using western blot analysis. (D) mRNA expression of the EMT markers E-cadherin and vimentin was detected in Huh-7 and Hep3B cells transfected with miR-300 mimic (miR-300) or miR-300 inhibitor using reverse transcription-quantitative polymerase chain reaction. (n=3; *P<0.05, **P<0.01). EMT, epithelial-to-mesenchymal transition; miR, microRNA.
LEF-1 is directly targeted by miR-300 in HCC
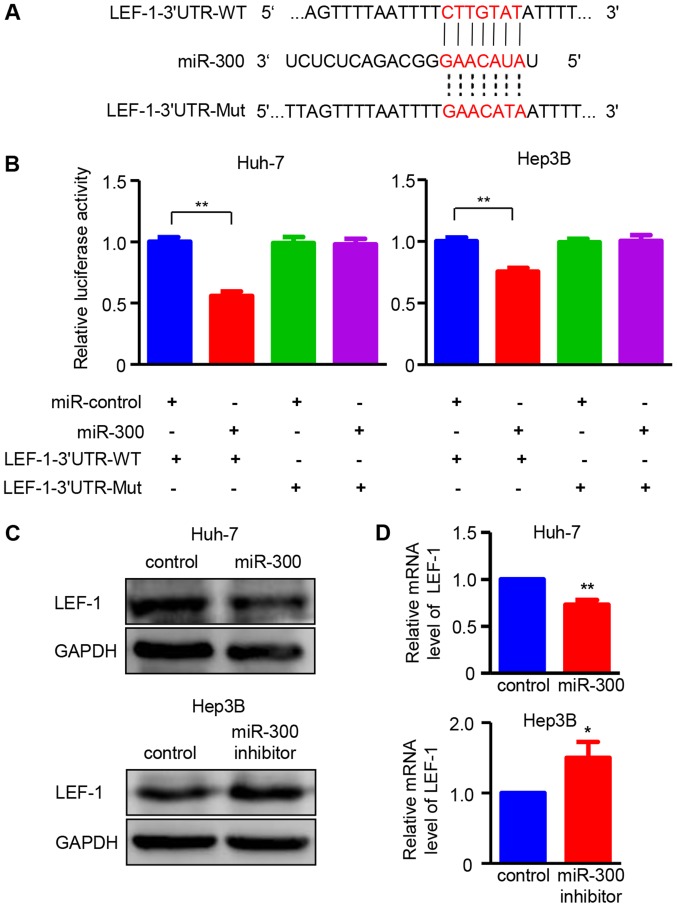
To elucidate the molecular mechanism underlying the effects of miR-300 on HCC cells, miR-300-targeted transcription factors were predicted using target prediction programs, including miRanda and miRDB. LEF-1 was identified as a potential target gene of miR-300, as the 3′UTR region of LEF-1 contained a complementary site for the seed region of miR-300 (Fig. 3A). To confirm this result, LEF-1 3′UTR and the mutant binding sequences were designed for use in a luciferase reporter assay. As demonstrated in Fig. 3B, miR-300 significantly reduced the relative luciferase activity in Huh-7 and Hep3B cells transfected with the wild-type LEF-1 3′UTR reported, indicating that miR-300 directly binds to the 3′UTR of LEF-1. Western blot and RT-PCR analyses were performed to verify the effect of miR-300 on LEF-1. The results demonstrated that overexpression of miR-300 significantly decreased the protein and mRNA expressions of LEF-1 in Huh-7 cells, whereas miR-300 inhibitor-transfected Hep3B cells exhibited increased LEF-1 expression at both the protein and mRNA level (Fig. 3C and D). Taken together, these results confirm that LEF-1 is a direct target gene of miR-300 in HCC cells.
Figure 3.
LEF-1 is directly targeted by miR-300 in hepatocellular carcinoma. (A) Bioinformatics analysis of miR-300 was performed to predict the binding sites in 3′UTR of the LEF-1 coding sequence. (B) Fluorescent reporter assay was conducted at 48 h after transfection with miR-300 mimic (miR-300) or miR-300 inhibitor in Huh7 and Hep3B cells. (n=3; **P<0.01). (C) The protein levels of LEF-1 were detected at 48 h after transfection with miR-300 mimic (miR-300) or miR-300 inhibitor in Huh-7 and Hep3B. (D) The mRNA levels of LEF-1 at 48 h after transfection with miR-300 mimic (miR-300) or miR-300 inhibitor in Huh-7 and Hep3B. (n=3; *P<0.05, **P<0.01 vs. control). LEF-1, lymphoid enhancer-binding factor 1; 3′UTR, 3′untranslated region; miR, microRNA; WT, wild-type; Mut, mutant.
LEF-1 promotes the proliferation, migration and invasion of HCC cells
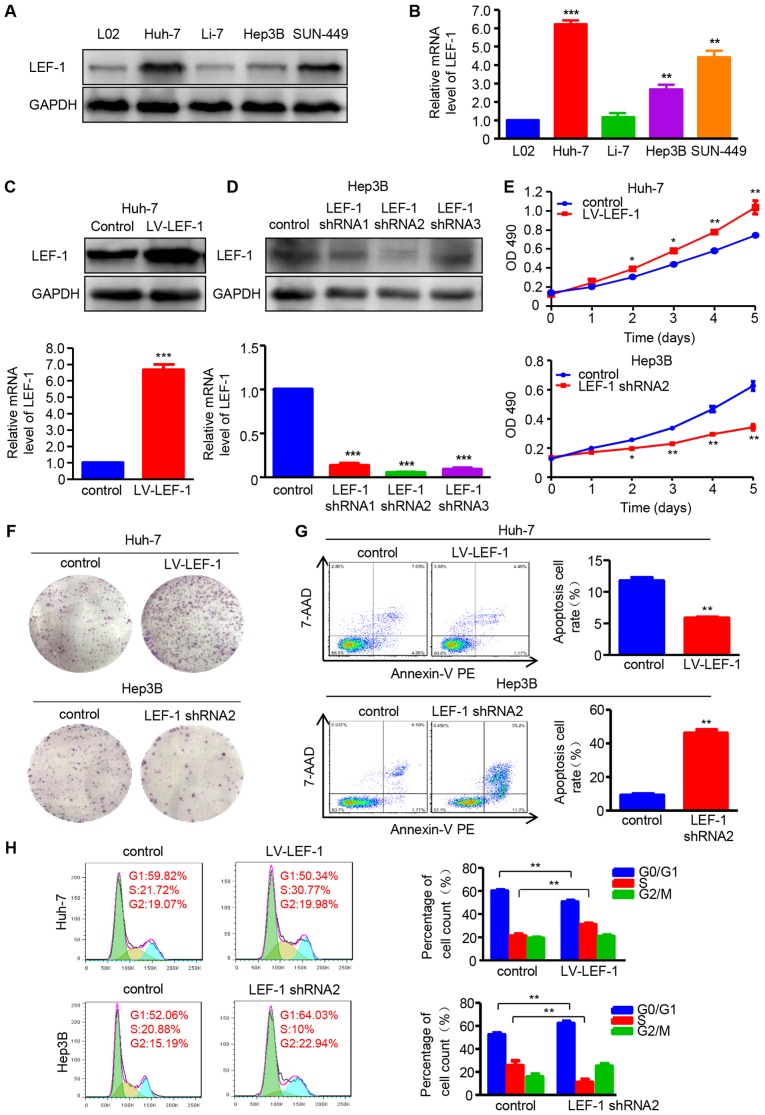
The role of LEF-1 in HCC cell proliferation, apoptosis and cell cycle arrest was investigated prior to determining whether LEF-1 mediated the regulatory effect of miR-300 on HCC cells. Contrary to miR-300 expression, the data demonstrated that LEF-1 protein and mRNA expression was increased in HCC cell lines (Huh-7, Hep3B and SNU-449) compared with that in the L02 cell line (Fig. 4A and B). Based on these data, the LV-LEF-1 vector and LEF-1 shRNA (shRNA1, shRNA2 and shRNA3) were designed and then transfected into Huh-7 or Hep3B cells to overexpress or silence LEF-1, respectively. Western blotting and RT-qPCR analysis demonstrated that LEF-1 mRNA and protein expression was upregulated by LV-LEF-1 in Huh-7 cells, whereas LEF-1 shRNA significantly decreased LEF-1 expression in Hep3B cells (Fig. 4C and D). MTT and colony formation assays were then performed, and the results suggested that cell viability and proliferation ability were enhanced by LEF-1 overexpression in Huh-7 cells and suppressed by LEF-1 silencing in Hep3B cells (Fig. 4E and F). Furthermore, the apoptosis rate was significantly increased in LEF-1 shRNA-transfected Hep3B cells, whereas Huh-7-LV-LEF-1 resulted in decreased cell apoptosis rate (Fig. 4G). In addition, as shown in Fig. 4H, the knockdown of LEF-1 significantly decreased the percentage of cells in the S phase and increased the percentage of cells in the G0/G1 phase of the cell cycle. By contrast, increased LEF-1 overexpression increased the percentage of cells in the S phase and decreased that in the G0/G1 phase.
Figure 4.
LEF-1 enhances cell proliferation and inhibits apoptosis and cell cycle arrest in HCC cells. (A) Western blotting was used to detect the LEF-1 protein expression in the HCC cell lines Huh-7, Li-7, Hep3B and SNU-449, and the normal hepatic cell line L02. (B) RT-qPCR was used to detect the LEF-1 mRNA expression in the HCC cell lines Huh-7, Li-7, Hep3B and SUN-449, and the normal hepatic cell line L02 (n=3; **P<0.01, ***P<0.001 vs. L02). (C) The protein and mRNA expression of LEF-1 was evaluated in the Huh-7 cells transfected with LV-LEF-1 using western blotting and RT-qPCR analysis, respectively (n=3; ***P<0.001 vs. control). (D) The protein and mRNA expression of LEF-1 was detected in the Hep3B cells transfected with three LEF-1 shRNAs using western blotting and RT-qPCR analysis, respectively (n=3; ***P<0.001 vs. control). (E) MTT assay was performed to detect the proliferation of Huh-7 cells transfected with LV-LEF-1 and Hep3B cells transfected with LEF-1 shRNA (n=3; *P<0.05, **P<0.01 vs. control). (F) The colony-forming ability was assessed using the colony formation assay. Representative images are shown. (G) Flow cytometric analysis was used to observe the apoptosis rate of Huh-7 and Hep3B cells following transfection with LV-LEF-1 or LEF-1 shRNA, respectively (n=3; **P<0.01 vs. control). (H) The DNA content was analyzed in Huh-7 and Hep3B cells transfected with LV-LEF-1 or LEF-1 shRNA using flow cytometry, and the percentage of cells in the G0/G1, S and G2/M phases of the cell cycle was calculated (n=3; **P<0.01). HCC, hepatocellular carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; LEF-1, lymphoid enhancer-binding factor 1; LV, lentivirus; shRNA, short hairpin RNA: 7-AAD, 7-aminoactinomycin D; PE, phycoerythrin.
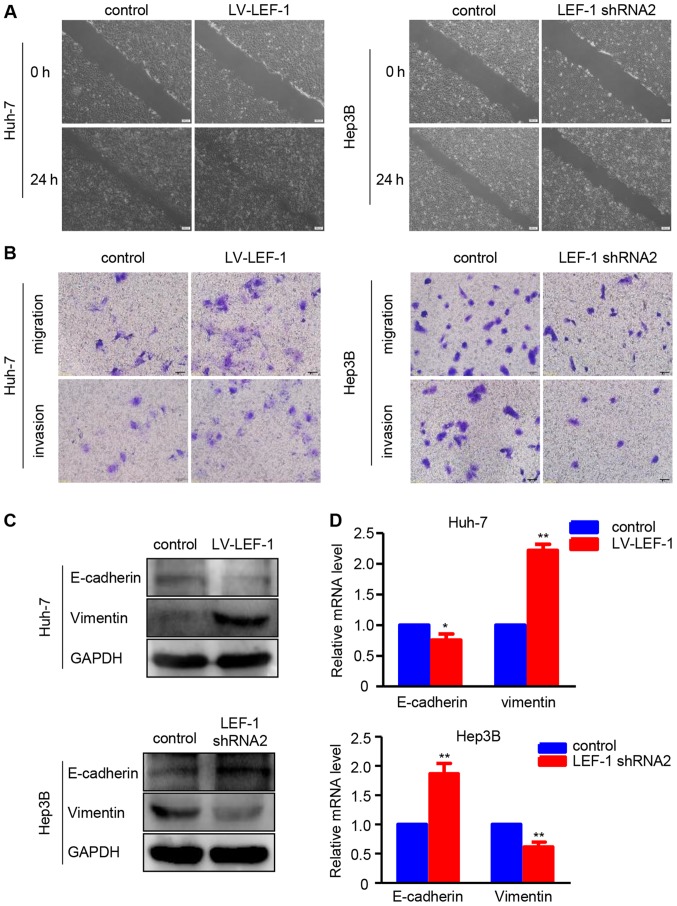
Furthermore, the association of LEF-1 with the regulation of migration and invasion in HCC cells was investigated. The wound healing assay revealed that the LEF-1 shRNA-transfected Hep3B cells exhibited a slower wound closure rate compared with the control group, whereas the LV-LEF-1-transfected Huh-7 cells exhibited an increased migratory ability compared with the control group (Fig. 5A). The migration and invasion assays demonstrated that LEF-1 overexpression increased the migration and invasion abilities of Huh7 cells, whereas LEF-1 knockdown decreased the migration and invasion abilities of Hep3B cells (Fig. 5B). Western blotting and RT-PCR analysis revealed that LEF-1 knockdown decreased the protein and mRNA levels of vimentin, but increased the protein and mRNA levels of E-cadherin in Hep3B cells. Conversely, LEF-1 overexpression increased the protein and mRNA levels of vimentin but decreased those of E-cadherin in Huh-7 (Fig. 5C and D).
Figure 5.
LEF-1 promotes migration, invasion and epithelial-to-mesenchymal transition in hepatocellular carcinoma cells. (A) The wound healing assay was performed to observe the cell migration ability at 24 h after transfection with LV-LEF-1 or LEF-1 shRNA in Huh-7 and Hep3B cells. Representative images are shown. Scale bars, 200 µm. (B) The Transwell migration assay and Transwell Matrigel invasion assay were performed to examine the migration and invasion ability of Huh-7 and Hep3B cells following transfection with LV-LEF-1 or LEF-1 shRNA. Representative images are shown. Scale bars, 50 µm. (C) The protein expression of E-cadherin and vimentin was detected in Huh-7 and Hep3B cells following transfection with LV-LEF-1or LEF-1 shRNA using western blot analysis. (D) The mRNA expression of E-cadherin and vimentin was detected in Huh-7 and Hep3B cells following transfection with LV-LEF-1 or LEF-1 shRNA using reverse transcription-quantitative polymerase chain reaction. (n=3; *P<0.05, **P<0.01 vs. control). LV, lentivirus; LEF-1, lymphoid enhancer-binding factor 1; shRNA, short hairpin RNA.
miR-300 affects proliferation, migration and invasion by regulating LEF-1 expression in HCC cells
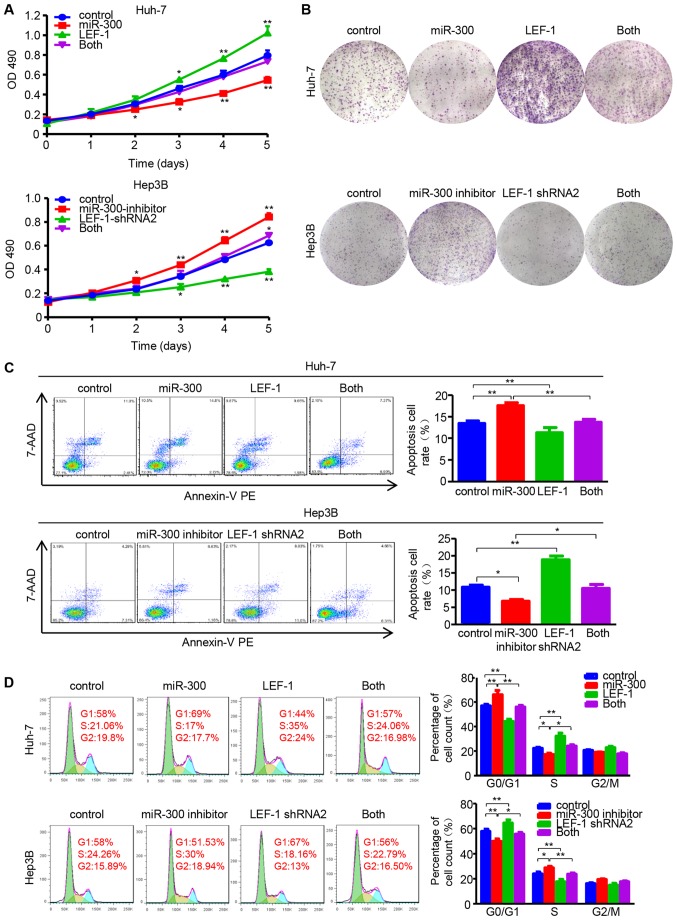
To determine whether miR-300 regulates cell proliferation and metastasis by targeting LEF-1, the LEF-1 vector and miR-300 mimic/LEF-1 shRNA and miR-300 inhibitor were co-transfected into Huh-7 or Hep3B cells, followed by evaluation of cell proliferation, migration and invasion abilities. The data demonstrated that the inhibitory effect of miR-300 mimic on cell viability and proliferation was partially rescued by the LEF-1 vector. In addition, LEF-1 shRNA suppressed the miR-300 inhibitor-induced cell growth (Fig. 6A and B). Similar to these results, LEF-1 and LEF-1 shRNA reversed the effects of miR-300 mimic and inhibitor on cell apoptosis and the cell cycle, respectively (Fig. 6C and D).
Figure 6.
miR-300 impacts on proliferation, apoptosis and cell cycle via regulating LEF-1 expression in hepatocellular carcinoma cells. (A) MTT assay was performed to detect the proliferation in Huh-7 cells co-transfected with LEF-1 vector (LEF-1) and miR-300 mimic (miR-300), as well as Hep3B cells co-transfected with LEF-1 shRNA and miR-300 inhibitor (n=3; *P<0.05, **P<0.01 vs. control). (B) The colony-forming ability was assessed using the colony formation assay. Representative images are shown. (C) The apoptosis rate was evaluated in Huh-7 and Hep3B cells by flow cytometric analysis following co-transfection with LEF-1 vector and miR-300/LEF-1 shRNA and miR-300 inhibitor (n=3; *P<0.05, **P<0.01). (D) The DNA content was analyzed using flow cytometry, and the percentage of cells in the G0/G1, S and G2/M phases of the cell cycle was calculated (n=3; *P<0.05, **P<0.01). OD, optical density; miR, microRNA; LEF-1, lymphoid enhancer-binding factor 1; shRNA, short hairpin RNA; 7-AAD, 7-aminoactinomycin D; PE, phycoerythrin.
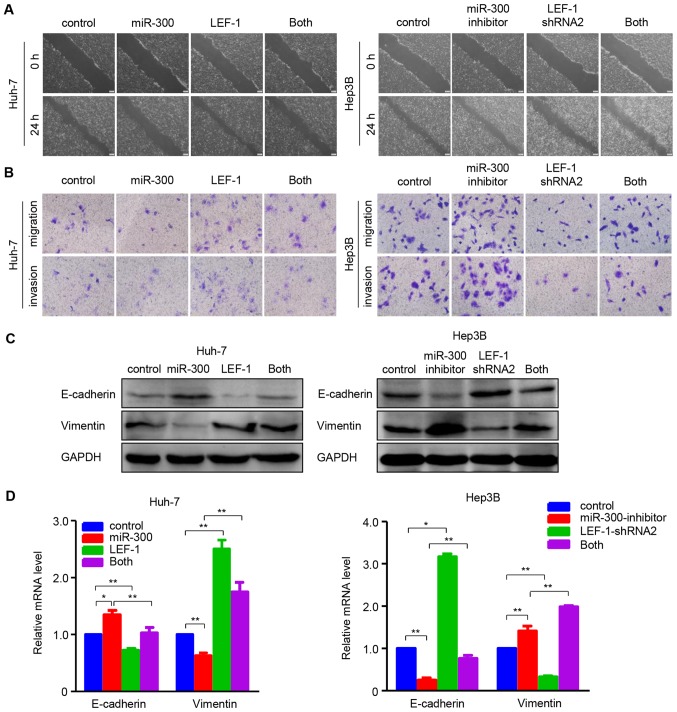
Furthermore, functional experiments were performed to determine whether LEF-1 mediated the effects of miR-300 on the migration and invasion in HCC cells. The wound healing and Transwell assays indicated that migration and invasion were inhibited in Huh-7 cells overexpressing miR-300, whereas they were restored when miR-300 mimic and LEF-1 vector were co-transfected into the cells. In the Hep3B cell line, miR-300 silencing-induced migration and invasion were inhibited by transfecting LEF-1 shRNA (Fig. 7A and B). In addition, the effects of miR-300 mimic/inhibitor on the mRNA and protein expression of EMT-associated factors were partially reversed by up- or downregulating LEF-1 expression using LEF-1 vector or LEF-1 shRNA, as shown by western blotting and RT-qPCR analysis (Fig. 7C and D). These data indicate that overexpressing/silencing miR-300 may suppress/promote cell proliferation, migration and invasion through down-/upregu-lating LEF-1 expression in HCC cells.
Figure 7.
miR-300 affects the migration, invasion and epithelial-to-mesenchymal transition via regulating LEF-1 expression in hepatocellular carcinoma cells. (A) The wound healing assay was performed to observe the cell migration ability at 24 h after co-transfection with LEF-1 vector and miR-300 mimic (Huh-7 cells) or LEF-1 shRNA and miR-300 inhibitor (Hep3B cells). Representative images are shown. Scale bars, 200 µm. (B) The Transwell migration and Transwell Matrigel invasion assays were performed to examine cell migration and invasion ability. Representative images are shown. Scale bars, 50 µm. (C) The protein expression of E-cadherin and vimentin was detected using western blot analysis. (D) Reverse transcription-quantitative polymerase chain reaction was used to detect the mRNA expression of E-cadherin and vimentin in Huh-7 cells following co-transfection with LEF-1 and miR-300, and in Hep3B cells following co-transfection with LEF-1 shRNA and miR-300 inhibitor (n=3; *P<0.05, **P<0.01). miR, microRNA; LEF-1, lymphoid enhancer-binding factor 1; shRNA, short hairpin RNA.
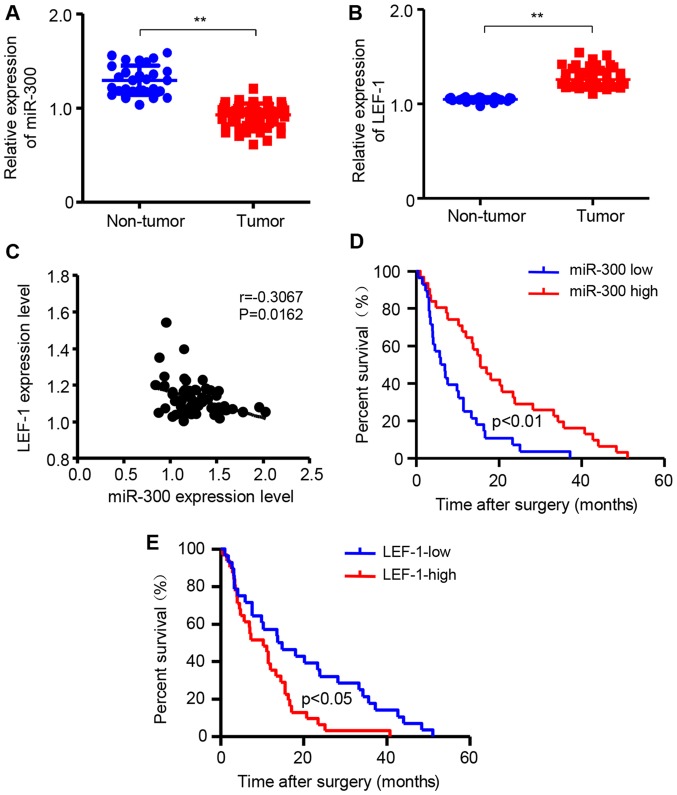
miR-300 is downregulated while LEF-1 is upregulated in HCC tissues
To further confirm that miR-300 and LEF-1 are associated with HCC development, the mRNA expression of miR-300 and LEF-1 was detected in HCC tissues and non-tumor liver tissues using RT-qPCR assay. The 62 patients with HCC were divided into two groups according to the median value of miR-300 or LEF-1 expression. The results demonstrated that miR-300 expression in HCC tissues was significantly decreased compared with that in non-tumor liver tissues (Fig. 8A). However, the mRNA expression of LEF-1 in HCC tissues was increased compared with that in non-tumor liver tissues (Fig. 8B). Additionally, the association between LEF-1 and miR-300 was investigated in patients with HCC and the expression level of LEF-1 was negatively correlated with the level of miR-300 mRNA (Fig. 8C). In addition, the association of miR-300 and LEF-1 with the overall survival time was investigated by Kaplan-Meier survival analysis. As shown in Fig. 8D and E, patients with HCC with low miR-300 levels had significantly shorter overall survival compared with those with high miR-300 levels. By contrast, the LEF-1 expression level was negatively associated with overall survival in patients with HCC. Based on those data, it may be concluded that low expression of miR-300 and high expression of LEF-1 in HCC tissues are associated with poor prognosis of patients with HCC, and that LEF-1 is a downstream target of miR-300 in HCC.
Figure 8.
miR-300 is downregulated and LEF-1 is upregulated in HCC tissues. Reverse transcription-quantitative polymerase chain reaction was performed to detect the mRNA expression of (A) miR-300 and (B) LEF-1 in HCC tissues (n=62) and non-tumor liver tissues (n=24; **P<0.01). (C) The expression of LEF-1 in patients with HCC was inversely correlated with the expression of miR-300. Kaplan-Meier survival analysis was used to evaluate the association between the expression of (D) miR-300 and (E) LEF-1 and overall survival in patients with HCC (n=62). The survival curve for patients with low and high expression of miR-300 and LEF-1 is depicted. HCC, hepatocellular carcinoma; miR, microRNA; LEF-1, lymphoid enhancer-binding factor 1.
Discussion
miR-300 was recently reported to be involved in the development and metastasis of different types of cancer (20,21). However, the effects of miR-300 on tumor cells and the underlying mechanism(s) remain unclear, particularly in HCC. The aim of the present study was to elucidate whether miR-300 affected the biological behavior of cancer cells and the possible mechanism of action. As demonstrated by the findings, miR-300 expression was low in HCC tissues and cell lines. Furthermore, miR-300 suppressed the proliferation, migration and invasion of HCC cells. In addition, LEF-1, which promotes the malignant behavior of HCC cells, was confirmed to mediate the effects of miR-300. Clinically, it was also observed that low miR-300 expression and high LEF-1 expression in HCC tissues were both associated with poor prognosis of patients with HCC. These finding suggest that miR-300 may be a potential treatment target and prognostic factor for HCC.
As mentioned above, accumulating evidence indicates that miR-300 is involved in the development and metastasis of several types of tumors; however, miR-300 may exert opposing effects on distinct types of human cancer. Zhou et al (22) demonstrated that miR-300 was significantly downregulated in glioblastoma tissues and cells (U87 and U251), and that the overexpression of miR-300 could suppress cell development and progression in vitro and in vivo, which was partially rescued by inhibiting Rho-associated protein kinase 1 expression. Similar to these results, Yu et al (23) confirmed that miR-300 inhibited cell invasion and metastasis by downregulating Twist-mediated EMT in human epithelial cancers. However, other studies demonstrated that miR-300 could promote cell growth in certain cancers. A previous study indicated that miR-300 upregulation in human gastric cancer tissues and cells promoted gastric cancer cell proliferation and invasion by targeting p53 (21). Xue et al (24) revealed that miR-300 acted as an oncogene in osteosarcoma, and demonstrated that increased expression of miR-300 promoted cell proliferation, invasion and EMT by suppressing bromodomain-containing protein 7; this discrepancy was attributed to differences in the tumor microenvironment. Only few studies on miR-300 have been reported in HCC, and only one study indicated that miR-300 was decreased in HCC and that this decrease was significantly associated with poor prognosis of patients with HCC (15). Similar to these results, miR-300 was downregulated in HCC tissues and cells, whereas miR-300 upregulation suppressed HCC cell growth, migration and invasion. These findings confirm that miR-300 is closely associated with the development and progression of HCC.
Most importantly, to the best of our knowledge, this study was the first to demonstrate that LEF-1 is a target gene of miR-300 and that it mediates the effects of miR-300 on HCC cell proliferation and metastasis. miR-300 has been confirmed to regulate cancer cell behavior by targeting downstream genes. Several target genes of miR-300 have been confirmed to date. For example, miR-300 was able to inhibit lung squamous cell carcinoma cell proliferation and invasion by targeting ROS1 (25). Furthermore, through targeting p53, miR-300 promotes the proliferation and EMT-mediated colorectal cancer cell migration and invasion, and desensitizes lung cancer cells to ionizing radiation by suppressing p53-dependent G2 cell cycle arrest, apoptosis and senescence (12,13). Notably, the present study identified a novel miR-300-targeted transcription factor, LEF-1, which has been confirmed to be a direct target gene of miR-300 by bioinformatics prediction and luciferase reporter assay. Furthermore, LEF-1 was revealed to be involved in the process of miR-300-mediated regulation of HCC cell behavior. These data provide a new direction for treatment of patients with HCC.
LEF-1 has been identified to be involved in the progression of HCC. A recent study demonstrated that Oct 4 promoted EMT by upregulating LEF-1 expression, which induced activation of β-catenin-dependent Wnt signaling (26). Another study demonstrated that LEF-1 binds the promoter of miR-HCC1 and activates its expression, thus contributing to the proliferation, migration and invasion of HCC cells (27). Similar results were observed in our study, with LEF-1 downregulated by miR-300, affecting the ability of proliferation and metastasis of HCC cells. The factors affecting the LEF-1 level in patients with HCC remain unclear. Hepatitis B virus (HBV) may induce LEF-1-mediated HCC cell growth. Tian et al (28) reported that the expression of LEF-1 was increased in HBsAg-positive HCC tissues, and hypothesized that HBsAg may stimulate the proliferation and functional modification of hepatocytes via LEF-1. A later study also revealed that HBV promoted the proliferation and metastasis of HCC cells through the LEF-1/miR-371a-5p/SRCIN1/pleiotrophin/Slug pathway (18). However, whether miR-300 and LEF-1 regulation of HCC progression is associated with HBV requires further investigation.
In summary, miR-300 was found to be frequently downregulated in HCC tissues and cells, which was significantly associated with poor patient prognosis. In addition, miR-300 overexpression inhibited the proliferation, migration and invasion of HCC cells. Furthermore, LEF-1 was identified as a direct target of miR-300, and mediated the effects of miR-300 on HCC cells. Collectively, these results suggest that miR-300 may represent a novel biomarker and promising therapeutic target for patients with HCC.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81572458), the University Outstanding Youth Talent Support Program of Anhui Province (grant no. gxyqZD2017065) and the Program for Graduate Research of Bengbu Medical College (grant no. Byycx1753).
Availability of data and materials
The datasets generated and analyzed during the present study are available from the corresponding author upon reasonable request.
Authors’ contributions
RW and QW participated in the design of the study, data interpretation and drafting of the manuscript. YC and JY performed the experiments. YG drafted the manuscript, and participated in analysis and interpretation of data. YL and JL participated in the clinical sample collection. All authors have read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The human tissue protocol utilized in this study was approved by the Ethics Committee of Bengbu Medical College (Bengbu, China). Informed consent was obtained from each patient registered in the study, in accordance with the institutional guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Mileo AM, Mattarocci S, Matarrese P, Anticoli S, Abbruzzese C, Catone S, Sacco R, Paggi MG, Ruggieri A. Hepatitis C virus core protein modulates pRb2/p130 expression in human hepatocellular carcinoma cell lines through promoter methylation. J Exp Clin Cancer Res. 2015;34:140. doi: 10.1186/s13046-015-0255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lombardi G, Perego S, Sansoni V, Banfi G. Circulating miRNA as fine regulators of the physiological responses to physical activity: Pre-analytical warnings for a novel class of biomarkers. Clin Biochem. 2016;49:1331–1339. doi: 10.1016/j.clinbiochem.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Zhou SL, Hu ZQ, Zhou ZJ, Dai Z, Wang Z, Cao Y, Fan J, Huang XW, Zhou J. miR-28-5p-IL-34-macrophage feedback loop modulates hepatocellular carcinoma metastasis. Hepatology. 2016;63:1560–1575. doi: 10.1002/hep.28445. [DOI] [PubMed] [Google Scholar]
- 7.Kumar A. MicroRNA in HCV infection and liver cancer. Biochim Biophys Acta. 18092011:694–699. doi: 10.1016/j.bbagrm.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang H, Zhang J, Yu Z, Ye L, Li K, Ding F, Feng X, Meng W. Mir-452-3p: A Potential Tumor Promoter That Targets the CPEB3/EGFR Axis in Human Hepatocellular Carcinoma. Technol Cancer Res Treat. 2017;16:1136–1149. doi: 10.1177/1533034617735931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao J, Qiu J, Wang X, Lu Z, Wang D, Feng H, Li X, Liu Q, Pan H, Han X, et al. Identification of microRNA-124 in regulation of Hepatocellular carcinoma through BIRC3 and the NF-κB pathway. J Cancer. 2018;9:3006–3015. doi: 10.7150/jca.25956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He C, Liu Z, Jin L, Zhang F, Peng X, Xiao Y, Wang X, Lyu Q, Cai X. lncRNA TUG1-Mediated Mir-142-3p Downregulation Contributes to Metastasis and the Epithelial-to-Mesenchymal Transition of Hepatocellular Carcinoma by Targeting ZEB1. Cell Physiol Biochem. 2018;48:1928–1941. doi: 10.1159/000492517. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Yu P. miR-300 promotes proliferation and EMT-mediated colorectal cancer migration and invasion by targeting p53. Oncol Rep. 2016;36:3225–3232. doi: 10.3892/or.2016.5193. [DOI] [PubMed] [Google Scholar]
- 13.He J, Feng X, Hua J, Wei L, Lu Z, Wei W, Cai H, Wang B, Shi W, Ding N, et al. miR-300 regulates cellular radiosensitivity through targeting p53 and apaf1 in human lung cancer cells. Cell Cycle. 2017;16:1943–1953. doi: 10.1080/15384101.2017.1367070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang JQ, Chen S, Gu JN, Zhu Y, Zhan Q, Cheng DF, Chen H, Deng XX, Shen BY, Peng CH. MicroRNA-300 promotes apoptosis and inhibits proliferation, migration, invasion and epithelial-mesenchymal transition via the Wnt/β-catenin signaling pathway by targeting CUL4B in pancreatic cancer cells. J Cell Biochem. 2018;119:1027–1040. doi: 10.1002/jcb.26270. [DOI] [PubMed] [Google Scholar]
- 15.Wang R, Yu Z, Chen F, Xu H, Shen S, Chen W, Chen L, Su Q, Zhang L, Bi J, et al. miR-300 regulates the epithelial-mesenchymal transition and invasion of hepatocellular carcinoma by targeting the FAK/PI3K/AKT signaling pathway. Biomed Pharmacother. 2018;103:1632–1642. doi: 10.1016/j.biopha.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Shang D, Bi R, Han T, Wang D, Tian Y, Liu Y. Expression and proliferation-promoting role of lymphoid enhancer-binding factor 1 in human clear cell renal carcinoma. Cancer Invest. 2014;32:368–374. doi: 10.3109/07357907.2014.919307. [DOI] [PubMed] [Google Scholar]
- 17.Xu S, Yang Z, Zhang J, Jiang Y, Chen Y, Li H, Liu X, Xu D, Chen Y, Yang Y, et al. Increased levels of β-catenin, LEF-1, and HPA-1 correlate with poor prognosis for acral melanoma with negative BRAF and NRAS mutation in BRAF exons 11 and 15 and NRAS exons 1–2. DNA Cell Biol. 2015;34:69–77. doi: 10.1089/dna.2014.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai PS, Hou P, Kong Y. Hepatitis B virus promotes proliferation and metastasis in male Chinese hepatocellular carcinoma patients through the LEF-1/miR-371a-5p/SRCIN1/pleiotrophin/Slug pathway. Exp Cell Res. 2018;370:174–188. doi: 10.1016/j.yexcr.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Zhang W, Jiang K, Chen B, Wang K, Lao L, Hou C, Wang F, Zhang C, Shen H. MicroRNA-300 regulates the ubiquitination of PTEN through the CRL4BDCAF13 E3 ligase in osteosarcoma cells. Mol Ther Nucleic Acids. 2018;10:254–268. doi: 10.1016/j.omtn.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Z, Li C, Zhang K, Yu W, Xiao H, Li B, Liu T. The up-regulation of miR-300 in gastric cancer and its effects on cells malignancy. Int J Clin Exp Med. 2015;8:6773–6783. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou F, Li Y, Hao Z, Liu X, Chen L, Cao Y, Liang Z, Yuan F, Liu J, Wang J, et al. MicroRNA-300 inhibited glioblastoma progression through ROCK1. Oncotarget. 2016;7:36529–36538. doi: 10.18632/oncotarget.9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Xie F, Bao X, Chen W, Xu Q. miR-300 inhibits epithelial to mesenchymal transition and metastasis by targeting Twist in human epithelial cancer. Mol Cancer. 2014;13:121. doi: 10.1186/1476-4598-13-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue Z, Zhao J, Niu L, An G, Guo Y, Ni L. Up-Regulation of MiR-300 promotes Proliferation and Invasion of Osteosarcoma by Targeting BRD7. PLoS One. 2015;10:e0127682. doi: 10.1371/journal.pone.0127682. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Ge W, Han C, Wang J, Zhang Y. MiR-300 suppresses laryngeal squamous cell carcinoma proliferation and metastasis by targeting ROS1. Am J Transl Res. 2016;8:3903–3911. [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Liu T, Zhang S, Guo K, Liu Y. Oct4 induces EMT through LEF1/β-catenin dependent WNT signaling pathway in hepatocellular carcinoma. Oncol Lett. 2017;13:2599–2606. doi: 10.3892/ol.2017.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y, Guo X, Wang J, Liu Y, Gao H, Fan H, Nong X, Yang X, Liu M, Li S, et al. A novel microRNA identified in hepatocellular carcinomas is responsive to LEF1 and facilitates proliferation and epithelial-mesenchymal transition via targeting of NFIX. Oncogenesis. 2018;7:22. doi: 10.1038/s41389-017-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Tian X, Li J, Ma ZM, Zhao C, Wan DF, Wen YM. Role of hepatitis B surface antigen in the development of hepatocellular carcinoma: Regulation of lymphoid enhancer-binding factor 1. J Exp Clin Cancer Res. 2009;28:58. doi: 10.1186/1756-9966-28-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the present study are available from the corresponding author upon reasonable request.