A network of nine MIR396 and 11 GRF genes delineates development of the crucial soybean cyst nematode feeding site, and homeostasis is necessary for successful infections.
Keywords: Growth-regulating factors, Heterodera glycines, miRNAs, miRNA396, plant-parasitic nematodes, post-transcriptional regulation, soybean, soybean cyst nematode, syncytium
Abstract
Heterodera glycines, the soybean cyst nematode, penetrates soybean roots and migrates to the vascular cylinder where it forms a feeding site called the syncytium. MiRNA396 (miR396) targets growth-regulating factor (GRF) genes, and the miR396–GRF1/3 module is a master regulator of syncytium development in model cyst nematode H. schachtii infection of Arabidopsis. Here, we investigated whether this regulatory system operates similarly in soybean roots and is likewise important for H. glycines infection. We found that a network involving nine MIR396 and 23 GRF genes is important for normal development of soybean roots and that GRF function is specified in the root apical meristem by miR396. All MIR396 genes are down-regulated in the syncytium during its formation phase while, specifically, 11 different GRF genes are up-regulated. The switch to the syncytium maintenance phase coincides with up-regulation of MIR396 and down-regulation of the 11 GRF genes specifically via post-transcriptional regulation by miR396. Furthermore, interference with the miR396–GRF6/8–13/15–17/19 regulatory network, through either overexpression or knockdown experiments, does not affect the number of H. glycines juveniles that enter the vascular cylinder to initiate syncytia, but specifically inhibits efficient H. glycines development to adult females. Therefore, homeostasis in the miR396–GRF6/8–13/15–17/19 regulatory network is essential for productive H. glycines infections.
Introduction
Cyst nematodes (Heterodera and Globodera spp.) are economically important, root-infecting, obligate biotrophs that form an elaborate feeding site within the vascular cylinder called the syncytium (Hussey and Grundler, 1998; Jones et al., 2013). The syncytium serves as the single source of nourishment throughout the life of the cyst nematode. The development of the feeding organ is initiated by migratory pre-parasitic second-stage juveniles (pre-J2) that enter the vascular cylinder and select a single cell that becomes enlarged and multinucleated [during parasitic (par)-J2 and early J3 stages] through cytoplasmic fusion of numerous nearby cortical or vascular parenchyma cells via cell wall dissolution. Syncytia are characterized by dense cytoplasm, reduced vacuoles, hypertrophied nuclei and nucleoli, and abundant endoplasmic reticulum, ribosomes, plastids, and mitochondria (Sobezak and Golinowski, 2009). This process of redirecting differentiated root cells into a novel developmental program ensues during a syncytium formation phase that involves immense transcriptional and post-transcriptional regulation of gene expression (Alkharouf et al., 2006; Ithal et al., 2007b; Klink et al., 2009; Szakasits et al., 2009; Hewezi and Baum, 2012; Hewezi et al., 2012). The fully formed syncytium then enters a maintenance phase (late in the J3 stage) where no additional cells are incorporated and, thus, has completed all major developmental changes for maintaining the function of feeding the developing nematode. Interestingly, much of this reprogramming of differentiated root cells involves the concerted action of small RNAs, in particular miRNAs and their target genes (Hewezi et al., 2008, 2012; Hewezi and Baum, 2012, 2015).
In plants, miRNAs are 20–24 nt endogenous molecules that are produced from their own MIRNA genes and function to suppress gene expression (Rogers and Chen, 2013). MIRNA genes are transcribed and produce a primary miRNA transcript that is first processed by DICER-LIKE 1 (DCL1) into a precursor (pre)-miRNA stem–loop structure (Bologna and Voinnet, 2014). The pre-miRNA is subsequently processed by DCL1 (if a 21 nt miRNA) into short dsRNAs consisting of miRNA guide and passenger (miRNA*) strands (Bologna and Voinnet, 2014). The miRNA/miRNA* duplex is 2'-O-methylated at the 3' ends for stabilization (Yu et al., 2005). Then, most commonly, the miRNA guide strand is loaded into the ARGONAUTE (AGO) component of the RNA-induced silencing complex (RISC) (Eamens et al., 2009). miRNA-loaded RISCs are then directed to target transcripts through miRNA/target complementarity and repress target transcripts most often through slicing or cleavage via AGO endonuclease activity (Mallory et al., 2008). miRNAs regulate the expression of transcription factors, proteins that mediate stress responses, and many other proteins that impact the development and physiology of plants (Rogers and Chen, 2013).
Various miRNAs change in expression in response to cyst and root-knot nematode infection (Hewezi et al., 2008; Li et al., 2012; Xu et al., 2014; Hewezi and Baum, 2015; Zhao et al., 2015; Cabrera et al., 2016). During infection of Arabidopsis by the beet cyst nematode Heterodera schachtii, many miRNAs are differentially expressed and are negatively correlated with target gene abundance (Hewezi et al., 2008). Of these differentially expressed miRNAs, miR396 was shown to be a master regulator of syncytium development (Hewezi et al., 2012). Also, an important role for miR390, TAS3 trans-acting short-interfering (tasi) RNAs, and their auxin response factor targets was demonstrated for root-knot nematode Meloidogyne javanica infection of Arabidopsis (Cabrera et al., 2016). Furthermore, the miR319–TCP4 module was shown to act as a responder and regulator of systemic defense signals, mediated by jasmonic acid, for resistance to the root-knot nematode Meloidogyne incognita in tomato (Solanum lycopersicum) (Zhao et al., 2015). These previous findings demonstrate that miRNAs are important regulatory factors during infection by cyst and root-knot nematodes (Hewezi and Baum, 2015).
Deep sequencing efforts have revealed that miRNAs in soybean (Glycine max) are differentially expressed during seed development, flowering time, and in the shoot apical meristem (Wong et al., 2011; Shamimuzzaman and Vodkin, 2012; Li et al., 2015). Soybean miRNAs are also differentially expressed during various abiotic (Kulcheski et al., 2011; Fang et al., 2013; Wang et al., 2013; Xu et al., 2013; Liu et al., 2017) and biotic stress conditions. These biotic stress conditions include rot and rust diseases (Guo et al., 2011; Kulcheski et al., 2011) and, interestingly, H. glycines infection (Li et al., 2012; Xu et al., 2014; Tian et al., 2017). However, the latter studies revealed only limited miRNA profiles. Collectively though, these deep sequencing efforts suggest that miRNAs are involved in a wide range of important processes in soybean, including H. glycines infection. Soybean miRNAs have been experimentally confirmed to be important for nodulation (H. Li et al., 2010; Turner et al., 2013; Yan et al., 2013, 2015, 2016; Wang et al., 2014, 2015) and low water availability (Liu et al., 2017).
miR396 targets the plant-specific growth-regulating factor (GRF) transcription factors characterized by the glutamine, leucine, glutamine (QLQ) protein interaction and tryptophan, arginine, cysteine (WRC) DNA-binding domains (Kim et al., 2003). The Arabidopsis miR396–GRF regulatory module is important for many developmental and stress-related processes (van der Knaap et al., 2000; Kim et al., 2003; Kim and Lee, 2006; Horiguchi et al., 2011; Hewezi and Baum, 2012; Hewezi et al., 2012; Kim et al., 2012; J. Liu et al., 2012; Bao et al., 2014; Debernardi et al., 2014; Liang et al., 2014; Liu et al., 2014; Pajoro et al., 2014; Rodriguez et al., 2015), but the most generalized function is the regulation of cell proliferation and expansion (Kim et al., 2003; Rodriguez et al., 2010; Horiguchi et al., 2011; Kim et al., 2012; Bao et al., 2014; Liang et al., 2014; Pajoro et al., 2014; Omidbakhshfard et al., 2015; Rodriguez et al., 2015). Homeostasis of the miR396–GRF regulatory module in both Arabidopsis and Medicago truncatula is important for normal root development (Hewezi et al., 2012; Bazin et al., 2013; Rodriguez et al., 2015). Recently, overexpression of soybean miR396 precursors in Arabidopsis gave an altered root phenotype (Liu et al., 2017). Interestingly, Arabidopsis GRF1- and GRF-3-mediated gene expression regulation probably accounts for almost 50% of the genes that are differentially expressed in the H. schachtii syncytium (Szakasits et al., 2009; Hewezi et al., 2012), and interfering with the miR396–GRF1/3 regulatory module resulted in decreased syncytium size and arrested nematode development (Hewezi et al., 2012). Thus, the miR396–GRF regulatory module may serve as a possible target for developing novel control measures against cyst nematodes.
Here we investigate whether a soybean miR396–GRF regulatory system is involved in, and necessary for, productive H. glycines infections. To the best of our knowledge, this is the first study that has attempted to extend findings made in the Arabidopsis–H. schachtii model system to the agronomically important interaction between soybean and H. glycines. Using a combination of molecular and genetic analyses, we first determine that a complex network involving nine MIR396 genes and 23 GRF genes operates in soybean roots. Interference with this regulatory network modifies the lateral root system, but the amount of root tissue available for H. glycines infections, and the number of J2 inside the vascular cylinder early on in infection are unaffected. Later on during H. glycines infection, we determine that a network involving all nine MIR396 and 11 different GRF genes delineates the syncytium formation phase, which begins with a miR396 down-regulation and a resulting GRF up-regulation. During the switch to the syncytium maintenance phase, a miR396 expression spike in the syncytium post-transcriptionally silences GRF genes. Furthermore, we indicate that interference with the homeostasis of this network prevents efficient H. glycines development to the adult female stage, showing an essential role for this regulatory network in productive H. glycines infections.
Materials and methods
Inoculation of whole plants
Soybean cultivar (cv.) Williams 82 seeds were surface sterilized with 10% sodium hypochlorite for 10 min and planted on seed germination paper (Anchor Paper). Ragdolls were incubated at 26 °C in the dark for 3 d. Seedlings were placed on circular steel blue seed germination blotter paper (Anchor paper) dampened with MES-buffered ddH2O, pH 6.5 in a circle with the radical tips facing towards the center (10 seedlings per plate). Each radical was inoculated with 500 surface-sterilized H. glycines line OP50 pre-J2s (Baum et al., 2000). Inoculated radicals were covered with dampened blotter paper, and infection chambers were incubated at 26 °C in the dark for 24 h. Four inoculated seedlings were acid fuchsin stained for H. glycines (Hussey, 1985) to ensure adequate infections. Infected seedlings were rinsed and placed back into ragdolls and incubated in a Percival growth chamber at 26 °C with a 14:10 h light dark cycle.
In silico analyses
Soybean pre-miR396 sequences from miRBase (Kozomara and Griffiths-Jones, 2014) were blastn-searched against the soybean genome at SoyBase using default parameters. Pre-miR396 stem–loops were modeled in silico using the Mfold Web Server (Zuker, 2003) with default settings. All GRF coding sequences (CDS) were submitted to the psRNATarget server (Dai and Zhao, 2011) with default parameters along with all miR396 molecules to evaluate for putative miR396 target sites.
Phylogenetic analysis
Multiple sequence alignments (MSAs) were generated with Clustal using default parameters in MEGA6 (Tamura et al., 2011). Poorly aligned regions were removed. Model selection analysis was performed in MEGA6 using default parameters to obtain the best-scoring model of nucleotide substitution. Phylogenetic analysis was performed in MEGA6 using bootstrapped Maximum Likelihood (ML) estimation with 100 bootstrap replications. Reported is the best scoring ML phylogenetic tree with bootstrap values indicated on the corresponding nodes.
Assessment of syncytial phases
Roots from five plants per time point were acid fuchsin stained for H. glycines (Hussey, 1985). For each plant, 100 H. glycines were observed with a stereo microscope (Zeiss) and each life stage was recorded. Pre-J2 have a slender body and tapered tail, and are most often found migrating through the cortex, while par-J2 have a swollen body and rounded tail, and are anteriorly attached in the vascular cylinder. J3 are much more swollen than even par-J2, and the rounded tail is more pronounced. J4/adult females are much larger than even J3, and have a near lemon shape. The acid fuchsin staining with thorough clearing in acidified glycerol allowed the stages to be easily distinguished with a stereo microscope. The average percentage of each life stage was calculated for each time point. This method was also used to compare the number of H. glycines penetrating J2s that infected the roots (i.e. reached the vascular cylinder) of the various mutants and empty vector (EV) controls.
RNA isolation and cDNA synthesis
Total RNA was isolated from 50 mg of ground root tissue using the NucleoSpin Kit (Clontech). Yields and purity were assessed with a NanoDrop, and integrity with agarose gel electrophoresis. Total RNA was polyadenylated and reverse transcribed using the Mir-X miRNA Kit (Clontech), which generates cDNA for both mature miRNAs and naturally polyadenylated transcripts, allowing quantitative real-time reverse transcription–PCR (qRT–PCR) analysis of pre-miR396, mature miR396, and GRF genes all from the same samples. cDNA was prepared from transgenic hairy root total RNA samples with qScript cDNA SuperMix (Quanta). In all cases, 1 μg of total RNA was used to prepare cDNA.
qRT–PCR
qRT–PCR was performed with iQ SYBR Green (Bio-Rad) on an iCycler iQ Real-Time PCR Detection System (Bio-Rad). For all reactions, cDNA consisted of 1/15th of the total reaction volume. Protocol: 95 °C for 3 min, 40 cycles of 95 °C for 15 s, and 60 °C for 30 s. Universal mRQ reverse primer (Mir-X miRNA Kit) was used along with miR396-specific oligonucleotides as forward primers. Forward primers specific to each pre-miR396 subfamily were used with mRQ reverse. Pre-miR396 had to be quantified as subfamilies because primers that attempted to quantify each subfamily member individually resulted in much lower primer efficiencies and poor melting curves. miR396-specific forward primers included two adenine nucleotides on the 3' ends to ensure binding to the poly(T) region of miR396 cDNAs and not to pre-miR396 (Gutierrez et al., 2009). U6 (Mir-X miRNA Kit) was used as a calibrator for normalization. For GRF genes, RNA levels were normalized to GmUBQ3 (GenBank: D28123.1). All primer sets were pre-validated on serially diluted soybean root cDNA with amplification efficiencies >90%. Amplification specificities were confirmed for all by melting curve analysis and agarose gel electrophoresis. Melting curve analysis protocol: 95 °C for 1 min, 55 °C for 10 s, and a slow temperature ramp from 55 °C to 95 °C . ddH2O and total RNA samples were included as negative controls with no amplification. Three biological replicates and four technical replicates were always used. Relative changes in gene expression levels were quantified using the 2−∆∆CT method (Livak and Schmittgen, 2001). Single statistical comparisons were made using the t-test, and multiple comparisons by ANOVA and Tukey–Kramer HSD post-hoc test in JMP Pro 11.
miRNA cleavage assays
miR396 cleavage sites were mapped with the FirstChoice RNA ligase-mediated (RLM)- RACE Kit (Ambion). Total RNA at 14 days post-inoculation (dpi) was poly(A)-selected with Dynabeads (Thermo) and ligated to the 5'-RACE RNA adaptor without calf intestine alkaline phosphatase treatment. cDNA synthesis was performed using GRF-specific outer primers. Subsequent steps followed the manufacturer’s instructions. RLM-RACE products were cloned into pGEM-T Easy (Promega) and sequenced at Iowa State University.
Vector construction
For promoter constructs, soybean Williams 82 genomic (g)DNA was isolated from a leaf of a 3-week-old plant according to Blin and Stafford (1976). From 1.4 kbp to 2.3 kbp of upstream regulatory DNA sequence in SoyBase was cloned for each promoter construct. PCR amplification was performed with Platinum Taq (Invitrogen). PCR products were cloned into pGEM-T Easy and restriction digest cloned into p4305.1 (GenBank: KT954098) (restriction enzyme sites are included on the primer sequences in Supplementary Table S1 at JXB online). For pre-miR396 overexpression constructs, pre-miR396 were PCR-amplified from soybean gDNA using Platinum Taq with primers exactly 20 nt 5' and 3' to the pre-miR396 sequences in SoyBase. PCR products were cloned into pGEM-T Easy and restriction digest cloned into the pG2XPRESS derivative of pG2RNAi2 (GenBank: KT954097); the GUS (β-glucuronidase) linker was restriction digested out (Noon et al., 2016). For RNAi, we PCR-amplified nucleotides 1–333 of the GRF9 CDS (GRF9i1–333) from soybean cDNA with Platinum Taq. PCR products were cloned into pGEM-T Easy and restriction digest cloned into pG2RNAi2 sense and antisense sites. For rGRF9 overexpression, we PCR-amplified the GRF9 CDS from cDNA, and synonymous mutations were introduced in the miR396 target site by overlap extension PCR (Ho et al., 1989). The rGRF9 PCR product was cloned into pGEM-T Easy and restriction digest cloned into pG2XPRESS. All empty vectors were sequenced, and then transformed into Agrobacterium rhizogenes strain K599.
Hairy root nematode infection assays
Transgenic hairy roots were generated and inoculated with surface-sterilized H. glycines (250 pre-J2s per root tip) similar to as described previously (Baum et al., 2000; S. Liu et al., 2012; Noon et al., 2016), but with some modifications. We used an inoculum of 250 pre-J2s for soybean hairy roots, as compared with 500 for whole roots, as the former tissue is much smaller/thinner than the latter. Each replicate consisted of 10 healthy root tips [white and with strong green fluorescent protein (GFP) fluorescence] transferred from a maintenance plate onto solid medium in a 150 mm×25 mm Petri dish. Root tips were inoculated 2–3 d after transfer. The number of J4/adult females was counted with a stereo microscope at 28 dpi. Statistical comparisons were made with the t-test in JMP Pro 11.
GUS histochemical staining
Transgenic hairy roots were inoculated with surface-sterilized H. glycines in 6-well plates (250 pre-J2s per well) similar to as described in Baum et al. (2000). Infected and uninfected roots were removed from the solid medium, the solid medium was removed, and then roots were placed back into the empty 6-well plates and subjected to histochemical staining for GUS according to Vitha et al. (1995). Substrate solution was vacuum infiltrated into the roots, and then roots were incubated at 37 °C for 1–4 h depending on the construct and efficiency of infiltration. All roots for all constructs (except EV control; always 4 h) were incubated in substrate until they reached maximum staining intensity, and before background was observed. Thus, end point staining intensity was purposely comparable for all constructs (except EV control; no staining), but the presence/absence of specific staining was what varied between promoter–GUS constructs. For the syncytium formation phase (J2 and J3 syncytia), infected roots were stained at 5 and 8 dpi. For the syncytium maintenance phase (J4/adult female syncytia), infected roots were stained at 15 dpi. Stained roots were mounted in ddH2O or glycerol and observed with a stereo microscope. Images were taken with an AxioCam HR 13 Megapixel Camera (Zeiss). Since there was no GUS staining in EV control roots, any specific staining in the tissue of interest was considered to be positive.
To measure promoter activity, only the healthy roots that grew inside the solid medium (white and with strong GFP fluorescence), as opposed to some root tips that can tend to grow upwards outside of the medium and dry out, were collected and stained. Thus, all of the roots included had the potential for staining. Many of these healthy roots were imaged. For uninfected roots, the percentage of n=20 of the healthy roots stained and imaged was scored for the presence or absence of GUS staining and averaged over three different experiments. For infected roots, the percentage of n=20 ‘healthy’ roots each with a particular H. glycines life stage with an observable syncytium was scored for the presence or absence of GUS staining and averaged over three different experiments. Multiple statistical comparisons were made by ANOVA and Tukey–Kramer HSD post-hoc test in JMP Pro 11.
Results
MIR396 and GRF gene families are active in young soybean roots
To begin, we first needed to identify the MIR396 and GRF gene families, and then determine if these genes are expressed in soybean roots. Eleven soybean pre-miR396 sequences were found in miRBase (pre-miR396a–k), and unique genomic co-ordinates were identified for all but pre-miR396d and g at Soybase (Schmutz et al., 2010) (Supplementary Table S2). Pre-miR396d and g are at the same genomic location and, thus, the latter was removed from our study. Modeled in silico stem–loop structures for all but pre-miR396h formed an miR396/miR396* duplex within the stems (Supplementary Fig. S1A). Thus, there are nine canonical MIR396 genes. Also, phylogenetic analysis (to inform primer design for expression analyses) identified four subfamilies (Supplementary Fig. S1B: subfamily 1, pre-miR396a/i; subfamily 2, pre-miR396e/h/j; subfamily 3, pre-miR396c/f; subfamily 4, pre-miR396b/d/k). Pre-miR396d and k within subfamily 4 are identical. Hence, there are eight unique pre-miR396 sequences. Moreover, pre-miR396 sequences within all four subfamilies were almost identical, with only a few nucleotide mismatches within the loops. Furthermore, three mature miR396 molecules were found to be produced by the eight unique pre-miR396, differentiated by the 3'-most nucleotide(s). Finally, qRT–PCR on RNA from roots of 10-day-old soybean cv. Williams 82 seedlings detected expression, albeit variable, of all pre-miR396 subfamilies and individual miR396 molecules (Supplementary Fig. S2A, B). Thus, there are eight unique, canonical pre-miR396 and three mature miR396 molecules, of which all four pre-miR396 subfamilies and mature miR396 molecules are expressed in young soybean roots.
Putative miR396 target sites were identified in all but one GRF mRNA sequence (GRF25; Supplementary Fig. S3). Thus, GRF1–24 have the potential for post-transcriptional regulation by miR396, consistent with the findings of Liu et al. (2017). GRF2 and 7 mRNAs were undetectable in 10-day-old soybean roots by qRT–PCR using as many as 40 cycles, while the remaining GRF mRNAs were detected, albeit with variable expression levels (Supplementary Fig. S2C). Also, GRF5/24, 9, 14, 18, 20–23, and 25 mRNAs resulted in much greater expression levels compared with the other 13 GRF genes. Thus, 23 GRF genes are active with varying levels of expression in young soybean roots, consistent with the expression of the MIR396 gene family.
Eleven GRF genes are up-regulated during the H. glycines syncytium formation phase
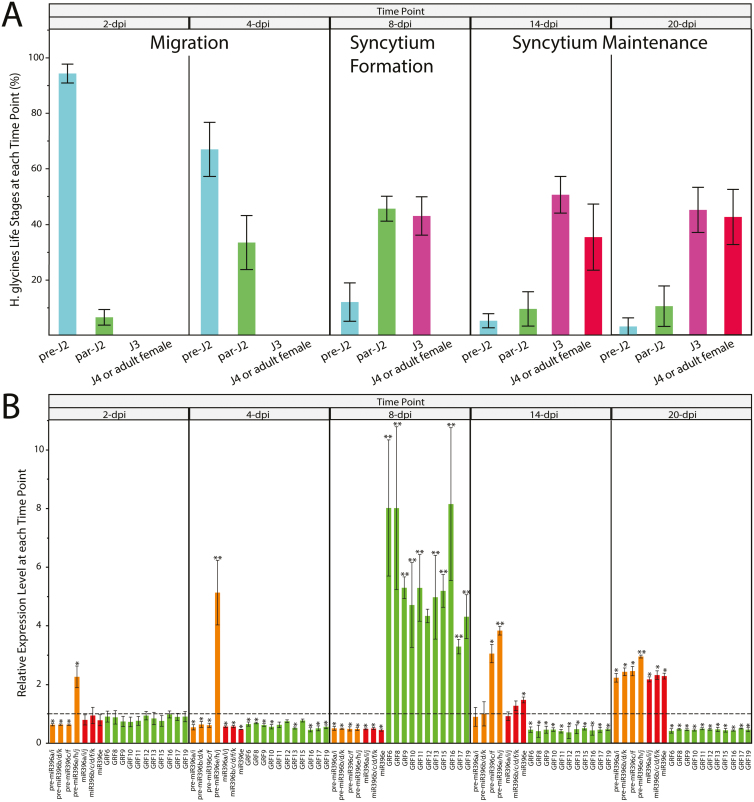
We next examined whether GRF genes change expression in response to H. glycines infection, specifically during the syncytium formation phase. However, since we always observe extensive variability in H. glycines life stages within each individual soybean root system, especially between 2 and 21 dpi, we first had to determine which time point cumulatively corresponded to the syncytium formation phase in our infection system. For this analysis, we inoculated soybean roots with H. glycines and, at 2, 4, 8, 14, and 20 dpi, evaluated in which life stage the majority of H. glycines juveniles were (Fig. 1A). Cumulatively, we determined <2 to 4 dpi as the migration phase, 5–13 dpi as the peak syncytium formation phase, and 14 to >20 dpi as the syncytium maintenance phase in our infection assay. Although the cumulative syncytium formation phase appears somewhat delayed compared with the findings of Ithal et al. (2007b), which could be due to both biological and technical differences, we did observe syncytia being formed as early as 2 dpi (5.9% par-J2), and more at 4 dpi (33.2% par-J2). However, cumulatively, the large majority of syncytium formation was occurring during the 8 dpi time point, with nearly 90% of H. glycines in either par-J2 or J3 stages. By 14 dpi, nearly 90% of H. glycines were in J3 or J4/adult female stages, indicating syncytium maintenance.
Fig. 1.
Time course expression analysis of the miR396–GRF6/8–13/15–17/19 regulatory network during H. glycines infection. (A) Assessment of syncytial phases in H. glycines-infected soybean roots (n=5 plants). At 2 dpi, an average of 94.1% of H. glycines were still in the migratory pre-parasitic (pre)-J2 stage while the few remaining were par-J2s. At 4 dpi, an average of 66.8% of H. glycines were in the pre-J2 stage and the remainder were par-J2s. Thus, we determined 2 and 4 dpi as early and late migration, respectively. By 8 dpi, an average of 45.4% and 42.8% of H. glycines were in par-J2 or early J3 stages, respectively, while the remaining few were pre-J2s. Thus, 8 dpi was designated as the syncytium formation phase. By 14 and 20 dpi, the majority of H. glycines were in late J3, J4, or adult female stages and, thus, these time points were designated as the syncytium maintenance phase. (B) Time course qRT–PCR analysis of pre-miR396 subfamilies, miR396 molecules, and the 11 GRF genes. Expression levels are relative to mock-inoculated; baseline expression is set to 1.0 and indicated with a dashed line. *P<0.05; **P<0.01. (A, B) Error bars represent ±1 SD from the mean.
A qRT–PCR screen was performed on RNA isolated from the 8 dpi roots for all 25 GRF genes. Interestingly, 11 GRF mRNAs resulted in significantly increased expression compared with mock-inoculated roots (between 3- and 8.5-fold increases, P<0.01), while 12 GRF mRNAs were unchanged and GRF2 and 7 remained undetected (Supplementary Fig. S4). The 11 up-regulated GRF genes were GRF6, 8, 9–13, 15–17, and 19. These results suggested that a network of 11 different GRF genes are involved in H. glycines infection.
The miR396–GRF6/8–13/15–17/19 regulatory network delineates the phases of the H. glycines syncytium
Having determined that the MIR396 gene family members are transcriptionally active in roots and that 11 GRF genes are up-regulated in response to H. glycines during the cumulative syncytium formation phase, it was of interest to examine the anticipated post-transcriptional silencing of these GRF genes by miR396 during various stages of infection. We used qRT–PCR to quantify the expression of the four pre-miR396 subfamilies, the three miR396 molecules, and the 11 GRF genes, at 2, 4, 8, 14, and 20 dpi, relative to mock-inoculated roots (Fig. 1B). With the exception of pre-miR396e/h/j, all pre-miR396 and miR396 as well as the GRF genes showed no significant changes or only a slight down-regulation during the cumulative migration time points (2–4 dpi). Interestingly, pre-miR396e/h/j showed significant up-regulation during the migration time points, but this up-regulation was not reflected by increased expression of the miR396 molecules, a possible indication of impaired miRNA maturation processing.
Strikingly, during the syncytium formation phase at 8 dpi, all pre-miR396 subfamilies and miR396 molecules showed significant down-regulation of >2-fold, and this down-regulation was accompanied by significant up-regulation of GRF genes showing between 3- and 8.5-fold mRNA increases (Fig. 1B). Furthermore, at 14 dpi (i.e. the switch to syncytium maintenance), pre-miR396c/f (subfamily 3) and pre-miR396e/h/j (subfamily 2) were significantly up-regulated between 3- and 4-fold, respectively, while pre-miR396a/i (subfamily 1) and pre-miR396 b/d/k (subfamily 4) were no longer down-regulated. Moreover, miR396e was significantly up-regulated >1.5-fold, while miR396a/i/j and miR396b/c/d/f/k were no longer down-regulated. Conversely, all 11 GRF genes were significantly down-regulated >2-fold. Furthermore, at 20 dpi, all pre-miR396 subfamilies and miR396 molecules were significantly up-regulated between 2.5- and 3.5-fold, while all 11 GRF genes remained significantly down-regulated >2-fold. The opposite expression patterns of miR396 and GRF genes pointed to post-transcriptional silencing of the 11 GRF genes by miR396 during H. glycines infection. These results indicated that the miR396–GRF6/8–13/15–17/19 regulatory network delineates the formation and maintenance phases of the H. glycines syncytium.
GRF6, 8, 9–13, 15–17, and 19 are post-transcriptionally regulated by miR396 during H. glycines infection
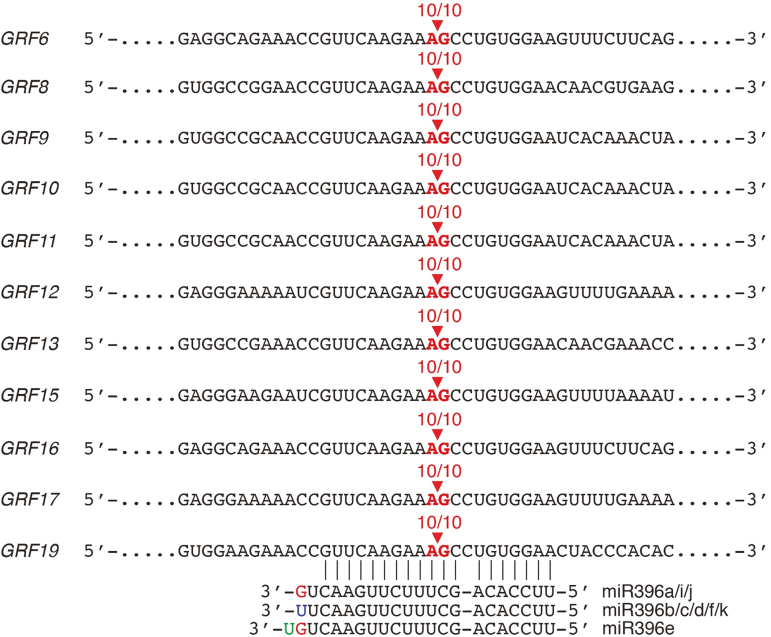
To determine whether the gene expression changes of the 11 H. glycines-responsive GRF genes are the results of their post-transcriptional regulation by miR396 during infection, we performed a 5' RLM-RACE assay on the 14 dpi RNA (i.e. during syncytium maintenance, the time point when down-regulated; see Fig. 1). Cloning and sequencing of the RLM-RACE clones indicated that the cleavage of all 11 GRF transcripts occurred within their miR396 target sites between positions 10 and 11 (Fig. 2, 10/10 clones). These results are consistent with previous reports for Arabidopsis GRF genes (Jones-Rhoades and Bartel, 2004; Hewezi et al., 2012) and confirmed that GRF mRNAs are post-transcriptionally regulated by miR396 in H. glycines-infected soybean roots, during the cumulative syncytium maintenance phase.
Fig. 2.
miRNA cleavage assays for GRF6, 8–13, 15–17, and 19. Ten different clones for each GRF degradation product were analyzed by DNA sequencing. The number of clones that resulted in the indicated cleavage positions within the miR396 target sites is indicated.
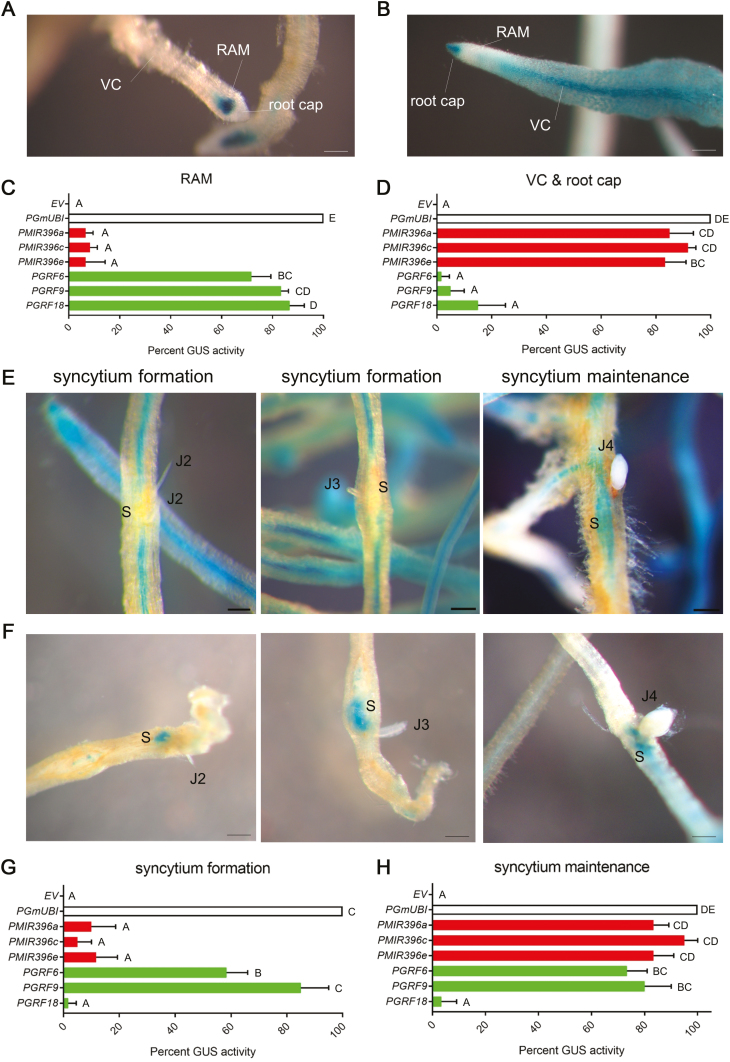
Then, we examined whether GRF genes may be post-transcriptionally regulated specifically in syncytia. We examined the spatiotemporal expression patterns of GRF6 and GRF9, which were two of the most highly up-regulated GRF genes during the cumulative syncytium formation phase (Fig. 1B), as well as GRF18 as a control that is highly expressed in roots (Supplementary Fig. S2C), but unresponsive to H. glycines infection (Supplementary Fig. S4), and MIR396a, MIR396c, and MIR396e to represent all miR396 molecules (see Supplementary Fig. S1A). We generated transgenic hairy roots expressing promoter:GUS fusion constructs for all six genes, and included EV and the constitutive soybean polyubiqutin promoter (PGmUBI; Hernandez-Garcia et al., 2009) as negative and positive controls, respectively. The histochemical localization of GUS activity directed by these promoters was assayed under both uninfected and H. glycines-infected conditions. Under uninfected conditions, PGRF6, PGRF9, and PGRF18 produced strong GUS staining within the root apical meristem (RAM) with minimal activity in the vascular cylinder and root cap (Fig. 3A, C, D). PMIR396a, PMIR396c, and PMIR396e produced consistent GUS activity in the vascular cylinder and in the root cap, but were mostly absent from the RAM (Fig. 3B, C, D). As controls, no GUS staining was observed in any EV roots, and all PGmUBI:GUS roots were strongly stained in all tissues (Fig. 3C, D).
Fig. 3.
GUS histochemical analyses for selected MIR396 and GRF promoters in uninfected and H. glycines-infected soybean roots. (A) Representative image for the native activity of GRF6, GRF9, and GRF18 promoters (shown is GRF9). (B) Representative image for the native activity of MIR396a, MIR396c, and MIR396e promoters (shown is MIR396c). (C) Percentage of GUS-stained roots (n=20 roots) in the root apical meristem (RAM) for MIR396a, MIR396c, MIR396e, GRF6, GRF9, and GRF18 promoters, with empty vector (EV) and constitutive PGmUBI included as negative and positive controls, respectively. (D) Percentage of GUS-stained roots (n=20 roots) in the vascular cylinder (VC) and root cap for MIR396a, MIR396c, MIR396e, GRF6, GRF9, and GRF18 promoters, with EV and constitutive PGmUBI included as negative and positive controls, respectively. (E) Representative images for the activity of MIR396a, MIR396c, and MIR396e promoters within the syncytium during formation (5 and 8 dpi) and maintenance (15 dpi) phases (shown is MIR396c). (F) Representative images for the activity of GRF6, GRF9, and GRF18 promoters within the syncytium during formation and maintenance phases (shown is GRF9). (G) Percentage of GUS-stained roots (n=20 infected roots) in the syncytium during the formation phase for MIR396a, MIR396c, MIR396e, GRF6, GRF9, and GRF18 promoters, with EV and constitutive PGmUBI included as negative and positive controls, respectively. (H) Percentage of GUS-stained roots (n=20 infected roots) in the syncytium during the maintenance phase as in (G). (A, B, E, F) Scale bars=0.5 mm. (C, D, G, H) Shown is the mean percentage GUS activity from three independent experiments. Error bars represent ±1 SD from the mean.
Under H. glycines-infected conditions, PMIR396a, PMIR396c, and PMIR396e showed clear down-regulation in the syncytia induced by J2 and early J3 nematodes (syncytium formation phase), but became very active in the syncytia of J4 nematodes (syncytium maintenance phase) (Fig. 3E, G, H). In contrast, PGRF6 and, in particular, PGRF9 showed sustained, high activation in the syncytia induced by J2, J3, and J4 nematodes (Fig. 3F–H). Interestingly, the strong activity of PGRF9 was observed in a significantly greater percentage of J2 and early J3 syncytia compared with PGRF6 (Fig. 3G), but this difference was no longer significant in syncytia of J4/adult females (Fig. 3H). Also, and as expected, consistent with the mRNA level (Supplementary Fig. S4), PGRF18 showed essentially no activity in syncytia of any H. glycines life stage (Fig. 3G, H). It is noteworthy that MIR396 and GRF promoter activities outside of syncytia at all time points evaluated were unchanged, closely mirroring the uninfected condition (Supplementary Fig. S5), strongly supporting that the changes in mRNA in response to H. glycines infection (Fig. 1B) occurred specifically in syncytia. Furthermore, as controls, and as in uninfected roots, no GUS staining was observed anywhere in any H. glycines-infected EV roots, and all H. glycines-infected PGmUBI:GUS roots were strongly stained in all tissues, including syncytia (Fig. 3G, H). Moreover, the strong activities of these three MIR396, GRF6, and GRF9 promoters in syncytia during the maintenance phase, in combination with the identified mRNA degradation products (Fig. 2), strongly support that the concomitant down-regulation of the respective GRF mRNAs (Fig. 1B) occurs post-transcriptionally from the miR396 expression spike.
Overexpression of pre-miR396 in soybean roots substantially reduces H. glycines development to adult females
All eight unique, canonical pre-miR396 were overexpressed in transgenic soybean hairy roots to determine whether interfering with the miR396–GRF6/8–13/15–17/19 regulatory network would modify susceptibility to H. glycines. PGmUBI was used for overexpression (Fig. 4A). Transgenic soybean cv. Williams 82 roots with high pre-miR396 overexpression (Fig. 4B) were selected for phenotyping. We used the same qRT–PCR primers as above to quantify the overexpression as this strategy gave optimal melting curves (as described in the Materials and methods).
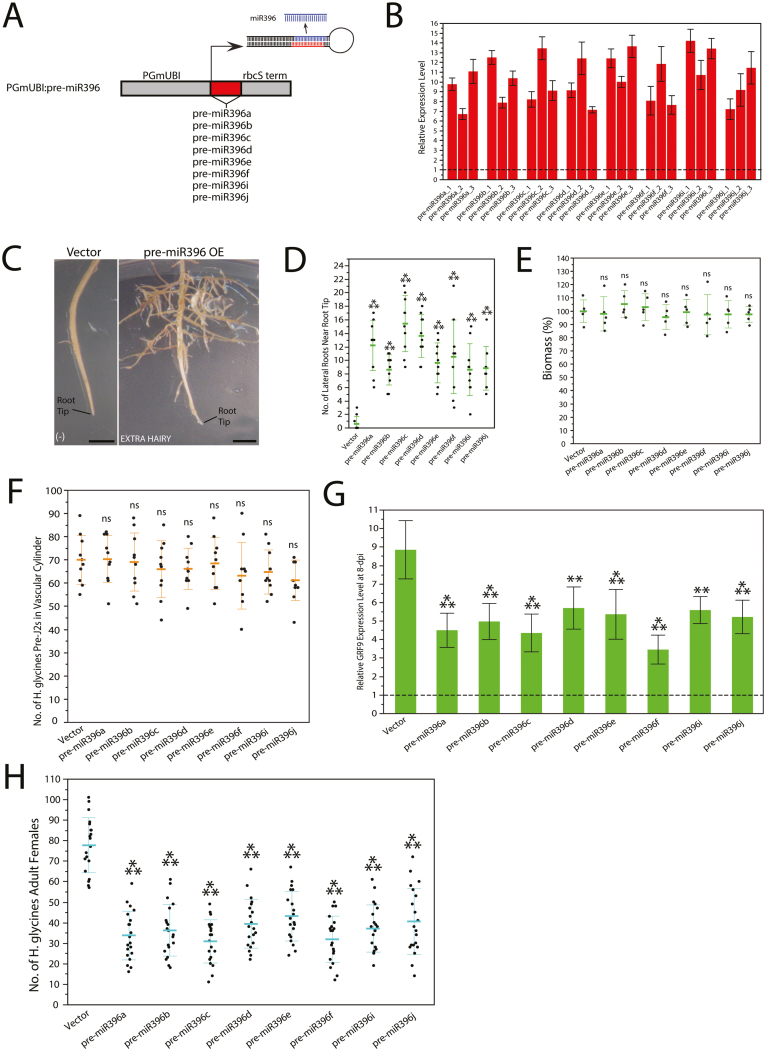
Fig. 4.
Overexpression of pre-miR396 in soybean roots. (A) Overexpression constructs. (B) qRT–PCR analysis on transgenic pre-miR396-overexpressing roots; three events per construct. Expression levels are relative to empty vector control; baseline expression is set to 1.0 and indicated with a dashed line. (C) EXTRA HAIRY phenotype caused by pre-miR396 overexpression. Scale bars=0.5 cm. (D) Comparisons between the number of lateral roots within 2.5 cm from the root tips for pre-miR396-overexpressing and empty vector control roots (n=10). (E) Comparisons between biomasses of pre-miR396-overexpressing and empty vector control roots (n=5) 1 week after transfer to new maintenance plates. Biomasses were measured as the percentage of dry root weight compared with empty vector control; empty vector control mean was set to 100%. (D, E) Data are representative of three independent experiments. (F) Comparisons between the number of H. glycines pre-J2s within the vascular cylinder of pre-miR396-overexpressing and empty vector control roots (n=10). (G) qRT–PCR analysis of GRF9 in H. glycines-infected pre-miR396-overexpressing and empty vector control roots at 8 dpi. Expression levels are relative to mock-inoculated roots for each construct; baseline expression is set to 1.0 and indicated with a dashed line. (H) Comparisons between the number of H. glycines adult females that developed on pre-miR396-overexpressing and empty vector control roots (n=20). (F, H) Data are representative of two independent experiments. (D–H) Error bars represent ±1 SD from the mean. **P<0.01; ***P<0.001; ns, not significant; all are statistically compared with empty vector control.
All pre-miR396-overexpressing roots resulted in an EXTRA HAIRY phenotype (Fig. 4C) characterized by more lateral roots within the first 2.5 cm from the root tips compared with the EV control (Fig. 4D). On the other hand, the overall biomasses (i.e. amount of root tissue generated) for pre-miR396-overexpressing roots were statistically similar to those of the EV control (Fig. 4E) and there were no differences in the number of penetrating H. glycines J2s that infected them at 4 dpi (Fig. 4F). In other words, the increased number of lateral roots near the root tips in the pre-miR396-overexpressing roots compared with EV control did not have any effect on the number of J2s that entered the vascular cylinder to initiate formation of syncytia.
At 8 dpi, all pre-miR396-overexpressing roots infected with H. glycines resulted in significantly reduced inductions of GRF9 during the syncytium formation phase compared with EV control (Fig. 4G). We selected GRF9 as a marker due to its particularly high promoter activity in the syncytium (Fig. 3F–H). Remarkably, in spite of the same number of J2s being inside the roots of pre-miR396 and EV control at 4 dpi (Fig. 4F), all pre-miR396-overexpressing roots resulted in highly significant reductions in the number of H. glycines adult females that developed by 28 dpi (Fig. 4H). Thus, overexpression of all canonical pre-miR396 substantially reduces susceptibility to H. glycines not by affecting the number of J2s that enter the vascular cylinder to initiate syncytia, but by inhibiting the development of the nematodes to the adult female stage, in association with silencing of GRF expression during the syncytium formation phase.
RNAi of GRF9 phenocopies pre-miR396 overexpression
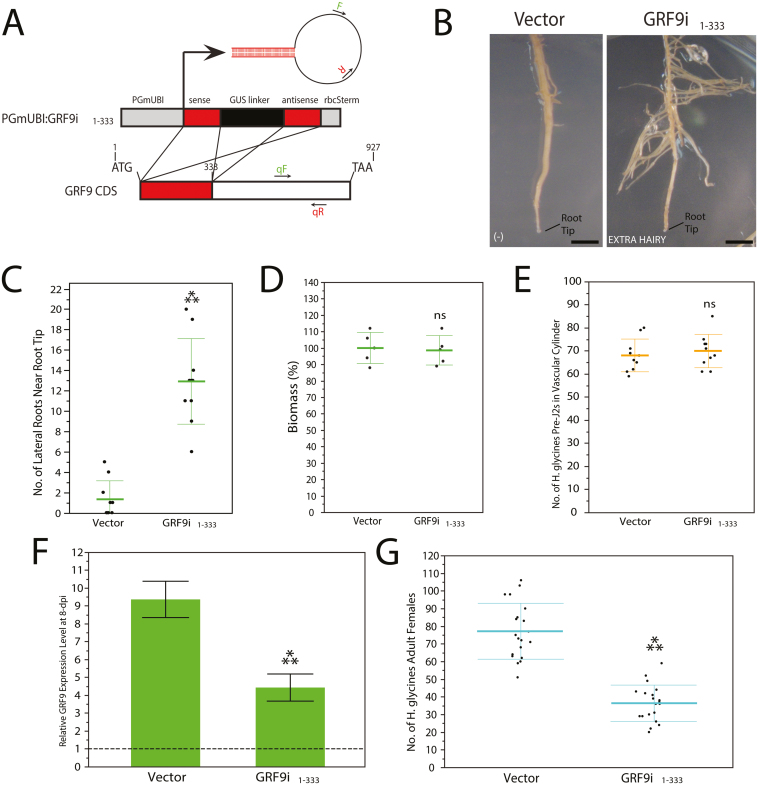
Due to GRF9 being among the most highly up-regulated GRF genes during the cumulative syncytium formation phase (Fig. 1B), having by far the highest steady-state mRNA levels among the 11 H. glycines responsive GRF genes (Supplementary Fig. S2C), and its particularly high promoter activity in syncytia (Fig. 3F–H), we next examined the effect of knocking down GRF9. An RNAi hairpin construct was generated for GRF9 and placed under the transcriptional control of the GmUBI promoter (Fig. 5A). Transgenic events that were determined to express GRF9i1–333 via RT–PCR were selected for phenotyping.
Fig. 5.
RNAi of GRF9 in soybean roots. (A) RNAi construct. Annealing sites for the primers used for RT–PCR diagnosis of transgene expression (F and R), and qRT–PCR analysis of GRF9 (qF and qR) are indicated. (B, C) EXTRA HAIRY phenotype for GRF9i1–333, as in Fig. 4C and D). (D) Comparison between biomasses of GRF9i1–333 and empty vector control roots, as in Fig. 4E. (E) Comparisons between the number of H. glycines pre-J2s within the vascular cylinder of GRF9i1–333 and empty vector control roots, as in Fig. 4F. (F) Comparison between GRF9 relative expression levels in H. glycines-infected GRF9i1–333 and empty vector control roots at 8 dpi, as in Fig. 4G. (G) Comparisons between the number of H. glycines adult females that developed on GRF9i1–333 and empty vector control roots, as in Fig. 4H.
Interestingly, and, at first, somewhat surprisingly, all GRF9i1–333 soybean roots phenocopied the EXTRA HAIRY phenotype that was observed for pre-miR396-overexpressing roots (Fig. 5B) resulting in similar numbers of lateral roots within the first 2.5 cm from the root tip (Fig. 5C). Also, as observed for pre-miR396-overexpressing roots, GRF9i1–333 roots showed no difference in root biomass (Fig. 5D) or in the number of penetrating H. glycines J2s that infected them at 4 dpi compared with EV control (Fig. 5E).
As expected, induction of GRF9 during the syncytium formation phase at 8 dpi was significantly reduced in the GRF9i1–333 roots (Fig. 5F) to a similar level to that at pre-miR396 overexpression. However, GRF9i1–333 also resulted in moderately reduced induction of the other 10 H. glycines-responsive GRF genes at 8 dpi, of which the GRF genes most similar to GRF9 (GRF10, GRF11,and GRF12; Supplementary Fig. S6) were significantly reduced (Supplementary Fig. S7). On the other hand, induction of GRF18 at 8 dpi was unaffected in GRF9i1–333 roots as it is unresponsive to H. glycines infection (Supplementary Fig. S7) and, thus, all of the silencing effects with GRF9i1–333 during infection are likely to be limited to the 11 H. glycines-responsive GRF genes, and GRF9 in particular. Strikingly, similar to pre-miR396 overexpression, GRF9i1–333 roots resulted in a highly significant reduction in the number of H. glycines that developed to adult females (Fig. 5G). Thus, RNAi-mediated down-regulation of GRF9, and to a moderate extent the other 10 H. glycines-responsive GRF genes, phenocopied the reduced susceptibility phenotypes caused by pre-miR396 overexpression. These results underscore the necessity of adequate expression of these 11 GRF genes, and in particular GRF9, during the syncytium formation phase for productive H. glycines infections.
Overexpression of miR396-resistant GRF9 resembles pre-miR396 overexpression and GRF9 RNAi
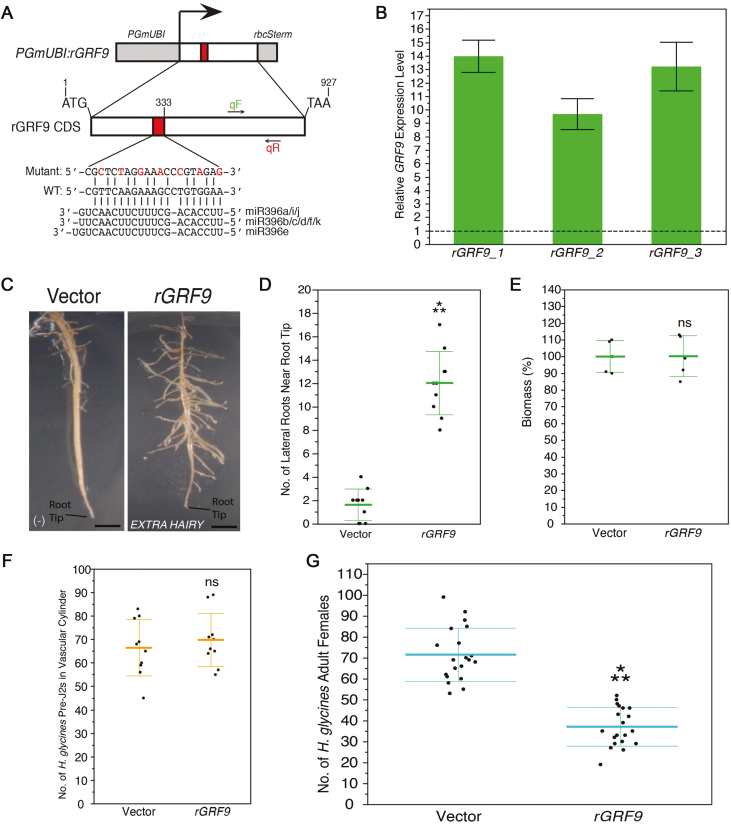
Finally, to demonstrate the necessary homeostasis in the miR396–GRF6/8–13/15–17/19 regulatory network during H. glycines infection, as observed in the Arabidopsis–H. schachtii model (Hewezi et al., 2012), we overexpressed a synonymous miR396-resistant (r)GRF9 mutant, under PGmUBI transcriptional control (Fig. 6A). GRF9 was selected for reasons explained above. Transgenic soybean roots determined to overexpress rGRF9 at high levels via qRT–PCR were selected for phenotyping (Fig. 6B).
Fig. 6.
Overexpression of an miR396-resistant mutant of GRF9 in soybean roots. (A) Overexpression construct for miR396-resistant GRF9 (rGRF9). An illustration of the synonymous mutations introduced into the miR396 target site is shown. Annealing sites for the primers used for qRT–PCR analysis of GRF9 levels are shown. (B) qRT–PCR analysis of GRF9 in transgenic rGRF9-overexpressing roots. Expression levels are relative to empty vector control; baseline expression is set to 1.0 and indicated with a dashed line. (C, D) EXTRA HAIRY developmental phenotype for rGRF9 overexpression, as in Fig. 4C and D. (E) Comparison between biomasses of rGRF9-overexpressing and empty vector control roots, as in Fig. 4E. (F) Comparisons between the number of H. glycines pre-J2s within the vascular cylinder of rGRF9-overexpressing and empty vector control roots, as in Fig. 4F. (G) Comparisons between the number of H. glycines adult females that developed on rGRF9-overexpressing and empty vector control roots, as in Fig. 4H.
Comparably with results from Rodriguez et al. (2015) and Hewezi et al. (2012) in Arabidopsis, rGRF9-overexpressing roots also showed the EXTRA HAIRY phenotype observed for pre-miR396 overexpression and GRF9i1–333 roots (Fig. 6C, D). Again, this manipulation did not alter the overall root biomass (Fig. 6E) or the number of penetrating H. glycines J2s that entered the vascular cylinder to initiate syncytium formation compared with EV control (Fig. 6F). However, overexpression of rGRF9 in soybean roots resulted in a highly significant reduction in the number of H. glycines that developed to adult females compared with EV control (Fig. 6G). As an additional control, in a separate experiment, overexpression of GUSPlus had no effect on the number of adult females (Supplementary Fig. S8). Thus, overexpression of rGRF9 highly resembled the reduced susceptibility phenotype caused by pre-miR396 overexpression and RNAi knockdown of GRF9 (and, to a moderate extent, the other H. glycines-responsive GRF gnes). Collectively, these in vivo studies indicated that homeostasis in the miR396–GRF6/8–13/15–17/19 regulatory network is essential for productive H. glycines infections.
Discussion
Previously, the miR396–GRF1/3 regulatory module was shown to be a master regulator of syncytium development in the model cyst nematode interaction between H. schachtii and Arabidopsis (Hewezi et al., 2012). Here, we investigated whether this syncytium post-transcriptional regulatory system is conserved in the interaction between H. glycines and soybean, which is of interest for at least two reasons: (i) for the obvious, potential translational benefit; and (ii) no study has addressed whether findings made in this model pathosystem are applicable to the H. glycines–soybean pathosystem. We have found that a network involving nine canonical MIR396 genes (eight unique) and, specifically, 11 GRF genes delineates the H. glycines syncytium formation and maintenance phases in soybean via transcriptional regulation of both MIR396 and GRF genes, and post-transcriptional regulation of GRF genes by miR396. Disrupting the balance in this network either by reducing GRF expression during the syncytium formation phase (either by pre-miR396 overexpression or RNAi knockdown) or by overexpression of rGRF9 dramatically inhibits the number of H. glycines that develop to adult females. Thus, although involving many more genes in soybean, the miR396–GRF regulatory network is clearly conserved between H. schachtii–Arabidopsis and H. glycines–soybean pathosystems, and a balanced network is essential for productive H. glycines infections.
Overexpression of all canonical pre-miR396 in soybean roots resulted in an EXTRA HAIRY phenotype (Fig. 4C, D) that was accompanied by GRF9 silencing during syncytium formation (Fig. 4G). Liu et al. (2017) generated Arabidopsis overexpression lines for all soybean pre-miR396, and all except pre-miR396d, f, and j gave similar mutant phenotypes. We suggest that lack of observed mutant phenotypes for soybean pre-miR396d, f, and j could have been due to the use of a surrogate transgenic system. Also, the number of lateral roots near the root tip, which we classified as an EXTRA HAIRY phenotype, was not scored in Liu et al. (2017), so it is not certain that soybean pre-miR396d, f, and j do not give this phenotype when overexpressed in Arabidopsis. Furthermore, the conclusion in Liu et al. (2017) that these soybean pre-miR396 do not function in the plant is not clear since they were capable of cleaving the consensus GRF target site in Arabidopsis protoplasts. In the present study, we have provided strong experimental evidence that these soybean pre-miR396 are functional in soybean roots.
Twenty-three GRF genes showed varying degrees of expression in young soybean roots, while only two GRF genes were undetected (Supplementary Fig. S2C). We compared these data with RNA sequencing (RNA-seq) data that are available for soybean root tips at the soybean functional genomics database (SFGD) (Yu et al., 2014). Even though these RNA-seq data were obtained from root tips as opposed to whole roots, and the plants were grown under different conditions from those in our experiments, we found that our qRT–PCR data were well correlated with the SFGD RNA-seq data (Supplementary Fig. S9; R2=0.48, P<0.001). Thus, this observation validates the accuracy of both our qRT–PCR data for GRF expression in whole soybean roots and the SFGD RNA-seq data for root tips. Also, MIR396 gene family members exhibited varying degrees of expression in young soybean roots (Supplementary Fig. S2A, B), which is consistent with previous findings (Li et al., 2012). However, Li et al. (2012) did not mention changes in miR396 abundance in response to H. glycines, but in their study soybean plants were grown in H. glycines-infested soil and they were only evaluated at a single time point long after infection was established. Tian et al. (2017) also did not mention changes in miR396 abundance in response to H. glycines, but, again, soybean plants were grown in H. glycines-infested soil, probably diluting out the phase-specific changes in developing syncytia (see Fig. 1B). We found that all canonical pre-miR396 and miR396 are down-regulated early during the syncytium formation phase, and up-regulated during the maintenance phase (Fig. 1B), mirroring MIR396 promoter activities in syncytia (Fig. 3E, G, H). Thus, our finding that expression patterns differ drastically at different time points (i.e. during different phases of syncytium development) during H. glycines infection probably explains why these previous studies did not mention miR396.
Liu et al. (2017) did not detect expression of the pre-miR396c/f subfamily or GRF genes 5, 6, 9, 12, 13, 16, 17, 22, and 24 (names according to this paper) in soybean roots, yet we detected them with relatively low Ct values in all biological replicates (Supplementary Fig. S2A, C). In fact, pre-miR396c/f was by far the most highly expressed subfamily (Supplementary Fig. S2A). Also, MIR396c (along with MIR396a and MIR396e), GRF6, and GRF9 promoters were clearly active in soybean roots (Fig. 3A–D). Moreover, Li et al. (2012) detected expression of all of these GRF genes in soybean root tips, which correlates well with our data (Supplementary Fig. S9). We suggest that the different growth conditions, RNA preparations, qRT–PCR parameters, and/or primers may be the cause for lack of detection of expression of these genes in soybean roots by Liu et al. (2017).
Eleven soybean GRF genes are specifically up-regulated during the cumulative syncytium formation phase (at 8 dpi), which in our infection system was between 5 and 13 dpi (Fig. 1A), while the other GRF genes do not change (Supplementary Fig. S4). Moreover, promoter analyses (Fig. 3F–H) indicate that this up-regulation is specific in syncytia during the formation phase as GRF promoters are, for the most part, specific to the RAM in uninfected roots, while syncytia are induced in the vascular cylinder. We also did not observe any changes in promoter activities for the respective MIR396 and GRF genes outside of syncytia (Supplementary Fig. S5). Microarray analysis was previously performed on laser capture-microdissected H. glycines syncytia at 2, 5, and 10 dpi (Ithal et al., 2007b). However, this study did not present any data on GRF genes, which is probably due to a number of factors. For instance, the only genes that were analyzed in the latter study were those that first changed in expression at 2 dpi, and then those genes were subsequently analyzed at 5 and 10 dpi. Thus, it is likely that at 2 dpi GRF genes are not yet up-regulated to the point of detection, which would be consistent with our results (Fig. 1B), and may explain why GRF genes were not mentioned in the latter study. Other microarray analyses were also performed on H. glycines-infected, whole soybean roots, but again no data were presented on GRF genes (Alkharouf et al., 2006; Ithal et al., 2007a). Lack of data presented for GRF genes in these studies could have also been due to insufficient representation on the GeneChip (Ithal et al., 2007a, b) or cDNA (Alkharouf et al., 2006) arrays, or possibly a combination of other factors. Also, many other microarray and RNA-seq studies have been performed on H. glycines-infected soybean roots, but the changes that are presented in those studies are representative of resistant reactions. It is noteworthy that our qR–PCR data for GRF expression changes during H. glycines infection (Fig. 1B) and promoter data in syncytia (Fig. 3F–H) are consistent with the previously published microarray, qRT–PCR, and promoter data for H. schachtii-infected Arabidopsis roots (Szakasits et al., 2009; Hewezi et al., 2012).
In the RAM, stem cell progeny undergo rapid cell division to ensure that there are enough cells for proper growth, and these rapidly dividing cells are called the transit-amplifying cells (TACs) (Rodriguez et al., 2015). In Arabidopsis roots, miR396 is abundant in the root cap and stem cell niche (SCN) formed by the quiescent center (QC) and adjacent stem cell initials, while GRF genes are abundant in TACs. GRFs promote rapid cell cycling within TACs, and miR396-mediated down-regulation of GRFs results in delayed cell cycling (Rodriguez et al., 2015). We found that soybean MIR396a, MIR396c, and MIR396e promoters are active within the root cap (most probably columella cells and the SCN) and the vascular cylinder leading up to the RAM (Fig. 3B–D), and that GRF6, GRF9, and GRF18 promoters are predominantly active within the RAM, most probably TACs (Fig. 5A, C, D). Thus, the function of the miR396–GRF regulatory network in soybean roots appears to be similar to that of other plant species (Bazin et al., 2013; Rodriguez et al., 2015), although we do not know if the other MIR396 and GRF promoters have the same patterns of activity throughout the root system during development, which was beyond the scope of our study. Importantly, GRF genes are up-regulated in the syncytium during the formation phase concomitant with the down-regulation of miR396 (Figs 1B, 3E–H). Conversely, during the syncytium maintenance phase, GRF genes are post-transcriptionally down-regulated by the de-repressed miR396 expression (Figs 1B, 3E–H, 4). We also found that all 11 GRF mRNAs are cleaved via miR396 during the syncytium maintenance phase (Fig. 2), but that GRF6 and GRF9 promoters remain highly active during this time (Fig. 3F–H), indicating post-transcriptional down-regulation by miR396. Hence, soybean GRF genes appear to be regulated in the H. glycines syncytium by miR396 in parallel to the RAM, suggesting that GRFs might function to maintain rapid cell cycling in the forming syncytium for proper organ development (Engler and Gheysen, 2013). It is noteworthy that GRF18, which we found as the most highly expressed GRF in young soybean roots (Supplementary Fig. S2C), specifically in the RAM (Fig. 3A, C, D), and does not change during H. glycines infection (Supplementary Fig. S4), has no promoter activity in H. glycines syncytia (Fig. 3G, H). Thus, the developmental program of developing H. glycines syncytia is clearly different from that of the RAM, involving select GRF genes.
When plants are under high pathogen stress, resources are devoted towards defense responses, while growth is stunted and development is delayed. This phenomenon is known as the growth–defense trade-off (Huot et al., 2014). GRFs have been implicated in various abiotic and biotic stress conditions (Liu et al., 2008; Y. Li et al., 2010; Hewezi et al., 2012; Kim et al., 2012; Casadevall et al., 2013; Liu et al., 2017), and regulate the expression of a wide range of genes involved in both developmental processes and defense responses (Liu et al., 2014). GRFs are thus hypothesized to co-ordinate the interactions between defense signaling and growth and developmental pathways (Liu et al., 2014). In this context, GRFs could be thought to promote growth by maintaining rapid cell cycles while simultaneously suppressing defense responses. We found that silencing GRF genes enhances soybean lateral root formation near the root tips (Figs 4C, D, 5B, C). This phenotype is probably reflected by a decreased elongation zone (Rodriguez et al., 2015), which may also explain the similar biomasses and numbers of H. glycines J2 that penetrated the vascular cylinder to initiate syncytia observed between control and mutant roots (Figs 4E, 5D), yet highly reduced susceptibility to H. glycines in mutant roots (Figs 4H, 5G), consistent with GRF genes promoting developmental processes and suppressing defenses. Interestingly, however, overexpression of rGRF9 gave an EXTRA HAIRY and highly reduced susceptibility phenotype similar to GRF silencing (Fig. 6). Thus, although GRF genes are required to maintain proper soybean lateral root numbers, and productive H. glycines infections, their precise expression levels, fine-tuned by miR396, are also required (probably too rapid and uncontrolled cell cycling in developing syncytia is also problematic), consistent with the Arabidopsis–H. schachtii model (Hewezi et al., 2012).
Feedback regulation of miRNAs by their transcription factor targets has been demonstrated in several studies (Gutierrez et al., 2009; Wang et al., 2009; Wu et al., 2009; Marin et al., 2010; Yant et al., 2010; Hewezi and Baum, 2012). Also, overexpression of rGRF1 and 3 in Arabidopsis not only down-regulates miR396, but also down-regulates other GRF genes as well as wild-type GRF1 and 3, respectively, in roots (Hewezi and Baum, 2012). Although some of the co-ordination between miR396 and GRF genes can be explained through PLETHORA (Rodriguez et al., 2015) and TCP4 (Rodriguez et al., 2010) transcription factors, it is clear that MIR396 and GRF genes are downstream targets that are negatively regulated by GRF genes in roots (Hewezi and Baum, 2012). This complex feedback loop ensures a precise transcriptional equilibrium. Thus, pre-miR396 overexpression, RNAi of GRF9 (and, to a lesser extent, the other 10 H. glycines-responsive GRF genes; Supplementary Fig. S6), and rGRF9 overexpression all resulting in comparable EXTRA HAIRY and highly reduced susceptibility phenotypes, consistent with the Arabidopsis–H. schachtii model (Hewezi et al., 2012), underscores the likely importance of this complex feedback loop to maintain such an equilibrium in soybean. Future studies that analyze the expression changes in miR396 and GRF genes in rGRF9-overexpressing roots, for example, will provide a more complete picture of the necessary feedback regulations within the miR396–GRF6/8–13/15–17/19 regulatory network during H. glycines infection.
In summary, we have investigated if a miR396–GRF regulatory system operates in the agronomically important interaction between H. glycines and soybean. Our results demonstrate that the miR396–GRF6/8–13/15–17/19 regulatory network delineates the phases of the H. glycines syncytium and that interfering in the homeostasis of this network inhibits productive H. glycines infections (i.e. greatly reduces the number of adult females that develop). As H. glycines is the most economically devastating soybean pathogen causing over US$1 billion in yield losses each year (Koenning and Wrather, 2010), control strategies more effective than the conventional measures (Conley et al., 2011) are urgently needed. Thus, by specifically interfering with this network in syncytia using an H. glycines-inducible promoter (thereby avoiding changes to the plant outside of syncytia), this network may serve as a potential target to develop soybean plants with novel, synthetic resistance to H. glycines.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The soybean MIR396 gene family.
Fig. S2. Native expression of pre-miR396, miR396, and GRF genes in young soybean roots.
Fig. S3. Putative miR396 target sites in GRF1–24.
Fig. S4. qRT–PCR screen of GRF genes in H. glycines-infected roots at 8 dpi.
Fig. S5. Activities of selected MIR396 and GRF promoters in H. glycines-infected soybean roots outside of syncytia.
Fig. S6. Multiple sequence (MUSCLE) alignment of soybean GRF genes.
Fig. S7. qRT–PCR analysis of H. glycines-responsive GRF genes in GRF9i1–333 roots.
Fig. S8. PGmUBI-driven expression of GUSPlus does not change susceptibility to H. glycines infection.
Fig. S9. Comparison between SFGD root tip RNA-seq data and our whole root qRT–PCR data for steady-state expression of soybean GRF genes.
Table S1. Complete list of primers used in our study.
Table S2. Soybase genome co-ordinates and gene calls for MIR396 and GRF genes.
Acknowledgements
This is a Journal Paper of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa, supported by Hatch Act and State of Iowa funds. This work was supported by grants from the Iowa Soybean Association. The authors would like to thank Tom Maier and Danielle Sill for technical assistance. The authors declare no conflict of interest.
References
- Alkharouf NW, Klink VP, Chouikha IB, Beard HS, MacDonald MH, Meyer S, Knap HT, Khan R, Matthews BF. 2006. Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224, 838–852. [DOI] [PubMed] [Google Scholar]
- Bao M, Bian H, Zha Y, Li F, Sun Y, Bai B, Chen Z, Wang J, Zhu M, Han N. 2014. miR396a-mediated basic helix–loop–helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant & Cell Physiology 55, 1343–1353. [DOI] [PubMed] [Google Scholar]
- Baum TJ, Wubben MJ, Hardyy KA, Su H, Rodermel SR. 2000. A screen for Arabidopsis thaliana mutants with altered susceptibility to Heterodera schachtii. Journal of Nematology 32, 166–173. [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Khan GA, Combier JP, et al. 2013. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. The Plant Journal 74, 920–934. [DOI] [PubMed] [Google Scholar]
- Blin N, Stafford DW. 1976. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Research 3, 2303–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O. 2014. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annual Review of Plant Biology 65, 473–503. [DOI] [PubMed] [Google Scholar]
- Cabrera J, Barcala M, García A, et al. 2016. Differentially expressed small RNAs in Arabidopsis galls formed by Meloidogyne javanica: a functional role for miR390 and its TAS3-derived tasiRNAs. New Phytologist 209, 1625–1640. [DOI] [PubMed] [Google Scholar]
- Casadevall R, Rodriguez RE, Debernardi JM, Palatnik JF, Casati P. 2013. Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. The Plant Cell 25, 3570–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley SP, Gaska JM, Pedersen P, Esker P. 2011. Soybean yield and Heterodera glycines response to rotation, tillage, and genetic resistance. Agronomy Journal 103, 1604–1609. [Google Scholar]
- Dai X, Zhao PX. 2011. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research 39, W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Mecchia MA, Vercruyssen L, Smaczniak C, Kaufmann K, Inze D, Rodriguez RE, Palatnik JF. 2014. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. The Plant Journal 79, 413–426. [DOI] [PubMed] [Google Scholar]
- Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM. 2009. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15, 2219–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler JdA, Gheysen G. 2013. Nematode-induced endoreduplication in plant host cells: why and how? Molecular Plant-Microbe Interactions 26, 17–24. [DOI] [PubMed] [Google Scholar]
- Fang XL, Zhao YY, Ma QB, Huang Y, Wang P, Zhang J, Nian H, Yang CY. 2013. Identification and comparative analysis of cadmium tolerance-associated miRNAs and their targets in two soybean genotypes. PLoS One 8, e81471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Ye WW, Wu XL, Shen DY, Wang YC, Xing H, Dou DL. 2011. Microarray profiling reveals microRNAs involving soybean resistance to Phytophthora sojae. Genome 54, 954–958. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C. 2009. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. The Plant Cell 21, 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Garcia CM, Martinelli AP, Bouchard RA, Finer JJ. 2009. A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Reports 28, 837–849. [DOI] [PubMed] [Google Scholar]
- Hewezi T, Baum TJ. 2012. Complex feedback regulations govern the expression of miRNA396 and its GRF target genes. Plant Signaling & Behavior 7, 749–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Baum TJ. 2015. Gene silencing in nematode feeding sites. Advanced Botanical Research 73, 221–239. [Google Scholar]
- Hewezi T, Howe P, Maier TR, Baum TJ. 2008. Arabidopsis small RNAs and their targets during cyst nematode parasitism. Molecular Plant-Microbe Interactions 21, 1622–1634. [DOI] [PubMed] [Google Scholar]
- Hewezi T, Maier TR, Nettleton D, Baum TJ. 2012. The Arabidopsis microRNA396–GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiology 159, 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Nakayama H, Ishikawa N, Kubo M, Demura T, Fukuda H, Tsukaya H. 2011. ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant & Cell Physiology 52, 112–124. [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. 2014. Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Molecular Plant 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey RS. 1985. Staining nematodes in plant tissue. In: Zuckerman BM, Mai WF, Harrison MB, eds. Plant nematology laboratory manual. Amherst, MA: The University of Massachusetts Agricultural Experiment Station, 197–199. [Google Scholar]
- Hussey RS, Grundler FMW. 1998. Nematode parasitism of plants. In: Perry RN, Wright DJ, eds. The physiology and biochemistry of free-living and plant-parasitic nematodes. Wallingford, Oxon: CAB International, 213–243. [Google Scholar]
- Ithal N, Recknor J, Nettleton D, Hearne L, Maier T, Baum TJ, Mitchum MG. 2007a. Parallel genome-wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Molecular Plant-Microbe Interactions 20, 293–305. [DOI] [PubMed] [Google Scholar]
- Ithal N, Recknor J, Nettleton D, Maier T, Baum TJ, Mitchum MG. 2007b. Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Molecular Plant-Microbe Interactions 20, 510–525. [DOI] [PubMed] [Google Scholar]
- Jones JT, Haegeman A, Danchin EG, et al. 2013. Top 10 plant-parasitic nematodes in molecular plant pathology. Molecular Plant Pathology 14, 946–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell 14, 787–799. [DOI] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H. 2003. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. The Plant Journal 36, 94–104. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee BH. 2006. GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. Journal of Plant Biology 49, 463–468. [Google Scholar]
- Kim JS, Mizoi J, Kidokoro S, et al. 2012. Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. The Plant Cell 24, 3393–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Hosseini P, Matsye P, Alkharouf NW, Matthews BF. 2009. A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode). Plant Molecular Biology 71, 525–567. [DOI] [PubMed] [Google Scholar]
- Koenning SR, Wrather JA. 2010. Suppression of soybean yield potential in the continental United States by plant diseases from 2006 to 2009. Plant Health Progress. doi: 10.1094/PHP-2010-1122-01-RS [DOI] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research 42, D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulcheski FR, de Oliveira LF, Molina LG, et al. 2011. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Deng Y, Wu T, Subramanian S, Yu O. 2010. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiology 153, 1759–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WB, Wang PP, Li YG, Zhang KX, Ding FQ, Nie TK, Yang X, Lv QX, Zhao L. 2015. Identification of microRNAs in response to different day lengths in soybean using high-throughput sequencing and qRT-PCR. PLoS One 10, e0132621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Wang X, Zhang SP, Liu DW, Duan YX, Dong W. 2012. Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS One 7, e9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM. 2010. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiology 152, 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Wang F, Yu D. 2014. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiology 164, 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. 2008. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA (New York, N.Y.) 14, 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hua W, Yang HL, Zhan GM, Li RJ, Deng LB, Wang XF, Liu GH, Wang HZ. 2012. The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. Journal of Experimental Botany 63, 3727–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rice JH, Chen N, Baum TJ, Hewezi T. 2014. Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PLoS One 9, e98477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kandoth PK, Warren SD, et al. 2012. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492, 256–260. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhou Y, Xiaowei L, et al. 2017. Tissue-specific regulation of Gma-miR396 family on coordinating development and low water availability responses. Frontiers in Plant Science 8, 1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Elmayan T, Vaucheret H. 2008. MicroRNA maturation and action—the expanding roles of ARGONAUTEs. Current Opinion in Plant Biology 11, 560–566. [DOI] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. 2010. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. The Plant Cell 22, 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon JB, Qi M, Sill DN, Muppirala U, Eves-van den Akker S, Maier TR, Dobbs D, Mitchum MG, Hewezi T, Baum TJ. 2016. A Plasmodium-like virulence effector of the soybean cyst nematode suppresses plant innate immunity. New Phytologist 212, 444–460. [DOI] [PubMed] [Google Scholar]
- Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B. 2015. Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Molecular Plant 8, 998–1010. [DOI] [PubMed] [Google Scholar]
- Pajoro A, Madrigal P, Muiño JM, et al. 2014. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biology 15, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Ercoli MF, Debernardi JM, Breakfield NW, Mecchia MA, Sabatini M, Cools T, De Veylder L, Benfey PN, Palatnik JF. 2015. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in arabidopsis roots. The Plant Cell 27, 3354–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. 2010. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. 2013. Biogenesis, turnover, and mode of action of plant microRNAs. The Plant Cell 25, 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Shamimuzzaman M, Vodkin L. 2012. Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genomics 13, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobezak M, Golinowski W. 2009. Structure of cyst nematode feeding sites. In: Berg RH, Taylor CG, eds. Plant cell monographs. Berlin: Springer, 153–187. [Google Scholar]
- Szakasits D, Heinen P, Wieczorek K, Hofmann J, Wagner F, Kreil DP, Sykacek P, Grundler FM, Bohlmann H. 2009. The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. The Plant Journal 57, 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Wang S, Todd TC, Johnson CD, Tang G, Trick HN. 2017. Genome-wide identification of soybean microRNA responsive to soybean cyst nematodes infection by deep sequencing. BMC Genomics 8, 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, Nizampatnam NR, Baron M, Coppin S, Damodaran S, Adhikari S, Arunachalam SP, Yu O, Subramanian S. 2013. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiology 162, 2042–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E, Kim JH, Kende H. 2000. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiology 122, 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, Benes K, Phillips JP, Gartland KMA. 1995. Histochemical GUS analysis. Methods in Molecular Biology 44, 185–193. [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li K, Chen L, et al. 2015. MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiology 168, 984–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang L, Zou Y, et al. 2014. Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. The Plant Cell 26, 4782–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Zhang CJ, Hao QN, Sha AH, Zhou R, Zhou XA, Yuan LP. 2013. Elucidation of miRNAs-mediated responses to low nitrogen stress by deep sequencing of two soybean genotypes. PLoS One 8, e67423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CE, Zhao YT, Wang XJ, Croft L, Wang ZH, Haerizadeh F, Mattick JS, Singh MB, Carroll BJ, Bhalla PL. 2011. MicroRNAs in the shoot apical meristem of soybean. Journal of Experimental Botany 62, 2495–2506. [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Liu Q, Chen LY, Kuang JB, Walk T, Wang JX, Liao H. 2013. Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genomics 14, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MY, Li YH, Zhang QX, Xu T, Qiu LJ, Fan YL, Wang L. 2014. Novel miRNA and phasiRNA biogenesis networks in soybean roots from two sister lines that are resistant and susceptible to SCN race 4. PLoS One 9, e110051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hossain MS, Arikit S, et al. 2015. Identification of microRNAs and their mRNA targets during soybean nodule development: functional analysis of the role of miR393j-3p in soybean nodulation. New Phytologist 207, 748–759. [DOI] [PubMed] [Google Scholar]
- Yan Z, Hossain MS, Valdés-López O, et al. 2016. Identification and functional characterization of soybean root hair microRNAs expressed in response to Bradyrhizobium japonicum infection. Plant Biotechnology Journal 14, 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hossain MS, Wang J, Valdés-López O, Liang Y, Libault M, Qiu L, Stacey G. 2013. miR172 regulates soybean nodulation. Molecular Plant-Microbe Interactions 26, 1371–1377. [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M. 2010. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. The Plant Cell 22, 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. 2005. Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhang Z, Wei J, Ling Y, Xu W, Su Z. 2014. SFGD: a comprehensive platform for mining functional information from soybean transcriptome data and its use in identifying acyl-lipid metabolism pathways. BMC Genomics 15, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Li Z, Fan J, Hu C, Yang R, Qi X, Chen H, Zhao F, Wang S. 2015. Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. Journal of Experimental Botany 66, 4653–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.