Abstract
Exposure to ultraviolet B (UVB) irradiation results in multitude of cellular responses including generation of reactive oxygen species and DNA damage and is responsible for non-melanoma skin cancers (NMSCs). Although genetic mutation is well documented, the epi-mutation, the alteration in epigenetics, remains elusive. In this study, we utilized CpG Methyl-seq to identify a genome-wide DNA CpG methylation, to profile the DNA methylation in UVB-irradiated SKH-1 mouse skin epidermis and non-melanoma skin papillomas at various stages. Methyl-seq and RNA-seq were performed to examine the methylation and corresponding transcriptome alterations. The methylation profiles in mouse epidermis were altered by UVB-irradiation as time progresses. Ingenuity Pathways Analysis (IPA) identified many cancer related pathways including PTEN, p53, Nrf2 and inflammatory signaling in UVB-irradiation induced carcinogenesis. Additionally, some novel genes involved in skin carcinogenesis that were not previously reported were differentially methylated, including Enf2, Mgst2, Vegfa, and Cdk4. Taken together, the current study provides novel profiles and insights of methylation and transcriptomic changes at different stages of carcinogenesis in UVB-irradiation induced NMSC and offers potential targets for prevention and treatment of NMSC at different stages of human skin cancer.
Keywords: Non-melanoma skin cancer, ultraviolet-B (UVB), Epigenetics, DNA methylation, RNA-seq, Methyl-seq
1. Introduction
Exposure to ultraviolet B (UVB) is one of the major causative factors for non-melanoma skin cancers (NMSCs). Long-term exposure to UVB radiation induces inflammation, oxidative stress, DNA mutation, and damage, which are involved in initiation, promotion and progression of NMSCs (1). The early UV-exposure induces inflammatory responses with the increased blood flow, vascular permeability, expression of cyclooxygenases-2 (COX-2), and production of prostaglandin (PG) metabolites (2). UVB-induced inflammation is an important event in all three stages of NMSCs (2), showing the importance of controlling the UVB-induced inflammation for prevention of skin cancers.
For many years cancer research has focused on genetic alterations in carcinogenesis, but during the last decade epigenetic deregulation has been increasingly recognized as a hallmark of cancer (3–7). Epigenomic alterations, including DNA methylation, histone modifications and miRNAs, are now well associated with cancer development and could become useful biomarkers and novel targets for prevention or treatment. DNA methylation can be altered by environmental influences and provides a mechanism to affect the phenotypes in skin aging and carcinogenesis (8). Currently, there are a few approaches commonly used with the next-generation sequencing (NGS) platforms to profile the genome-wide DNA methylation. We have previous applied protein-affinity enrichment of methylated regions in two selected representative carcinogenesis models (9). This method reports the varying enrichment CpG density but would not provide the base-pair resolution of methylated cytosines (10,11). An unbiased method to detect methylated CpG sites at base-pair resolution is the whole genome bisulfite sequencing. However, only 70% −80% of the sequenced reads provide useful DNA methylation information (12), and a more cost-effective method is to detect the methylation at base-pair resolution without bias of CpG dense and poor regions for the genome-wide DNA methylation. Our current study describes the application of Methyl-Seq for the mouse methylome and it reliably detects the DNA methylation in different stages of skin carcinogenesis. This study will provide insights in the understanding of the alterations of the genome-wide DNA methylation signatures in UVB-induced skin carcinogenesis mouse model.
In this study, we used UVB-induced skin cancer mouse models to examine the transcriptomic and epigenomic changes during different stages of skin cancer from imitation, promotion, to later progression. The results from the study will contribute to the development of safe and efficient biomarkers by natural phytochemicals chemopreventive compounds to prevent skin cancers and to identify potential transcriptomic and epigenomic biomarkers during skin carcinogenesis to provide novel therapeutic strategies.
2. Materials and methods
2.1. Chemicals and reagents
Acetone (HPLC grade) and 10% phosphate-buffered formalin were obtained from Fisher Scientific (Hampton, NH, USA). UV lamps that emit UVB (280 – 320 nm; 75–80% of total energy) and UVA (320–375 nm; 20–25% of total energy), as described in previous studies (13). These UV lamps (FS72T12-UVB-HO; National Biological Corp., Twinsburg, OH, USA) emit little or no radiation < 280 nm or > 375 nm. The lamps emit UVB (280–320 nm; 75–80% of total energy) and UVA (320–375 nm; 20–25% of total energy), as described in our previous studies (14,15). The dose of UVB was quantified using a UVB Spectra 305 dosimeter (Daavlin Co., Bryan, OH, USA). The radiation was calibrated with an IL-1700 research radiometer/photometer from International Light Inc. (Newburyport, MA, USA).
2.2. UV-induced skin carcinogenesis model
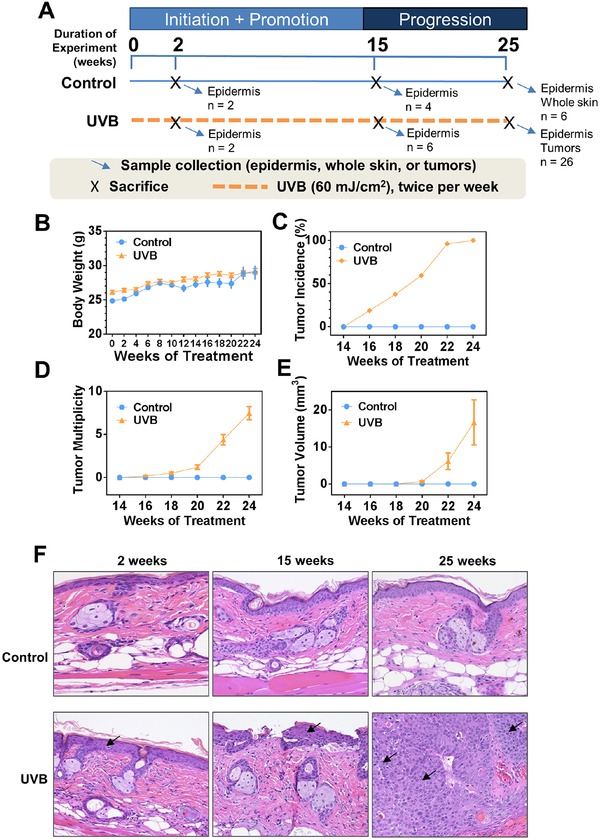
The UV-induced model was generated as previously described (16) and the procedure is presented in Figure 1A. Six weeks old female SKH-1 hairless mice were randomly assigned into two groups and the tattoo of mouse ID number was placed on the tail of each mouse. Starting at the age of eight weeks, mice were applied 60 mJ/cm2 UVB-irradiation twice per week for 25 weeks. Body weight was measured biweekly. The health condition was monitored every three days, especially the skin condition including UVB-induced epidermal hyperplasia, and the actinic keratosis as pre-cancers.
Figure 1. UVB-irradiation induces non-melanoma skin carcinogenesis.
(A) Experimental design of the animal study. Mice were 8 weeks old when study began (week 0). (B) The biweekly recording of body weight during the experiment. (C-E) Measurements of tumor incidence, tumor multiplicity and tumor volume during the animal study. Note that tumor volume was calculated with formula V = (L*W*W)/2 only for tumors that had diameter greater than 2 mm. (F) Histopathological examination of epidermal tissues at ×200 magnification. Arrows indicate epidermis and tumor for early (2 and 15 weeks) and late (25 weeks) stages, respectively.
2.3. Animals and sample preparation
Female SKH-1 hairless mice were purchased from Charles River Laboratories (Wilmington, MA, USA), as described in previous studies (17). Mice were housed at the Rutgers Animal Facility, maintained under 12 hours light and dark cycles, and provided ad libitum access to food and water. These mice were housed in the animal facility for at least one week before experiments. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC; protocol number: PROTO999900171) of Rutgers University. Mice were sacrificed followed by immediately extraction of skin epidermis. For Methyl-Seq experiments, 2 biological replicates of these tissues were used. For subsequent pyrosequencing sequencing validation, 2 identical specimens of these tissues were used. DNA extraction was performed using the DNA extraction kit (QIAGEN Cat. No. 80204) as described in the manufacturer’s protocol and previously (18).
2.4. Histopathological analysis
The histopathological analysis was performed as described previously (19). Tissue blocks were serially sectioned (4 μm) and mounted on glass slides. The sections were stained with hematoxylin and eosin and were carefully evaluated by a histopathologist. Images of H&E stained sections were captured at 200x total magnification.
2.5. Methyl-Seq library preparation
Methyl-seq library preparation was performed using Agilent SureSelect Methyl-Seq kit (Cat. No. G9651A) as described in the manufacturer’s protocol (Methyl-seq protocol, Version X, August 2017). 3 μg of DNA was used in the library preparation. 550 ng of adaptor ligated DNA was used for the hybridization capture and the final concentration of indexed library was 8 to 15 nM. The following changes were made to minimize the loss of DNA in the process of enzymatic reactions. AMPure XP beads (Cat. No. A63880, Beckman Coulter, USA) were incubated with DNA reaction mix for 10 min at room temperature prior to pelleting by magnetization. AMPure XP beads were then washed twice with 80% ethanol and dried at 37°C for 5 min. DNA was dissolved for 10 min at 37°C. AMPure beads were retained in the solution for adenylation and end-repair reaction. An equal volume of binding buffer was added to this reaction mix at 1:1 ratio to enable the AMPure XP beads to rebind to DNA, incubated at room temperature for 10 min. DNA was further purified as described above. Concentration and size of DNA fragments were determined by Agilent Bioanalyzer 2100.
2.6. Bisulfite conversion and next-generation sequencing
Bisulfite conversion was performed using EZ DNA Methylation-Gold kit (Zymo Research, USA) as described in the manufacturer’s protocol. Sequencing was performed on an Illumina HiSeq 2000 platform with 75 bp paired-end reads as described in the manufacturer’s protocol.
2.7. Bioinformatics analyses of SureSelect Methyl-seq
The reads were aligned to the in silico bisulfite-converted mouse genome (mm10) with the Bismark (version 0.15.0) alignment algorithm (20). After alignment, DMRfinder (version 0.1) was used to extract methylation counts and cluster CpG sites into DMRs (21). Each DMR contains at least three CpG sites. Methylation differences greater than 0.10 and with a P value smaller than 0.05 were considered significant. Genomic annotation was performed with ChIPseeker (version 1.10.3) in R (version 3.4.0) (22).
2.8. Bisulfite Pyrosequencing
The bisulfite-treated DNA was amplified by PCR using Platinum PCR Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA) with the forward and reverse primers listed in Supplementary Table 1. Specifically, the reverse primers were biotinylated at the 5’ end. The PCR product was separated by agarose gel electrophoresis and was visualized by ethidium bromide staining using a Gel Documentation 2000 system (Bio-Rad, Hercules, CA, USA) to ensure purity of the PCR products. Later, the biotinylated PCR product was captured using streptavidin-coated beads (GE Healthcare, Piscataway, NJ, USA). After annealing with the sequencing primer at 80°C for 5 min, the single-stranded PCR product was pyrosequenced on a PyroMark Q24 advanced instrument (Qiagen).
2.9. RNA extraction, library preparation, and next-generation sequencing
Total RNA was extracted from snap-frozen skin tissue and/or tumor samples from the control and experimental groups using the AllPrep DND/RNA Mini Kit (Qiagen, Valencia, CA, USA). The quality and quantity of the extracted RNA samples were determined with an Agilent 2100 Bioanalyzer. The library was constructed using the Illumina TruSeq RNA preparation kit (Illumina, San Diego, CA, USA) according to the manufacturer’s manual. Samples were sequenced on the Illumina NextSeq 500 instrument with 75 bp paired-end reads, to a minimum depth of 25 – 30 million reads per sample.
2.10. Computational analyses of RNA-seq data
The reads were aligned to the mouse genome (mm10) with TopHat v2.0.9 (23). Reference gene annotations from UCSC were supplied to TopHat (-G genes.gtf0; otherwise, default parameters were used. The Cufflinks v2.2.1 (24) program cuffdiff was used to calculate expression levels, using the UCSC gene annotations and default parameters, as previously described (25).
2.11. Ingenuity pathway analysis (IPA)
Genes that exhibited a log2 fold change greater than 1 and a false detection rate (FDR) adjusted p value (q value)less than 0.01 were subjected to Ingenuity Pathway Analysis (IPA 4.0, Ingenuity Systems, www.Ingenuity.com). The input genes were mapped to IPA’s knowledge bases, and the relevant biological functions, networks, and pathways related to the treatment of UA were identified.
2.12. Statistical analysis
The data are presented as means ± SD. Comparisons of multiple groups were analyzed using one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test, and simple comparisons between two groups were analyzed using Student’s t-test. Tumor incidence was examined by Fisher’s exact test. Methylation differences were analyzed by Mann-Whitney U test. For qPCR and pyrosequencing data, a P value less than 0.05 was considered statistically significant unless otherwise indicated.
3. Results
3.1. Non-melanoma Skin Carcinogenesis by UVB irradiation
The animal study was carried out according to scheme shown in Figure 1A. We initiated the study with 8-week-old female SKH-1 mice (denoted as week 0). Body weight, tumor incidence, tumor multiplicity, and tumor volume were measured every two weeks. Designated numbers of mice from both groups were sacrificed at three time points: week 2, 15, and 25. Only epidermis were collected for the first two time points while both epidermis and tumor (UVB group) or whole skin (Control group) were collected for the last time point. During the experimental period, we did not observe noticeable body weight loss or sickness in UVB-irradiated mice (Figure 1B). Starting from week 16, noticeable tumors were observed in the UVB group (Figures 1C–D). For tumors with diameter greater than 2 mm, we calculated the volume with equation V = (L*W*W)/2, where V is the tumor volume, L is tumor length, and W is tumor width. The average tumor volume in UVB group reached 17 mm3 at week 24 (Figure 1E). H/E stained slides show that UVB irradiation increased the thickness of epidermis from two layers of cells to 4~6 layers, starting as early as two weeks after UVB exposure (Figure 1F, lower panel, left and middle images). By week 22, UVB exposure greatly increased tumor incidence attaining almost 100%, and with further increased in tumor multiplicity and tumor volume at weeks 25 (Fig 1C–E).
3.2. DNA methylation changes in UVB-irradiation induced non-melanoma carcinogenesis
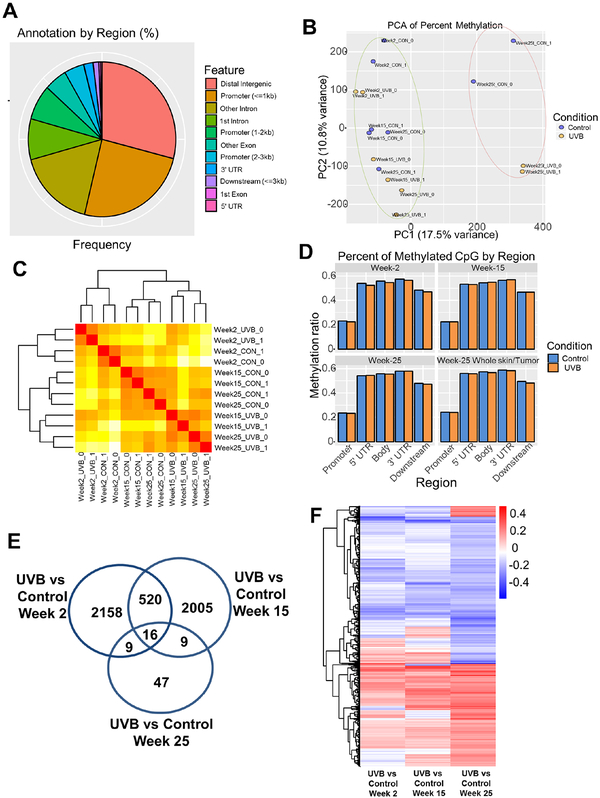
To identify DNA methylation changes in UVB-induced non-melanoma skin carcinogenesis, we performed single base-pair resolution Methyl-seq with DNA samples from all three time points. For week-2 and week-15 groups, DNA was extracted from epidermis while for week-25 group, both epidermis and whole skin (in Control group) and tumor (in UVB group) were used for DNA extraction (Figure S1A). A total of 16 DNA samples (n = 2 per group) were subjected to Agilent SureSelect Mouse Methyl-seq library preparation then sequenced on Illumina Hiseq 2000 platform. Sequencing reads were aligned to in silico C-T converted mouse genome (mm10) and deduplicated. Individual CpG sites were clustered into DMRs according to the default settings in the DMRfinder package (21). Specifically, each DMR has at least three CpG sites and has a maximum length of 500 bp with no more than 100 bp between any two CpG sites. Average methylation ratio for each DMR was calculated as aggregated counts of 5-mC (as C in bisulfite converted sequencing) divided by the aggregated counts of 5-mC and C (as either C or T in bisulfite converted sequencing) for all CpG dinucleotides in that DMR. We then collected DNA methylation data for all 16 samples with a total of 237,904 DMRs. These DMRs were further annotated with gene features using ChIPseeker (v1.14.2). As shown in Figure 2A, most of the DMRs are located in the distal intergenic (> 3 kb upstream transcription start site; TSS or downstream 3’ untranslated region; UTR) regions and the promoters. All these 16 samples were clustered by Euclidean distances of the methylation levels and it shows that DNA methylation of whole skin versus tumor samples at week-25 are clearly separated from the other epidermis samples (Figure S1B). Principal component analysis (PCA) also shows the same finding for these samples (Figure 2B). To further identify the methylation changes by UVB irradiation and/or aging effects, Euclidean distance clustering was performed with epidermis samples. As shown in Figure 2C, irrespective of UVB irradiation, week-2 samples are clustered separately from week-15 and week-25 samples. When comparing all samples at week-15 and week-25, UVB irradiation has a stronger impact on methylation changes than aging effect (Fig 2C, dendrogram). These results suggest that both UVB irradiation and aging effects can alter DNA methylation profiles of mouse epidermal cells. We next compared the DNA methylation level of samples from UVB group versus those samples from Control group. As shown in Figure 2D, no significant methylation difference was observed from these two groups of samples. However, CpG methylation in the promoters was much lower than in other regions for both groups. We next focused on gene promoters (<3 kb) containing DMRs in comparison of UVB versus Control for all three time points, using a cutoff for the methylation ratio difference of greater than or equal to 0.1 and a P-value of less than or equal to 0.05. The comparisons of UVB versus Control at time points of Week-2 and Week-15 showed the greatest differences with 2703 and 2550 DMRs, whereas only 81 DMRs were observed with time point Week-25. The number of common DMRs between these comparisons are shown in a Venn Diagram (Figure 2E). MA plots of methylation change in comparison of UVB versus Control for all three time points are shown in Figure S1C–E. When looking at DMRs for all genomic locations except distal intergenic regions with a methylation cutoff level of 0.1 and a P-value of 0.05, the number of commonly shared DMRs across the three comparisons increased from 16 to 974. The methylation changes of these 974 DMRs for all three comparisons are shown in a heatmap (Figure 2F). About half of the DMRs were hypomethylated (upper half heatmap, blue) by UVB irradiation while the other half were hypermethylated (lower half heatmap, red).
Figure 2. DNA methylation alteration by aging and UVB-irradiation in mouse epidermal cells.
(A) Distribution of annotated differentially methylated regions (DMRs) by gene feature. Each DMR has at least three CpG sites. (B) Principal component analysis (PCA) on methylation profiles of the 16 samples. (C) Dendrogram clustering by Euclidean distance on methylation profiles of 12 epidermis samples. It shows that the samples are first clustered by time point and within each time point, samples are separated by condition. (D) Average methylation levels of DMRs based on gene regions for samples in Control and UVB groups. (E) Venn Diagram showing number of significantly changed DMRs in promoter region in comparison of UVB versus Control for all three time points. The cutoff was 0.1 for methylation ratios and 0.05 for q value. (F) Heatmap showing DMRs in promoter region and gene body with significant changes between UVB and Control groups for all three time points. A total of 974 DMRs were identified with cutoff of methylation ratios at 0.1 and P value at 0.05.
3.3. DNA methylation changes during aging and in different stages of carcinogenesis
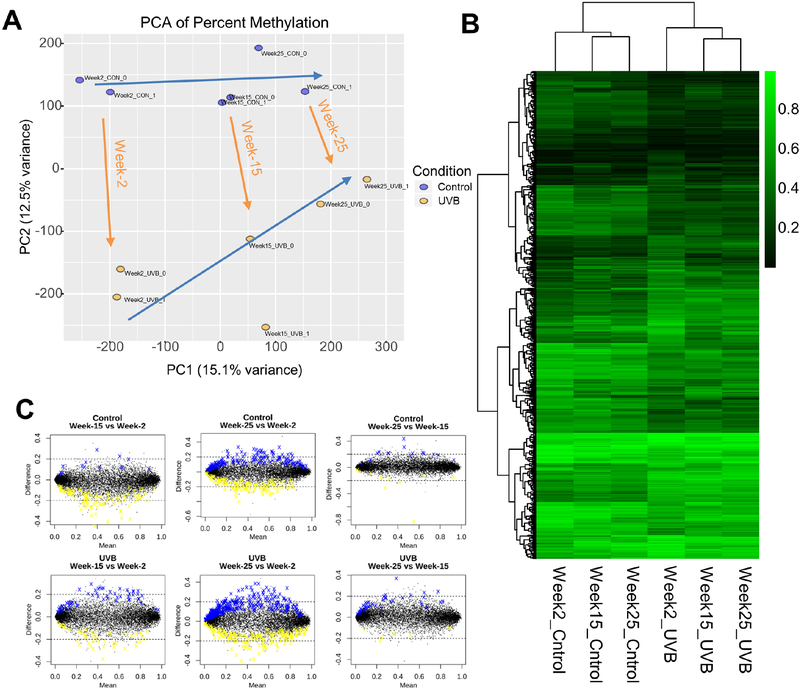
To further dissect the UVB-irradiation induced methylation changes at different stages of non-melanoma skin carcinogenesis, we performed principal component analysis (PCA) on the 12 epidermis samples. As shown in Figure 3A, all six groups are clustered separately with aging effects shifting right (blue arrows) and UVB induced carcinogenesis shifting down (orange arrows). When clustering the 974 DMRs (identified in Section 3.2 above) with Euclidean distance, as shown in the dendrogram in Figure 3B, samples in the Control groups for all three time points are clustered together and are separated from samples in the UVB groups for all three time points. And within these two conditions, Week-15 and Week-25 are clustered together and are separate from Week-2. Using a cutoff of methylation level of 0.1 and a P-value of 0.05, we also compared and contrasted the methylation changes between timepoints for each condition, i.e., Control and UVB. As shown in Figure 3C, substantial number of DMRs are hyper- (blue) or hypo- (yellow) when comparing late stages (Week-15 and Week-25) versus the early stage (Week-2), while only a few DMRs are changed when comparing Week-15 with Week-25. Similar trends are also observed for both the Control and the UVB conditions, which is consistent with the findings in Figure 3B (Dendrogram).
Figure 3. DNA methylation changes at different stages of carcinogenesis.
(A) Principal component analysis (PCA) on the same 12 samples. Two blue arrows showing the shifting trends of age and orange arrows showing the shifting trends of UVB irradiation. Note the two blue arrows approach each other as age progresses. (B) Heatmap showing average methylation ratio of 974 DMRs from Control and UVB conditions for all three time points. These 974 DMRs are the same as identified in Figure 2F. All six groups are from epidermis samples. (C) MA plots showing methylation changes between timepoints for Control condition (upper three plots) and UVB condition (lower three plots).
3.4. Gene expression changes in UVB-irradiation induced non-melanoma carcinogenesis
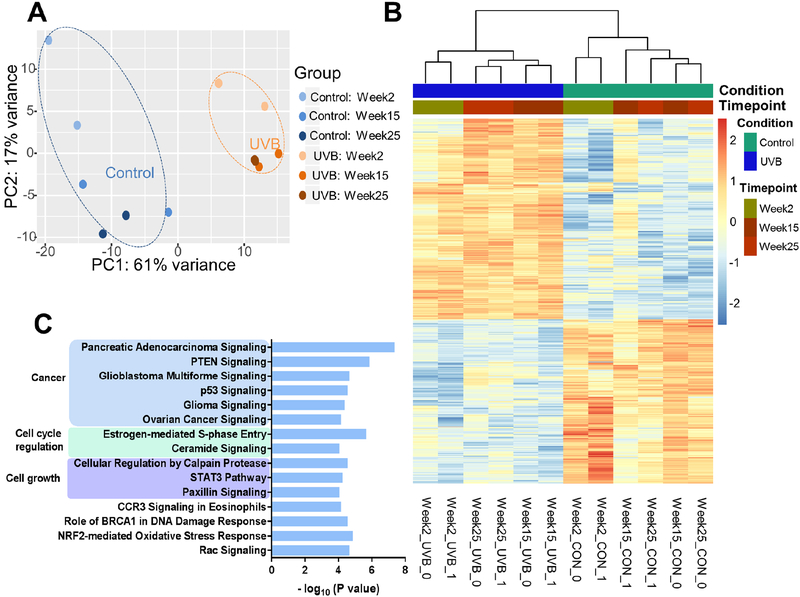
Matching RNA-seq for the samples mentioned above in Methyl-seq was performed (Figure S2A). Principal component analysis (PCA) reveals that, similar to the methyl-seq analysis above, tumor and whole skin samples from week-25 UVB group are clustered separately from the epidermis samples from all time points (Figure S2B). Dendrogram and heatmap for the top 1,000 most regulated genes across all 16 samples show that similar trends of results as shown in PCA plot (Figure S2C). Additionally, the dendrogram shows that the epidermis samples are separated by UVB-exposed condition rather than aging time point. These findings suggest that UVB irradiation dominates over aging effects in the regulation of RNA expression in mouse epidermal cells. Due to the large differences between epidermis samples and tumor or whole skin samples, we next focused on comparing the RNA expression between UVB and Control groups in epidermis samples only. Principal component analysis (PCA) of these 12 samples shows that samples in UVB groups are clustered separately from the samples in the Control groups and the difference between time points in UVB appears to be minimal (Figure 4A). When comparing the RNA expression of UVB groups to Control groups, a list of 2,301 genes was obtained with false discovery rate (FDR) adjusted p value (q value) of less than 0.01, among which 569 genes had at least two-fold change in RNA expression. The relative RNA expression of these genes is shown in a heatmap (Figure 4B). Of these 569 genes, 323 were upregulated and 246 were down-regulated in UVB groups when compared to Control groups. Ingenuity Pathway Analysis (IPA) on these 569 genes identified a list of 56 significantly regulated signaling pathways (p < 0.01, Table S2). The top 15 significant pathways are shown in Figure 4C. These pathways are primarily clustered in three categories: Cancer, Cell cycle regulation, and cell growth. When comparing the RNA expression of week-25 tumor samples (UVB group) with that of week-25 whole skin samples, we obtained a list of 50 significantly (q < 0.01) expressed genes, with 41 of them had at least two-fold change in RNA expression. These genes are listed in Table S3. We also performed IPA analysis with a much looser cutoff (p < 0.05) on input genes and obtained a list of 29 significantly regulated pathways (Table S4).
Figure 4. Gene expression change by UVB-irradiation induced non-melanoma skin carcinogenesis.
(A) Principal component analysis (PCA) on RNA expression of the same 12 samples as mentioned in Figure 2. RNA expression profiles of samples in the Control groups for all time points are discrete (blue oval), whereas all samples in the UVB groups are clustered together (orange oval), suggesting UVB-irradiation has prevailing effects over aging on gene expression in mouse epidermal cells. (B) Dendrogram clustering by Euclidean distance and heatmap showing top 569 regulated genes by UVB-irradiation with cutoff q < 0.01 and log2(Fold Change) > 1 or < −1. The dendrogram shows the samples are first clustered by treatment condition then by time point, which is consistent with the PCA in Figure A. (C) Top 15 regulated pathways that were regulated by UVB-irradiation. Pathways were identified by Ingenuity Pathway Analysis (IPA) with the list of 569 regulated genes in Figure B. These pathways have P values smaller than 0.0001 (shown as – log (P values) on x-axis).
3.5. Correlations between DNA methylation and RNA expression in UVB-irradiation induced non-melanoma skin carcinogenesis
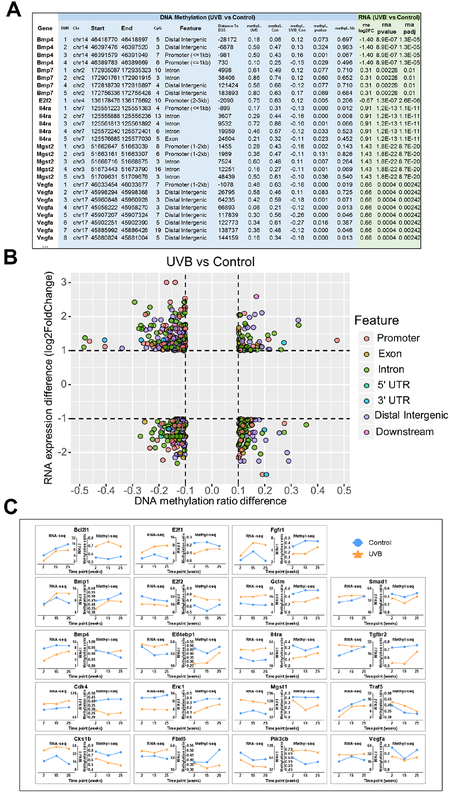
One of the current fundamental biological questions is the correlation or lack of correlation between CpG methylation and RNA expression. Previous DNA methylation analysis and RNA expression analysis showed that UVB-irradiation had profound effects over aging. We next combined DNA methylation profiles with RNA expression profiles in the comparison of UVB vs Control and obtained a list of 6,357 DMRs with corresponding RNA expression data. A representative image of the list is shown in Figure 5A. DNA methylation data are shown on the left columns (in light blue background) that include the DMR positions, gene features, distance to transcription start site (TSS), absolute methylation ratios, methylation differences between the Control and UVB groups, and statistics. RNA expression data are shown on the right columns (in light green background) including log2 fold-change between UVB and Control groups and statistics. When filtering the list by a cutoff of 0.1 for DNA methylation ratio change and a cutoff of two-fold change for RNA expression change, 502 DMRs with changes in both DNA methylation and corresponding RNA expression change were identified. The DNA methylation and RNA expression profiles of these 502 DMRs are shown in Figure 5B. Each dot represents a DMR and their corresponding features are indicated by different colors. These DMRs were separated into two lists with one containing 296 DMRs that have an inverse relationship between DNA methylation and RNA expression and the other one containing 206 DMRs that have no such inverse correlation (Tables S5 and S6). Of these DMRs, we further focused on the DMRs and genes that were involved in the previously identified 56 pathways (Table S1) that were regulated by UVB-irradiation to finally obtain a list of 19 genes. The RNA-seq data and Methylseq data of these genes are shown in Figure 5C. Of these 19 genes, 18 have an inverse relationship between DNA methylation and RNA expression, which is consistent with the dogma of suppressive effect of DNA CpG methylation on gene expression. However, with Enc1, both decreased RNA expression and decreased CpG methylation were observed.
Figure 5. Correlations between DNA methylation and RNA expression.
(A) A representative image of the 6357 DMRs that have corresponding RNA expression data in comparison of UVB vs Control. Gene names are shown in the first column. Each gene has several coupling DMRs as shown in the second column. DMR information including location, number of CpG sites and methylation ratios in Control and UVB groups are shown in blue background and RNA expression data including log2(Fold Change) and statistics are shown in green background. (B) Scatter plot showing 502 DMRs with cutoff 0.1 for DNA methylation and 2-fold change for RNA expression. DMR locations (gene features) are indicated by colors. (C) A list of genes that were involved in the pathway regulations and their methylation and expression profiles. Time points are shown on x-axis. Control and UVB conditions are shown with blue and orange lines, respectively.
3.6. Validation of key genes regulated in UVB-irradiation induced non-melanoma skin carcinogenesis
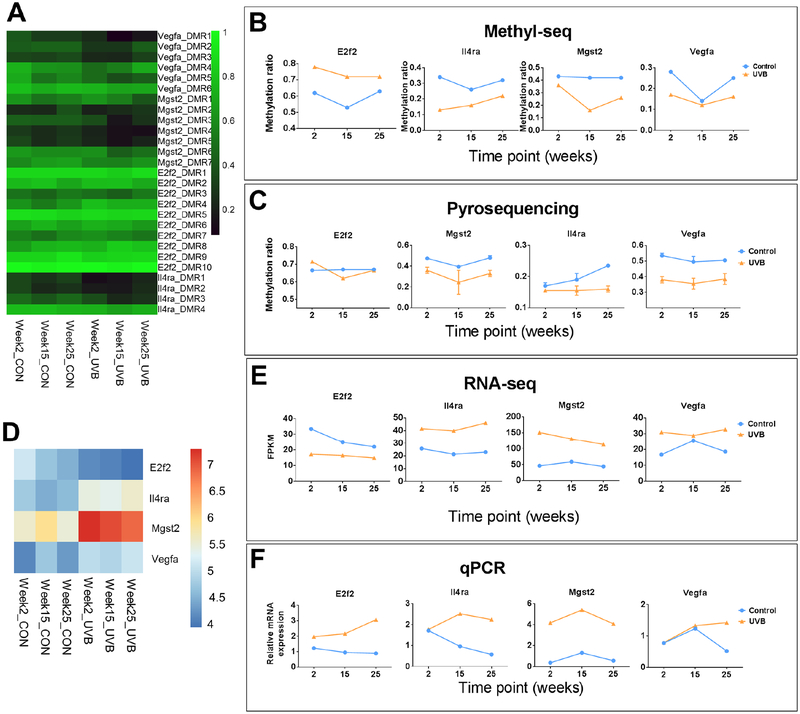
We next performed pyrosequencing and qPCR on E2f2, Il4ra, Mgst2 and Vegfa to validate the Methyl-seq and RNA-seq results. The oligo primers used for validation are listed in Table S1. Each gene has several DMRs and the methylation ratio are shown in the heatmap in Figure 6A. The average methylation ratios of all DMRs for each gene are shown in Figure 6B. Methylation ratios of one DMR in promoter region for each gene are shown in Figure 6C. Due to the limitation that pyrosequencing can only practically cover less than 200 bp of DNA, the selected regions for validation in Figure 6C are not the same as any DMRs in Figure 6A. Methylation ratio of individual CpG sites for these genes are shown in Figure S3. These results show that data from pyrosequencing showed similar methylation changes by UVB irradiation for these four validated genes, although the trends among the three time points are not exactly the same. RNA-seq data for these four validated genes are shown in a heatmap (figure 6D) and the values are plotted in Figure 5E, upper half. qPCR data are shown in Figure 5F. Both RNA-seq data and qPCR data showed similar expression change by UVB irradiation except for the E2f2 gene, where qPCR data were not in line with RNA-seq data. These results suggest that UVB-irradiation induced DNA methylation change and RNA expression change were mostly accurately measured by NGS approaches.
Figure 6. Validation of 4 genes by pyrosequencing and qPCR.
(A) Heatmap showing methylation ratio of the DMRs for these 4 genes from Methyl-seq data. Note each gene has several coupling DMRs, which are shown to the right of the heatmap. (B) Average methylation levels of DMRs for each gene. (C) The met+hylation ratio of one DMR in promoter region for each gene was determined by pyrosequencing. (D) Heatmap showing gene expression from RNA-seq (log2FPKM) of these for genes. These values are plotted in Figure E. Values from qPCR are plotted in Figure F.
4. Discussion
Non-melanoma skin cancers, including basal cell carcinomas (BCCs) and squamous cell carcinomas (SCCs), account for approximately 80% and 16% of all skin cancers, respectively, while malignant melanomas account for only 4% of all skin cancers (26). BCCs and SCCs develop primarily on sun-exposed areas of the body such as the head and neck. The process of non-melanoma carcinogenesis is generally divided into three stages — initiation, promotion and progression. The initiation process involves generation of reactive oxygen species and UVB-irradiation induced DNA damage, which can further introduce mutations to genome, including tumor-suppressor genes such as Trp53. Both UVA and UVB irradiation can impact on mouse skin models as a complete carcinogen. UVB light has been shown to initiate the benign tumors called papillomas in mouse-skin studies (27,28).
The present study provides unique new insights into the alterations induced by UVB-irradiation for both DNA Methylation and gene expression and more specifically, how different stages of DNA CpG methylation could regulate gene expression. We observed skin tumor development as early as 16 weeks after the first dose of UVB irradiation, which is consistent with findings reported by other investigators (29). In this study, we got a list of 502 DMRs that had at least 0.1 (10%) in methylation ratio change and 2-fold change in RNA expression after UVB irradiation. About 60% of these DMRs showed inversive relationship between DNA methylation and RNA expression, i.e., hypermethylation coupled with suppression of transcription or hypomethylation coupled with promotion of transcription, while the other 40% of these DMRs did not show such relationship. we have identified 19 genes with altered CpG methylation in the different stages of UVB-induced skin carcinogenesis’s progression and 18 of these showed inverse relationship between DNA methylation and RNA expression (Figure 5C). For instance, cyclin-dependent kinase 4 (Cdk4), a gene involved in cell cycle regulation (30), was up-regulated by UVB irradiation and was also CpG demethylated. Ming et al, reported that PTEN pathway was involved in the survival of epidermal keratinocytes upon UVB irradiation (31), here we also found that a number of genes in the PTEN pathway were regulated by UVB irradiation and many of them, including Tgfbr2, Fgfr1, Bcl2l1, and Pik3cb had inverse DNA methylation-RNA expression patterns, which would be a new discovery for the first time in an in vivo skin cancer model. While UVB irradiation could induce generation of ROS, Nrf2-mediated oxidative stress response was also activated by UVB for removal of excessive ROS. Specifically, Nrf2-mediated anti-oxidative stress genes Gclm, Mgst2, Fgfr1, and Pik3cb were regulated by UVB irradiation, implicating potential regulation by DNA methylation in the promoter regions of these genes. Lahtz et al. reported that UVB irradiation does not directly induce detectable changes of DNA methylation in human keratinocytes (32), the discrepancy between their study and our current study could be due to different responses between animal and cell line models and/or different coverage of CpG sites in these studies. As the Agilent SureSelect Kit targets about 3 million CpG sites in the mouse genome and we only identified about 1% of that CpG sites/DMRs (2,703 DMRs in Figures 2E versus 237,904 DMRs in Section 3.2) were differently methylated by UVB irradiation, the microarray system, which targeted the 3KB promoter region of 27,728 RefSeq genes, could miss most of the DNA CpG sites and islands that were regulated by UVB irradiation in Lahtz’s study.
In general, UVB irradiation can trigger the activation of several pathways/genes including Tumor Necrosis Factor Receptor (Tnfr), Epidermal Growth Factor Receptor (EGFR) and mitogen-activated protein kinases (MAPKs) as a response to UVB-induced ROS and or DNA damage. Kim et al, reported that matrix metalloproteinase genes were activated in human keratinocyte cells (33,34), however, we did not observe such inductions of these genes in our current in vivo study. The discrepancy could be due in part to different responses between human and mouse cells and/or difference between the in vivo and in vitro cell culture model systems. Chitsazzadeh et al. performed a similar animal study and found that matrix metalloproteinase genes were overexpressed in UV irradiation-induced cutaneous squamous cell carcinoma samples (35). However, the transcriptomic analysis in that study was performed with whole skin and tumor samples, which are different from (mainly) epidermis samples in our study. One of the advantages in Chitsazzadeh’s study is that mutations were detected in tumor suppressor genes such as p53. Similar approaches could be utilized in our future studies on UVB irradiation and skin cancer.
In summary, we have utilized the latest Methyl-seq and RNA-seq approaches to dissect the epigenomic CpG methylation changes and gene expression changes in different stages of UVB-induced non-melanoma skin carcinogenesis. DNA methylation was altered by both UVB irradiation and aging, while RNA expression was mainly affected by UVB irradiation. RNA-seq data revealed a list of regulated pathways by UVB irradiation and the top regulated pathways were cancer, cell cycle regulation and cell growth. A list of key genes involved in cancer and cell cycle regulations were also identified with the concomitant epigenetic CpG modifications of these genes’ promoters and or gene body by UVB irradiation. Taken together, our current findings could benefit future studies in targeting these genes for prevention and treatment of skin cancers, focusing on the epigenetically regulated genes and pathways.
Supplementary Material
Highlights.
DNA methylation was altered by both UVB irradiation and aging, while RNA expression was mainly affected by UVB irradiation.
A list of key genes involved in UVB irradiation induced carcinogenesis were identified with concomitant DNA CpG methylations.
This study provides novel profiles of CpG methylation and transcriptomic changes at different stages of UVB irradiation induced skin cancer and offers potential targets for prevention/treatment of human skin cancer.
Acknowledgements
We thank all members of Dr. Ah-Ng Tony Kong’s lab for helpful discussions and preparation of this manuscript.
Funding
This study was supported in part by R01 CA200129, from the National Cancer Institute to Dr. Ah-Ng Tony Kong.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes
Transcript sequencing data and Bisulfite methyl sequencing data have been deposited under Gene Expression Omnibus (GEO) accession GSE122227.
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Katiyar SK, Matsui MS, Mukhtar H. Kinetics of UV light-induced cyclobutane pyrimidine dimers in human skin in vivo: an immunohistochemical analysis of both epidermis and dermis. Photochemistry and photobiology 2000;72:788–93 [DOI] [PubMed] [Google Scholar]
- 2.Mukhtar H, Elmets CA. Photocarcinogenesis: mechanisms, models and human health implications. Photochemistry and photobiology 1996;63:356–7 [DOI] [PubMed] [Google Scholar]
- 3.Nakayama M, Gonzalgo ML, Yegnasubramanian S, Lin X, De Marzo AM, Nelson WG. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J Cell Biochem 2004;91:540–52 [DOI] [PubMed] [Google Scholar]
- 4.Nelson WG, Yegnasubramanian S, Agoston AT, Bastian PJ, Lee BH, Nakayama M, et al. Abnormal DNA methylation, epigenetics, and prostate cancer. Front Biosci 2007;12:4254–66 [DOI] [PubMed] [Google Scholar]
- 5.Nelson WG, De Marzo AM, Yegnasubramanian S. Epigenetic alterations in human prostate cancers. Endocrinology 2009;150:3991–4002. Epub 2009 Jun 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeronimo C, Henrique R. Epigenetic biomarkers in urological tumors: A systematic review. Cancer letters 2011. [DOI] [PubMed] [Google Scholar]
- 7.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev 2012;22:50–5 [DOI] [PubMed] [Google Scholar]
- 8.Perez RF, Tejedor JR, Bayon GF, Fernandez AF, Fraga MF. Distinct chromatin signatures of DNA hypomethylation in aging and cancer. Aging Cell 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung KL, Lee JH, Khor TO, Wu TY, Li GX, Chan J, et al. Nrf2 knockout enhances intestinal tumorigenesis in Apc(min/+) mice due to attenuation of anti-oxidative stress pathway while potentiates inflammation. Mol Carcinog 2014;53:77–84 [DOI] [PubMed] [Google Scholar]
- 10.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 2010;28:1097–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, Strbenac D, et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 2011;6:34–44 [DOI] [PubMed] [Google Scholar]
- 12.Guan C, Barron AB, He XJ, Wang ZL, Yan WY, Zeng ZJ. A comparison of digital gene expression profiling and methyl DNA immunoprecipitation as methods for gene discovery in honeybee (Apis mellifera) behavioural genomic analyses. PLoS One 2013;8:e73628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lou YR, Peng QY, Li T, Medvecky CM, Lin Y, Shih WJ, et al. Effects of high-fat diets rich in either omega-3 or omega-6 fatty acids on UVB-induced skin carcinogenesis in SKH-1 mice. Carcinogenesis 2011;32:1078–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu YP, Lou YR, Yen P, Mitchell D, Huang MT, Conney AH. Time course for early adaptive responses to ultraviolet B light in the epidermis of SKH-1 mice. Cancer Res 1999;59:4591–602 [PubMed] [Google Scholar]
- 15.Saw CL, Huang MT, Liu Y, Khor TO, Conney AH, Kong AN. Impact of Nrf2 on UVB-induced skin inflammation/photoprotection and photoprotective effect of sulforaphane. Mol Carcinog 2011;50:479–86 [DOI] [PubMed] [Google Scholar]
- 16.Lu YP, Lou YR, Xie JG, Peng QY, Liao J, Yang CS, et al. Topical applications of caffeine or (−)-epigallocatechin gallate (EGCG) inhibit carcinogenesis and selectively increase apoptosis in UVB-induced skin tumors in mice. Proc Natl Acad Sci U S A 2002;99:12455–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu YP, Lou YR, Peng QY, Nghiem P, Conney AH. Caffeine decreases phospho-Chk1 (Ser317) and increases mitotic cells with cyclin B1 and caspase 3 in tumors from UVB-treated mice. Cancer prevention research 2011;4:1118–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, Wu R, Gaspar JM, Sargsyan D, Su ZY, Zhang C, et al. DNA Methylome and Transcriptome Alterations and Cancer Prevention by Curcumin in Colitis-accelerated Colon Cancer in Mice. Carcinogenesis 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Liu Y, Zhang C, Su ZY, Li W, Huang MT, et al. The epigenetic effects of aspirin: the modification of histone H3 lysine 27 acetylation in the prevention of colon carcinogenesis in azoxymethane- and dextran sulfate sodium-treated CF-1 mice. Carcinogenesis 2016;37:616–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaspar JM, Hart RP. DMRfinder: efficiently identifying differentially methylated regions from MethylC-seq data. BMC Bioinformatics 2017;18:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015;31:2382–3 [DOI] [PubMed] [Google Scholar]
- 23.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Y, Su ZY, Zhang C, Gaspar JM, Wang R, Hart RP, et al. Mechanisms of colitis-accelerated colon carcinogenesis and its prevention with the combination of aspirin and curcumin: Transcriptomic analysis using RNA-seq. Biochem Pharmacol 2017;135:22–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowden GT. Prevention of non-melanoma skin cancer by targeting ultraviolet-B-light signalling. Nat Rev Cancer 2004;4:23–35 [DOI] [PubMed] [Google Scholar]
- 27.Husain Z, Pathak MA, Flotte T, Wick MM. Role of ultraviolet radiation in the induction of melanocytic tumors in hairless mice following 7,12-dimethylbenz(a)anthracene application and ultraviolet irradiation. Cancer Res 1991;51:4964–70 [PubMed] [Google Scholar]
- 28.Uberoi A, Yoshida S, Frazer IH, Pitot HC, Lambert PF. Role of Ultraviolet Radiation in Papillomavirus-Induced Disease. PLoS Pathog 2016;12:e1005664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bair WB 3rd, Hart N, Einspahr J, Liu G, Dong Z, Alberts D, et al. Inhibitory effects of sodium salicylate and acetylsalicylic acid on UVB-induced mouse skin carcinogenesis. Cancer Epidemiol Biomarkers Prev 2002;11:1645–52 [PubMed] [Google Scholar]
- 30.Rane SG, Dubus P, Mettus RV, Galbreath EJ, Boden G, Reddy EP, et al. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat Genet 1999;22:44–52 [DOI] [PubMed] [Google Scholar]
- 31.Ming M, Han W, Maddox J, Soltani K, Shea CR, Freeman DM, et al. UVB-induced ERK/AKT-dependent PTEN suppression promotes survival of epidermal keratinocytes. Oncogene 2010;29:492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lahtz C, Kim SI, Bates SE, Li AX, Wu X, Pfeifer GP. UVB irradiation does not directly induce detectable changes of DNA methylation in human keratinocytes. F1000Res 2013;2:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim C, Ryu HC, Kim JH. Low-dose UVB irradiation stimulates matrix metalloproteinase-1 expression via a BLT2-linked pathway in HaCaT cells. Exp Mol Med 2010;42:833–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ujfaludi Z, Tuzesi A, Majoros H, Rothler B, Pankotai T, Boros IM. Coordinated activation of a cluster of MMP genes in response to UVB radiation. Sci Rep 2018;8:2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chitsazzadeh V, Coarfa C, Drummond JA, Nguyen T, Joseph A, Chilukuri S, et al. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun 2016;7:12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.