Abstract
The dopamine transporter (DAT) is a transmembrane protein that terminates dopamine signaling in the brain by driving rapid dopamine reuptake into presynaptic nerve terminals. Several lines of evidence indicate that DAT dysfunction is linked to neuropsychiatric disorders such as attention-deficit/hyperactivity disorder (ADHD), bipolar disorder (BPD), and autism spectrum disorder (ASD). Indeed, individuals with these disorders have been found to express the rare, functional DAT coding variant Val559, which confers anomalous dopamine efflux (ADE) in vitro and in vivo. To elucidate the impact of the DAT Val559 variant on membrane diffusion dynamics, we implemented our antagonist-conjugated quantum dot (QD) labeling approach to monitor the lateral mobility of single particle-labeled transporters in transfected HEK-293 and SK-N-MC cells. Our results demonstrate significantly higher diffusion coefficients of DAT Val559 compared to those of DAT Ala559, effects likely determined by elevated N-terminal transporter phosphorylation. We also provide pharmacological evidence that PKCβ- mediated signaling supports enhanced DAT Val559 membrane diffusion rates. Additionally, our results are complimented with diffusion rates of phosphomimicked and phosphorylation-occluded DAT variants. Furthermore, we show DAT Val559 has a lower propensity for membrane clustering, which may be caused by a mutation-derived shift out of membrane microdomains leading to faster lateral membrane diffusion rates. These findings further demonstrate a functional impact of DAT Val559 and suggest that changes in transporter localization and lateral mobility may sustain ADE and contribute to alterations in dopamine signaling underlying multiple neuropsychiatric disorders.
For Table of Contents Use Only
Introduction
The catecholamine neurotransmitter dopamine is central to the modulation of neuronal pathways that control diverse behaviors including those linked to movement, reward, mood, attention, and cognition.(1,2) Disrupted dopamine signaling is associated with multiple brain disorders such as Parkinson’s disease (PD), schizophrenia, bipolar disorder (BPD), attention-deficit/hyperactive disorder (ADHD), and addiction.(3–6) The presynaptic Na+/Cl- coupled dopamine transporter (DAT) determines dopamine signaling amplitude and duration by actively clearing synaptic dopamine following vesicular release.(7–9) Importantly, genetic polymorphisms of the human DAT gene (DAT 1, SLC6A3) have been identified in cases of ADHD, BPD, autism spectrum disorder (ASD), PD, and juvenile dystonia.(10–15) DAT endocytic trafficking at presynaptic terminals is likely a major regulatory mode of synaptic strength in dopamine neurons,(16–19) a process that can be referred to as vesicle trafficking, wherein DAT proteins are moved into and out of the plasma membrane from intracellular compartments. Consequently, constitutive and regulated vesicle trafficking is considered to be the principal determinant of functional DAT availability, though engagement of these mechanisms appears to be region dependent.(20,21) DAT proteins can also engage cell surface trafficking or lateral membrane diffusion that can be impacted by DAT-associated proteins and disease-associated mutations revealed by total internal reflection fluorescence (TIRF) microscopy,(22,23) fluorescence recovery after photobleaching (FRAP),(24) and single particle tracking (SPT) techniques.(25,26)
Single molecule imaging offers information such as kinetics and dynamics of molecules in real-time, which would be lost in conventional ensemble measurements. The study of lateral diffusion at a single protein level requires an approach that uses bright probes, such as quantum dots (QD), to achieve signal-to-noise ratios suitable for high spatiotemporal resolution. QDs exhibit unique photophysical properties that make them an attractive first choice for single molecule imaging applications,(27) which many groups have employed to investigate the diffusion dynamics of single transmembrane, neurotransmitter receptors, and transporter proteins (e.g., GABA receptors, glycine receptors, serotonin transporters, DAT).(26,28–32) First, QDs offer a prolonged photostability required for imaging acquisition times on the order of minutes.(27) Second, QDs have broad absorption spectra and size-tunable narrow Gaussian photoluminescence profiles that permit simultaneous multicolor tracking with little to no spectral bleedthrough. As a product of high quantum yields and large absorption cross sections, QDs are also very bright upon laser irradiation.(33) Together, these properties have enabled the detection of single proteins in living cells targeted by antagonist- and antibody-conjugated biocompatible QDs.(27,34) Because surface trafficking is believed to be a critical posttranslational regulatory mechanism,(35–37) our group developed an antagonist-conjugated QD labeling approach to monitor individual membrane proteins in live cells (Figure 1).(38–41) Subsequently, we reported that the ADHD-associated DAT Cys615 coding variant exhibited significantly increased membrane mobility and a pronounced lack of dynamic response to lipid raft disruption and amphetamine (AMPH) stimulation.(26)
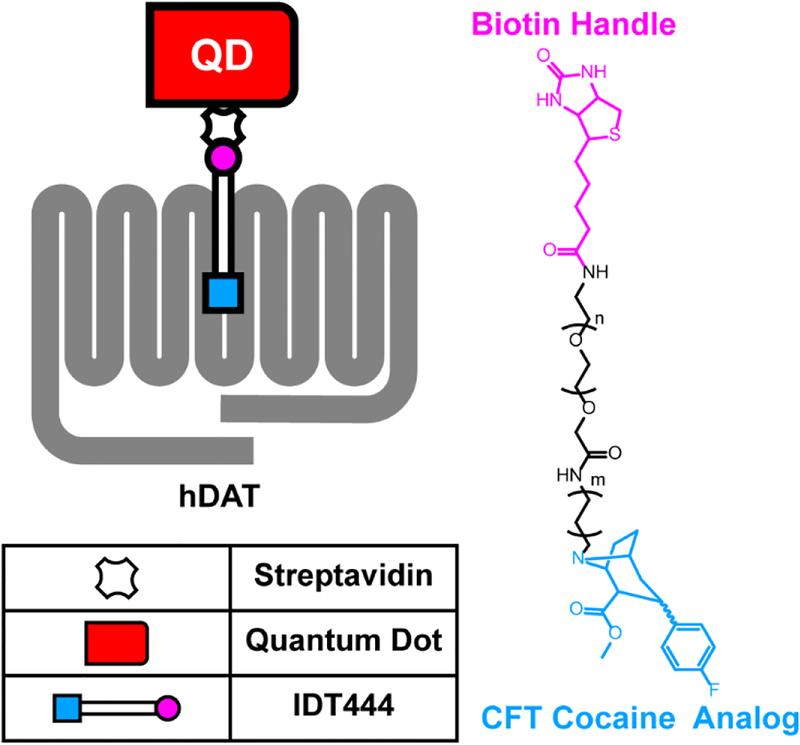
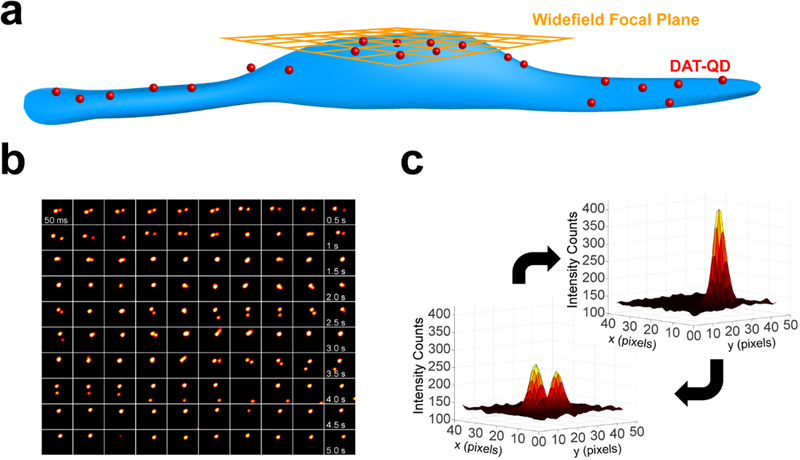
Figure 1: Schematic outlining single QD-DAT labeling architecture and chemical structure of the DAT-specific IDT444 affinity tool.
Along with Cys615 as one of the multiple genetic DAT variants, a second variant, Val559, has been identified in subjects with distinct disorders associated with dopamine signaling dysfunction. The Val559 mutation was first identified in a female proband presenting with BPD(11) followed by its detection in two brothers with ADHD(10) and subsequently in two unrelated adolescent males with ASD.(42) Studies with mice expressing the DAT Val559 variant demonstrate elevated extracellular dopamine levels, altered biochemical and behavioral responses to psychostimulants, and changes in behaviors linked to reward and impulsivity circuits.(43,44) In reduced preparations, the mutant transporter displays multiple, striking phenotypes. Mazei-Robison and colleagues demonstrated that the Val559 mutation, though not impacting DAT surface expression or dopamine uptake, induces anomalous dopamine efflux (ADE), whereby mutant transporters spontaneously move dopamine from the cytosol to the extracellular space.(45) DAT Val559 also demonstrates elevated levels of N-terminal phosphorylation at distal Ser residues.(42) Mutation of these sites eliminates ADE, suggesting that N- terminal phosphorylation plays an essential role in sustaining dopamine reverse transport. Whether ADE is induced directly by transporter phosphorylation or is a consequence of changes in membrane distribution, lateral membrane trafficking, and/or the spatiotemporal organization of DAT with membrane partners is unclear. Here, we implemented our dynamic QD-based DAT monitoring approach to examine the impact of the DAT Val559 mutation on DAT membrane diffusion dynamics. We demonstrate that DAT exhibits increased lateral mobility in transiently transfected HEK-293 and SK-N-MC cells, movements that are also sensitive to mutations and pharmacological approaches impacting DAT phosphorylation. Using tracking and localization microscopy (TALM) and an intensity-based clustering analysis we developed, we demonstrate that the mutant targets to surface membrane clusters of HEK-293 cells to a lesser degree than the wild- type transporter. Our findings support the idea that disruption of normal DAT spatiotemporal organization may impose elevated risk for neuropsychiatric disorders linked to perturbed dopamine signaling.
Results and Discussion
Single QD Tracking Analysis Reveals DAT Val559 Has Aberrant Membrane Diffusion Dynamics
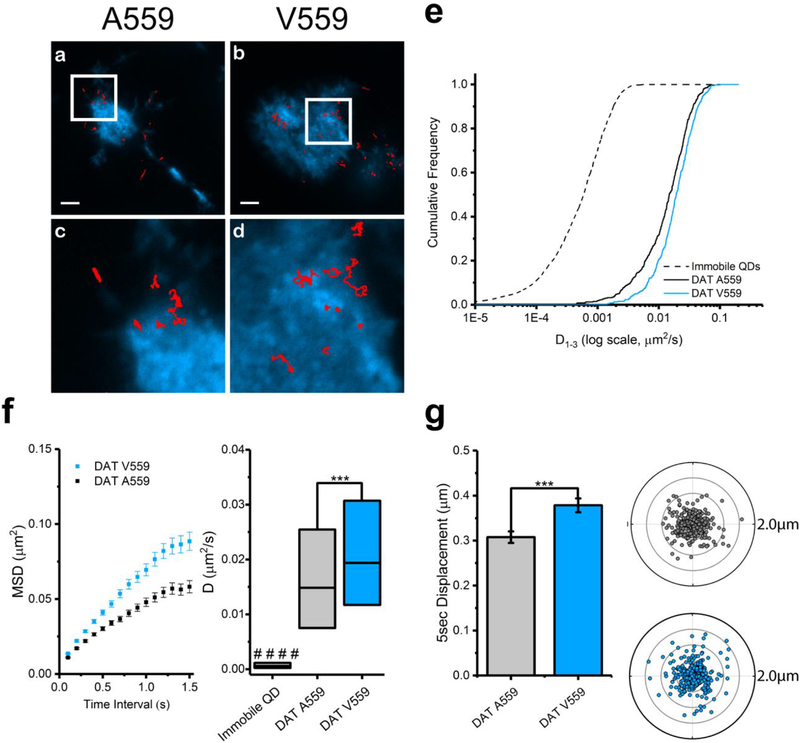
DAT Val559 has been reported to display altered vesicle trafficking in vitro and in vivo.(21,42) To assess the impact of the DAT Val559 mutation on lateral membrane trafficking, we targeted DATs with DAT antagonist-conjugated QDs.(40) Figure 1 illustrates the chemical structure of IDT444, the DAT-specific ligand used in our labeling paradigm, which makes use of the high-affinity carbomethoxy-fluorophenyl cocaine analog β-CFT (also known as WIN 35,428), an 11-carbon alkyl spacer to allow the antagonist to access its DAT binding site, and a PEG linker connected to a biotin molecule to provide for streptavidin-mediated QD binding.(40) This QD-IDT444 labeling strategy has proven successful in single QD tracking of DAT proteins.(26,40) At concentrations of 100 nM IDT444 and <0.1 nM QD, we previously demonstrated that nonspecific binding was virtually eliminated.(26,40) Because DAT Val559 is similar to wild-type DAT Ala559 in both β-CFT binding affinity and cocaine inhibition of [3H]-DA uptake,(46,47) we expect IDT444 labeling to be comparable across cells expressing DAT Ala559 or Val559. Because dopamine D2 receptors (D2Rs) physically associate with DAT and promote DAT Val559-induced ADE,(48–50) we pursued studies of multiple DAT variants (e.g., A559V, YFPDAT, YFPDAT S/D, DAT S53A, DAT S53D, A559V + S53A, A559V + S53D) in HEK- 293 cells that endogenously express D2Rs.(50) QD-labeled human DAT Ala559 and human DAT Val559 were studied in live, transiently transfected HEK-293 cells and imaged using TIRF microscopy at 10 hz (see Movie S1 for a representative time series). Representative trajectories for both DAT Ala559 and DAT Val559 coding variants are shown in Figure 2a and b. IDT307 (4-(4-dimethylamino)phenyl-1-methylpyridinium, APP+),(51,52) a DAT-specific fluorescent analog of 1-methyl-4-phenylpyridinium (MPP+), was used to outline the cell boundaries (Figure 2a–d). Diffusion coefficients (D1–3, see Materials and Methods for details on calculations and analysis) were determined for populations of DAT Ala559 and DAT Val559 trajectories. Cumulative probability distributions (Figure 2e) of D1–3 values reveal a significant increase in DAT Val559 diffusion rates under basal conditions as compared to DAT Ala559. Averaged mean square displacement (MSD) curves show confined motion for both DAT Ala559 and DAT Val559, though the Val559 slope is significantly larger than the wild-type. DAT Val559 D1–3 interquartile ranges (25–75%) are also significantly greater than DAT Ala559 (Figure 2f). In addition to diffusion coefficients, 5 s radial displacement vectors were calculated by obtaining QD particle distance and direction traveled in 5 s (50 frames at 10 Hz) normalized to the particle position at the first frame (see Materials and Methods). Averaged 5 s displacements for DAT Ala559 and DAT Val559 under basal conditions proved to be significantly different (Figure 2g). To increase the physiological relevance of our studies, we repeated our experiments using catecholaminergic neuroblastoma cells (SK-N-MC) derived from the human brain. Results in Figure S1 demonstrate findings similar to those in our HEK-293 studies using SK-N-MC cells expressing DAT Ala559 and DAT Val559.
Figure 2. DAT Val559 exhibits faster mobility compared to that of DAT Ala559 transiently expressed in HEK-293 cells.
(a,b) Representative trajectories collected over 60 s of QD-bound DAT and DAT Val559 coding variants superimposed to the IDT307 channel (scale bar = 5 μm). (c,d) Images at 4× magnification of images in a and b, respectively. (e) Cumulative frequency distributions of diffusion coefficients (D1–3) of immobile QDs, DAT Ala559, and DAT Val559 (Kolmogorov–Smirnov 2-sample test, p < 0.0001). (f) Averaged mean square displacement (MSD) plots (mean ± S.E.M.) and diffusion coefficient box plots (median, 25% and 75% interquartiles, one-way ANOVA followed by Bonferroni’s multiple comparison test, ***p < 0.001, ####p < 0.001 comparing data sets to immobilized QDs as control) of trajectories analyzed for DAT Ala559 and DAT Val559. (g) A 5 s displacement bar graph (mean ± S.E.M., unpaired Student’s t test, ***p < 0.001) and polar plots (outer radius limit = 2 μm) of single DAT Ala559 (gray) and DAT Val559 (blue). DAT displacements are normalized to their spatial origin. N (trajectories) are provided in Table S1.
DAT Val559 Membrane Mobility Is Insensitive to PMA-Triggered PKC Activation but Can Be Diminished by PKCβ Inhibition
DAT-mediated dopamine efflux and DAT surface density are impacted by protein kinase C (PKC) signaling.(5) To examine whether faster DAT Val559 mobility is associated with transporter phosphorylation status, we examined the effects of general PKC activation on DAT Ala559 and DAT Val559 mobility using phorbol-12-myristate-13- acetate (PMA), a diester that binds to the catalytic C1 domain of PKC, which leads to stimulation of PKC activity. HEK-293 cells were preincubated with 100 nM PMA for 30 min prior to QD labeling. PKC activation induced an increase in DAT Ala559 D1–3 and 5 s displacements. However, these effects of PMA were not observed for DAT Val559 (Figure 3b, c). Similar results to those observed in HEK-293 cells were obtained with transfected SK-N-MC cells, demonstrating consistency across cell lines (Figure S2).
Figure 3. DAT Val559 diffusion is unresponsive to protein kinase C (PKC) activation and is attenuated by PKCβ inhibition in HEK-293 cells.
(a) Schematic of DAT with PKC activation by Pm A highlighted in blue and PKCβ inhibition by enzastaurin (Enz) highlighted in red. The dashed line denotes the general region of phosphorylation mediated by PKC activation. (b) Diffusion coefficient box plot (median, 25% and 75% interquantiles, one-way ANOVA followed by Bonferroni’s multiple comparison test, NS p > 0.05, ***p < 0.001) of trajectories analyzed for DAT Ala559 and DAT Val559 under basal, stimulated (+PMA) conditions, and inhibited (+Enz) conditions. (c) A 5 s displacement bar graph (mean ± S.E.M., unpaired Student’s t test, NS p> 0.05 *p < 0.05, **p < 0.01, ***p < 0.001) of trajectories analyzed for DAT Ala559 and DAT Val559 under basal, stimulated (+PMA), and inhibited (+Enz) conditions. N (trajectories) are provided in Table S1.
PMA activates multiple PKC isoforms that can regulate DAT.(5) Because PKCβ regulates dopamine efflux,(53) PKCβ activity is elevated in DAT Val559 expressing HEK- 293 cells,(42) and antagonism of PKCβ restores AMPH-induced DAT internalization,(54) we focused further studies on this PKC isoform. Specifically, we tested the ability of the PKCβ-specific inhibitor enzastaurin, previously shown to attenuate dopamine efflux in vivo,(55) to determine whether PKCβ activity supports enhanced DAT Val559 membrane diffusion. DAT Ala559 and DAT Val559 transfected cells were preincubated with 1 μM enzastaurin for 30 min prior to PMA treatment and QD labeling. Both DAT and DAT Val559 responded to PKCβ inhibition by reduced D1–3 and reduced 5 s displacements (Figure 3b, c). Similar results were obtained using SK-N-MC cells as well (Figure S2). These findings indicate that PKCβ tone exists in our cell models that supports basal DAT Ala559 and DAT Val559 lateral mobility and that PMA can further enhance wild-type but not mutant DAT lateral mobility in a PKCβ-dependent manner. Our findings are also consistent with a model whereby the elevated PKCβ activity reported in DAT Val559 transfected cells leads to elevated DAT phosphorylation and increased lateral mobility,(42) effects mimicked by treating DAT Ala559 expressing cells with PMA.
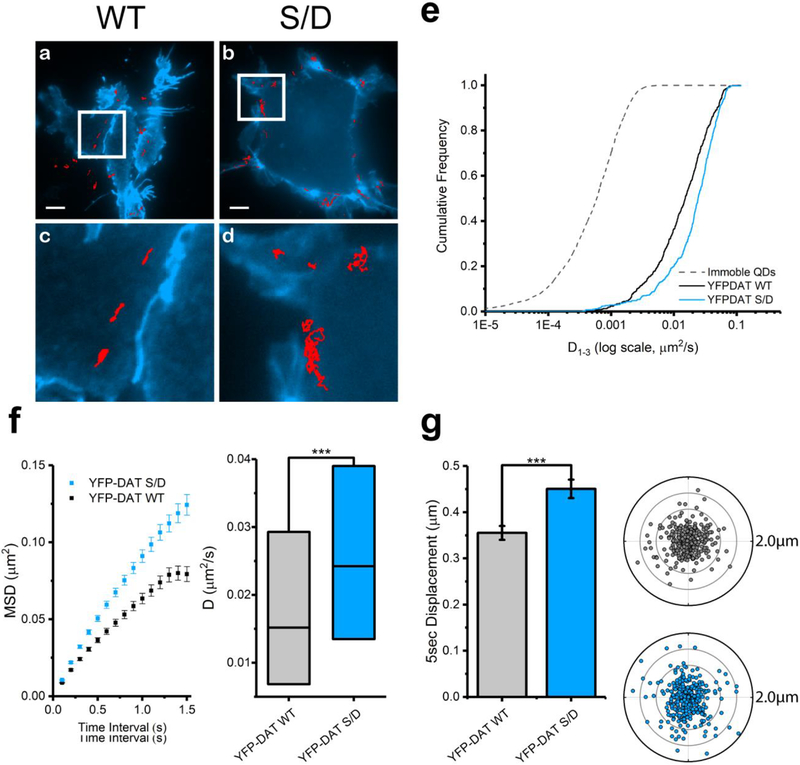
DAT Val559 has been shown to exhibit elevated phosphorylation of multiple N- terminal Ser residues, which is essential for DAT Val559 ADE.(50) We hypothesized that elevated Ser phosphorylation of the DAT Val559 N-terminus might also be involved in the enhanced lateral mobility of this variant. To test this idea, we evaluated the impact of phosphomimetic mutations of the N-termimal Ser residues on lateral mobility using our antagonist-coupled QD approach. Here, Ser residues were mutated to Asp residues (S/D), which are negatively charged at pH 7, thus mimicking a phosphorylated state. We chose to use YFP-DAT and YFP-DAT S/D available for purchase from Addgene (see Materials and Methods) considering GFP and YFP moieties have been reported to induce no adverse effects on DAT function.(56,57) Representative trajectories of QD-labeled YFP-DAT S/D demonstrate a greater area explored compared to QD-labeled YFP-DAT (Figure 4a–d). In Figure 4e, the cumulative probability distribution plot of D1–3 clearly demonstrates an increase in YFP-DAT S/D diffusion rate compared to that of YFP-DAT. Complementing the mobility of DAT Val559, YFP-DAT S/D has elevated D1–3 and 5 s displacements compared to those of YFP-DAT. Results using SK-N-MC are in agreement with these data (Figure S3).
Figure 4. Phosphomimetic YFP-DAT S/D exhibits faster membrane mobility than that of wild-type YFP-DAT in HEK-293 cells.
(a,b) Representative trajectories collected over 60 s of QD-bound YFP-DAT WT and YFP-DAT S/D superimposed to cell membranes outlined by YFP (scale bar = 5 μm). (c,d) Images at 4× magnification of images in a and b, respectively. (e) Cumulative frequency distributions of diffusion coefficients (D1–3) of immobile QDs, YFP-DAT WT, and YFP-DAT S/D (Kolmogorov– Smirnov 2-sample test, p< 0.0001). (f) Averaged mean square displacement (MSD) plots (mean ± S.E.M.) and a diffusion coefficient box plot (median, 25% and 75% interquantiles, one-way ANOVA followed by Bonferroni’s multiple comparison test, ***p < 0.001) of trajectories analyzed for YFP-DAT WT and YFP-DAT S/D. (g) A 5 s displacement bar graph (mean ± S.E.M., unpaired Student’s t test, ***p < 0.001) and polar plots (outer radius limit = 2 μm) of single YFP-DAT WT (gray) and YFP-DAT S/D (blue). DAT-QD displacements are normalized to their spatial origin. N (trajectories) are provided in Table S1.
A559V-Induced Ser53 Phosphorylation Increases Membrane DAT Mobility
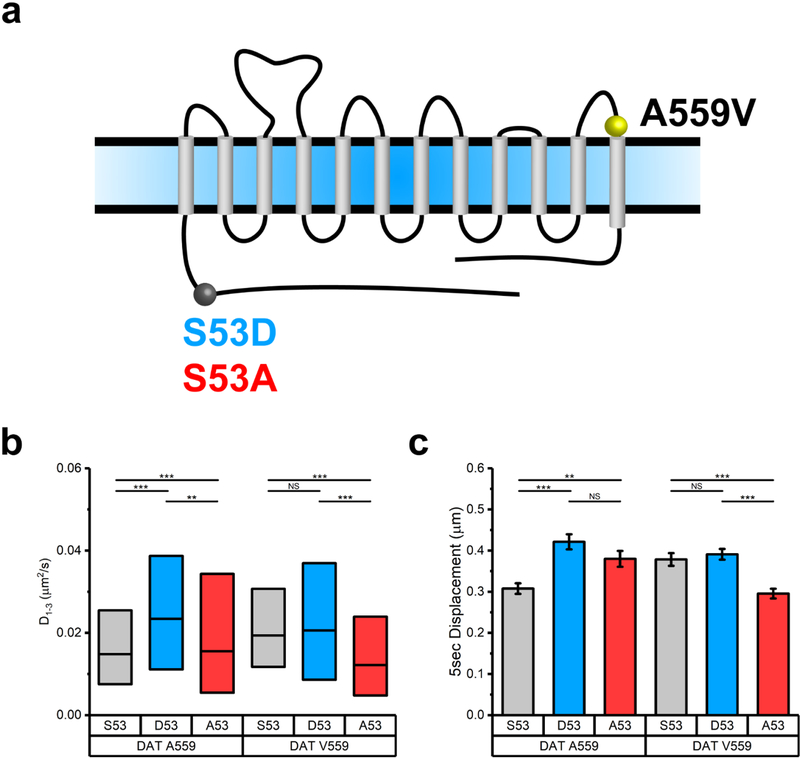
In addition to the phosphorylation of N-terminal Ser residues, juxtamembrane DAT residue Thr53 (Ser53 in humans) accounts for a portion of basal DAT phosphorylation in transfected cells and in vivo.(58) Phosphorylation of Thr53 has been reported to impact dopamine uptake and AMPH-induced efflux,(58) whereas spatiotemporal effects of Thr53 phosphorylation on DAT lateral mobility remain unexplored. Intriguingly, elevated DAT Thr53 phosphorylation is evident in DAT Val559 knock-in mice(21) in concert with elevated DAT Val559 surface expression and ADE. Thus, we explored a possible contribution of Ser53 phosphorylation to DAT membrane mobility. Upon generating diffusion profiles for DAT Ala559 and DAT Val559 expressing phosphorylation-occluded (S53A) and phosphomimetic (S53D) mutations, we observed a significant increase in D1–3 and 5 s displacements comparing DAT Ala559,Asp53 to DAT Ala559 populations, whereas DAT Val559,Asp53 exhibited no significant difference in D1–3 or 5 s displacements compared to those of DAT Val559 populations (Figure 5b, c).
Figure 5. S53 phosphorylation alters m obility of DAT and DAT Val559. (a) Schematic of DAT and positions of variants tested.
The A559V site is highlighted in yellow; S53A is highlighted in blue, and S53A is highlighted in red. (b) Diffusion coefficient box plot (median, 25% and 75% interquantiles, one-way ANOVA followed by Bonferroni’s multiple comparison test, NS p > 0.05, **p < 0.01 ***p < 0.001) of trajectories analyzed for Ser53, Asp53, and Ala53 under DAT Ala559 and DAT Val559 backgrounds. (c) A 5 s displacement bar graph (mean ± S.E.M., unpaired Student’s t test, NS p > 0.05, **p < 0.01, ***p < 0.001) of trajectories analyzed for Ser53, Asp53, and Ala53 under DAT Ala559 and DAT Val559 backgrounds. N (trajectories) are provided in Table S1.
Additionally, we observed that the DAT Val559,Ala53 mutant exhibited lower D1–3 and 5 s displacements compared to those of DAT Val559. Unexpectedly, the DAT Ala559,Ala53 mutant exhibited higher D1–3 and faster 5 s displacements than those of DAT Ala559, which may reflect a nonspecific impact of the Ala53 substitution on the membrane transporter diffusion. Nonetheless, occluding Ser53 phosphorylation in DAT Val559 with the Ala53 substitution resulted in diffusion coefficients comparable to those of DAT Ala559, consistent with a model where elevated phosphorylation at Ser53 in DAT Val559 is essential to the mutant’s increased lateral mobility. Given that phosphorylation at distal N-terminal Ser residues as well as at Ser53 appears required for increased lateral mobility of DAT Val559, we suggest that these sites may “communicate” with each other through either transmitted conformational changes in the N-terminus or through changes in protein associations that impact transporter lateral mobility. In this regard, a number of DAT-associated proteins interact with the N-terminus including D2Rs, syntaxin 1A, and kappa opioid receptors.(49,59,60) Future studies of DAT Val559 lateral mobility should explore contributions of disrupted associations of one or more of these proteins.
DAT Val559 Exhibits Reduced Clustering at the Apical Surface of HEK-293 Cells
Several groups demonstrated that DAT undergoes oligomerization via various biochemical and optical approaches.(61–65) A more recent superresolution microscopy study demonstrated that DAT proteins are organized into functional, cholesterol- dependent nanodomains in both transfected CAD cells and dopamine neurons.(66) Moreover, transmembrane domain 12 (TM12), where the Ala559Val mutation is located, has been suggested to support dimer formation in DAT via in silico experiments as well as serotonin transporter (SERT) proteins via in vitro and in silico studies.(67–69) To explore the possibility of the Ala559Val mutation altering DAT clustering, we evaluated DAT Val559 clustering at the apical surface of HEK-293 cells by widefield epifluorescence microscopy. Consistent with our tracking experiments, we treated cells expressing DAT Ala559 and DAT Val559 with 100 nM IDT444 and <0.1 nM QD. Briefly, we analyzed clusters by an in house-developed intensity-based algorithm (see Materials and Methods, Figure 6a) where integrated density (ID) values were obtained by integrating point spread functions (PSF) from acquired images (Figure 6b, c). Higher ID values indicate the presence of clustered DAT-QDs (Figure 6c). For the best visualization of clusters, TALM reconstructed maps are provided (Figure 7a) for both DAT Ala559 and DAT Val559. We fit ID distributions with a lognormal function and used quantile-quantile (Q-Q) plots to compare ID values that fall outside of the fit with a 99% confidence value. DAT Ala559 ID values clearly indicate a distinct population of density values that fall outside the lognormal fit in the cluster regime, unlike DAT Val559 ID values that deviate less from the reference line (Figure 7b). These findings indicate that, in addition to an increase in lateral mobility, DAT Val559 proteins appear to cluster less on the apical surface of HEK-293 cells with other labeled transporters. These changes may preclude interactions with other DAT regulators that support normal dopamine influx/efflux bias and proper regulation of the transporter by cell signaling mechanisms. Notably, studies in the literature are mixed with respect to whether cocaine impacts multimer formation with differences possibly related to expression systems, levels of DAT expressed, and methods for multimer capture.(63,64,70–72) Although we used low concentrations of IDT444-conjugated QDs to afford labeling of a small number of targets, additional studies are needed to know whether the binding of probes to these transporters has effects on multimer formation probability on its own. Studies using TIRF approaches with GFP-tagged transporters in the presence and absence of IDT444 present one possible route to explore this possibility.(73)
Figure 6. Intensity-based widefield imaging analysis reveals clusters at the apical cell surface.
(a) Cartoon outlining the experimental widefield focal plane (orange) and DAT-QD (red) localization. (b) Representative montage of widefield epifluorescence micrographs of two DAT-QD PSFs interacting over a period of 5 s. (c) 3D surface plots of representative PSFs for single DAT-QDs and DAT-QD clusters.
Figure 7. DAT Val559 has a lower propensity to reside in clusters at the apical surface of HEK-293 cells.
(a) TALM reconstructions of DAT Ala559-QD and DAT Val559-QD PSFs from 1000-frame time series (scale bars = 2 μm). Q-Q plots (99% confidence intervals, black) of background-subtracted integrated intensity values for DAT Ala559 and DAT Val559. Upper and lower percentiles are calculated with a 99% confidence. NA559 = 1871, NV559 = 1864, 26 cells analyzed.
In the model suggesting that alterations in cholesterol may indirectly influence DAT Val559 engagement in multi-transporter clusters, conformational changes and/or steric clashes that DAT Val559 imparts on TM12 may shift the equilibrium from a stabilized clustered state in membrane rafts toward a more mobile, efflux-prone dissociated state. This would agree with the idea explored by Sitte and Freissmuth in which oligomerization acts as an important factor in reverse dopamine transport.(74) Such modification in the DAT membrane lipid environment could affect the efficacy of DAT regulation by endogenous G protein coupled receptors, such as D2R, that result in dysfunctional DAT Val559 phenotypes. Another component that could be affecting DAT clustering is phosphatidylinositol 4,5-biphosphate (PIP2) interaction with the N-terminus of Val559. Interestingly, an unphosphorylated DAT N-terminus interacts with PIP2 insilico,(75) which the Galli group reported enables consequent AMPH-induced efflux.(76) The possibility of impaired efflux as a result of Val559 mistargeting PIP2 pools cannot be easily reconciled with the observation of PIP2 electrostatic interaction with the N-terminus being necessary for AMPH-induced efflux.(76) However, considering the phosphorylation status of DAT is important in PIP2 binding and direct PIP2 binding has been shown to facilitate membrane oligomerization of SERT,(73) we cannot exclude the possibility of impaired interaction between the DAT Val559 and PIP2.
Conclusions
Neuropsychiatric disease-derived DAT missense mutations demonstrate both trafficking-dependent and -independent DAT modes of transporter dysfunction, reinforcing perturbed synaptic dopamine homeostasis as a risk determinant for neuropsychiatric disease. Although the underlying molecular mechanisms of DAT dysregulation in neuropsychiatric disorders remain to be fully elucidated, recent evidence suggests that disrupted DAT membrane microdomain compartmentalization is potentially a common culprit of DAT-mediated dopamine pathology.(26,77) Previously, we demonstrated that the ADHD-associated DAT coding Cys615 demonstrates alterations in membrane lateral mobility. Here, we provide the first single molecule tracking evidence for the DAT Val559 variant, a mutation found in subjects with ADHD, ASD, and BPD. Like DAT Cys615, DAT Val559 is more mobile compared to DAT under basal conditions. Longstanding evidence indicating that PKC mediates N-terminal phosphorylation of DAT at Ser and Thr residues,(5) and our observation of the DAT Val559 variant exhibiting altered diffusion rates, led us to perform pharmacological investigation of requirements for PKC-mediated phosphorylation. We demonstrate that DAT Val559 mobility is unresponsive to PKC activation but is attenuated by a potent and selective PKCβ inhibitor. Furthermore, DAT phosphomimicked at distal serine residues is more mobile, supporting N-terminal phosphorylation as one determinant of DAT membrane diffusion. Adding to the growing appreciation of the role of Ser/Thr53 phosphorylation in DAT function, we provide evidence that phosphorylation at this site mobilizes DAT for increased lateral diffusion, effects (phosphorylation and mobilization) instituted constitutively in the DAT Val559 variant. We report here the first experimental evidence of reduced DAT Val559 clustering related to aberrant membrane mobility.
Several groups have reported PKC modulation of DAT endocytic trafficking observed in heterologous cells to be absent in cultured midbrain neurons.(24,78–80) In contrast, Gabriel et al. recently reported PKC-mediated action on DAT trafficking observed intact striatal slices.(23) It is possible that some of this discrepancy may derive from culture versus slice approaches or the study of circuits that differentially support regulated DAT trafficking. For instance, Gowrishankar et al. found in studies of acute brain slices that D2R-dependent DAT trafficking occurs in dorsal but not ventral striatum.(21) Currently, the degree to which Val559 variant proteins diffuse and cluster in vivo is unknown, though this is a goal for our efforts moving forward.
As the ADHD-associated DAT Cys615 variant also demonstrates increased lateral mobility,(26) disrupted membrane localization may be a common attribute of disease- associated changes in neurotransmitter transporter availability and/or function. Beyond this more general conclusion, our results reveal previously unreported perturbations of the membrane dynamics DAT Val559 variant implicating PKCβ activation as an important determinant of transporter lateral mobility. Future studies are needed to dissect whether altered DAT Val559 membrane dynamics are a consequence of disrupted interactions between membrane domains enriched with cholesterol and/or PIP2. Resolution of this question may lead to new opportunities for neuropsychiatric disease intervention.
Materials and Methods
Materials
AmpQD-655, SavQD-605, and SavQD655 were purchased from Invitrogen. DMEM, fetal bovine serum (Gibco), phosphate buffer saline (PBS, w/o Ca2+, Mg2+), penicillin/streptomycin, and Lipofectamine 3000 were purchased from ThermoFisher. EMEM was purchased from VWR. MatTek dishes were purchased from MatTek directly. Laminin (Millipore), phorbol-12-myristate-13-acetate (PMA) (Sigma), and poly-D-lysine (Sigma) were purchased from Millipore Sigma. IDT307 (Blakely et al., 2011, U.S. Patent Number 7947255 B2) and IDT444 were synthesized in the Rosenthal Lab by Dr. Ian D. Tomlinson.(37)
Coding Variant Constructs
pcDNA3.1(+) DAT and pcDNA3.1(+) DAT Val559 constructs were previously detailed.(15) yfpsyndat (Addgene plasmid # 19991) and YFP-synDAT-S/D (Addgene plasmid # 48793) were gifts from Jonathan Javitch, Ph.D. at Columbia University. Q5 site directed mutagenesis (NEB Inc.) was used to generate hDAT A53 and D53 on both the hDAT and hDAT A559V plasmid with forward primer RB5576 5’- CCCGCGGCAGgccCCCGTGGAGG-3’ and RB5578 5’- CCCGCGGCAGgacCCCGTGGAGG-3’, respectively, and reverse primer RB5577 5’- TTGGTGAGGGTGGAGCTGG-3’.
Cell Culture
HEK-293 cells were a kind gift from Eva Harth, Ph.D. at Vanderbilt University, and SK-N- MC cells were kindly provided by Jerry Chang, Ph.D. at the Rockefeller University Laboratory of Molecular and Cellular Neuroscience. HEK-293 and SK-N-MC cells were maintained in DMEM and EMEM, respectively, with 2 mM/L glutamine, 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin. Both cell cultures were transiently transfected 24 h prior to labeling at 1 μg DNA:3 μL Lipofectamine 3000 ratio.
QD Labeling
DAT-expressing HEK-293 and SK-N-MC cells were plated on MatTek No. 1.5 coverslips coated with poly-D-lysine and laminin, respectively. QD labeling was employed following a two-step protocol previously outlined.(81) Briefly, cells in 2 mL of full growth medium were spiked with 20 μL of 10 μM IDT444 suspended in PBS (w/o Ca2+, Mg2+) and incubated at 37 °C and 5% CO2 for 10 min. Three washes with warm DMEM Fluorobrite were performed prior to treating cells with 10 pM SavQD 2% dialyzed FBS in DMEM Fluorobrite. The QD-IDT444 DAT-labeled cells were washed three times with warm DMEM Fluorobrite. Activation of PKC and inhibition of PKCβ were performed by administering 100 nM PMA 30 min prior to washing and 1 μM Enz prior to PMA treatment, respectively. PMA- and Enz-treated cells were washed an additional 3 times with warm DMEM Fluorobrite. At the microscope, IDT307 was added to the MatTek dish prior to imaging.
Microscopy
Time-series images were generated by TIRF and widefield epifluorescence microscopy on a Nikon Eclipse Ti-E inverted microscope equipped with an Andor Zyla 4.2 PLUS sCMOS camera and viewed with an Apo TIRF 60×/1.49 NA oil objective. Excitation at 488 nm was sourced by a Nikon LU-NV laser unit. YFP-DAT and IDT307 emissions were collected with a 525 ± 18 nm emission filter. SavQD-605 and SavQD-655 signals were collected with 603 ± 15 nm and 655 ±15 nm emission filters, respectively. For SPT experiments, time series were produced at a 10 Hz frame rate. For clustering experiments, time series were produced at a 17 Hz frame rate.
Single Particle Tracking Analysis
Raw TIFF stacks were extracted from Nikon Elements ndl files in Fiji, an ImageJ distribution (National Institutes of Health, Bethesda, MD). T rajectories were compiled from these raw data given the conditions that (i) the particle emission is intermittent to ensure discrimination of single particles, (ii) the blinking gap is less than 10 frames, (iii) the PSF is located within a 3 × 3 pixel area surrounding the PSF location from the previous frame, and (iv) the trajectory persists at least 50 frames. Mean square displacement, ˂r2(nδt)˃, values were calculated for each of the trajectories collected for time intervals of 0.1–1.5 s in 0.1 s intervals via
where δt is the temporal resolution, (x(jδt), x(jδt)) is the coordinate at t = jδt, and N is the number of total frames recorded during a single trajectory.
Diffusion coefficients (D1–3) were determined from the linear fits of first three MSD values in the following algorithm:
where δt is the temporal resolution. The uncertainty, dependent on the signal-to-noise ratio (SNR) and limited by the diffraction limit using visible light, was estimated by Δσ≈ ω/SNR, where w is approximately the widefield mode diffraction limit and SNR values for QD emitters exited by an evanescent field in TIRF mode range between 20 and 30. D1–3 values were populated, and a significant difference between distributions was determined by one-way ANOVA followed by Bonferroni’s multiple comparison test. 5 s displacement vectors were obtained by indexing particle coordinates after 50 frames and normalizing to the particle origin. Populated 5 s displacement values were analyzed by unpaired Student’s t test. All analysis was performed and automated by MATLAB codes. For extensive detail regarding microscopy and analysis in SPT experiments, see Chang and Rosenthal et al.(82)
Cluster Analysis
Raw TIFF stacks were extracted from Nikon Elements ndl files in Fiji. PSF centroids were subsequently identified and indexed. For the intensity of each PSF to be quantified, ID values were calculated by integrating raw intensity values in a 5 × 5 pixel matrix and normalized to the particle centroid coordinate. Because background in widefield mode is heterogeneous due to emission from QDs outside of the focal plane, background values were calculated by averaging intensity counts of a 9 × 9 pixel parameter centered around each centroid. Raw IDs were then corrected by their assigned backgrounds. All analysis was performed and automated by an in-house MATLAB script. The QuickPALM plugin in Fiji was used to generate TALM images.
Supplementary Material
Acknowledgements
Imaging was performed in the VUMC Cell Imaging Resource Center (supported by NIH grants CA68485, DK20593, DK20953, DK58404, DK59637, and EY08126). LBT was supported by the NIH Chemistry-Biology Interface training grant (NIH T32GM065086–14). RDB was supported by NIH grant MH105094.
Footnotes
Supporting Information
The authors declare no competing financial interest.
References
- 1.Giros B; Caron MG, Molecular characterization of the dopamine transporter. Trends in Pharmacological Sciences 1993, 14 (2), 43–49. [DOI] [PubMed] [Google Scholar]
- 2.Palmiter RD, Dopamine signaling in the dorsal striatum Is essential for motivated behaviors. Annals of the New York Academy of Sciences 2008, 1129 (1), 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannon MJ; Michelhaugh SK; Wang J; Sacchetti P, The human dopamine transporter gene: gene organization, transcriptional regulation, and potential involvement in neuropsychiatric disorders. European Neuropsychopharmacology 11 (6), 449–455. [DOI] [PubMed] [Google Scholar]
- 4.Howes O; McCutcheon R; Stone J, Glutamate and dopamine in schizophrenia: An update for the 21st century. Journal of Psychopharmacology 2015, 29 (2), 97–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bermingham DP; Blakely RD, Kinase-dependent regulation of monoamine neurotransm itter transporters. Pharmacological Reviews 2016, 68 (4), 888–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nutt DJ; Lingford-Hughes A; Erritzoe D; Stokes PRA, The dopamine theory of addiction: 40 years of highs and lows. Nature Reviews Neuroscience 2015, 16, 305. [DOI] [PubMed] [Google Scholar]
- 7.Giros B; Jaber M; Jones SR; Wightman RM; Caron MG, Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature 1996, 379, 606. [DOI] [PubMed] [Google Scholar]
- 8.Gainetdinov RR; Jones SR; Caron MG, Functional hyperdopaminergia in dopamine transporter knock-out mice. Biological Psychiatry 1999, 46 (3), 303–311. [DOI] [PubMed] [Google Scholar]
- 9.Ralph RJ; Paulus MP; Fumagalli F; Caron MG; Geyer MA, Prepulse inhibition deficits and perseverative m otor patterns in dopamine transporter knock-out mice: Differential effects of D1 and D2 receptor antagonists. The Journal of Neuroscience 2001, 21 (1), 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazei-Robison MS; Couch RS; Shelton RC; Stein MA; Blakely RD, Sequence variation in the human dopamine transporter gene in children with attention deficit hyperactivity disorder. Neuropharmacology 2005, 49 (6), 724–736. [DOI] [PubMed] [Google Scholar]
- 11.Grunhage F; Schulze TG; Muller DJ; Lanczik M; Franzek E; Albus M; Borrmann-Hassenbach M; Knapp M; Cichon S; Maier W; Rietschel M; Propping P; Nothen MM, Systematic screening for DNA sequence variation in the coding region of the human dopamine transporter gene (DAT1). Molecular Psychiatry 2000, 5, 275. [DOI] [PubMed] [Google Scholar]
- 12.Hahn MK; Blakely RD, The functional impact of SLC6 transporter genetic variation. Annual Review of Pharmacology and Toxicology 2007, 47 (1), 401–441. [DOI] [PubMed] [Google Scholar]
- 13.Hansen FH; Skj0rringe T; Yasmeen S; Arends NV; Sahai MA; Erreger K; Andreassen TF; Holy M; Hamilton PJ; Neergheen V; Karlsborg M; Newman AH; Pope S; Heales SJR; Friberg L; Law I; Pinborg LH; Sitte HH; Loland C; Shi L; Weinstein H; Galli A; Hjermind LE; M0ller LB; Gether U, Missense dopamine transporter mutations associate with adult parkinsonism and ADHD. The Journal of Clinical Investigation 2014, 124 (7), 3107–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurian MA; Gissen P; Smith M; Heales SJR; Clayton PT, The monoamine neurotransm itter disorders: an expanding range of neurological syndromes. The Lancet Neurology 2011, 10 (8), 721–733. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton PJ; Campbell NG; Sharma S; Erreger K; Herborg Hansen F; Saunders C; Belovich AN; Consortium NAAS; Sahai MA; Cook EH; Gether U; McHaourab HS; Matthies HJG; Sutcliffe JS; Galli A, De novo m utation in the dopamine transporter gene associates dopamine dysfunction w ith autism spectrum disorder. Molecular Psychiatry 2013, 18, 1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S; Fagan RR; Uttamapinant C; Lifshitz LM; Fogarty KE; Ting AY; Melikian HE, The dopamine transporter recycles via a retrom er-dependent postendocytic mechanism: Tracking studies using a novel fluorophore-coupling approach. The Journal of Neuroscience 2017, 37 (39), 9438–9452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S; Bellve KD; Fogarty KE; Melikian HE, Ack1 is a dopamine transporter endocytic brake that rescues a trafficking-dysregulated ADHD coding variant. Proceedings of the National Academy of Sciences 2015, 112 (50), 15480–15485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler DS; Underhill SM; Stolz DB; Murdoch GH; Thiels E; Romero G; Amara SG, Amphetamine activates Rho GTPase signaling to mediate dopamine transporter internalization and acute behavioral effects of amphetamine. Proceedings of the National Academy of Sciences 2015, 112 (51), E7138–E7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu S; Zhao C; Wu Y; Yang Q; Shao A; Wang T; Wu J; Yin Y; Li Y; Hou J; Zhang X; Zhou G; Gu X; Wang X; Bustelo XR; Zhou J, Identification of a Vav2-dependent mechanism for GDNF/Ret control of mesolimbic DAT trafficking. Nature Neuroscience 2015, 18, 1084. [DOI] [PubMed] [Google Scholar]
- 20.Block ER; Nuttle J; Balcita-Pedicino JJ; Caltagarone J; Watkins SC; Sesack SR; Sorkin A, Brain region-specific trafficking of the dopamine transporter. The Journal of Neuroscience 2015, 35 (37), 12845–12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gowrishankar R; Gresch PJ; Davis GL; Katamish RM; Riele JR; Stewart AM; Vaughan RA; Hahn MK; Blakely RD, Region-specific regulation of presynaptic dopamine homeostasis by D2 autoreceptors shapes the In Vivo impact of the neuropsychiatric disease-associated DAT variant Val559. The Journal of Neuroscience 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furman CA; Chen R; Guptaroy B; Zhang M; Holz RW; Gnegy M, Dopamine and amphetamine rapidly increase dopamine transporter trafficking to the surface: Live-cell imaging using total internal reflection fluorescence microscopy. The Journal of Neuroscience 2009, 29 (10), 3328–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel LR; Wu S; Kearney P; Bellve KD; Standley C; Fogarty KE; Melikian HE, Dopamine transporter endocytic trafficking in striatal dopaminergic neurons: differential dependence on dynamin and the actin cytoskeleton. The Journal of Neuroscience 2013, 33 (45), 17836–17846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eriksen J; Rasmussen SGF; Rasmussen TN; Vaegter CB; Cha JH; Zou M-F; Newman AH; Gether U, Visualization of dopamine transporter trafficking in live neurons by use of fluorescent cocaine analogs. The Journal of Neuroscience 2009, 29 (21), 6794–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorkina T; Caltagarone J; Sorkin A, Flotillins regulate membrane m obility of the dopamine transporter but are not required fo r its protein kinase C dependent endocytosis. Traffic 2013, 14 (6), 709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovtun O; Sakrikar D; Tomlinson ID; Chang JC; Arzeta-Ferrer X; Blakely RD; Rosenthal SJ, Single-quantum-dot tracking reveals altered membrane dynamics of an attention-deficit/hyperactivity-disorder-derived dopamine transporter coding variant. ACS chemical neuroscience 2015, 6 (4), 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenthal SJ; Chang JC; Kovtun O; McBride JR; Tomlinson ID, Biocompatible quantum dots for biological applications. Chemistry & Biology 2011, 18 (1), 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahan M; Levi S; Luccardini C; Rostaing P; Riveau B; Triller A, Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science 2003, 302 (5644), 442–445. [DOI] [PubMed] [Google Scholar]
- 29.Bouzigues C; Morel M; Triller A; Dahan M, Asymmetric redistribution of GABA receptors during GABA gradient sensing by nerve growth cones analyzed by single quantum dot imaging. Proceedings of the National Academy of Sciences 2007, 104 (27), 11251–11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frischknecht R; Heine M; Perrais D; Seidenbecher CI; Choquet D; Gundelfinger ED, Brain extracellular matrix affects AMPA receptor lateral m obility and short-term synaptic plasticity. Nature neuroscience 2009, 12 (7), 897–904. [DOI] [PubMed] [Google Scholar]
- 31.Chang JC; Tomlinson ID; Warnement MR; Ustione A; Carneiro AMD; Piston DW; Blakely RD; Rosenthal SJ, Single molecule analysis of serotonin transporter regulation using antagonist-conjugated quantum dots reveals restricted, p38 MAPK-dependent mobilization underlying uptake activation. The Journal of neuroscience : the official journal of the Society for Neuroscience 2012, 32 (26), 8919–8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey DM; Catron MA; Kovtun O; Macdonald RL; Zhang Q; Rosenthal SJ, Single quantum dot tracking reveals serotonin transporter diffusion dynamics are correlated w ith cholesterol- sensitive threonine 276 phosphorylation status in primary midbrain neurons. ACS Chemical Neuroscience 2018. [DOI] [PubMed] [Google Scholar]
- 33.McBride J; Treadway J; Feldman LC; Pennycook SJ; Rosenthal SJ, Structural basis for near unity quantum yield core/shell nanostructures. Nano Letters 2006, 6 (7), 1496–1501. [DOI] [PubMed] [Google Scholar]
- 34.Kovtun O; Tomlinson ID; Bailey DM; Thal LB; Ross EJ; Harris L; Frankland MP; Ferguson RS; Glaser Z; Greer J; Rosenthal SJ, Single quantum dot tracking illuminates neuroscience at the nanoscale. Chemical Physics Letters 2018, 706, 741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choquet D; Triller A, The role of receptor diffusion in the organization of the postsynaptic membrane. Nature Reviews Neuroscience 2003, 4, 251. [DOI] [PubMed] [Google Scholar]
- 36.Triller A; Choquet D, New concepts in synaptic biology derived from single-molecule imaging. Neuron 2008, 59 (3), 359–374. [DOI] [PubMed] [Google Scholar]
- 37.Kahlig KM; Lute BJ; Wei Y; Loland CJ; Gether U; Javitch JA; Galli A, Regulation of dopamine transporter trafficking by intracellular amphetamine. Molecular Pharmacology 2006, 70 (2), 542–548. [DOI] [PubMed] [Google Scholar]
- 38.Gussin HA; Tomlinson ID; Cao D; Qian H; Rosenthal SJ; Pepperberg DR, Quantum dot conjugates of GABA and muscimol: Binding to a ip 2 y2 and p i GABAA receptors. ACS Chemical Neuroscience 2013, 4 (3), 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gussin HA; Tomlinson ID; Little DM; W arnement MR; Qian H; Rosenthal SJ; Pepperberg DR, Binding of muscimol-conjugated quantum dots to GABAC receptors. Journal of the American Chemical Society 2006, 128 (49), 15701–15713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovtun O; Tomlinson ID; Sakrikar DS; Chang JC; Blakely RD; Rosenthal SJ, Visualization of the cocaine-sensitive dopamine transporter w ith ligand-conjugated quantum dots. ACS Chemical Neuroscience 2011, 2 (7), 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenthal SJ; Tomlinson I; Adkins EM; Schroeter S; Adams S; Swafford L; McBride J; Wang Y; DeFelice LJ; Blakely RD, Targeting cell surface receptors w ith ligand-conjugated nanocrystals. Journal of the American Chemical Society 2002, 124 (17), 4586–4594. [DOI] [PubMed] [Google Scholar]
- 42.Bowton E; Saunders C; Reddy IA; Campbell NG; Hamilton PJ; Henry LK; Coon H; Sakrikar D; Veenstra-VanderWeele JM; Blakely RD; Sutcliffe J; Matthies HJG; Erreger K; Galli A, SLC6A3 coding variant Ala559Val found in tw o autism probands alters dopamine transporter function and trafficking. Transi Psychiatry 2014, 4, e464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mergy MA; Gowrishankar R; Gresch PJ; Gantz SC; Williams J; Davis GL; Wheeler CA; Stanwood GD; Hahn MK; Blakely RD, The rare DAT coding variant Val559 perturbs DA neuron function, changes behavior, and alters in vivo responses to psychostimulants. Proceedings of the National Academy of Sciences 2014, 111 (44), E4779–E4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis GL; Stewart A; Stanwood GD; Gowrishankar R; Hahn MK; Blakely RD, Functional coding variation in the presynaptic dopamine transporter associated w ith neuropsychiatric disorders drives enhanced m otivation and context-dependent impulsivity in mice. Behavioural Brain Research 2018, 337, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazei-Robison MS; Bowton E; Holy M; Schmudermaier M; Freissmuth M; Sitte HH; Galli A; Blakely RD, Anomalous dopamine release associated w ith a human dopamine transporter coding variant. The Journal of Neuroscience 2008, 28 (28), 7040–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazei-Robison MS; Blakely RD, Expression studies of naturally occurring human dopamine transporter variants identifies a novel state of transporter inactivation associated w ith Val382Ala. Neuropharmacology 2005, 49 (6), 737–749. [DOI] [PubMed] [Google Scholar]
- 47.Herborg F; Andreassen TF; Berlin F; Loland CJ; Gether U, Neuropsychiatric disease- associated genetic variants of the dopamine transporter display heterogeneous molecular phenotypes. Journal of Biological Chemistry 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolan EA; Kivell B; Jaligam V; Oz M; Jayanthi LD; Han Y; Sen N; Urizar E; Gomes I; Devi LA; Ramamoorthy S; Javitch JA; Zapata A; Shippenberg TS, D2 receptors regulate dopamine transporter function via an extracellular signal-regulated kinases 1 and 2-dependent and phosphoinositide 3 kinase-independent mechanism. Molecular Pharmacology 2007, 71 (5), 1222–1232. [DOI] [PubMed] [Google Scholar]
- 49.Lee FJ; Pei L; Moszczynska A; Vukusic B; Fletcher PJ; Liu F, Dopamine transporter cell surface localization facilitated by a direct interaction w ith the dopamine D2 receptor. The EMBO Journal 2007, 26 (8), 2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowton E; Saunders C; Erreger K; Sakrikar D; Matthies HJ; Sen N; Jessen T; Colbran RJ; Caron MG; Javitch JA; Blakely RD; Galli A, Dysregulation of dopamine transporters via dopamine D2 autoreceptors triggers anomalous dopamine efflux associated w ith attention-deficit hyperactivity disorder. The Journal of Neuroscience 2010, 30 (17), 6048–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blakely RD; Mason JN; Tomlinson ID; Rosenthal SJ, Fluorescent substrates for neurotransm itter transporters. Google Patents: 2011. [Google Scholar]
- 52.Mason JN; Farmer H; Tomlinson ID; Schwartz JW; Savchenko V; DeFelice LJ; Rosenthal SJ; Blakely RD, Novel fluorescence-based approaches for the study of biogenic amine transporter localization, activity, and regulation. Journal of Neuroscience Methods 2005, 143 (1), 3–25. [DOI] [PubMed] [Google Scholar]
- 53.Johnson LAA; Guptaroy B; Lund D; Shamban S; Gnegy ME, Regulation of amphetamine- stimulated dopamine efflux by protein kinase C p. Journal of Biological Chemistry 2005, 280 (12), 1091410919. [DOI] [PubMed] [Google Scholar]
- 54.Chen R; Furman CA; Zhang M; Kim MN; Gereau RW; Leitges M; Gnegy ME, Protein kinase C p is a critical regulator of dopamine transporter trafficking and regulates the behavioral response to amphetamine in mice. Journal of Pharmacology and Experimental Therapeutics 2009, 328 (3), 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zestos AG; Mikelman SR; Kennedy RT; Gnegy ME, PKCΒ inhibitors attenuate amphetamine-stimulated dopamine efflux. ACS Chemical Neuroscience 2016, 7 (6), 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daniels GM; Amara SG, Regulated trafficking of the human dopamine transporter: Clatherin- mediated internalization and lysosomal degradation in response to phorbol esters. Journal of Biological Chemistry 1999, 274 (50), 35794–35801. [DOI] [PubMed] [Google Scholar]
- 57.Carvelli L, Moron JA, Kahlig KM, Ferrer JV, Sen N, Lechleiter JD, Leeb-Lundberg LM, Merrill G , Lafer EM, Ballou LM, Shippenberg TS, Javitch JA, Lin RZ and Galli A, PI 3-kinase regulation of dopamine uptake. Journal of Neurochemistry 2002, 81 (4), 859–869. [DOI] [PubMed] [Google Scholar]
- 58.Foster JD; Yang J-W; Moritz AE; ChallaSivaKanaka S; Smith MA; Holy M; Wilebski K; Sitte HH; Vaughan RA, Dopamine transporter phosphorylation site threonine 53 regulates substrate reuptake and amphetamine-stimulated efflux. Journal of Biological Chemistry 2012, 287 (35), 29702–29712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Binda F; Dipace C; Bowton E; Robertson SD; Lute BJ; Fog JU; Zhang M; Sen N; Colbran RJ; Gnegy ME; Gether U; Javitch JA; Erreger K; Galli A, Syntaxin 1A interaction w ith the dopamine transporter promotes amphetamine-induced dopamine efflux. Molecular Pharmacology 2008, 74 (4), 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kivell B; Uzelac Z; Sundaramurthy S; Rajamanickam J; Ewald A; Chefer V; Jaligam V; Bolan E; Simonson B; Annamalai B; Mannangatti P; Prisinzano TE; Gomes I; Devi LA; Jayanthi LD; Sitte HH; Ramamoorthy S; Shippenberg TS, Salvinorin A regulates dopamine transporter function via a kappa opioid receptor and ERK1/2-dependent mechanism. Neuropharmacology 2014, 86, 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torres GE; Carneiro A; Seamans K; Fiorentini C; Sweeney A; Yao W-D; Caron MG, Oligomerization and trafficking of the human dopamine transporter: M utational analysis identifies critical domains im portant for the functional expression of the transporter. Journal of Biological Chemistry 2003, 278 (4), 2731–2739. [DOI] [PubMed] [Google Scholar]
- 62.Sorkina T; Doolen S; Galperin E; Zahniser NR; Sorkin A, Oligomerization of dopamine transporters visualized in living cells by fluorescence resonance energy transfer microscopy. Journal of Biological Chemistry 2003, 278 (30), 28274–28283. [DOI] [PubMed] [Google Scholar]
- 63.Hastrup H; Sen N; Javitch JA, The human dopamine transporter forms a tetram er in the plasma membrane: Cross-linking of a cysteine in the fourth segment is sensitive to cocaine analogs. Journal of Biological Chemistry 2003, 278 (46), 45045–45048. [DOI] [PubMed] [Google Scholar]
- 64.Chen N; Reith MEA, Substrates dissociate dopamine transporter oligomers. Journal of Neurochemistry 2008, 105 (3), 910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Egana LA; Cuevas RA; Baust TB; Parra LA; Leak RK; Hochendoner S; Pena K; Quiroz M; Hong WC; Dorostkar MM; Janz R; Sitte HH; Torres GE, Physical and functional interaction between the dopamine transporter and the synaptic vesicle protein synaptogyrin-3. The Journal of Neuroscience 2009, 29 (14), 4592–4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rahbek-Clemmensen T; Lycas MD; Erlendsson S; Eriksen J; Apuschkin M; Vilhardt F; Jorgensen TN; Hansen FH; Gether U, Super-resolution microscopy reveals functional organization of dopamine transporters into cholesterol and neuronal activity-dependent nanodomains. Nature communications 2017, 8 (1), 740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Periole X; Zeppelin T; Schiott B, Dimer interface of the human serotonin transporter and effect of the membrane composition. Scientific Reports 2018, 8 (1), 5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Just H; Sitte HH; Schmid JA; Freissmuth M; Kudlacek O, Identification of an additional interaction domain in transmembrane domains 11 and 12 that supports oligomer form ation in the human serotonin transporter. Journal of Biological Chemistry 2004, 279 (8), 6650–6657. [DOI] [PubMed] [Google Scholar]
- 69.Jayaraman K; Morley AN; Szollosi D; Wassenaar TA; Sitte HH; Stockner T, Dopamine transporter oligomerization involves the scaffold domain, but spares the bundle domain. PLOS Computational Biology 2018, 14 (6), e1006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderluh A; Klotzsch E; Reismann AWAF; Brameshuber M; Kudlacek O; Newman AH; Sitte HH; Schutz GJ, Single molecule analysis reveals coexistence of stable serotonin transporter monomers and oligomers in the live cell plasma membrane. Journal of Biological Chemistry 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siciliano CA; Saha K; Calipari ES; Fordahl SC; Chen R; Khoshbouei H; Jones SR, Amphetamine reverses escalated cocaine intake via restoration of dopamine transporter conformation. The Journal of Neuroscience 2018, 38 (2), 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorkina T; Ma S; Larsen MB; Watkins SC; Sorkin A, Small molecule induced oligomerization, clustering and clathrin-independent endocytosis of the dopamine transporter. eLife 2018, 7, e32293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anderluh A; Hofmaier T; Klotzsch E; Kudlacek O; Stockner T; Sitte HH; Schutz GJ, Direct PIP2 binding mediates stable oligomer form ation of the serotonin transporter. Nature Communications 2017, 8, 14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sitte HH; Freissmuth M, Amphetamines, new psychoactive drugs and the monoamine transporter cycle. Trends in Pharmacological Sciences 2015, 36 (1), 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khelashvili G; Stanley N; Sahai MA; Medina J; LeVine MV; Shi L; De Fabritiis G; Weinstein H, Spontaneous inward opening of the dopamine transporter is triggered by PIP2-regulated dynamics of the N-terminus. ACS Chemical Neuroscience 2015, 6 (11), 1825–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamilton PJ; Belovich AN; Khelashvili G; Saunders C; Erreger K; Javitch JA; Sitte HH; Weinstein H; Matthies HJ; Galli A, PIP 2 regulates psychostimulant behaviors through its interaction w ith a membrane protein. Nature chemical biology 2014, 10 (7), 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson SB; Hardaway JA; Hardie SL; W right J; Glynn RM; Bermingham DP; Han Q; Sturgeon SM; Freeman P; Blakely RD, Sequence determinants of the Caenhorhabditis elegans dopamine transporter dictating in vivo axonal export and synaptic localization. Molecular and Cellular Neuroscience 2017, 78, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sorkina T; Miranda M; Dionne KR; Hoover BR; Zahniser NR; Sorkin A, RNA interference screen reveals an essential role of nedd4–2 in dopamine transporter ubiquitination and endocytosis. The Journal of Neuroscience 2006, 26 (31), 8195–8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rao A; Simmons D; Sorkin A, Differential subcellular distribution of endosomal compartments and the dopamine transporter in dopaminergic neurons. Molecular and Cellular Neuroscience 2011, 46 (1), 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rao A; Richards TL; Simmons D; Zahniser NR; Sorkin A, Epitope-tagged dopamine transporter knock-in mice reveal rapid endocytic trafficking and filopodia targeting of the transporter in dopaminergic axons. The FASEB Journal 2012, 26 (5), 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thal LB; Bailey DM; Kovtun O; Rosenthal SJ, Quantum dot toolbox in membrane neurotransm itter transporter research Chemical and Synthetic Approaches in Membrane Biology. Springer Protocols Handbooks. Humana Press, New York, NY Humana Press: Totowa, NJ, 2017; pp 1–12. [Google Scholar]
- 82.Chang JC; Rosenthal SJ, Real-time quantum dot tracking of single proteins In Biomedical Nanotechnology: Methods and Protocols, Hurst SJ, Ed. Humana Press: Totowa, NJ, 2011; pp 51–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.