Abstract
Objective and Background:
The expression of periodontitis, including age of onset, extent, and severity is considered to represent an interaction of the individual’s oral microbiome and host response to the microbial challenge that is modified by both genetics and environmental factors. The aim of this study was to determine the distribution of periodontitis in a population of nonhuman primates, to document features of familial distribution that could reflect heritability and transmission of microbes with enhanced virulence.
Methods:
This report presents our findings from evaluation of periodontal disease bone defects in skulls from 569 animals (5–31 years of age) derived from the skeletons of the rhesus monkeys (Macaca mulatta) of Cayo Santiago derived from 8 matrilines over 6–9 generations. The distance from the base of alveolar bone to the cemento-enamel junction on 1st/2nd premolars and 1st/2nd molars from all 4 quadrants was evaluated as a measure of periodontal disease. Additionally, we documented the presence of periodontitis in 79 living descendants within these matrilines.
Results:
The results demonstrated an increased extent and severity of periodontitis with aging across all matrilines. Extensive heterogeneity in disease expression was observed among the animals and this was linked to specific periodontitis susceptible matrilines. Moreover, we identified some matrilines in which the members appeared to show some resistance to more severe disease, even with aging.
Conclusions:
Linking these disease variations to multi-generational matriarchal family units supported familial susceptibility of periodontitis. This familial disease relationship was reinforced by the distribution of naturally-occurring periodontitis in the living descendants.
Keywords: nonhuman primates, periodontitis, familial risk
INTRODUCTION
Studies in humans have continued to provide some evidence regarding the contribution of genetics to the expression of periodontal disease(s). These include an array of genome wide association studies (GWAS) and more recently studies suggesting epigenetic changes that associate with periodontitis (1, 2). These modern approaches to genetic markers of disease risk are built on historical data indicating an increased prevalence of early onset disease in families (3, 4) and findings of various single nucleotide polymorphisms (SNPs) occurring in various populations of chronic adult periodontitis (5). However in chronic complex diseases, such as periodontitis, these types of cross-sectional associational studies generally require very large populations, with challenges existing in the studies related to a range of confounders in the population and a recognition that many individual genes likely each demonstrating a small contribution to the variance in the population (6).
Existing evidence supports a familial tendency for this disease, particularly with members exhibiting more generalized severe disease (6–11). Data derived from international populations also demonstrates the familial distribution of periodontitis in younger subjects that has been shown to have a genetic predisposition based upon specific immune abnormalities (10–14). This genomic impact is also linked to the virtually universal occurrence of a particular periodontal pathogen, Aggregatibacter actinomycetemcomitans within the affected individuals in these families (15–19).
Substantial research in chronic adult periodontitis, which represents the vast majority of the global disease, has been less definitive on a genetic contribution. Various studies have examined immune response gene polymorphisms (5, 20), genome-wide association studies (GWAS) have provided some potential gene contributors (21–23), and more recently studies describing epigenetic changes that could present as a risk factor for this disease (2). However, beyond early twin studies of periodontitis (24, 25) the fundamental concept of the magnitude of heritability of this disease remains to be determined. As importantly, in the human model of this disease, familial genetic studies are usually limited to one or 2 generations (vertical) and few family members in the same generation (horizontal) in attempting to define heritability.
Nearly all species of nonhuman primates (e.g. M. fascicularis, M. mulatta, M. nemestrina, S. scuireus, P. anubis) have been shown to express naturally-occurring periodontitis that increases with aging, as observed in humans (26–28). We have collaborated with the Caribbean Primate Research Center in Sabana Seca, Puerto Rico (CPRC) to examine aging effects on the immune system in gingival tissues and the relationship to the oral microbiome with initiation and progression of periodontitis (29–31). As we have reported previously (32), these animals were derived from a large, free-ranging colony of rhesus monkeys that was created in 1938 on “Cayo Santiago” off the coast of Puerto Rico (33).
The creation and description of this colony over 80 years allowed the definition of extensive pedigrees for the colony over 8–9 generations (34, 35). The natural death of the animals has enabled establishment of a “library” of full skeletons from over 2000 animals with defined matrilines (34, 36–38). Using this skeleton collection we hypothesized that periodontitis will be increased in members of selected matriline families that could be defined as a heritable feature of disease susceptibility or resistance. This report provides documentation of the distribution of periodontitis in this large nonhuman primate colony and the relationship of this distribution to the matrilineage of the cohort.
METHODS
Skull collection and matrilines
The CPRC Skeletal collection contains 569 specimens that were evaluated derived from 8 founding mothers, 065, 22, 116, 022, 073, 076, DM, and 004. Skeletons from these matrilines were assessed through 6–9 generations. The skulls were selected among animals ranging from 5–31 years of age. An extensive pedigree history of the nonhuman primates enabled determination of the approximate age of each animal at the time of death. Thus, the “population” could be stratified based upon age of the sample. Detailed analysis of the alveolar bone characteristics were acquired throughout the maxillary and mandibular quadrants (32). These periodontal characteristics were related to age and matriline of each animal.
No animals were sacrificed for this study. Rather, material from a pre-existing skeletal collection served as the database. Institutional Animal Care and Use Committee (IACUC) review as not required for analysis of the skeletons, as this is defined as an exempt protocol. Living rhesus monkeys (n=79; 42 females and 37 males) housed at the Caribbean Primate Research Center (CPRC) at Sabana Seca, Puerto Rico, who were members of the 8 matrilines were also used in this study. A protocol approved by the IACUC of the University of Puerto Rico, enabled anesthetized animals to be examined for clinical measures of periodontal health including probing pocket depth (PPD), and bleeding on probing (BOP), as we have described previously (31). Periodontitis was defined as mean mouth values of PPD≥3mm and BOP≥1. The nonhuman primates were typically fed a 20% protein, 5% fat, and 10% fiber commercial monkey diet (diet 8773, Teklad NIB primate diet modified: Harlan Teklad). The diet was supplemented with fruits and vegetables, and water was provided ad libitum in an enclosed corral setting. This research adhered to the American Society of Primatologists Principles for the Ethical Treatment of Nonhuman primates.
Periodontal bone defect measures in skulls
Periodontal disease was evaluated using a standard UNC-15 periodontal probe by determining the distance from the deepest extent of the vertical bone to the cemento-enamel junction (CEJ) on 1stand 2nd premolars and 1st and 2nd molars from all 4 quadrants (32). The alveolar bone levels were measured on both mesiobuccal and distobuccal sites for each tooth since periodontitis is frequently developed in these sites by non-human primates. Data was assessed for total bone loss defects (mm; BD), as well as frequency of sites with loss that was greater than 4 mm and 5 mm. Missing teeth that demonstrated alveolar bone loss were assigned a value of 10 based upon the maximal magnitude of bone loss that was detected across the population for existing diseased teeth.
Statistical analyses
Differences among the matrilines in bone defect levels and frequency of sites with bone defects were evaluated using an ANOVA with post hoc testing of groups using Tukey’s HSD method (SigmaStat, Systat Software, Inc., Richmond, CA). Differences in frequency of affected animals used a X2 test for 2x2 contingency analysis with Yates correction. Data with an alpha <0.05 (after being adjusted for the multiple comparisons) were accepted as statistically significant using a two-tailed analysis. Correlation analyses were conducted using a Pearson correlation coefficient for linear relationships between the age and bone defects.
RESULTS
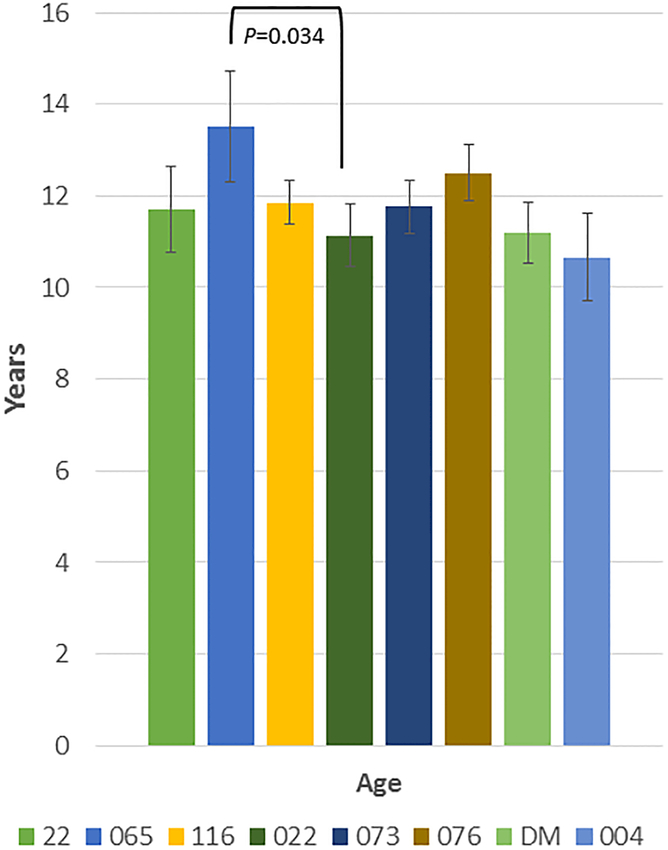
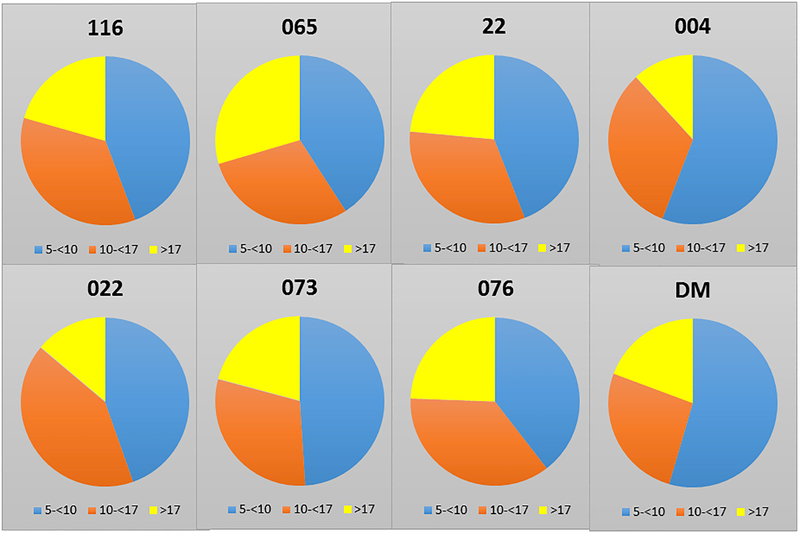
Table 1 summarizes the demographics of the population that was examined with 569 skulls from the 8 matrilines over 6–9 generations being evaluated. Fig. 1A shows that only the age range between matriline 065 and 022 were significantly different among all the groups. Figure 1B provides additional information on the proportions of animals from each matriline across the age range. No differences were noted in the distribution of ages among any of the matrilines.
Table 1:
Demographics of the population and matrilines
| Matriline | N | % Male | Generations | Age range (yrs.) |
|---|---|---|---|---|
| 22 | 32 | 46.8 | 9 | 5.12–23.50 |
| 065 | 44 | 43.5 | 7 | 5.10–31.44 |
| 116 | 126 | 34.6 | 8 | 5.81–24.92 |
| 022 | 64 | 24.8 | 9 | 5.02–23.87 |
| 073 | 96 | 37.9 | 7 | 5.00–25.19 |
| 076 | 86 | 42.6 | 6 | 5.22–28.95 |
| DM | 87 | 19.4 | 8 | 5.04–26.26 |
| 004 | 34 | 45.8 | 8 | 5.08–22.64 |
Figure 1:
(A) Age of the members of each matriline whose skulls were evaluated. Bars denote mean age ± SD (vertical brackets) (B) Distribution of animals from different age groups in each of the matrilines.
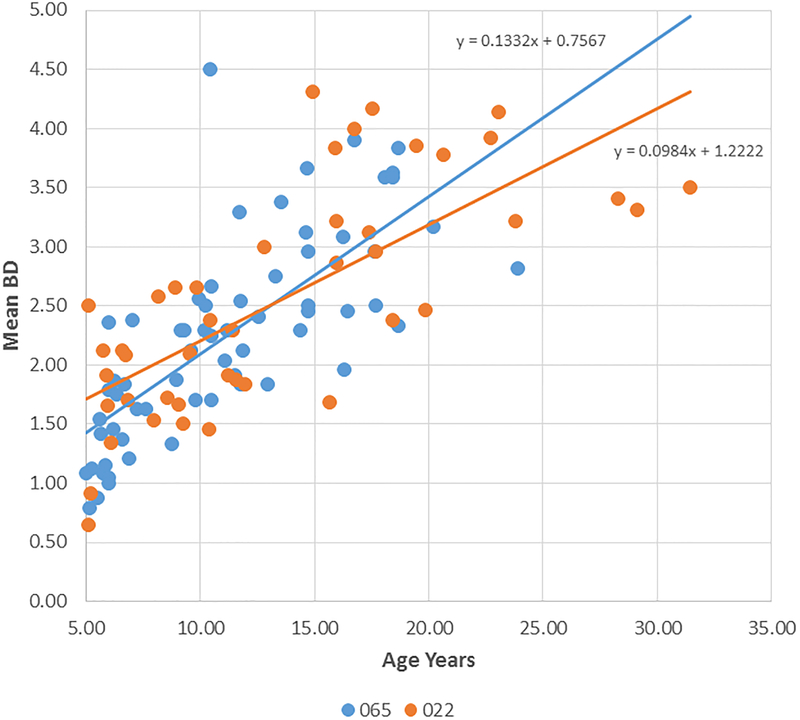
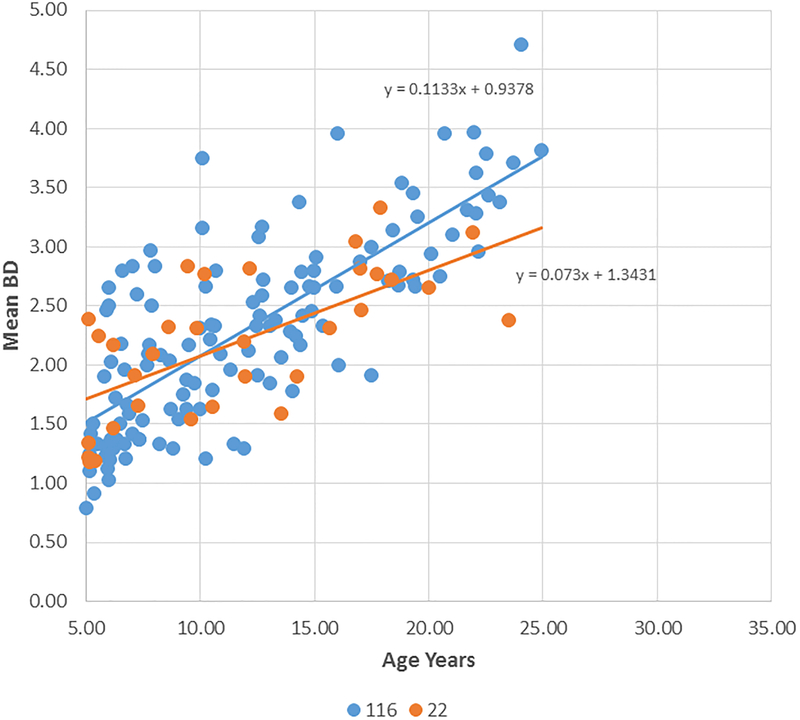
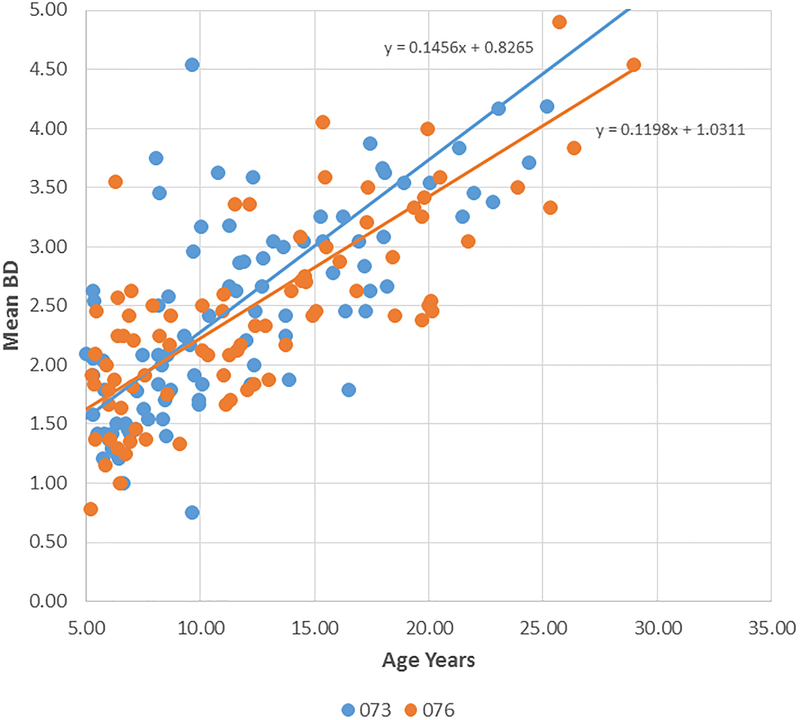
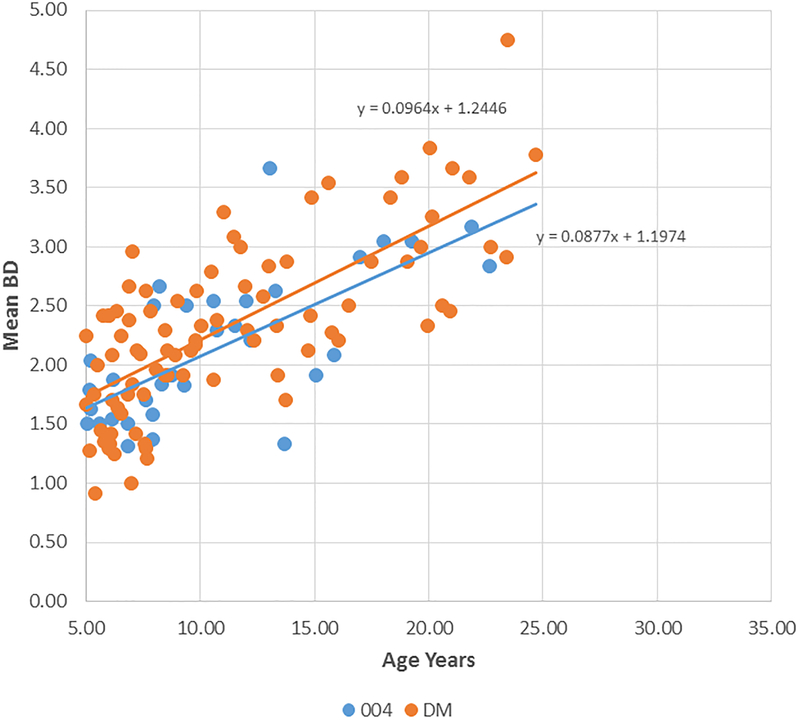
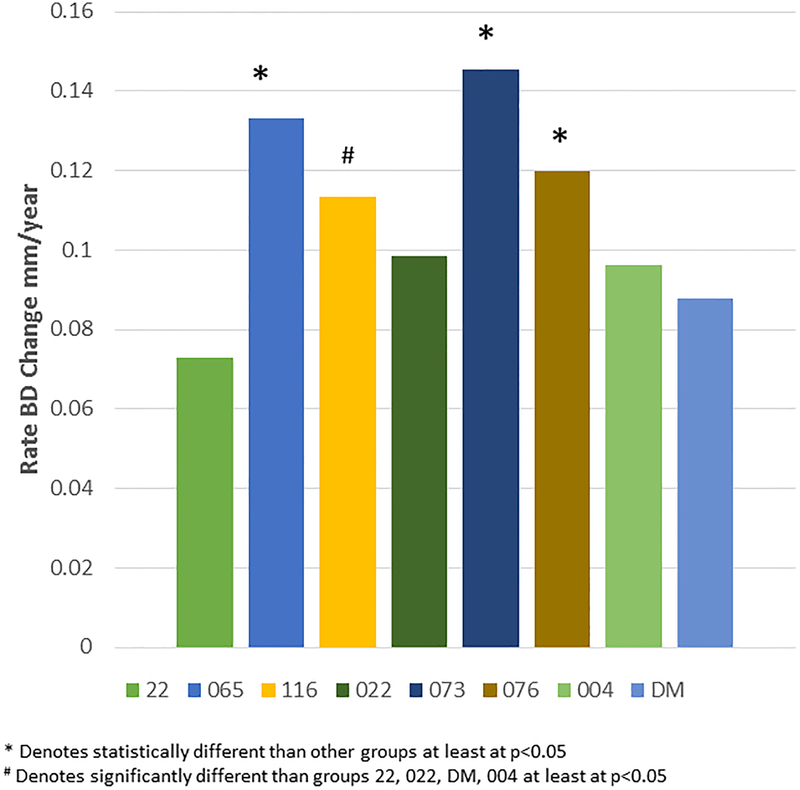
Figure 2A-D provide summaries of the relationship of the bone loss data depicting mean bone defect (mm) for individual animals across the entire age range in each of the 8 matrilines. There was a clear increase in disease with aging in each of the family units, although most of the matrilines demonstrated rather similar levels of bone defects early in life (i.e. 5 years; comparable to approximately 20 human years). Fig. 2E summarizes these differences and shows significantly increased rate of bone defect formation with aging in matrilines 065, 073, and 076 compared to all other groups. Additionally, matriline 116 animals showed a significantly increased rate of bone defects compared to the other 4 matrilines (22, 022, DM, 004).
Figure 2:
Correlation analysis of bone defects (BD) with age for all matrilines. (A-D) demonstrate the correlation of mean bone defect measures on maxillary and mandibular premolars and molars for each skull in the various matrilines, with linear regression noted. (E) The mean rate of bone defect formation for each matriline over the entire age of the members is shown. Asterisk (*) denotes significantly different than other groups at least a p<0.05 and the hashtag (#) denotes the group that as significantly different from 22, 022, 004 and DM at least at p<0.05.
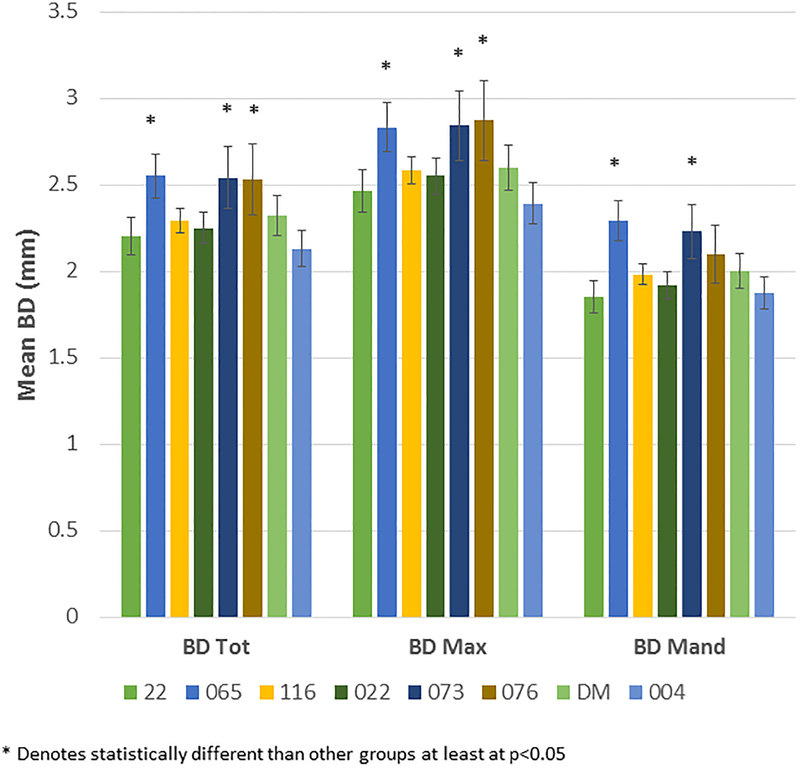
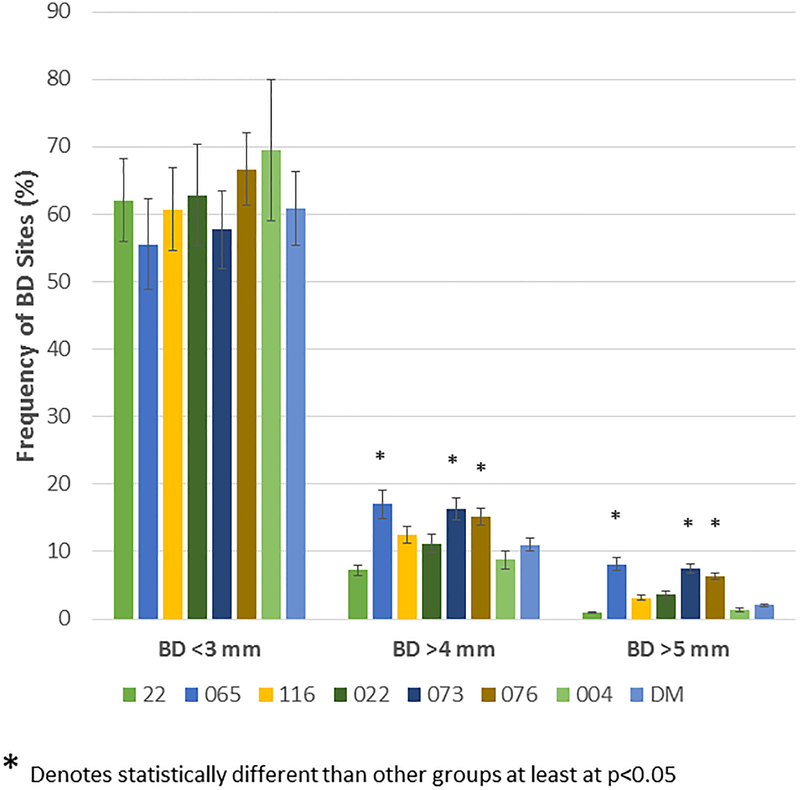
Figure 3A demonstrates the mean bone defect for the entire mouth (BD Tot), for the maxillary quadrants (BD Max) and mandibular quadrants (BD Mand). These data show, as was suggested by the age progression data, that the 065, 073 and 076 matriline animals exhibited significantly greater disease than the other groups, generally in both the maxilla and mandible. Extending this information, Figure 3B shows the severity of disease in the animals as a percentage of sites with greater levels of alveolar bone loss. The 065, 073, and 076 animals demonstrated a greater frequency of sites with bone defects >4mm and >5mm compared to the other groups. Figure 3C summarizes the frequency of more severe disease identified at the animal level across all animals in each of the 8 matrilines and demonstrates a heightened level of disease severity in a greater number of animals in the 065, 073, and 076 matrilines.
Figure 3:
(A) Bars denote the mean bone defect for the matrilines for total mouth (BD Tol), maxillary (BD Max) and mandibular (BD Mand) quadrants. Vertical brackets enclose 1 SD. (B) Bars denote the mean % of sites with bone measures of < 3, >4, and >5 mm in each skull stratified by matriline. Asterisk (*) denotes significantly different than other groups at least a p<0.05. (C) Bars signify the frequency of skulls (animals) in each matriline that demonstrated bone defects of >5 mm. Asterisk (*) denotes significantly different than other groups at least a p<0.05.
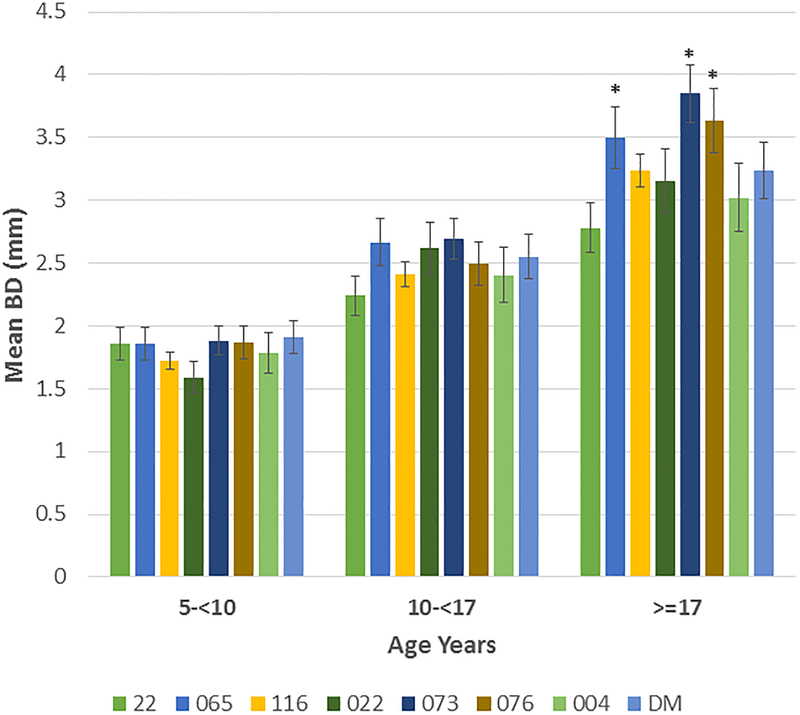
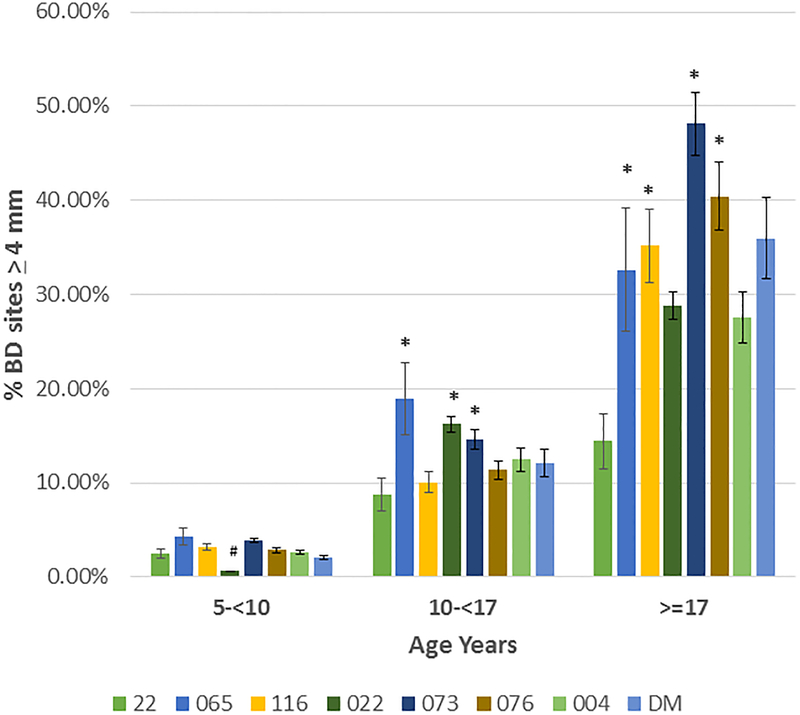
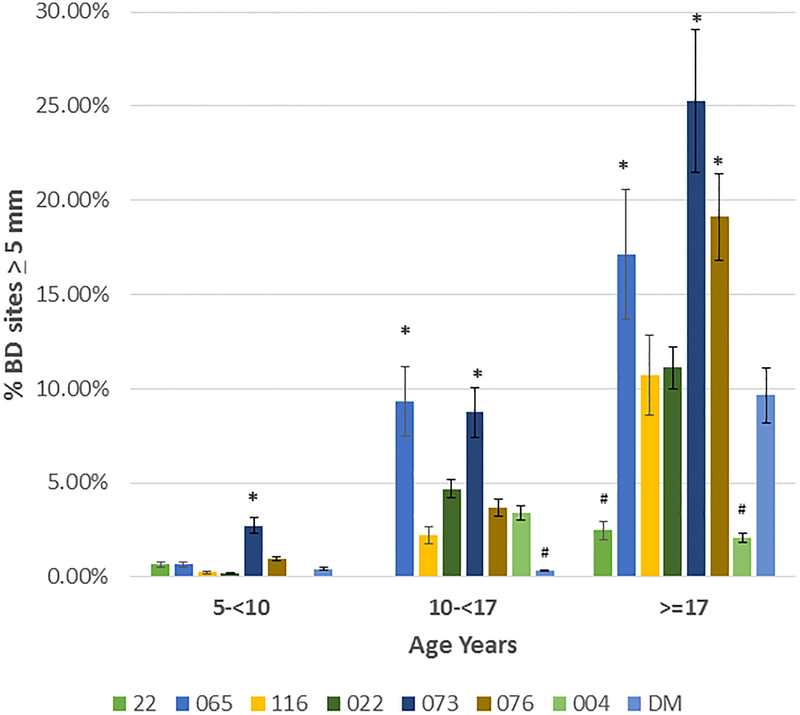
Based upon the correlation of aging and bone defect expression across all of the matrilines, Figure 4A depicts the data describing the expression of disease stratified by age category and analyzed within the 8 matrilines. In younger animals, no differences were observed in mean bone defect across all sites evaluated. However, increases in disease appeared in the 10–17 year old animals from the 065, 073, and 076 matrilines compared to each of the other groups. Within the population greater than17 years of age, elevated disease levels were seen in the 065, 073 and 076 matrilines. Figure 4B&C summarize the severity data stratified with age across the matrilines. Members of 065, 073, and 076 families demonstrate more severe disease in the subset >10 years of age compared to the other groups for bone defects ≥4 mm and ≥5 mm. Of interest was that older members of families 116 and DM demonstrated increased disease, the early onset of more severe disease in the 073 matriline, and matrilines 22 and 004 members that appeared to be relatively resistant to severe disease even in older animals.
Figure 4:
(A) demonstrates the mean bone defect for the skulls in each matriline stratified according to age of the animal at the time of death. (B) description of frequency of sites in skulls of each matriline stratified on age. (C) description of frequency of sites in skulls of each matriline stratified on age. Bars denote group mean and vertical brackets enclose 1 SD. Asterisk (*) denotes significantly different than other groups at least a p<0.05. Hashtag (#) denotes the groups with significantly less disease compared to all other matrilines at least at p<0.05.
Table 2 provides a summary that links the multi-generational data derived from the skulls within the matrilines to the presentation of naturally-occurring periodontitis in living descendants of these families. The findings represent results from animals who were randomly selected and periodontal disease evaluated for participation in ongoing studies of aging effects on periodontitis and mucosal immune responses (39–46). Within the 3 matrilines demonstrating the least periodontal disease in the skulls, few living animals showed any naturally-occurring disease, with <30% presenting with any disease even >10 years of age (~35 human years). In contrast, the susceptible matrilines demonstrated increases in expression of periodontitis in both younger and older animals (p<0.06) that were 2–5-fold elevated in frequency.
Table 2:
Characteristics of prevalence of periodontitis in living descendants of the matrilines. Data reflect the numbers of animals for each matriline and at each age determined to be periodontally healthy or demonstrating periodontitis. Asterisks denotes different from high risk matriline members ≥10 years of age group.
| Matriline | Total | Healthy | Periodontitis | |||
|---|---|---|---|---|---|---|
| <10 yr. | ≥10 yrs | <10 yr. | ≥10 yrs | <10 yr. | ≥10 yrs | |
| 004 | 0 | 2 | 0 | 1 | 0 | 1 (50.0%) |
| 022 | 17 | 11 | 16 | 9 | 1 (5.9%) |
2 (22.2%) |
| DM | 1 | 4 | 1 | 2 | 0 | 2 (50.0%) |
| Total | 18 | 17 | 17 | 12 | 1 (5.6%) |
5* (29.4%) |
| 065 | 0 | 7 | 0 | 4 | 0 | 3 (42.9%) |
| 073 | 9 | 10 | 7 | 4 | 2 (22.2%) |
6 (60.0%) |
| 076 | 6 | 7 | 4 | 2 | 2 (33.3%) |
5 (71.4%) |
| Total | 15 | 24 | 11 | 10 | 4 (26.7%) |
14* (583%) |
| 116 | 1 | 4 | 1 | 3 | 0 | 1 (25.0%) |
DISCUSSION
Periodontitis affects primarily adults on a global scale that negatively impacts quality of life, workplace productivity, and social interactions (47, 48). The disease represents a polymicrobial infection that results in a chronic inflammatory response resulting in soft and hard tissue damage and is the primary reason for tooth loss in adults (7, 49).
Numerous disease phenotypes, including periodontitis, are quantitative under natural conditions, with a complex etiology, encompassing multiple environmental and genetic triggers. Since we have been documenting the clinical, microbiological, and immunologic aspects of periodontitis in nonhuman primates for many decades, this investigation tested the hypothesis that periodontitis will be increased in specific nonhuman primate pedigrees in which the multigenerational families demonstrate variations in susceptibility to the expression of this disease. The results demonstrated a clear familial relationship of the onset and severity of disease expression in the matrilines of nonhuman primates comprising the population of animals at Cayo Santiago.
We evaluated a large population of rhesus monkeys spanning 6–9 generations via an historic skeleton collection across 8 matrilines in animals aged 5–31 years. The results demonstrated that within this population, animals derived from the 3 matrilines (065, 073, 076) appeared to show an increased susceptibility to periodontitis with 29–45% of the family members demonstrating bone defects of ≥5 mm and mean mouth bone defects significantly greater than the other 5 matrilines. This susceptibility was also reflected by the rates of disease expression with the amount of bone loss was similar to the other matrilines at a youngest age, but each showed a significantly more rapid increase in disease extent and severity with aging. Distinctive features within the matrilines was that 073 members showed more severe disease sites at a younger age, and both 065 and 073 matrilines demonstrated more severe disease presentation even within the 10-<17 year old grouping equivalent to approximately 35–60 year old humans). Additionally, we identified that matrilines 22, 022, 004 and DM showed less bone loss in extent and severity at all ages. Both matriline 22 and 004 members actually showed minimal severe disease, even in the oldest age group ≥17 years. Finally, matriline 116, 073, 076 and DM members ≥17 years demonstrated a substantial increase in more severe disease (3–3.5-fold), compared to the data from the 10-<17 year subset, which suggests that there could be differences in the age-related onset of disease between matrilines.
Based upon the resources of the Caribbean Primate Research Center and the colony of animals derived from Cayo Santiago, we had the ability to evaluate periodontitis in living descendants of these various matrilines. Our hypothesis was that living members of the susceptible matrilines would demonstrate an elevated prevalence of naturally-occurring periodontitis. The results showed that, in fact, this was true, as well as the matrilines that appeared to show some resistance to disease even with aging had less periodontal disease at the individual monkey level.
While substantial information is available on tooth wear and aging and diet effects on dentition, only a single report (50) described the dentition from about 180 wild-shot great apes related to potential periodontitis. These researcher’s findings suggested no change in alveolar bone height from young adulthood to old age. They interpreted their findings that with increasing tooth wear there is a compensatory eruption to maintain a constant height of tooth tissue, and that chronic pulpo/periodontal infections were the basis of alveolar bone and tooth loss in these great apes. Although, our measurement approach for the skulls could have been impacted somewhat due to compensatory eruption, the frequency and severity of bone defects that were detected suggested potential differences in aging effects on macaques versus great apes, as well as differences in a more controlled environment (eg. Cayo Santiago) compared to free-ranging wild animals.
This retrospective study allowed us to track the expression of periodontitis over 9 generations with a family, as well as having numerous members of each family in any given generation (data not shown); however, the underlying etiology for these differences across the matrilines cannot be discerned at this point. The current paradigm of human periodontitis extent and severity focuses on the interaction of an altered (dysbiotic) oral microbial ecology and a dysregulated host response attempting to control this chronic infection that appears to reflect both genetic predisposition and environmental modulation of these responses (51–54). While the decedents in this matrilines provided us some novel insights of a human model of periodontitis, the expanded opportunities to use this nonhuman primate model are based upon the descendants of each of these matrilines that still exist within the population. The initial data presented in this report support that these living animals demonstrate some conservation of susceptibility or resistance to the individual matrilines. Thus, prospective studies can be performed on these animals using a ligature-induced model of disease to assess the interactions of the microbiome and host responses during the initiation and progression of periodontitis to better understand these factors as explanatory variables in disease extent and severity in the human population. Furthermore, recent studies have demonstrated similarities and overlap in SNPs between humans and macaques (55–58), as well as clear epigenetic evidence for altered biological functions in these nonhuman primates (59, 60). Thus, this model may also provide access to a better understanding of the genetic contributors to periodontitis susceptibility.
ACKNOWLEDGMENTS
This work was supported by P20 GM103538 from the National Institute of General Medical Sciences, the National Center for Research Resources (NCRR) and the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health (NIH) through Grant Number 5P40OD012217 to the Caribbean Primate Research Center. Infrastructure support was provided, in part, by grants from the National Center for Research Resources G12RR003051 (National Center for Research Resources) and G12MD007600 (National Institute on Minority Health and Health Disparities) from the National Institutes of Health. The authors acknowledge no conflict of interest with the support for this research project.
REFERENCES
- (1).Rhodin K, Divaris K, North KE, et al. Chronic periodontitis genome-wide association studies: gene-centric and gene set enrichment analyses. J Dent Res 2014; 93: 882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Larsson L Current Concepts of Epigenetics and Its Role in Periodontitis. Current oral health reports 2017; 4: 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Genco RJ, Borgnakke WS. Risk factors for periodontal disease. Periodontol 2000 2013; 62: 59–94. [DOI] [PubMed] [Google Scholar]
- (4).Demmer RT, Papapanou PN. Epidemiologic patterns of chronic and aggressive periodontitis. Periodontol 2000 2010; 53: 28–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Yoshie H, Kobayashi T, Tai H, Galicia JC. The role of genetic polymorphisms in periodontitis. Periodontol 2000 2007; 43: 102–132. [DOI] [PubMed] [Google Scholar]
- (6).Kinane DF, Shiba H, Hart TC. The genetic basis of periodontitis. Periodontol 2000 2005; 39: 91–117. [DOI] [PubMed] [Google Scholar]
- (7).Armitage GC, Cullinan MP. Comparison of the clinical features of chronic and aggressive periodontitis. Periodontol 2000 2010; 53: 12–27. [DOI] [PubMed] [Google Scholar]
- (8).Carlsson G, Wahlin YB, Johansson A, et al. Periodontal disease in patients from the original Kostmann family with severe congenital neutropenia. J Periodontol 2006; 77: 744–751. [DOI] [PubMed] [Google Scholar]
- (9).Hodge PJ, Teague PW, Wright AF, Kinane DF. Clinical and genetic analysis of a large North European Caucasian family affected by early-onset periodontitis. J Dent Res 2000; 79: 857–863. [DOI] [PubMed] [Google Scholar]
- (10).Marazita ML, Burmeister JA, Gunsolley JC, Koertge TE, Lake K, Schenkein HA. Evidence for autosomal dominant inheritance and race-specific heterogeneity in early-onset periodontitis. J Periodontol 1994; 65: 623–630. [DOI] [PubMed] [Google Scholar]
- (11).Page RC, Vandesteen GE, Ebersole JL, Williams BL, Dixon IL, Altman LC. Clinical and laboratory studies of a family with a high prevalence of juvenile periodontitis. J Periodontol 1985; 56: 602–610. [DOI] [PubMed] [Google Scholar]
- (12).Kinane DF, Hodge P, Eskdale J, Ellis R, Gallagher G. Analysis of genetic polymorphisms at the interleukin-10 and tumour necrosis factor loci in early-onset periodontitis. J Periodontal Res 1999; 34: 379–386. [DOI] [PubMed] [Google Scholar]
- (13).Waldrop TC, Anderson DC, Hallmon WW, Schmalstieg FC, Jacobs RL. Periodontal manifestations of the heritable Mac-1, LFA-1, deficiency syndrome. Clinical, histopathologic and molecular characteristics. J Periodontol 1987; 58: 400–416. [DOI] [PubMed] [Google Scholar]
- (14).Hart TC, Hart PS, Michalec MD, et al. Localisation of a gene for prepubertal periodontitis to chromosome 11q14 and identification of a cathepsin C gene mutation. Journal of medical genetics 2000; 37: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Dogan B, Kipalev AS, Okte E, Sultan N, Asikainen SE. Consistent intrafamilial transmission of Actinobacillus actinomycetemcomitans despite clonal diversity. J Periodontol 2008; 79: 307–315. [DOI] [PubMed] [Google Scholar]
- (16).Kilian M, Frandsen EV, Haubek D, Poulsen K. The etiology of periodontal disease revisited by population genetic analysis. Periodontol 2000 2006; 42: 158–179. [DOI] [PubMed] [Google Scholar]
- (17).Papapanou PN. Population studies of microbial ecology in periodontal health and disease. Annals of periodontology / the American Academy of Periodontology 2002; 7: 54–61. [DOI] [PubMed] [Google Scholar]
- (18).Schenkein HA, Van Dyke TE. Early-onset periodontitis: systemic aspects of etiology and pathogenesis. Periodontol 2000 1994; 6: 7–25. [DOI] [PubMed] [Google Scholar]
- (19).Zambon JJ, Christersson LA, Slots J. Actinobacillus actinomycetemcomitans in human periodontal disease. Prevalence in patient groups and distribution of biotypes and serotypes within families. J Periodontol 1983; 54: 707–711. [DOI] [PubMed] [Google Scholar]
- (20).Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Critical reviews in oral biology and medicine: an official publication of the American Association of Oral Biologists 2003; 14: 430–449. [DOI] [PubMed] [Google Scholar]
- (21).Divaris K, Monda KL, North KE, et al. Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Human molecular genetics 2013; 22: 2312–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Divaris K, Monda KL, North KE, et al. Genome-wide association study of periodontal pathogen colonization. J Dent Res 2012; 91: 21S–28S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Feng P, Wang X, Casado PL, et al. Genome wide association scan for chronic periodontitis implicates novel locus. BMC oral health 2014; 14: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Michalowicz BS. Genetic and heritable risk factors in periodontal disease. J Periodontol 1994; 65: 479–488. [DOI] [PubMed] [Google Scholar]
- (25).Michalowicz BS. Genetic risk factors for the periodontal diseases. Compendium 1994; 15: 1036, 1038, 1040 passim. [PubMed] [Google Scholar]
- (26).Oz HS, Puleo DA. Animal models for periodontal disease. Journal of biomedicine & biotechnology 2011; 2011: 754857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Struillou X, Boutigny H, Soueidan A, Layrolle P. Experimental animal models in periodontology: a review. Open Dent J 2010; 4: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Schou S, Holmstrup P, Kornman KS. Non-human primates used in studies of periodontal disease pathogenesis: a review of the literature. J Periodontol 1993; 64: 497–508. [DOI] [PubMed] [Google Scholar]
- (29).Gonzalez OA, Novak MJ, Kirakodu S, et al. Comparative analysis of gingival tissue antigen presentation pathways in ageing and periodontitis. J Clin Periodontol 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Gonzalez OA, Stromberg AJ, Huggins PM, Gonzalez-Martinez J, Novak MJ, Ebersole JL. Apoptotic genes are differentially expressed in aged gingival tissue. J Dent Res 2011; 90: 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ebersole JL, Steffen MJ, Gonzalez-Martinez J, Novak MJ. Effects of age and oral disease on systemic inflammatory and immune parameters in nonhuman primates. Clin Vaccine Immunol 2008; 15: 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gonzalez OA, Orraca L, Kensler TB, Gonzalez-Martinez J, Maldonado E, Ebersole JL. Familial periodontal disease in the cayo santiago rhesus macaques. American journal of primatology 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Kessler MJ, Berard JD. A brief description of the Cayo Santiago rhesus monkey colony. Puerto Rico health sciences journal 1989; 8: 55–59. [PubMed] [Google Scholar]
- (34).Rothschild BM, Hong N, Turnquist JE. Skeletal survey of Cayo Santiago rhesus macaques: osteoarthritis and articular plate excrescences. Seminars in arthritis and rheumatism 1999; 29: 100–111. [DOI] [PubMed] [Google Scholar]
- (35).Hernandez-Pacheco R, Rawlins RG, Kessler MJ, et al. Demographic variability and density-dependent dynamics of a free-ranging rhesus macaque population. American journal of primatology 2013; 75: 1152–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wang Q, Opperman LA, Havill LM, Carlson DS, Dechow PC. Inheritance of sutural pattern at the pterion in Rhesus monkey skulls. The anatomical record Part A, Discoveries in molecular, cellular, and evolutionary biology 2006; 288: 1042–1049. [DOI] [PubMed] [Google Scholar]
- (37).Wang Q, Strait DS, Dechow PC. Fusion patterns of craniofacial sutures in rhesus monkey skulls of known age and sex from Cayo Santiago. American journal of physical anthropology 2006; 131: 469–485. [DOI] [PubMed] [Google Scholar]
- (38).Wang Q, Dechow PC, Hens SM. Ontogeny and diachronic changes in sexual dimorphism in the craniofacial skeleton of rhesus macaques from Cayo Santiago, Puerto Rico. Journal of human evolution 2007; 53: 350–361. [DOI] [PubMed] [Google Scholar]
- (39).Gonzalez OA, Novak MJ, Kirakodu S, et al. Comparative analysis of gingival tissue antigen presentation pathways in ageing and periodontitis. J Clin Periodontol 2014; 41: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ebersole JL, Kirakodu S, Novak MJ, et al. Cytokine gene expression profiles during initiation, progression and resolution of periodontitis. J Clin Periodontol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Gonzalez OA, John Novak M, Kirakodu S, et al. Effects of aging on apoptosis gene expression in oral mucosal tissues. Apoptosis 2013; 18: 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Gonzalez OA, Kirakodu S, Novak MJ, et al. Comparative analysis of microbial sensing molecules in mucosal tissues with aging. Immunobiology 2018; 223: 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Gonzalez OA, Nagarajan R, Novak MJ, et al. Immune system transcriptome in gingival tissues of young nonhuman primates. J Periodontal Res 2016; 51: 152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ebersole JL, Kirakodu SS, Novak MJ, et al. Transcriptome Analysis of B Cell Immune Functions in Periodontitis: Mucosal Tissue Responses to the Oral Microbiome in Aging . Frontiers in immunology 2016; 7: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ebersole JL, Kirakodu S, Novak MJ, et al. Effects of aging in the expression of NOD-like receptors and inflammasome-related genes in oral mucosa. Mol Oral Microbiol 2016; 31: 18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Gonzalez OA, Novak MJ, Kirakodu S, et al. Differential Gene Expression Profiles Reflecting Macrophage Polarization in Aging and Periodontitis Gingival Tissues. Immunological investigations 2015; 44: 643–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Richards D Oral diseases affect some 3.9 billion people. Evidence-based dentistry 2013; 14: 35. [DOI] [PubMed] [Google Scholar]
- (48).Marcenes W, Kassebaum NJ, Bernabe E, et al. Global burden of oral conditions in 1990–2010: a systematic analysis. J Dent Res 2013; 92: 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Armitage GC, Robertson PB. The biology, prevention, diagnosis and treatment of periodontal diseases: scientific advances in the United States. J Am Dent Assoc 2009; 140 Suppl 1: 36S–43S. [DOI] [PubMed] [Google Scholar]
- (50).Dean M, Jones M, Pilley J. The natural history of tooth wear, continuous eruption and periodontal disease in wild shot great apes. Journal of human evolution 1992; 22: 23–39. [Google Scholar]
- (51).Kornman KS, Polverini PJ. Clinical application of genetics to guide prevention and treatment of oral diseases. Clinical genetics 2014; 86: 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wade WG. The oral microbiome in health and disease. Pharmacol Res 2013; 69: 137–143. [DOI] [PubMed] [Google Scholar]
- (53).Ebersole JL, Dawson DR 3rd, Morford LA, Peyyala R, Miller CS, Gonzalez OA. Periodontal disease immunology: ‘double indemnity’ in protecting the host. Periodontol 2000 2013; 62: 163–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hajishengallis G, Krauss JL, Liang S, McIntosh ML, Lambris JD. Pathogenic microbes and community service through manipulation of innate immunity. Adv Exp Med Biol 2012; 946: 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Rogers J The behavioral genetics of nonhuman primates: Status and prospects. American journal of physical anthropology 2018; 165 Suppl 65: 23–36. [DOI] [PubMed] [Google Scholar]
- (56).Xue C, Raveendran M, Harris RA, et al. The population genomics of rhesus macaques (Macaca mulatta) based on whole-genome sequences. Genome research 2016; 26: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Haus T, Ferguson B, Rogers J, et al. Genome typing of nonhuman primate models: implications for biomedical research. Trends in genetics: TIG 2014; 30: 482–487. [DOI] [PubMed] [Google Scholar]
- (58).Kanthaswamy S, Johnson Z, Trask JS, et al. Development and validation of a SNP-based assay for inferring the genetic ancestry of rhesus macaques (Macaca mulatta). American journal of primatology 2014; 76: 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Maegawa S, Lu Y, Tahara T, et al. Caloric restriction delays age-related methylation drift. Nature communications 2017; 8: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Cain CE, Blekhman R, Marioni JC, Gilad Y. Gene expression differences among primates are associated with changes in a histone epigenetic modification. Genetics 2011; 187: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]