Abstract
In addition to the development of beta amyloid plaques and neurofibrillary tangles, Alzheimer's disease (AD) involves the loss of connecting structures including degeneration of myelinated axons and synaptic connections. However, the extent to which white matter tracts change longitudinally, particularly in the asymptomatic, preclinical stage of AD, remains poorly characterized. In this study we used a novel graph wavelet algorithm to determine the extent to which microstructural brain changes evolve in concert with the development of AD neuropathology as observed using CSF biomarkers. A total of 118 participants with at least two diffusion tensor imaging (DTI) scans and one lumbar puncture for CSF were selected from two observational and longitudinally followed cohorts. CSF was assayed for pathology specific to AD (Aβ42 and phosphorylated-tau), neurodegeneration (total-tau), axonal degeneration (neurofilament light chain protein; NFL), and synaptic degeneration (neurogranin). Tractography was performed on DTI scans to obtain structural connectivity networks with 160 nodes where the nodes correspond to specific brain regions of interest (ROIs) and their connections were defined by DTI metrics (i.e., fractional anisotropy (FA) and mean diffusivity (MD)). For the analysis, we adopted a multi-resolution graph wavelet technique called Wavelet Connectivity Signature (WaCS) which derives higher order representations from DTI metrics at each brain connection. Our statistical analysis showed interactions between the CSF measures and the MRI time interval, such that elevated CSF biomarkers and longer time were associated with greater longitudinal changes in white matter microstructure (decreasing FA and increasing MD). Specifically, we detected a total of 17 fiber tracts whose WaCS representations showed an association between longitudinal decline in white matter microstructure and both CSF p-tau and neurogranin. While development of neurofibrillary tangles and synaptic degeneration are cortical phenomena, the results show that they are also associated with degeneration of underlying white matter tracts, a process which may eventually play a role in the development of cognitive decline and dementia.
Keywords: DTI tractography, CSF biomarker, Longitudinal brain connectivity, Alzheimer's disease pathology, Multi-resolution analysis
Highlights
-
•
Derives novel representation of brain connectivity identifying associations between changes in white matter tracts and CSF
-
•
Demonstrates statistically significant associations between white matter microstructure and CSF at the asymptomatic stage
-
•
Shows that biomarkers of both neurofibrillary tangles and synaptic dysfunction are associated with altered connectivity
1. Introduction
Loss of white matter connectivity is an increasingly recognized feature of Alzheimer's disease (AD) and other dementias (Jung et al., 2014; Mahoney et al., 2015; Mascalchi et al., 2017; Meijboom et al., 2017; Racine et al., 2017b). Post mortem, biochemical, and brain imaging studies show extensive white matter degeneration in AD (Balthazar et al., 2009; Baxter et al., 2006; Chaim et al., 2007; Im et al., 2008; Racine et al., 2017b; Salat et al., 2009; Teipel et al., 1998; Vermersch et al., 1994). However, the temporal emergence of white matter disconnection abnormalities and their interactions with cortical pathology are not well defined. Studies using diffusion tensor imaging (DTI) acquired in concert with cerebrospinal fluid (CSF) measures of AD pathology show that greater concentration of AD neuropathology is associated with altered microstructure among individuals with mild cognitive impairment (MCI) and AD (Li et al., 2014; Stenset et al., 2011), as well as among cognitively asymptomatic middle-aged and older adults (Bendlin et al., 2012; Gold et al., 2014; Molinuevo et al., 2014). The observed white matter changes may be an independent but co-occurring feature of AD, or it may be that amyloidogenic and neurodegenerative processes drive white matter changes very early in the disease course.
Longitudinal in vivo measurements of white matter microstructure are expected to shed light on how white matter alterations evolve in disease. Furthermore, given that changes in cognitively asymptomatic populations may be subtle, highly sensitive approaches are essential. In our recent work, we showed that using a multi-resolution analysis can significantly improve sensitivity to subtle alterations in white matter connectivity, in addition to providing a method for maintaining sensitivity in the face of multiple comparisons which are common in neuroimaging studies (Kim et al., 2013; 2015a). Considering a brain connectivity as a graph that consists of nodes and edges, we previously proposed a Wavelet Connectivity Signature (WaCS) that enables a multi-resolution view (i.e., coarse-to-fine details) of measures for connectivity strength between regions of interest (ROIs). In the traditional Euclidean setting (e.g.1-D or 2-D), multi-resolution view of a signal is obtained by performing band-pass filter operations in the frequency space. In our case where the domain is a graph, we leverage spectral graph theory to define its dual space (i.e., an analogue of the frequency space) and perform band-pass filtering in different bandwidths to obtain coarse-to-fine details of a signal on the graph. This operation yields the multi-resolution characteristics to define WaCS, which is essentially a higher-order representation that takes into account the connectivity strength of the specific fiber bundle concurrently with the connectivity strength of other fiber bundles in a graph theoretic sense (Kim et al., 2015a). Utilizing WaCS has shown improved sensitivity for detecting group differences and brain alterations present in neuroimaging data over the use of raw measurements only (Hwang et al., 2018a; Kim et al., 2013; 2015a; 2012; 2015b; 2014).
To investigate whether development of AD pathology is associated with progressively impaired connectivity of white matter, we examined the relationship between changes in WaCS based representations over an average of 2.7 years and CSF biomarkers of amyloid (Aβ42), neurofibrillary tangles (p-tau), axonal degeneration (neurofilament light chain protein; NFL), and synaptic dysfunction (neurogranin). Participants were late middle aged and cognitively asymptomatic, although several participants harbored increased risk for AD due to parental family history and APOE ε4 genotype. We hypothesized that biomarker levels indicative of greater AD pathology would be associated with decreased white matter connectivity strength captured in the multi-resolution WaCS representations, measured over time. This would suggest that elevated AD pathology is at least one driver of white matter degeneration, serving as a possible biological substrate underlying progressive cognitive impairment. Our paradigm also allowed us to test the utility of WaCS based signatures for detecting subtle changes in the longitudinal setting.
2. Materials and methods
2.1. Participants
We combined data collected from cognitively asymptomatic participants enrolled in two longitudinal cohort studies: 1) the Wisconsin Registry for Alzheimer's Prevention (WRAP) study and 2) the Wisconsin Alzheimer's Disease Research Center (ADRC). The WRAP is a longitudinal observational study of middle-aged adults who were cognitively normal at study entry, and approximately 70% of the cohort has a parental history of AD. The Wisconsin ADRC clinical core comprises participants at various AD stages ranging from cognitively asymptomatic to having dementia due to AD; to investigate early white matter changes, we limited our analysis to participants who were non-demented. In sum, the present investigation included 118 participants who were non-demented at cohort entry, completed at least two DTI scans, and underwent CSF collection by lumbar puncture (Racine et al., 2016). Among the 118 participants, n = 11 were Wisconsin ADRC subjects (mean age 55.57 years, σ = 6.62) and n = 107 were WRAP subjects (mean age 60.41 years, σ = 6.31). The mean age for the total sample (40 males and 78 females) was 59.8 (σ = 6.16) and the mean time period between the two DTI visits was 2.73 years (σ = 1.29). The University of Wisconsin Institutional Review Board approved all study procedures, each participant provided signed informed consent before participation, and all research was completed in accordance with the Helsinki Declaration. Also, the data that support the findings of this study are available from the corresponding author upon reasonable request without any information that may indicate specific participant in the study. The full demographics are reported in Table 1.
Table 1.
Demographics of the Wisconsin Registry for Alzheimer's Prevention (WRAP) and the Wisconsin Alzheimer's Disease Research Center (ADRC).
| Demographics | WRAP | Wisconsin ADRC | Total |
|---|---|---|---|
| Number of subjects | 107 | 11 | 118 |
| Sex (M/F) | 39/68 | 1/10 | 40/78 |
| Age (mean/σ) | 60.42/6.31 | 55.57/6.62 | 59.80/6.16 |
| Ptau in pg/ml (mean/σ) | 43.07/13.23 | 40.72/10.85 | 42.85/13 |
| Neurogranin in pg/ml (mean/σ) | 386.24/165.56 | 404.61/151.71 | 387.96/163.8 |
2.2. MRI acquisition
Participants were imaged on two identical General Electric 3.0 Tesla Discovery MR750 (Waukesha, WI) MRI systems fitted with an 8-channel head coil and using parallel imaging (ASSET). All participants in the W-ADRC dataset were imaged on one scanner, while all WRAP participants were imaged on a second, identical scanner. For both cohorts, Diffusion Tensor Imaging (DTI) was acquired using a diffusion-weighted, spin-echo, single-shot, echo planar imaging (EPI) pulse sequence in 40 encoding directions at b = 1300 s/mm2, with eight non-diffusion weighted (b = 0) reference images. The cerebrum was covered using contiguous 2.5 mm thick axial slices, FOV = 24 cm, TR = 8000 ms, TE = 67.8 ms, matrix = 96 × 96, resulting in isotropic 2.5 mm3 voxels. High order shimming was performed prior to the DTI acquisition to optimize the homogeneity of the magnetic field across the brain and to minimize EPI distortions. We employed a robust processing pipeline, based on methods in (Adluru et al., 2014). The eddy current correction, field map correction and brain mask segmentation were performed using tools from FSL, and the tensors were estimated using non-linear least squares method.
2.3. Deriving brain connectivity networks
Brain connectivity networks were derived using a 3-step process.
2.3.1. Coordinate system
Image registration was achieved following recommended procedures for deriving an unbiased coordinate system for 4D data (Keihaninejad et al., 2013). This approach reduces interpolation asymmetry that can arise if one were to select only one time point in a longitudinal dataset when generating the population/study level coordinate system as described in recent works (Keihaninejad et al., 2013; Yushkevich et al., 2010). We first estimated a subject-specific average that is temporally unbiased. The subject specific averages were then used to generate an unbiased population level average template space. Image registration was achieved using the Diffusion Tensor Imaging Toolkit (DTI-TK, http://dti-tk.sourceforge.net/pmwiki/pmwiki.php), an empirically validated software that supports affine and diffeomorphic transformations while allowing tensor interpolation and reorientation.
2.3.2. Nodes
The second step involved definition of node regions in gray matter. This was achieved using the gray matter atlas defined on a publicly available Illinois Institute of Technology-Diffusion Tensor Image (IIT-DTI) template (Varentsova et al., 2014) which provides 160 distinct brain regions (discarded regions labelled as ‘unknown’ and ‘cerebellum’). These regions (i.e., nodes) varies in their size and defined based on anatomical prior, and they share partial volume with the white matter around their boundaries due to limited MRI resolution. The white matter fiber tracts end in these regions due to stopping criteria used in the tracking algorithm.
The fractional anisotropy (FA) map of the IIT-DTI template provided a co-registered FA image with its gray matter atlas. Since our population atlas provide FA images and not T1-weighted images, we performed FA to FA registration by employing advanced normalization tools (ANTS, http://picsl.upenn.edu/software/ants). Here, the FA map was the fixed image and the IIT-DTI-FA image was the moving image. This FA to FA registration produced accurate registration between the IIT atlas and our population average FA image. The resulting transformation was then applied to the integer valued gray-matter atlas map with nearest neighbor interpolation to provide the 160 regions in the population coordinate system.
2.3.3. Edges/links in the connectivity graph
In the third step, we performed tractography to derive brain edges representing the structural networks/graph. The edges are composed of fiber bundles connecting pairs of gray matter regions, and they have rich information such as their tract curvatures and torsions (i.e., geometric shape information) as well as DTI measures defined along these tracts (i.e., functional values). Data were fit with a diffusion tensor model. The principal eigenvector of the diffusion tensor in a voxel provides a proxy to the predominant orientation of the white matter fibers in that voxel. A key factor in tractography algorithms is error accumulation. The two key techniques we employed to avoid the error issue were bootstrapping and tensor deflection (TEND) interpolation. Using this information, we repeated probabilistic tractography 20 times with implementations available in (Cook et al., 2006). Since the goal of this study is to identify fiber pathways connecting pairs of specific gray matter regions, we started the streamlines from each node region. For each pair of nodes, we merged the region-wise fibers and then filtered them to include only those that pass through both the regions. During the tractography processing, we found this approach produced qualitatively better fiber connections (i.e., lower false positives) compared to the streamlines that starts from the whole brain as a seed file and applying pairs of gray matter regions as end-point filters. The other criteria for tractography (for reproducibility) were as follows: (i) Curvature threshold of 45 degrees checked every 10 mm for stopping, and (ii) Euler integration step size of 0.1 mm for streamline generation. The stopping criteria did not involve any FA thresholding since the tracts are filtered using the gray matter regions. For tracking purpose, the gray matter regions were dilated twice but this was not for filtering. We used Camino (Cook et al., 2006) package to implement our protocol. Once the tracts connecting the pairwise gray matter ROIs were reconstructed, mean diffusivity (MD) and average FA in these tracts then served as the edge strength between the nodes.
2.4. Cerebrospinal fluid (CSF) biomarkers
CSF samples were collected after a minimum 4-hour fast and processed in the same manner as in (Starks et al., 2015). CSF t-tau, p-tau181 and Aβ42 were assayed using commercially available enzyme-linked immunosorbent assays (ELISA) (INNOTEST, Fujirebio, Ghent Belgium). Neurogranin concentration was measured using an in house ELISA and NFL was measured using the NF-Light kit from UmanDiagnostics (Umeå, Sweden), as previously described in (Mattsson et al., 2016). Lower levels of CSF Aβ42 indicate greater amyloid burden in the brain (Racine et al., 2016). High levels of NFL, neurogranin and p-tau respectively indicate axonal degeneration, synaptic degeneration and neurofibrillary tangle burden. CSF assays were completed in two separate batches. Due to minor batch-related differences in the components of research-grade assays, the values of a CSF analyte in one batch are not necessarily directly comparable to that same CSF analyte assayed in a different batch (Zhou et al., 2018). We therefore corrected for batch using a simple linear regression (SLR) model as described previously in (Racine et al., 2016), with the exception of NFL which was comparable between the two batches. Additionally, a square root transform was applied to neurogranin to correct for heteroscedasticity. These CSF measures together with the DTI metrics has been previously assessed in a longitudinal analysis as in (Racine et al., 2017a).
2.5. Multi-resolution connectivity signature using wavelet transform on graphs
The goal of this study was to identify brain connections that show close associations between longitudinal changes in structural brain connectivity and AD neuropathology as indexed by the CSF measures. To increase the sensitivity of our analysis, we adopted WaCS methodology introduced in (Kim et al., 2013; 2015a) which derives multi-resolution (higher order) descriptors on edges of a graph. WaCS obtains context information in multiple resolutions on graph edges based on the connectivity information of a graph. Such a multi-resolution strategy is the intuition behind feature descriptor matching schemes widely used in computer vision, where an image is considered as a continuous signal (Lowe, 2004; Morel and Yu, 2009; Shen and Ip, 1999). These traditional approaches for multi-resolution are not directly applicable on a complex domain such as brain connectivity graphs since they work only for signals in Euclidean spaces. Fortunately, wavelet transforms for graphs recently became available (Coifman and Maggioni, 2006; Hammond et al., 2011) which enable one to derive multi-resolution from signals defined in non-Euclidean spaces.
As described in (Kim et al., 2015a), in traditional wavelet approaches, a wavelet basis ψs, a(x) (i.e., a mother wavelet), an oscillating function at a specific scale s (i.e., dilation) centred at a specific location a, is first defined over the domain x in a Euclidean space. Given a signal f(x), this pre-defined basis function ψs, a(x) at multiple scales is used for the wavelet transform which derives the multi-resolution perspective of the original signal. However, when it comes to a graph, it is difficult to define such a basis function over the graph nodes n because the concepts of scale and localization become ambiguous with arbitrary structure of a graph. Spectral graph wavelet transform (SGWT) in (Hammond et al., 2011) overcame this difficulty by defining a wavelet in a 1D dual space which is an analogue of the frequency space and localizing it in the original space. To define the dual space, eigenvalues and eigenvectors pairs of a graph Laplacian (λl, χl), l = 0, 1, ⋯, N is used to define graph Fourier transform that acts on a signal f(n) as
where the is the resultant graph Fourier coefficient. Then, the wavelet basis is defined in the graph Fourier space as a kernel function g(λl) as
where the g() is a band-pass filter that represents a traditional wavelet basis in the frequency space. Here, the shape of the kernel function g() determines the shape of the wavelet basis ψs, n in the original graph space. With a mother wavelet ψs, n at hand, the spectral graph wavelet transform of a function f(n) is defined as
yielding a wavelet coefficient Wf(s, n) at scale s at centered n. The wavelet coefficients are the key ingredient to derive a multi-resolution view of a signal at different scales which encode coarse to fine details of the original signal at different scales.
To obtain WaCS, we need to take one step further. First, a brain network is considered as a graph G ∈ {V, E, ω} where the gray matter regions of interest (ROIs) become the nodes V and the edges E denote their connections/relations where the strength of the connections is described by the edge weights ω. Since the signal that we want to analyze is on the edges and not on the nodes, the SGWT described above cannot be applied directly. We therefore utilize a concept of a dual graph, i.e., a line graph L(G) of G that simply exchanges the roles of vertices and edges (Harary, 1969). Together with the SGWT, we obtain multi-resolution view of the edge signal (i.e., ω) via wavelet coefficients W(s, e) at each edge e at scale s as described in (Kim et al., 2013; 2015a) as
where S = {s0, s1, s2, ⋯, sJ} and J is the total number of scales without s0.
Our recent work have shown the utility of the WaCS for characterizing brain connectivity graphs by demonstrating its sensitivity in detecting subtle group differences in bipolar disease (Kim et al., 2013) and in family history of AD from preclinical AD data (Kim et al., 2015a). The strength of the approach is that by comparing local context of signals at each location, essentially higher order interaction terms, it is possible to achieve much greater reliability and accuracy versus a simple comparison of values at one resolution (which explains the broad use of this idea in feature matching (Lowe, 1999, Lowe, 2004; Morel and Yu, 2009; Shen and Ip, 1999; Won et al., 2013)).
For the derivation of the WaCS, we used the Multi-resolution Brain Connectivity Analysis (MBCA) toolbox (Kim et al., 2015a) together with the Spectral graph wavelet toolbox (SGWT) (Hammond et al., 2011). We obtained J = 5 scales of WaCS at each connection, then selected s = 2, 3, 4 scales which contained the most information based on observation of the eigenspectrum of L(G). The distribution of eigenvalues of graph Laplacian of L(G) demonstrated very small number of eigenvalues in scales corresponding to s = {0, 1},and we removed s = 5 (i.e., high frequency components) considering it as a noise component. The mother wavelet that was used in this study was a spline wavelet, which is the default wavelet provided by SGWT toolbox.
2.6. Statistical analysis
Since WaCS is a multi-scale descriptor which aggregates information from other fiber bundles neighboring to the fiber bundle of interest, we performed an analysis based on a multivariate general linear model (MGLM) at each connection to determine whether longitudinal connectivity changes in white matter connectivity are influenced by AD pathology or neural injury as indexed by each CSF biomarker. Under Gaussian assumption, our model included the covariates of age, sex, time interval between the longitudinal imaging visits (∆Time, 2.73 years in average), time interval between CSF collection (i.e., lumbar puncture) and baseline MRI (CSFtoMRI, 0.35 years in average), the cohort (WRAP or Wisconsin ADRC) from which the participant was recruited (Cohort) and the interaction term CSF × ∆Time. Because it was generally the case that WRAP participants were scanned on one GE 3 T scanner, and ADRC participants on an identical but separate GE 3 T scanner, we did not add an additional covariate for scanner (this was largely accounted for by the “cohort” covariate). The primary outcome was longitudinal changes in WaCS at each scale (i.e., ∆WaCS) which is multivariate, and the primary predictor of interest was the interaction CSF × ∆Time, where the variable CSF was an individual CSF biomarker as expressed by:
which reflects a hypothesis that ∆WaCS is independent from the interaction CSF × ∆Time. We chose to examine the interaction CSF × ∆Time rather than a main effect of each CSF biomarker because we expected there to be greater detectable changes in WaCS over longer intervals between DTI scans. This interaction term indicates that CSF biomarker level in addition to interval is important for predicting changes in connectivity over time. This hypothesis test was applied to WaCS of all existing connections using MGLM, and Wilks's λ was used to compute p-values. After running MGLM for WaCS of all connections, Bonferroni correction at α = 0.05 was used to correct for multiple comparisons. Effects surviving the correction for multiple comparisons (i.e., showing lower p-values than the correction threshold) provide a strong statistical evidence that the CSF biomarker together with the MRI interval are associated with the longitudinal strength changes observed in the WaCS representation of that connection. Since the connectivity matrix for each brain used in our experiment was sparse (i.e., only a few ROIs are structurally connected to each other) and symmetric, the total number of tests performed over the whole brain connectivity for a single CSF measure was 2964.
We tested the described model for each CSF biomarker of interest and ΔWaCSFA derived from FA and ΔWaCSMD derived from MD. Additionally, in order to investigate whether WaCS is more sensitive than conventional analysis of simple DTI metrics, we performed the same models as described above but substituted ΔFA or ΔMD in place of ∆WaCS. We again inferred significance of the model by examining the CSF × ∆Time predictor using the Bonferroni correction.
In the next step, we extracted FA and MD from tracts that were identified as significant in the WaCS model for further interrogation. While WaCS provides a sensitive approach to identify statistical associations, the underlying directional relationship between predictors and outcomes is not observable in the WaCS representation due to its multi-scale representation. Therefore, inferences about the directional association between the CSF biomarkers and the underlying white matter microstructure were determined by Pearson correlations between the CSF biomarker levels and the extracted simple MD and FA values from the connectivities identified in the WaCS analyses. The Pearson correlations are reported to facilitate interpretation of our primary findings using WaCS, and corresponding scatter plots are demonstrated in the Appendix.
3. Results
No significant brain connectivity edges were found between longitudinal simple MD/FA (i.e., raw measurements) and CSF measures of Aβ42, t-tau and NFL even with our multi-resolution framework using WaCS. However, we detected WaCS representations derived from several brain connections that were statistically significant and meaningful with p-tau and neurogranin together with longitudinal WaCS from MD and FA, which are demonstrated in the following subsections.
3.1. Mean diffusivity and CSF biomarkers
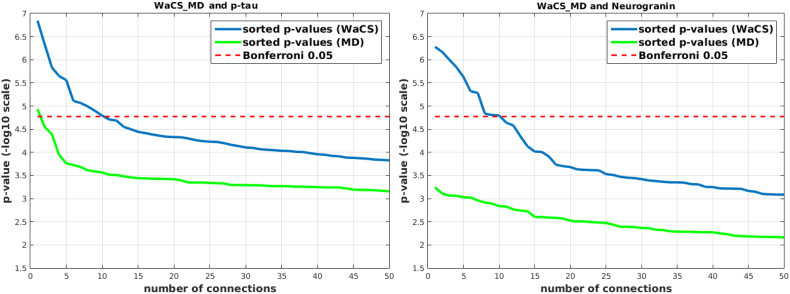
We identified statistically significant associations between the interaction between some CSF biomarkers (i.e., p-tau and neurogranin) and time (i.e., CSF × ∆Time) and ΔWaCSMD; the associated p-values are reported in Table 2, Table 3. The left and middle panels in Fig. 1 displays the p-values from ΔWaCSMD (blue) together with p-values from simple MD analysis (green) and the Bonferroni threshold (dashed red) in −log10 scale, where the connections above the threshold are statistically significant. Significant associations were identified with p-tau and neurogranin, but none with other CSF biomarkers (i.e., Aβ42, t-tau, and NFL). In contrast to the WaCS analyses, we detected only one connection for the p-tau analysis when we used the simple MD values as a response variable. Of note, this connection was also identified in ΔWaCSMD analyses.
Table 2.
Connections identified using ptau × ∆Time as a predictor for longitudinal changes in WaCSMD (multiple comparisons corrected using Bonferroni at 0.05) sorted by corresponding p-values (column 1–3), and the correlation between simple MD changes and ptau × ∆Time on the identified connections (column 4). Two ROIs (i.e., ROI 1 and ROI 2) represent one connection and the ROI indices corresponds to their labels in the IIT template.
| ROI 1 (IIT index) | ROI 2 (IIT index) | p-Value (1e-4) (WaCSMD) | Correlation (simple MD) |
|---|---|---|---|
| ctx_rh_G_and_S_frontomargin (12101) | ctx_rh_S_intrapariet_and_P_tran (12157) | 0.0014 | 0.3157 |
| ctx_rh_G_and_S_frontomargin (12101) | ctx_lh_G_and_S_transv_frontopol (11105) | 0.0046 | 0.0711 |
| Right-Hippocampus (53) | ctx_rh_G_and_S_frontomargin (12101) | 0.0147 | 0.3671 |
| ctx_rh_G_and_S_frontomargin (12101) | ctx_rh_G_front_middle (12115) | 0.0225 | 0.2554 |
| Right-Amygdala (54) | ctx_rh_G_cingulum-Post-ventral (12110) | 0.0274 | 0.3026 |
| Left-Caudate (11) | Right-Amygdala (54) | 0.0776 | 0.3071 |
| ctx_rh_G_and_S_frontomargin (12101) | ctx_rh_S_precentral-sup-part (12170) | 0.0859 | 0.2507 |
| Right-Caudate (50) | ctx_rh_G_and_S_frontomargin (12101) | 0.1022 | 0.4041 |
| Right-Hippocampus (53) | Left-Amygdala (18) | 0.1270 | 0.3483 |
| Left-Caudate (11) | Right-Hippocampus (53) | 0.1611 | 0.2017 |
ctx: cortex, lh: left hemisphere, rh: right hemisphere, S: sulcus, G: gyrus, sup: superior, inf: inferior, post: posterior, frontomargin: frontomarginal, pariet: parietal, tran/transv: transverse, frontopol: frontopolar.
Table 3.
Connections identified using neurogranin × ∆Time as a predictor for longitudinal changes in WaCSMD (multiple comparisons corrected using Bonferroni at 0.05) sorted by corresponding p-values (column 1–3), and the correlation between simple MD changes and neurogranin × ∆Time on the identified connections (column 4). Two ROIs (i.e., ROI 1 and ROI 2) represent one connection and the ROI indices corresponds to their labels in the IIT template.
| ROI 1 (IIT index) | ROI 2 (IIT index) | p-Value (1e-4) (WaCSMD) | Correlation (simple MD) |
|---|---|---|---|
| ctx_rh_G_and_S_frontomargin (12101) | ctx_rh_S_intrapariet_and_P_tran (12157) | 0.0053 | 0.3180 |
| ctx_rh_G_and_S_frontomargin (12101) | ctx_lh_G_and_S_transv_frontopol (11105) | 0.0068 | 0.0884 |
| ctx_rh_G_and_S_frontomargin (12101) | ctx_rh_G_front_middle (12115) | 0.0099 | 0.2637 |
| ctx_rh_G_and_S_frontomargin (12101) | ctx_rh_G_insular_short (12118) | 0.0145 | 0.0986 |
| ctx_rh_G_and_S_frontomargin (12101) | ctx_rh_G_pariet_inf-Angular (12125) | 0.0236 | 0.2277 |
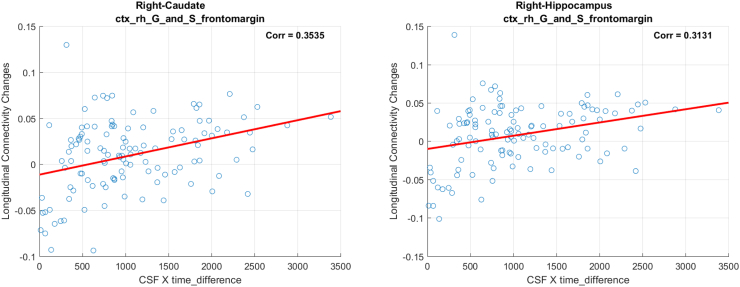
| Right-Hippocampus (53) | ctx_rh_G_and_S_frontomargin (12101) | 0.0481 | 0.3131 |
| Right-Caudate (50) | ctx_rh_G_and_S_frontomargin (12101) | 0.0519 | 0.3535 |
| Right-Hippocampus (53) | ctx_lh_G_parietal_sup (11127) | 0.1462 | 0.2751 |
| ctx_rh_G_and_S_frontomargin (12101) | ctx_rh_S_precentral-sup-part (12170) | 0.1564 | 0.2460 |
| ctx_rh_G_and_S_frontomargin (12101) | ctx_rh_G_and_S_paracentral (12103) | 0.1597 | 0.1183 |
ctx: cortex, lh: left hemisphere, rh: right hemisphere, S: sulcus, G: gyrus, sup: superior, inf: inferior, post: posterior, frontomargin: frontomarginal, pariet: parietal, tran/transv: transverse, frontopol: frontopolar.
Fig. 1.
Sorted resultant p-values from analyses using WaCSMD(blue) and simple MD (green) with CSF biomarkers (i.e., p-tau and neurogranin). Bonferroni threshold (dashed red) at α = 0.05 in −log10 scale are shown together. The connections surviving (above) the multiple comparisons threshold are the connections significantly associated with CSF biomarkers. Left: WaCSMD and p-tau, Right: WaCSMD and neurogranin.
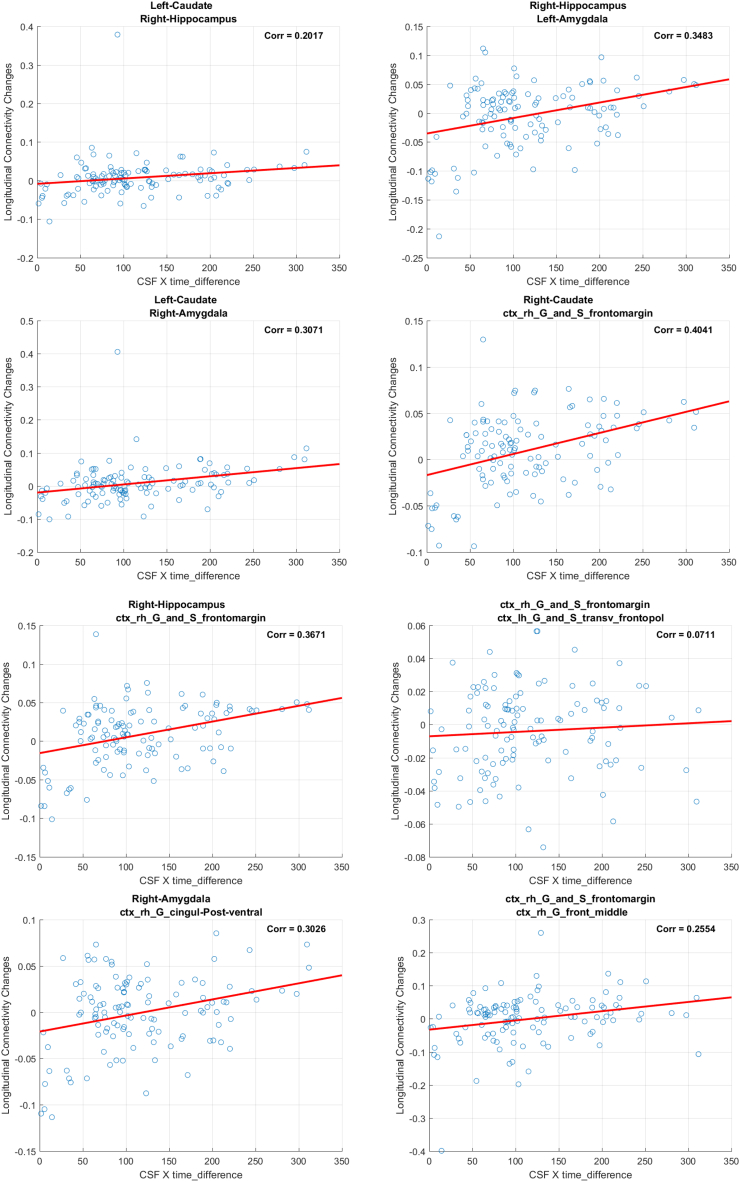
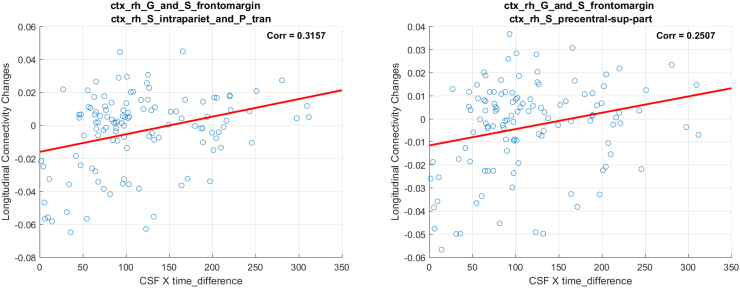
3.1.1. Results from ΔWaCSMD and P-tau analysis
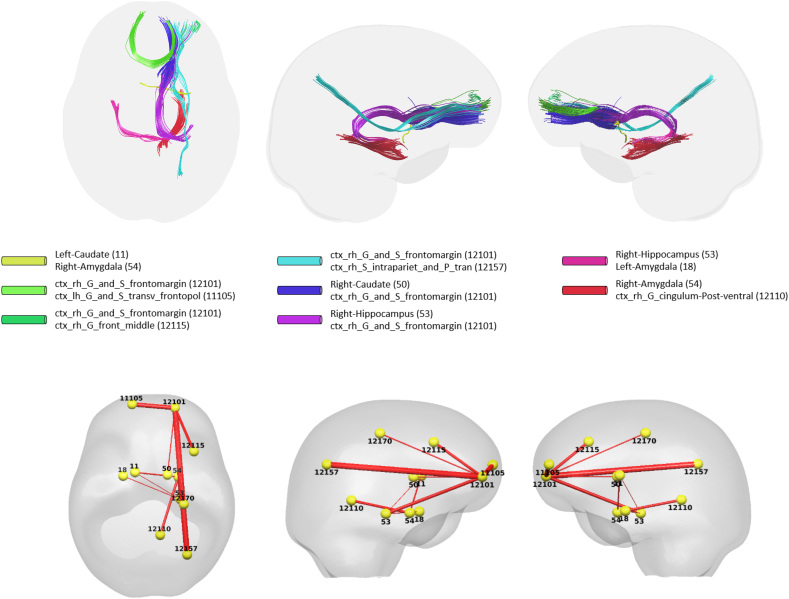
We identified 10 connections using ptau × ∆Time as in Fig. 1 (left), which are listed in Table 2 and visualized in Fig. 2. The top row in Fig. 2 shows the fiber tracts reflecting the 10 connections derived from tractography and the bottom row shows straight connections between the centers of the ROIs where the thickness of the connections represents the corresponding p-values (thicker connections denote lower p-values, i.e. a more robust association). These connections span over 11 different ROIs (three on the left and eight on the right hemisphere) including left and right amygdala, left and right caudate, left transverse frontopolar gyrus/sulcus, right hippocampus, right frontomarginal gyrus/sulcus, right posterior ventral cingulate gyrus, right middle frontal gyrus, right intraparietal sulcus and right precentral superior regions. The significant connections tended to show moderate positive (Pearson's) correlations between the simple MD changes and the interaction (i.e., ptau × ∆Time), ranging from 0.071 to 0.4041, indicating that increase in p-tau levels is associated with increase in MD over time, where higher MD indicates more diffusion possibly due to neurodegeneration.
Fig. 2.
Visualization of identified brain connections whose longitudinal MD changes are dependent on p-tau (i.e. ptau × ∆Time). Top: visualization of tractography generated tracts from the identified connectivity where the tracts in the same color belong to the same connectivity, Bottom: lines representing the identified connections with thickness corresponding to the p-values, where thicker line corresponds to lower p-value and ROI indices. Left: top view, Middle: left view, Right: right view.
Among the gray matter regions that were associated with altered connections, right frontomarginal gyrus/sulcus had the greatest number of associated connections with six, followed by right hippocampus with three connections. Right amygdala and left caudate were each associated with two altered connections. Among the identified connections, three interconnected the two hemispheres: 1) the connection between right frontomarginal gyrus/sulcus and left transverse frontopolar gyrus/sulcus, 2) the left caudate and right amygdala, 3) the left amygdala and the right hippocampus and 4) the left caudate and right hippocampus. Note that while this analysis shows the associations between structural connectivity and p-tau, since WaCS is a multi-resolution representation, the association between p-tau may not necessarily be driven by the specific fiber bundle, entirely by itself.
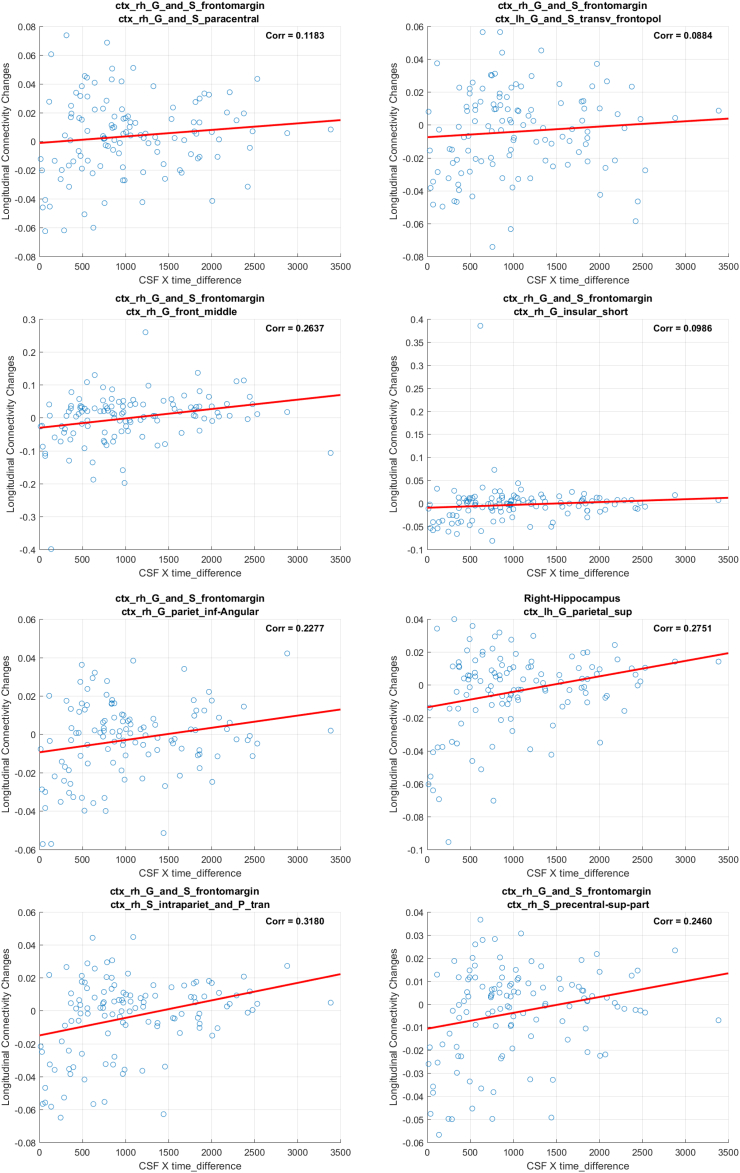
3.1.2. Results from ΔWaCSMD and neurogranin analysis
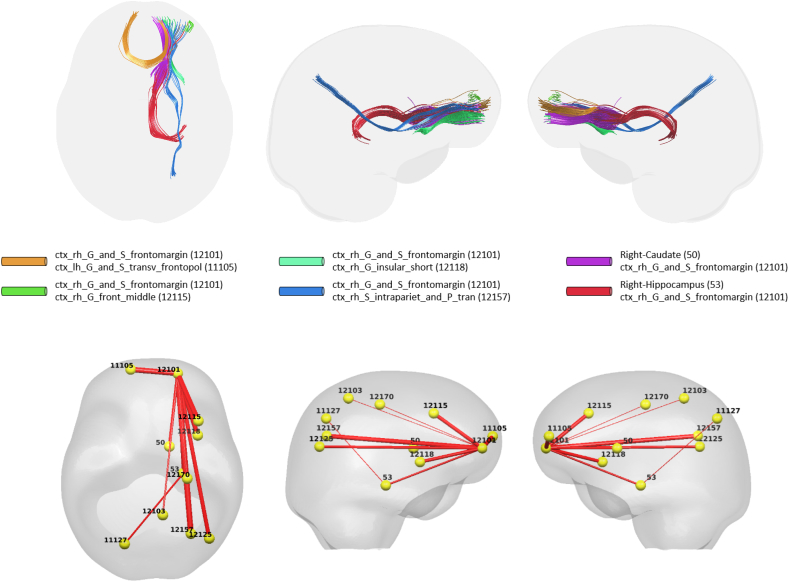
Using the interaction between neurogranin and MRI time interval (i.e., neurogranin × ∆Time) as the main predictor for the longitudinal changes in ΔWaCSMD, we also found 10 connections (listed in Table 3) with low p-values surviving Bonferroni correction at α = 0.05. The p-values and correction threshold in −log10 scale are shown in Fig. 1 (right). The 10 identified connections, as visualized in Fig. 3, were present across 11 different ROIs (two on the left and nine on the right hemisphere). Interestingly, most of the significant connections (nine out of 10) were associated with right frontomarginal gyrus/sulcus and two connections were found with right hippocampus. The hippocampus is well documented as a key region affected in all stages of Alzheimer's disease. Studies have also consistently found atrophy within the frontal lobes in Alzheimer's disease, including ROIs overlapping with the frontomarginal gyrus (Burgmans et al., 2009; Du et al., 2007; Singh et al., 2006). The correlations between the longitudinal change in simple MD and the interaction neurogranin × ∆Time also showed small to moderate positive correlations ranging from 0.0884 to 0.3535, indicating that as neurogranin increases, there is a corresponding increase in MD in these tracts. Of course, similar to the associations with p-tau, since WaCS is a multi-resolution representation, we cannot conclude that the associations are entirely driven by the specific fiber bundle, entirely by itself.
Fig. 3.
Visualization of identified brain connections whose longitudinal MD changes are dependent on neurogranin (i.e., neurogranin × ∆Time). Top: visualization of tractography generated tracts from the identified connectivity where the tracts in the same color belong to the same connectivity, Bottom: lines representing the identified connections with thickness corresponding to the p-values. Thicker line corresponds to lower p-value. Left: top view, Middle: left view, Right: right view.
Results of the analysis of p-tau and analysis of neurogranin revealed 6 shared connections: 1) right frontomarginal gyrus/sulcus and right intraparietal sulcus, 2) right frontomarginal gyrus/sulcus and right transverse frontopolar gyrus/sulcus, 3) right frontomarginal gyrus/sulcus and right frontal middle, 4) right frontomarginal gyrus/sulcus and right hippocampus, 5) right frontomarginal gyrus/sulcus and right precentral sulcus and 6) right frontomarginal gyrus/sulcus and right caudate. Of note, all the common connections (in terms of their WaCS representations) were associated with right frontomarginal gyrus and sulcus.
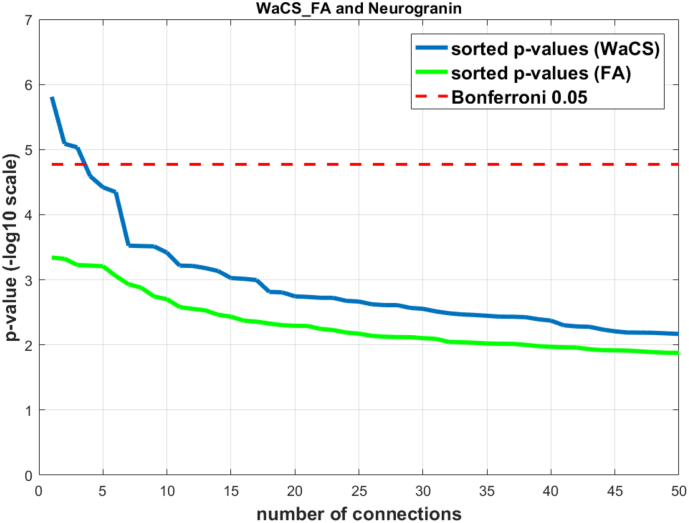
3.2. Fractional anisotropy and CSF biomarkers
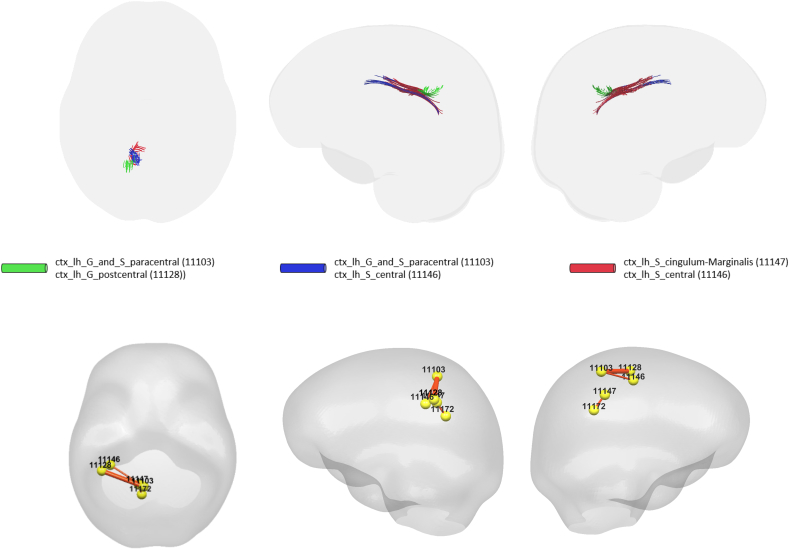
Using ΔWaCSFA and neurogranin × ∆Time, three out of 2964 connections were statistically associated with neurogranin as listed in Table 4; we did not find any significant connections using other CSF biomarkers. In this experiment, fewer connections survived the multiple comparisons correction using Bonferroni at 0.05. The p-values for the identified connections are shown in Fig. 4 with Bonferroni correction thresholds, and the identified connections are visualized in Fig. 5. As shown in Fig. 4, no significant associations were identified using the simple FA values. Five different ROIs were involved in the identified connections: left paracentral gyrus/sulcus, left postcentral gyrus, left cingulate Marginalis sulcus, left subparietal sulcus, left central sulcus. While most of the identified connections in the previous ΔWaCSMD analyses were found on the right hemisphere, the ΔWaCSFA analysis yielded connections only in the left hemisphere. The only ROI in common between WaCSMD and WaCSFA analyses was left paracentral gyrus/sulcus.
Table 4.
Connections identified using neurogranin × ∆Time as a predictor for longitudinal changes in WaCSFA (multiple comparisons corrected using Bonferroni at 0.05) sorted by corresponding p-values (column 1–3), and the correlation between simple FA changes and neurogranin × ∆Time on those identified connections (column 4). Two ROIs (i.e., ROI 1 and ROI 2) represent one connection and the ROI indices corresponds to their labels in the IIT template.
| ROI 1 (IIT index) | ROI 2 (IIT index) | p-Value (1e-4) (WaCSFA) | Correlation (simple FA) |
|---|---|---|---|
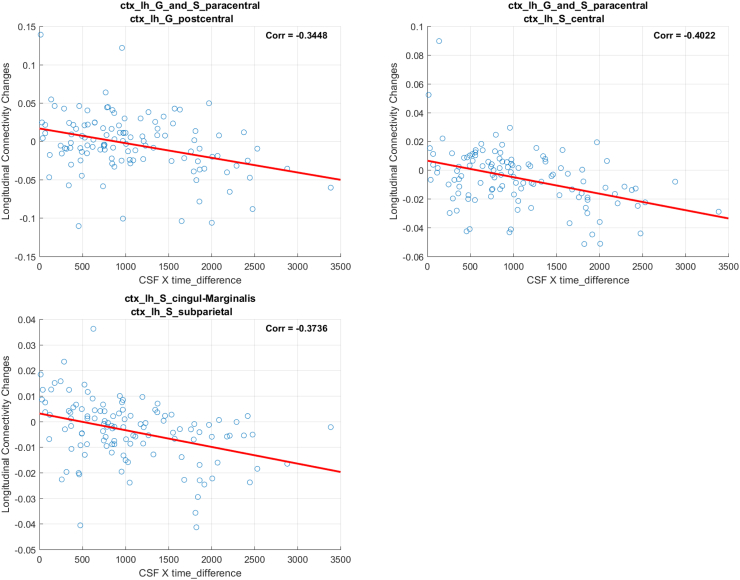
| ctx_lh_G_and_S_paracentral (11103) | ctx_lh_G_postcentral (11128) | 0.0155 | −0.3448 |
| ctx_lh_S_cingulum-Marginalis (11147) | ctx_lh_S_subparietal (11172) | 0.0815 | −0.3736 |
| ctx_lh_G_and_S_paracentral (11103) | ctx_lh_S_central (11146) | 0.0924 | −0.4022 |
ctx: cortex, lh: left hemisphere, rh: right hemisphere, S: sulcus, G: gyrus, sup: superior, inf: inferior, post: posterior, frontomargin: frontomarginal, pariet: parietal, tran/transv: transverse, frontopol: frontopolar.
Fig. 4.
Sorted resultant p-values from analysis using WaCSFA (blue) and simple FA (green) with neurogranin. Bonferroni threshold at α = 0.05 in −log10 scale is shown in dotted red. The connections surviving (above) the multiple comparisons threshold are the connections significantly associated with neurogranin.
Fig. 5.
Visualization of the identified brain connections whose longitudinal FA changes are dependent on neurogranin (i.e., neurogranin × ∆Time). Top: visualization of tractography generated tracts from the identified connectivity where the tracts in the same color belong to the same connectivity, Bottom: lines representing the identified connections with thickness corresponding to the p-values. Thicker line corresponds to lower p-value. Left: top view, Middle: left view, Right: right view.
Again, we computed the correlation between the changes in simple FA and the interaction neurogranin × ∆Time. The correlations were mostly moderate ranging between −0.4022 and −0.3448. As expected given the inverse relationship between MD and FA, the correlations were negative which means that the changes in FA decrease (i.e., directional coherence of the white matter fiber tract decreases) with increases in neurogranin × ∆Time.
4. Discussion
Several neuroimaging studies confirm that white matter degeneration is a feature of AD and other dementias, and that degeneration occurs across disease development (Acosta-Cabronero et al., 2010; Liu et al., 2017; Steketee et al., 2016; Zhuang et al., 2010). However, few longitudinal studies have been carried out, especially studies in cognitively asymptomatic individuals. In the current study, we examined longitudinal change and employed a novel multi-resolution graph analysis method which offers very high sensitivity for identifying associations between connectivity graphs (in terms of hybrid local-global representations) and neuropathology. We found that elevated CSF p-tau and neurogranin predicted changes in white matter connection strength after correction for multiple comparisons. Using a conventional approach for analysing FA and MD with GLM did not yield sufficient sensitivity to detect subtle changes at a statistically significant threshold across the entire brain.
The development of AD from the preclinical to the dementia stage involves progressive accumulation of cortical pathology. In addition to development of amyloid plaques and neurofibrillary tangles, additional disease features include neural injury, resulting in loss of synaptic connections and axonal loss which results in a disconnection of more distal brain regions (Perez-Nievas et al., 2013). Of the AD-specific and neural injury biomarkers, only p-tau and neurogranin were significant predictors of change in connectivity strength. It is intriguing that these two markers were associated with loss of connectivity, given several reports that tau pathology may spread via synaptically connected circuits (De Calignon et al., 2012; Liu et al., 2012). Neurogranin, a postsynaptic protein enriched in the brain and primarily localized in the cortex, hippocampus, and amygdala, is thought to reflect synaptic loss and several studies indicate that CSF neurogranin levels are elevated in patients with AD and MCI compared to controls (Hellwig et al., 2015; Kvartsberg et al., 2015; Thorsell et al., 2010). Elevated neurogranin also appears specific to AD compared to other neurodegenerative diseases (Portelius et al., 2018; Wellington et al., 2016). There was considerable overlap between the p-tau and neurogranin findings, particularly for connections within the frontal lobe, and connections between the frontal lobe and the hippocampus as well as parietal ROIs, possibly reflecting propagation of tau pathology via axonally connected regions. Hippocampal involvement in AD is present in Braak stages II and III of the disease process, and in our study, hippocampus showed 2 abnormal connections in each of the p-tau and neurogranin analyses. Tangle development in high order association cortices — including the frontal and parietal cortices detected in this study — do not occur until Braak stage IV; however, prior to dense tangle accumulation, tau abnormalities appear in axons, and at least one mouse study has shown that abnormalities can be detected using diffusion weighted imaging, even at the early stage of disease development prior to severe tauopathy (Sahara et al., 2014). Further interrogation of temporal ordering of connectivity loss and regional neurofibrillary tangle development will require regional mapping of tau pathology in conjunction with diffusion weighted imaging (Hwang et al., 2018b). A caveat of this interpretation in our study is that while hyperphosphorylation of tau protein is known to begin within the axon, we did not observe a relationship between NFL and connectivity strength. CSF NFL levels (a marker of axonal injury) are elevated in AD and predict faster disease progression (Zetterberg et al., 2016). NFL is consistently elevated among individuals with more advanced pathology, so it may be the case that an association between NFL and connectivity change is not observed until later disease stages.
While WaCS provides a sensitive tool for detecting significant differences or associations, it does not provide the direction of the association. To accomplish this, we extracted FA and MD values from regions that were detected using WaCS and computed correlations between the FA/MD values and the interaction between CSF biomarkers and time intervals. The correlations between p-tau and neurogranin and WaCSMD connections were all moderately positive, indicating that more tangle pathology and synaptic degeneration are associated with weakened connectivity strength over time. CSF neurogranin, but not p-tau, was also a significant predictor of WaCSFA. While fewer connections overall reached statistical significance, as expected, these correlations were all negative with higher neurogranin being associated with weaker connection strength. Separately, since WaCS is a multi-resolution representation, localizing a significant association to a fiber bundle by itself is problematic since the signal at other adjacent fiber bundles (in the graph sense) may nominally contribute to the derived WaCS representation.
At an uncorrected threshold, CSF biomarkers predicted change in simple FA and MD in a similar pattern to that found using WaCS. This consistent pattern demonstrates that FA and MD still remain useful measures for assessing microstructure, particularly in analyses that involve an a priori driven hypothesis for which there are few multiple comparisons. WaCS, however, provides a sensitive method for detecting subtle effects across the entire brain, which requires correction for multiple comparisons given the thousands of hypothesis tests. WaCS captures subtle variations that are represented in multiple bandwidths in the frequency space, which may not manifest in the original space taking into account signal at multiple resolutions. This approach can be particularly useful for detecting changes in the preclinical stages of disease. Indeed, this framework could be adopted for the analysis of other preclinical diseases, not just AD. No relationships were observed with markers of amyloid. Thus, it may be the case that our findings represent early AD-related connectivity changes that are independent of amyloid. Alternatively, p-tau levels and associated white matter degeneration observed in the population studied here may reflect a process independent of AD, such as suspected non-Alzheimer's pathology (SNAP) (Knopman et al., 2013) or primary age-related tauopathy (PART) (Crary et al., 2014). Elevated neurogranin could also reflect age-related rather than AD-related loss of synapses and plasticity. Because our study population is a preclinical cohort and the course of AD occurs simultaneously with age-related or other disease-related brain changes, it is likely that both AD and non-AD pathological changes may underlie some of the observed white matter alterations as demonstrated in this study.
We find that the multi-resolution analysis using WaCS outperforms traditional approaches with the raw measures for various reasons. First, the multi-resolution view of a signal on each data point (e.g., pixel in an image or node/edge on a graph) yields its relationship with the signal on its neighboring data points (i.e., local context) by applying a filter at different scales. Such a technique is well known in computer vision and image processing to yield robust comparisons (Morel and Yu, 2009). Also, the idea of the multi-resolution shares similarities with feature extraction using convolution neural network (CNN), which derives features by applying multiple filters with different parameters to achieve novel representation of an image. In the end, our multi-resolution approach makes the true signal stronger in the locations where the p-values with the raw measure are low but not enough to survive multiple comparisons corrections due to small sample size or small effect size, without increasing false positives, by aggregating information based on the topology of the graph. Although not reported in this manuscript for simplicity, we empirically tested the uni-resolution version of WaCS based on using only a single scale which turned out to be ineffective in the current study as well as other previous studies (Kim et al., 2015a; 2014). This scheme returned improved results but quite similar to only using FA or MD values directly.
Note that since WaCS is derived using graph wavelets, it is leveraging various properties of well-studied concepts from physics such as diffusion or heat equation. Diffusion has been shown to be closely related to wavelets in earlier papers (Coifman and Maggioni, 2006). Separate from this body of work, diffusion or heat equation based operators have been used to better understand the physical process of pathology propagation (Raj et al., 2012; 2015) by essentially treating the structural connectivity network as physical “pathways” to explain the spatial distribution of pathology at a subsequent time point. This suggests that ideas related to our multi-resolution WaCS may provide efficient ways to investigate pathology propagation, if an appropriate imaging modality (such as amyloid PET) is available for the participants. A preliminary attempt to investigate this hypothesis is presented in (Hwang et al., 2018b).
4.1. Limitations
WaCS is a sensitive and effective way for characterizing connection strength of white matter tracts in a multi-resolution (hybrid global-local) manner; however, it is not without its limitations. Multi-resolution strategy deriving WaCS involves a quadratic increase of computation depending on the number of connections (i.e., O(|E|2)) during the transformation between G and L(G). When the given brain connectivity (i.e., graph) is dense, this can pose a problem, however the number of ROIs studied here was relatively small (<200) and their connections were sparse. Specifying parameters (i.e., number of scales J, choice of scales and choice of a type of wavelet) also requires a heuristic input from the user, which in turn may affect the statistical power of the analysis. It is also important to note that WaCS is a representation of the connectivity that embeds the strength of the connections in a high dimensional space where the direction of the associations become ambiguous. Therefore, additional analysis is required to make further inferences about the underlying microstructure. Finally, association studies using WaCS provide useful information regarding structural connectivity captured in a global-local sense in the context of one or more predictors. However, the association may not necessarily be limited to a specific fiber bundle due to the inherent multi-resolution representation. Aside from the technical issues with WaCS, an additional limitation of this study was the use of DTI model for diffusion MRI, which may not capture the crossing white matter fibers in the white matter connectivity construction that may have been affected by some of the CSF measures. Also, the lateralization of results where associations between CSF and MD were primarily observed in the right cortex and with FA in the left cortex is an interesting finding to note but remains unclear and need to be further investigated as an important future study. Finally, while this study is an improvement over cross-sectional analyses, our study included only two time-points over a relatively short interval (~3 years on average). Considering that pre-clinical pathology may begin decades prior to clinical impairment, longer follow-up is needed.
5. Conclusion
While white matter alteration is an increasingly recognized feature of neurodegeneration, the relationship between cortical pathology and degeneration of underlying white matter tracts remains poorly defined. The present study demonstrated that elevated levels of biomarkers of both neurofibrillary tangles and synaptic dysfunction are associated with altered connectivity over several years. Using techniques that provide superior sensitivity to connectivity changes such as WaCS, are expected to facilitate the understanding of the earliest stages of disease.
Funding
This research was supported by the National Institutes of Health (R01AG040396, R01AG037639, R01AG021155, R01AG027161, P50AG033514), by a Clinical and Translational Science Award (UL1RR025011) to the University of Wisconsin, Madison, the Center for Predictive Computational Phenotyping; and by the Swedish Research Council, the Swedish Brain Foundation, the Knut and Alice Wallenberg Foundation, and Torsten Söderberg's Foundation to the University of Gothenburg. Portions of this research were supported by a NSF CAREER Award to Singh and the Veterans Administration including facilities and resources at the Geriatric Research Education and Clinical Center of the William S. Middleton Memorial Veterans Hospital, Madison, WI.
Conflict of interests
Kaj Blennow and Henrik Zetterberg are co-founders of Brain Biomarker Solutions in Gothenburg AB, a GU Venture-based platform company at the University of Gothenburg. Kaj Blennow has served as a consultant or at advisory boards for IBL International, Roche Diagnostics, Eli Lilly, Fujirebio Europe, and Novartis. The other authors declare that they have no conflict of interest.
Appendix A
In this appendix, we demonstrate figures of scatter plots using the longitudinal connectivity strength changes (i.e., FA and MD measures) and the main predictor (i.e. interaction between a CSF measure and longitudinal imaging visits) on the brain connectivities that we identified in our study.
A.1. MD and p-tau analysis
Our analysis on WaCS from MD and p-tau identified 10 connectivities that showed statistically significant associations. In the following, we demonstrate the scatter plots using simple MD measure and interaction term (i.e., ptau × ∆Time) from each of the identified connections. In each of the figure, the title shows the ROIs associated with each connection, the x-axis is the interaction and the y-axis reflects the longitudinal MD changes.
A.2. MD and neurogranin analysis
Our analysis on WaCS from MD and neurogranin identified 10 connectivities that showed statistically significant associations. In the following, we demonstrate the scatter plots using simple MD measure and interaction term (i.e., neurogranin×∆Time) from each of the identified connections. In each of the figure, the title shows the ROIs associated with each connection, the x-axis is the interaction and the y-axis reflects the longitudinal MD changes.
A.3. FA and neurogranin analysis
Our analysis on WaCS from FA and neurogranin identified 3 connectivities that showed statistically significant associations after Bonferroni correction. In the following, we demonstrate the scatter plots using simple FA measure and interaction term (i.e., neurogranin×∆Time) from each of the identified connections. In each of the figure, the title shows the ROIs associated with each connection, the x-axis is the interaction and the y-axis reflects the longitudinal MD changes.
References
- Acosta-Cabronero J., Williams G.B., Pengas G., Nestor P.J. Absolute diffusivities define the landscape of white matter degeneration in Alzheimer's disease. Brain. 2010;133:529–539. doi: 10.1093/brain/awp257. [DOI] [PubMed] [Google Scholar]
- Adluru N., Destiche D.J., Lu S.Y.-F., Doran S.T., Birdsill A.C., Melah K.E., Okonkwo O.C., Alexander A.L., Dowling N.M., Johnson S.C. White matter microstructure in late middle-age: effects of apolipoprotein E4 and parental family history of Alzheimer's disease. NeuroImage. 2014;4:730–742. doi: 10.1016/j.nicl.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balthazar M., Yasuda C., Pereira F., Pedro T., Damasceno B., Cendes F. Differences in grey and white matter atrophy in amnestic mild cognitive impairment and mild Alzheimer's disease. Eur. J. Neurol. 2009;16:468–474. doi: 10.1111/j.1468-1331.2008.02408.x. [DOI] [PubMed] [Google Scholar]
- Baxter L.C., Sparks D.L., Johnson S.C., Lenoski B., Lopez J.E., Connor D.J., Sabbagh M.N. Relationship of cognitive measures and gray and white matter in Alzheimer's disease. J. Alzheimers Dis. 2006;9:253–260. doi: 10.3233/jad-2006-9304. [DOI] [PubMed] [Google Scholar]
- Bendlin B.B., Carlsson C.M., Johnson S.C., Zetterberg H., Blennow K., Willette A.A., Okonkwo O.C., Sodhi A., Ries M.L., Birdsill A.C. CSF T-Tau/Aβ42 predicts white matter microstructure in healthy adults at risk for Alzheimer's disease. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0037720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgmans S., Van Boxtel M., Smeets F., Vuurman E., Gronenschild E., Verhey F., Uylings H., Jolles J. Prefrontal cortex atrophy predicts dementia over a six-year period. Neurobiol. Aging. 2009;30:1413–1419. doi: 10.1016/j.neurobiolaging.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Chaim T.M., Duran F.L., Uchida R.R., Périco C.A., de Castro C.C., Busatto G.F. Volumetric reduction of the corpus callosum in Alzheimer's disease in vivo as assessed with voxel-based morphometry. Psychiatry Res. Neuroimaging. 2007;154:59–68. doi: 10.1016/j.pscychresns.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Coifman R.R., Maggioni M. Diffusion wavelets. Appl. Comput. Harmon. Anal. 2006;21:53–94. [Google Scholar]
- Cook P., Bai Y., Nedjati-Gilani S., Seunarine K., Hall M., Parker G., Alexander D. 14th Scientific Meeting of the International Society for Magnetic Resonance in Medicine. Seattle WA; USA: 2006. Camino: open-source diffusion-MRI reconstruction and processing. [Google Scholar]
- Crary J.F., Trojanowski J.Q., Schneider J.A., Abisambra J.F., Abner E.L., Alafuzoff I., Arnold S.E., Attems J., Beach T.G., Bigio E.H. Primary age-related tauopathy (PART): a common pathology associated with human aging. Acta Neuropathol. 2014;128:755–766. doi: 10.1007/s00401-014-1349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Calignon A., Polydoro M., Suárez-Calvet M., William C., Adamowicz D.H., Kopeikina K.J., Pitstick R., Sahara N., Ashe K.H., Carlson G.A. Propagation of tau pathology in a model of early Alzheimer's disease. Neuron. 2012;73:685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du A.-T., Schuff N., Kramer J.H., Rosen H.J., Gorno-Tempini M.L., Rankin K., Miller B.L., Weiner M.W. Different regional patterns of cortical thinning in Alzheimer's disease and frontotemporal dementia. Brain. 2007;130:1159–1166. doi: 10.1093/brain/awm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B.T., Zhu Z., Brown C.A., Andersen A.H., Ladu M.J., Tai L., Jicha G.A., Kryscio R.J., Estus S., Nelson P.T. White matter integrity is associated with cerebrospinal fluid markers of Alzheimer's disease in normal adults. Neurobiol. Aging. 2014;35:2263–2271. doi: 10.1016/j.neurobiolaging.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond D.K., Vandergheynst P., Gribonval R. Wavelets on graphs via spectral graph theory. Appl. Comput. Harmon. Anal. 2011;30:129–150. [Google Scholar]
- Harary F. 1969. Graph Theory. Addison-Wesley, Reading, MA. [Google Scholar]
- Hellwig K., Kvartsberg H., Portelius E., Andreasson U., Oberstein T.J., Lewczuk P., Blennow K., Kornhuber J., Maler J.M., Zetterberg H. Neurogranin and YKL-40: independent markers of synaptic degeneration and neuroinflammation in Alzheimer's disease. Alzheimers Res. Ther. 2015;7:74. doi: 10.1186/s13195-015-0161-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S., Adluru N., Kim W., Johnson S., Bendlin B., Singh V. Associations between PET Amyloid Pathology and DTI Brain Connectivity in Preclinical Alzheimer's Disease. Brain Connect. 2018 doi: 10.1089/brain.2018.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S.J., Ravi S., Adluru N., Bendlin B.B., Johnson S.C., Singh V. 2018. Data-Driven Propagation Modeling of PET-Derived Alzheimer's Pathology in a Preclinical Cohort. Alzheimer's Association International Conference. [Google Scholar]
- Im K., Lee J.-M., Seo S.W., Kim S.H., Kim S.I., Na D.L. Sulcal morphology changes and their relationship with cortical thickness and gyral white matter volume in mild cognitive impairment and Alzheimer's disease. NeuroImage. 2008;43:103–113. doi: 10.1016/j.neuroimage.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Jung W.B., Mun C.W., Kim Y.H., Park J.M., Lee B.D., Lee Y.M., Moon E., Jeong H.J., Chung Y.I. Cortical atrophy, reduced integrity of white matter and cognitive impairment in subcortical vascular dementia of Binswanger type. Psychiatry Clin. Neurosci. 2014;68:821–832. doi: 10.1111/pcn.12196. [DOI] [PubMed] [Google Scholar]
- Keihaninejad S., Zhang H., Ryan N.S., Malone I.B., Modat M., Cardoso M.J., Cash D.M., Fox N.C., Ourselin S. An unbiased longitudinal analysis framework for tracking white matter changes using diffusion tensor imaging with application to Alzheimer's disease. NeuroImage. 2013;72:153–163. doi: 10.1016/j.neuroimage.2013.01.044. [DOI] [PubMed] [Google Scholar]
- Kim W.H., Pachauri D., Hatt C., Chung M.K., Johnson S., Singh V. Advances in Neural Information Processing Systems. 2012. Wavelet based multi-scale shape features on arbitrary surfaces for cortical thickness discrimination; pp. 1241–1249. [PMC free article] [PubMed] [Google Scholar]
- Kim W.H., Adluru N., Chung M.K., Charchut S., Gadelkarim J.J., Altshuler L., Moody T., Kumar A., Singh V., Leow A.D. Springer; 2013. Multi-Resolutional Brain Network Filtering and Analysis Via Wavelets on Non-Euclidean Space. International Conference on Medical Image Computing and Computer-Assisted Intervention; pp. 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.H., Singh V., Chung M.K., Hinrichs C., Pachauri D., Okonkwo O.C., Johnson S.C., Initiative A.s.D.N. Multi-resolutional shape features via non-Euclidean wavelets: applications to statistical analysis of cortical thickness. NeuroImage. 2014;93:107–123. doi: 10.1016/j.neuroimage.2014.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.H., Adluru N., Chung M.K., Okonkwo O.C., Johnson S.C., Bendlin B.B., Singh V. Multi-resolution statistical analysis of brain connectivity graphs in preclinical Alzheimer's disease. NeuroImage. 2015;118:103–117. doi: 10.1016/j.neuroimage.2015.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.H., Singh V., Chung M.K., Adluru N., Bendlin B.B., Johnson S.C. Multi-resolution statistical analysis on graph structured data in neuroimaging. Biomedical Imaging (ISBI), 2015 IEEE 12th International Symposium on. IEEE. 2015:1548–1551. doi: 10.1109/ISBI.2015.7164173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopman D.S., Jack C.R., Wiste H.J., Weigand S.D., Vemuri P., Lowe V.J., Kantarci K., Gunter J.L., Senjem M.L., Mielke M.M. Brain injury biomarkers are not dependent on β-amyloid in normal elderly. Ann. Neurol. 2013;73:472–480. doi: 10.1002/ana.23816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvartsberg H., Portelius E., Andreasson U., Brinkmalm G., Hellwig K., Lelental N., Kornhuber J., Hansson O., Minthon L., Spitzer P. Characterization of the postsynaptic protein neurogranin in paired cerebrospinal fluid and plasma samples from Alzheimer's disease patients and healthy controls. Alzheimers Res. Ther. 2015;7:40. doi: 10.1186/s13195-015-0124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Li T.Q., Andreasen N., Wiberg M., Westman E., Wahlund L.O. The association between biomarkers in cerebrospinal fluid and structural changes in the brain in patients with Alzheimer's disease. J. Intern. Med. 2014;275:418–427. doi: 10.1111/joim.12164. [DOI] [PubMed] [Google Scholar]
- Liu L., Drouet V., Wu J.W., Witter M.P., Small S.A., Clelland C., Duff K. Trans-synaptic spread of tau pathology in vivo. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liang P., Yin L., Shu N., Zhao T., Xing Y., Li F., Zhao Z., Li K., Han Y. White matter abnormalities in two different subtypes of amnestic mild cognitive impairment. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0170185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe D.G. Object recognition from local scale-invariant features. Computer vision, 1999. The proceedings of the seventh IEEE international conference on. IEEE. 1999:1150–1157. [Google Scholar]
- Lowe D.G. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004;60:91–110. [Google Scholar]
- Mahoney C.J., Simpson I.J., Nicholas J.M., Fletcher P.D., Downey L.E., Golden H.L., Clark C.N., Schmitz N., Rohrer J.D., Schott J.M. Longitudinal diffusion tensor imaging in frontotemporal dementia. Ann. Neurol. 2015;77:33–46. doi: 10.1002/ana.24296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascalchi M., Pantoni L., Giannelli M., Valenti R., Bianchi A., Pracucci G., Orsolini S., Ciulli S., Tessa C., Poggesi A. Diffusion tensor imaging to map brain microstructural changes in CADASIL. J. Neuroimaging. 2017;27:85–91. doi: 10.1111/jon.12374. [DOI] [PubMed] [Google Scholar]
- Mattsson N., Insel P.S., Palmqvist S., Portelius E., Zetterberg H., Weiner M., Blennow K., Hansson O., Initiative A.s.D.N. Cerebrospinal fluid tau, neurogranin, and neurofilament light in Alzheimer's disease. EMBO Mol. Med. 2016;8:1184–1196. doi: 10.15252/emmm.201606540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijboom R., Steketee R., de Koning I., Osse R.J., Jiskoot L., De Jong F.J., van der Lugt A., van Swieten J., Smits M. Functional connectivity and microstructural white matter changes in phenocopy frontotemporal dementia. Eur. Radiol. 2017;27:1352–1360. doi: 10.1007/s00330-016-4490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinuevo J.L., Ripolles P., Simó M., Lladó A., Olives J., Balasa M., Antonell A., Rodriguez-Fornells A., Rami L. White matter changes in preclinical Alzheimer's disease: a magnetic resonance imaging-diffusion tensor imaging study on cognitively normal older people with positive amyloid β protein 42 levels. Neurobiol. Aging. 2014;35:2671–2680. doi: 10.1016/j.neurobiolaging.2014.05.027. [DOI] [PubMed] [Google Scholar]
- Morel J.-M., Yu G. ASIFT: a new framework for fully affine invariant image comparison. SIAM J. Imag. Sci. 2009;2:438–469. [Google Scholar]
- Perez-Nievas B.G., Stein T.D., Tai H.C., Dols-Icardo O., Scotton T.C., Barroeta-Espar I., Fernandez-Carballo L., Munain E.L., Perez J., Marquie M., Serrano-Pozo A., Frosch M.P., Lowe V., Parisi J.E., Petersen R.C., Ikonomovic M.D., López O.L., Klunk W., Hyman B.T., Gómez-Isla T. Dissecting phenotypic traits linked to human resilience to Alzheimer's pathology. Brain. 2013 Aug;136(Pt 8):2510–2526. doi: 10.1093/brain/awt171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portelius E., Olsson B., Höglund K., Cullen N.C., Kvartsberg H., Andreasson U., Zetterberg H., Sandelius Å., Shaw L.M., Lee V.M. Cerebrospinal fluid neurogranin concentration in neurodegeneration: relation to clinical phenotypes and neuropathology. Acta Neuropathol. 2018:1–14. doi: 10.1007/s00401-018-1851-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine A.M., Koscik R.L., Nicholas C.R., Clark L.R., Okonkwo O.C., Oh J.M., Hillmer A.T., Murali D., Barnhart T.E., Betthauser T.J. Cerebrospinal fluid ratios with Aβ 42 predict preclinical brain β-amyloid accumulation. Alzheimer's Dementia. 2016;2:27–38. doi: 10.1016/j.dadm.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine A., Merluzzi A., Adluru N., Norton D., Koscik R., Clark L., Berman S., Nicholas C., Asthana S., Alexander A. Association of longitudinal white matter degeneration and cerebrospinal fluid biomarkers of neurodegeneration, inflammation and Alzheimer's disease in late-middle-aged adults. Brain Imag. Behav. 2017 doi: 10.1007/s11682-017-9732-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine A.M., Merluzzi A.P., Adluru N., Norton D., Koscik R.L., Clark L.R., Berman S.E., Nicholas C.R., Asthana S., Alexander A.L. Association of longitudinal white matter degeneration and cerebrospinal fluid biomarkers of neurodegeneration, inflammation and Alzheimer's disease in late-middle-aged adults. Brain Imag. Behav. 2017:1–12. doi: 10.1007/s11682-017-9732-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Kuceyeski A., Weiner M. A network diffusion model of disease progression in dementia. Neuron. 2012;73:1204–1215. doi: 10.1016/j.neuron.2011.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Locastro E., Kuceyeski A., Tosun D., Relkin N., Weiner M., Initiative A.s.D.N. Network diffusion model of progression predicts longitudinal patterns of atrophy and metabolism in Alzheimer's disease. Cell Rep. 2015;10:359–369. doi: 10.1016/j.celrep.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara N., Perez P.D., Lin W.-L., Dickson D.W., Ren Y., Zeng H., Lewis J., Febo M. Age-related decline in white matter integrity in a mouse model of tauopathy: an in vivo diffusion tensor magnetic resonance imaging study. Neurobiol. Aging. 2014;35:1364–1374. doi: 10.1016/j.neurobiolaging.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat D.H., Greve D.N., Pacheco J.L., Quinn B.T., Helmer K.G., Buckner R.L., Fischl B. Regional white matter volume differences in nondemented aging and Alzheimer's disease. NeuroImage. 2009;44:1247–1258. doi: 10.1016/j.neuroimage.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Ip H.H. Discriminative wavelet shape descriptors for recognition of 2-D patterns. Pattern Recogn. 1999;32:151–165. [Google Scholar]
- Singh V., Chertkow H., Lerch J.P., Evans A.C., Dorr A.E., Kabani N.J. Spatial patterns of cortical thinning in mild cognitive impairment and Alzheimer's disease. Brain. 2006;129:2885–2893. doi: 10.1093/brain/awl256. [DOI] [PubMed] [Google Scholar]
- Starks E.J., Patrick O'Grady J., Hoscheidt S.M., Racine A.M., Carlsson C.M., Zetterberg H., Blennow K., Okonkwo O.C., Puglielli L., Asthana S. Insulin resistance is associated with higher cerebrospinal fluid tau levels in asymptomatic APOEɛ4 carriers. J. Alzheimers Dis. 2015;46:525–533. doi: 10.3233/JAD-150072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steketee R.M., Meijboom R., de Groot M., Bron E.E., Niessen W.J., van der Lugt A., van Swieten J.C., Smits M. Concurrent white and gray matter degeneration of disease-specific networks in early-stage Alzheimer's disease and behavioral variant frontotemporal dementia. Neurobiol. Aging. 2016;43:119–128. doi: 10.1016/j.neurobiolaging.2016.03.031. [DOI] [PubMed] [Google Scholar]
- Stenset V., Bjørnerud A., Fjell A.M., Walhovd K.B., Hofoss D., Due-Tønnessen P., Gjerstad L., Fladby T. Cingulum fiber diffusivity and CSF T-tau in patients with subjective and mild cognitive impairment. Neurobiol. Aging. 2011;32:581–589. doi: 10.1016/j.neurobiolaging.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Teipel S.J., Hampel H., Alexander G., Schapiro M., Horwitz B., Teichberg D., Daley E., Hippius H., Möller H.-J., Rapoport S. Dissociation between corpus callosum atrophy and white matter pathology in Alzheimer's disease. Neurology. 1998;51:1381–1385. doi: 10.1212/wnl.51.5.1381. [DOI] [PubMed] [Google Scholar]
- Thorsell A., Bjerke M., Gobom J., Brunhage E., Vanmechelen E., Andreasen N., Hansson O., Minthon L., Zetterberg H., Blennow K. Neurogranin in cerebrospinal fluid as a marker of synaptic degeneration in Alzheimer's disease. Brain Res. 2010;1362:13–22. doi: 10.1016/j.brainres.2010.09.073. [DOI] [PubMed] [Google Scholar]
- Varentsova A., Zhang S., Arfanakis K. Development of a high angular resolution diffusion imaging human brain template. NeuroImage. 2014;91:177–186. doi: 10.1016/j.neuroimage.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermersch P., Scheltens P., Barkhof F., Steinling M., Leys D. Evidence for atrophy of the corpus callosum in Alzheimer's disease. Eur. Neurol. 1994;34:83–86. doi: 10.1159/000117014. [DOI] [PubMed] [Google Scholar]
- Wellington H., Paterson R.W., Portelius E., Törnqvist U., Magdalinou N., Fox N.C., Blennow K., Schott J.M., Zetterberg H. Increased CSF neurogranin concentration is specific to Alzheimer disease. Neurology. 2016;86:829–835. doi: 10.1212/WNL.0000000000002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won Hwa Kim, Moo K. Chung, Vikas Singh. Multi-resolution shape analysis via non-euclidean wavelets: Applications to mesh segmentation and surface alignment problems. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2013:2139–2146. doi: 10.1109/CVPR.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P.A., Avants B.B., Das S.R., Pluta J., Altinay M., Craige C., Initiative A.s.D.N. Bias in estimation of hippocampal atrophy using deformation-based morphometry arises from asymmetric global normalization: an illustration in ADNI 3 T MRI data. NeuroImage. 2010;50:434–445. doi: 10.1016/j.neuroimage.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg H., Skillbäck T., Mattsson N., Trojanowski J.Q., Portelius E., Shaw L.M., Weiner M.W., Blennow K. Association of cerebrospinal fluid neurofilament light concentration with Alzheimer disease progression. JAMA Neurol. 2016;73:60–67. doi: 10.1001/jamaneurol.2015.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.H., Singh V., Johnson S.C., Wahba G. Statistical tests and identifiability conditions for pooling and analyzing multisite datasets. Proc. Natl. Acad. Sci. 2018 doi: 10.1073/pnas.1719747115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L., Wen W., Zhu W., Trollor J., Kochan N., Crawford J., Reppermund S., Brodaty H., Sachdev P. White matter integrity in mild cognitive impairment: a tract-based spatial statistics study. NeuroImage. 2010;53:16–25. doi: 10.1016/j.neuroimage.2010.05.068. [DOI] [PubMed] [Google Scholar]