Abstract
Manual quantification of the hippocampal atrophy state and rate is time consuming and prone to poor reproducibility, even when performed by neuroanatomical experts. The automation of hippocampal segmentation has been investigated in normal aging, epilepsy, and in Alzheimer's disease. Our first goal was to compare manual and automated hippocampal segmentation in ischemic stroke and to, secondly, study the impact of stroke lesion presence on hippocampal volume estimation. We used eight automated methods to segment T1-weighted MR images from 105 ischemic stroke patients and 39 age-matched controls sampled from the Cognition And Neocortical Volume After Stroke (CANVAS) study. The methods were: AdaBoost, Atlas-based Hippocampal Segmentation (ABHS) from the IDeALab, Computational Anatomy Toolbox (CAT) using 3 atlas variants (Hammers, LPBA40 and Neuromorphometics), FIRST, FreeSurfer v5.3, and FreeSurfer v6.0-Subfields. A number of these methods were employed to re-segment the T1 images for the stroke group after the stroke lesions were masked (i.e., removed). The automated methods were assessed on eight measures: process yield (i.e. segmentation success rate), correlation (Pearson's R and Shrout's ICC), concordance (Lin's RC and Kandall's W), slope ‘a’ of best-fit line from correlation plots, percentage of outliers from Bland-Altman plots, and significance of control−stroke difference. We eliminated the redundant measures after analysing between-measure correlations using Spearman's rank correlation. We ranked the automated methods based on the sum of the remaining non-redundant measures where each measure ranged between 0 and 1. Subfields attained an overall score of 96.3%, followed by AdaBoost (95.0%) and FIRST (94.7%). CAT using the LPBA40 atlas inflated hippocampal volumes the most, while the Hammers atlas returned the smallest volumes overall. FIRST (p = 0.014), FreeSurfer v5.3 (p = 0.007), manual tracing (p = 0.049), and CAT using the Neuromorphometics atlas (p = 0.017) all showed a significantly reduced hippocampal volume mean for the stroke group compared to control at three months. Moreover, masking of the stroke lesions prior to segmentation resulted in hippocampal volumes which agreed less with manual tracing. These findings recommend an automated segmentation without lesion masking as a more reliable procedure for the estimation of hippocampal volume in ischemic stroke.
Keywords: Hippocampus, Magnetic resonance imaging, Automatic segmentation, Manual segmentation, Stroke
Highlights
-
•
FreeSurfer–Subfields segmentation is favoured for hippocampal volume estimation in healthy and ischemic stroke participants
-
•
Subfields agreed most with manual tracing of the hippocampus performed in accordance with the EADC-ADNI harmonized protocol
-
•
Masking (removal) of stroke lesions in MRI images negatively impact on the performance of automated hippocampal segmentation
-
•
Manual and automated segmentation revealed a significantly compromised hippocampal state in ischemic stroke patients
1. Introduction
Brain atrophy is associated with both normal aging and with brain pathology, especially neurodegenerative disorders. At the age of 65 years, a third of men and half of women are at risk of having a stroke or developing dementia (Hénon et al., 2006). Stroke is one of the major causes of death and disability worldwide (Aerts et al., 2016; Maier et al., 2015; Strong et al., 2007) and a third of stroke survivors develop post-stroke dementia (PSD) months (early-onset) to years (late-onset) after the initial stroke incident (Mok et al., 2017). Vascular dementia, including PSD, is the second most common cause of cognitive decline, with only Alzheimer's disease (AD) being more prevalent (Seshadri and Wolf, 2007). Yet, the regional atrophy patterns in PSD are not understood.
Quantitative magnetic resonance imaging (MRI) studies have revealed changes in brain structures in normal aging and in neurodegenerative dementias. Accelerated hippocampal volume loss has been described in many neurological and psychiatric disorders including temporal lobe epilepsy, Huntington's disease, AD, mild cognitive impairment, schizophrenia, major depression, post-traumatic stress disorder, chronic alcoholism and panic disorder (Geuze et al., 2005). In AD, for instance, hippocampal atrophy is considered a distinguishing feature (Scheltens et al., 1992) and a hallmark of this disease. In conditions such as schizophrenia, post-traumatic stress disorder and major depression, several studies have reported smaller hippocampal volumes (Apfel et al., 2011; Frodl et al., 2006; Meisenzahl et al., 2009; Woon et al., 2010). Smaller hippocampal sizes have also been reported in subcortical ischemic vascular dementia (Fein et al., 2000). In major stroke, hippocampal atrophy has been associated with cognitive decline in survivors (Gemmell et al., 2012; Kliper et al., 2013).
Manual tracing of the hippocampus is the accepted gold standard among neuroanatomical experts (Boccardi et al., 2015; Frisoni et al., 2015). However, manual segmentation is laborious and very time consuming. An expert rater may need up to two hours to trace the hippocampus (Morey et al., 2009) and results may be influenced by rater bias (Colon-Perez et al., 2016). For large sets, manual tracing of the hippocampus is restricted by time and cost. For this, automated segmentation was proposed as a reliable alternative to human manual tracing, and its application is now widely performed in large datasets of patients with conditions such as AD (Morra et al., 2010) and temporal lobe epilepsy (Kim et al., 2012; Pardoe et al., 2009). In AD, computational segmentation techniques have shown good reproducibility and are comparable in accuracy to manual tracing (Fischl et al., 2002; Fischl et al., 2004b; Pantel et al., 2000). The high reproducibility of automated methods reduces bias and facilitates the replication of findings between studies.
Approaches to computerised segmentation include multi-atlas–based registration and propagation (Aljabar et al., 2009; Heckemann et al., 2006; Klein et al., 2005; Wang et al., 2013) and machine learning–based clustering (Maglietta et al., 2016; Morra et al., 2010; Patenaude et al., 2011). For instance, Morra et al. (2010) evaluated FreeSurfer and three machine learning classifiers (hierarchical AdaBoost, support vector machines (SVM) with manual feature selection, and hierarchical SVM with automated feature selection (Ada-SVM)) for normal and AD populations. AdaBoost and Ada-SVM compared more favourably with manual segmentation. Also, a novel strategy for hippocampal segmentation was proposed by Maglietta et al. (2016) using the RUSBoost classifier, which correlated the most to manual tracing when compared to random forest and FreeSurfer.
Available automated methods were not designed to segment MRI images in stroke populations and very little is known about the influence of stroke lesions on their performance. The stroke lesions, often considerably large, appear hypo-intense on T1-weighted images and are hard to discriminate from the gray matter. In this study, we investigated the performance of several automated segmentation tools in two populations: healthy controls and participants with ischemic stroke. We compared their performance against human manual tracing of the hippocampus with and without the masking of stroke lesions. We hypothesized, based on findings from prior studies on healthy and other disease groups, that:
-
1.
Automated methods and manual tracing would return comparable hippocampal volumes for control and stroke populations on both group and individual levels; especially for methods using protocols similar to the EADC-ADNI harmonized protocol (Boccardi et al., 2015; Frisoni et al., 2015) used here for manual tracing.
-
2.
Automated segmentation methods and manual tracing would comparably characterize atrophy states of control and stroke groups from data acquired at equivalent timepoints.
-
3.
Given that lesions in our stroke population occurred remotely outside the hippocampus, their presence would not significantly and negatively impact on the estimation of hippocampal volume by the automated methods.
2. Materials and methods
-
2.1
CANVAS study
It is an observational longitudinal case control study in which 135 ischaemic stroke patients – confirmed clinically and radiologically – and 40 age-matched healthy controls are followed over five years. The CANVAS study is described in detail elsewhere (Brodtmann et al., 2014). Briefly, participants were recruited from three sites in Melbourne (Austin Health, Eastern Health, and Melbourne Health), with all imaging sessions held at The Florey, Melbourne Brain Centre, Austin Hospital campus. Ethical approval was granted by each hospital's human research ethics committee and all participants gave informed consent. Participants completed an interview, MRI scans and neuropsychological assessments, at five timepoints: baseline (within six weeks of index stroke), three months, one year, three years, and at five years. They also provided a blood sample to determine their apolipoprotein E epsilon-4 status. Patients diagnosed with primary hemorrhagic stroke, transient ischemic attack (TIA), venous infarction, or significant medical comorbidities were excluded from participation. Age-matched healthy controls with no history of stroke or TIA were recruited from the local community, from family members of stroke participants and from volunteers who had previously undertaken MRI research at the Florey Institute of Neuroscience and Mental Health. All participants had no pre-existing dementia, neurodegenerative disorders, major psychiatric illnesses or substance abuse problems.
2.1. Participants
We used data from 105 stroke patients (age: 67.4 (mean) ± 11.9 (SD) years, 33 women) and 39 healthy controls (age: 69.1 ± 5.7 years, 15 women) at the 3-month timepoint when most of stroke participants were recruited to the CANVAS study. Among the stroke patients, 60 (57.1%) had right hemispheric stroke, 42 (40%) had left stroke, and three (2.9%) had bilateral stroke. Overall, 60 patients had infarcts in the anterior and middle cerebral artery territories, 17 (16.2%) had posterior cerebral artery infarcts, 13 (12.4%) had stroke in the brainstem, 11 (10.5%) had stroke in the cerebellum, and four (3.8%) had infarcts in multiple locations and vascular territories.
2.2. MRI acquisition
Whole brain images were acquired on a 3 T Siemens Tim Trio Scanner with a 32-channel head coil (Siemens, Erlangen, Germany). The MR images were obtained using a T1-weighted 3D magnetization-prepared rapid gradient sequence: 160 coronal slices, repetition time RT = 1900 ms, echo time TE = 2.6 ms, inversion time TI = 900 ms, flip angle = 9°, matrix size = 256 × 256, slice thickness = 1 mm, voxel size = 1 × 1 × 1 mm3.
A high-resolution, 3D sampling perfection with application-optimized contrasts using different flip angle evolutions (SPACE)-fluid-attenuated inversion recovery (FLAIR) images were also acquired: 160 sagittal slices, 1 mm thick, RT = 6000 ms, TE = 380 ms, 120° flip angle, and 256 × 256 acquisition matrix. The FLAIR images were used to manually outline the stroke lesions.
2.3. Hippocampal volumetry
Manual and automated hippocampal volumes for each subject in this study were averaged across the left and right hippocampi and were expressed in mm3. The hippocampal volumes estimated by manual tracing were assumed to be closest to true measures.
2.3.1. Manual tracing
Hippocampi for the 144 participants were manually traced by an expert rater (MSK) following guidelines described in the EADC-ADNI harmonized protocol for manual hippocampal segmentation (Boccardi et al., 2015; Frisoni et al., 2015). Special attention was paid to the determination of the most rostral and caudal slices, exclusion of the choroid plexus, inclusion of the alveus/fimbria, and exclusion of internal CSF pools within the hippocampus. Coronal slices were used to trace the hippocampi on 3D Slicer 4.5.0.1 (Fedorov et al., 2012) available from http://www.slicer.org. Images in sagittal views were consulted to confirm hippocampal boundaries. A subset made of 101 participants (22 controls and 79 strokes) was separately traced by a second expert rater (QL). The manual tracing of the hippocampi progressed blindly based on the subject ID alone.
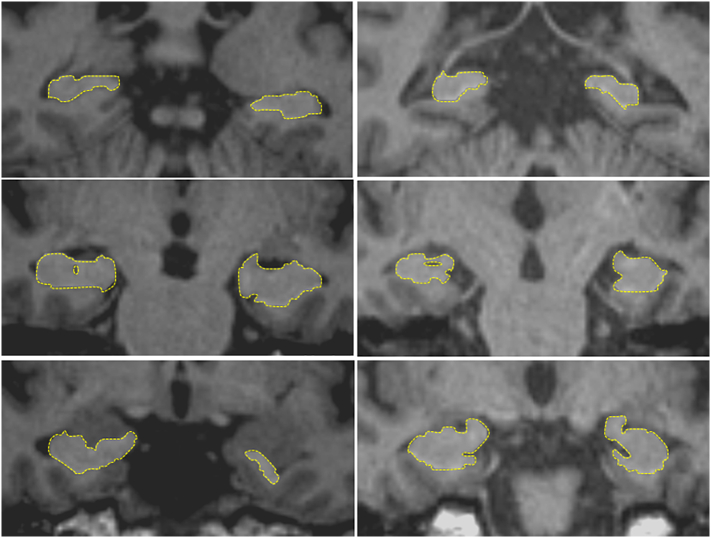
The intra-class correlation coefficient (ICC) (Shrout and Fleiss, 1979) was used to measure inter-rater reliability for raters MSK and QL. ICCs were 0.86 for controls, 0.9 for stroke participants and 0.9 for the two groups together. For comparison with automated methods, manual tracing by MSK was used. A sample representation of hippocampal manual tracing of control and stroke participants is shown in Fig. 1.
Fig. 1.
Sample representation of hippocampal manual tracing of a control (left) and a stroke participant (right).
2.3.2. AdaBoost
AdaBoost is a machine learning based segmentation method. It produced accurate, high-quality automated segmentations in 400 participants from the Alzheimer's Disease Neuroimaging Initiative (Morra et al., 2008). The AdaBoost-based method classifies voxels as belonging to the hippocampus based on thousands of features, which are learned from a training set of manually delineated data. The implementation of AdaBoost used in the current study, applied by one of the authors (AR) using the neuGRID platform (Redolfi et al., 2013), was trained on the EADC-ADNI harmonized protocol for hippocampal segmentation. Adaboost corrects images through N3 bias field correction algorithm and then registers brain to the standard ICBM-152 template through affine registration using FSL FLIRT algorithm.
2.3.3. Atlas-based Hippocampal Segmentation (ABHS)
We obtained hippocampal masks and volumes using an adaptation of a multi-atlas based segmentation technique (Aljabar et al., 2009) specifically for the hippocampus. We created 100 atlas brains with carefully hand-segmented hippocampal masks according to the EADC-ADNI harmonization protocol (Frisoni and Jack 2011; Frisoni et al., 2015). To segment the hippocampi of an individual brain, each of the atlas brains is non-linearly registered to the target image. Then we use a “voting” protocol, in which the cross-correlation match score of each deformed atlas to the target weights the vote of each atlas brain. The voting takes account of the deformed locations of the atlas masks and a priori tissue segmentation of the target image to arrive at a final determination of voxel locations in the native hippocampus. Finally, we use human visual inspection (and clean-up as necessary) for quality control. ABHS is available from ‘http: //idealab.ucdavis.edu’.
2.3.4. Computational Anatomy Toolbox (CAT)
CAT (Gaser and Dahnke, 2016), available from ‘http://dbm.neuro.uni-jena.de/cat12’, is an extension toolbox to SPM12 (www.fil.ion.ucl.ac.uk/spm/software/spm12) that provides easy-to-use tools that cover diverse morphometric methods, such as voxel-based morphometry (VBM), surface-based morphometry (SBM), deformation-based morphometry (DBM), and region or label-based morphometry (RBM).
The T1 images were normalized to the standard ICBM template space and segmented into gray matter, white matter and cerebrospinal fluid. By default, the SPM12 tissue probability maps (TPMs) were used for the affine spatial registration. The segmentation approach in CAT is based on an Adaptive Maximum a Posterior (AMAP) technique without the need for a priori information about tissue probabilities. Tissue probability maps are not used constantly in the sense of the classical unified segmentation approach (Ashburner and Friston, 2005), but only for spatial normalization and for initial skull-stripping. AMAP is adaptive in the sense that local variations (in mean and variance) are modelled as slowly varying spatial functions (Rajapakse et al., 1997). This not only accounts for intensity inhomogeneities, but also for other local variations of intensity. Additionally, the segmentation approach uses a Partial Volume Estimation (PVE) with a simplified mixed model of a maximum of two tissue types (Tohka et al., 2004). Segmentation starts initially with three pure classes (gray matter, white matter, and cerebrospinal fluid) and is followed by a PVE of two additional mixed classes: gray-white matter and gray- cerebrospinal fluid. This results in an estimation of the amount of each pure tissue type present in every voxel. CAT estimates of hippocampal volume were based on the Hammers (Gousias et al., 2008), LPBA40 (LONI Probabilistic Brain Atlas) (Shattuck et al., 2008) and Neuromorphometics (Caviness Jr et al., 1999) brain atlases. Here, we refer to the CAT methods by their respective atlas names: Hammers, LPBA40 and Neuro (short for Neuromorphometics).
2.3.5. FIRST-FSL
FIRST, part of the FSL 5.0.8 analysis library (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki), is a model-based segmentation tool (Patenaude et al., 2011). The shape and appearance models used in FIRST are constructed from manually segmented images provided by the Center for Morphometric Analysis, Boston MA, United States. The manual labels are parameterized as surface meshes and modelled as a point distribution model. Deformable surfaces are used to automatically parameterize the volumetric labels in terms of meshes. The deformable surfaces are constrained to preserve vertex correspondence across the training data. Furthermore, normalized intensities along the surface normals are sampled and modelled. The shape and appearance model is based on multivariate Gaussian assumptions. FIRST searches through linear combinations of shape modes of variation for the most probable shape instance given the observed intensities in a T1-weighted image.
The T1-weighted volumes were RAS-oriented and centred using the alignment script “acpcdetect_v2.0” (https://www.nitrc.org/frs/?group_id=90&release_id=3772). Centring was based on a midpoint between anterior and posterior commissures. Then, the images were registered using “first_flirt”. This script runs two-stage affine registration to MNI152 space at 1 mm resolution. The first stage is a standard 12 degrees of freedom registration to the template. The second stage applies 12 degrees of freedom registration using an MNI152 sub-cortical mask to exclude voxels outside the sub-cortical regions.
2.3.6. FreeSurfer v5.3
Referred to here as just ‘FreeSurfer’, this method used the cross-sectional pipeline (recon-all) in FreeSurfer v5.3, which is freely available from ‘http://surfer.nmr.mgh.harvard.edu/’. Briefly, FreeSurfer segmentation includes 1) motion correction and averaging (Reuter et al., 2010) of multiple images when available, 2) removal of non-brain tissue (Segonne et al., 2004), 3) Talairach transformation (affine transform from the original volume to the MNI305 atlas), 4) segmentation of subcortical white matter and deep gray matter structures including hippocampus, amygdala, caudate, putamen, ventricles (Fischl et al., 2002; Fischl et al., 2004a), 5) intensity normalization (Sled et al., 1998), 6) tessellation of the gray matter-white matter boundary, 7) topology correction (Fischl et al., 2001; Segonne et al., 2007), and 8) surface deformation following intensity gradients (Dale et al., 1999; Fischl and Dale, 2000). FreeSurfer morphometric procedures show good test-retest reliability across scanner manufacturers and field strengths (Reuter et al., 2012; Han et al., 2006).
2.3.7. FreeSurfer v6.0 − Subfields
Referred to here as ‘Subfields’, this algorithm (recon-all -hippocampal-subfields-T1) generates an automated segmentation of the hippocampal subfields based on a statistical atlas built primarily upon ultra-high resolution (~0.1 mm isotropic) ex vivo MRI data (Iglesias et al., 2015). In this study, we used the sum of all subfield volumes to represent whole hippocampal volume with this segmentation method, which solves a number of limitations of the in vivo atlas that was distributed with FreeSurfer v5.3. Namely, these limitations were: a) lower resolution of the training images forcing the human labellers to heavily rely on geometric criteria to trace boundaries; b) the delineation protocol did not include the “molecular layer” which corresponds to the stratum radiatum, lacunosum moleculare, hippocampal sulcus and molecular layer of the dentate gyrus; and c) the delineation protocol of the in vivo atlas was designed for the hippocampal body and did not translate well to the hippocampal head or tail.
2.3.8. Lesion masking
To assess the influence of stroke lesions on the automated segmentation of the hippocampus, modified T1 images were created then re-segmented. The stroke lesions were initially identified by stroke neurologist (AB) and subsequently traced by MSK using the FLAIR images to create the lesion masks. The T1 images were first registered to the FLAIR images using FLIRT (Jenkinson et al., 2002; Jenkinson and Smith, 2001) then the created lesion masks were projected onto the registered T1 images. Finally, the intensities of all voxels on T1 images corresponding to the projected masks were zeroed to create the modified T1 versions.
2.3.9. Quality control
The registered images were visually checked to confirm that the orientation and size of the subject brain corresponded with that of the template and to verify that the subcortical structures were appropriately situated. The segmented images were inspected to verify that the segmentation of the hippocampus didn't overlap with that of another adjacent structure such as the amygdala and that no part of the hippocampus was left out during segmentation.
2.4. Performance measures
We utilized eight measures to quantify the performance of the automated segmentation methods described in the previous section. The methods ranked differently depending on which measure was used. Since different measures portrait different aspects of performance (e.g., reliability, agreement, efficacy), we ranked the methods according to the sum (overall score) of the non-redundant (non-correlated), equally weighted, measures. Next, we briefly describe these measures.
2.4.1. Process yield: segmentation success rate
Process yield was measured as the percentage of T1-weighted scans that successfully completed segmentation and where both left and right hippocampal volumes could be estimated.
2.4.2. Measures of concordance
We used two measures to quantify concordance between hippocampal volumes obtained through manual tracing and each of the automated segmentation methods. These were Lin's concordance correlation coefficient (RC) (Lin, 1989) and Kendall's coefficient of concordance (W) (Kendall and Smith, 1939).
Lin's RC is the concordance between a new measurement and an existing “gold standard” measurement. This statistic quantifies the agreement between two measures on the same observation. RC ranges from −1 to 1 with perfect agreement at 1. RC is a measure of absolute agreement in the ratings and can be legitimately calculated on as few as 10 observations.
Kendall's coefficient of concordance (also known as Kendall's W) is a non-parametric statistic. It is a normalization of the statistic of the Friedman test, and can be used for assessing agreement among raters. Kendall's W ranges from 0 for no agreement to 1 for complete agreement.
2.4.3. Measures of correlation
For correlation between manual and automated hippocampal volumes, we computed Pearson's correlation coefficient (R) and Shrout's intra-class correlation coefficient (ICC) (Shrout and Fleiss, 1979).
Pearson's correlation measured the degree by which the automated and manual hippocampal volumes were associated. High correlation does not automatically imply that there is good agreement between compared sets as correlation evaluates only the linear association of observations in these sets (Morey et al., 2009).
In the computation of ICC, no generalization to a larger population of raters was assumed. Since we were interested in comparing specific segmentations at hand, the methods were assumed fixed. When we assumed them to be random, we found ICC to be remarkably similar to Lin's RC.
2.4.4. Bland-Altman plots
Bland–Altman (BA) plots (Bland and Altman, 1999) offer another approach to assessing agreement between two quantitative measurements. Bland–Altman analyses are of value in clinical settings: they provide information about interchangeability of two measures without assuming that either is the gold standard (Morey et al., 2009). Agreement between measures is quantified by constructing limits of agreement (LOA). We calculated these statistical limits using the mean (d) and the standard deviation (s) of the percent difference between manual hippocampal volumes and volumes estimated by each of the automated segmentation methods. The percent volume difference was computed by dividing the difference between two compared volumes by their average and then plotted against this average. We used the average of the two compared volumes in the calculation of percent volume difference since true volumes were unknown and the average was the best estimate there was (Giavarina, 2015).
Distributions of percent volume differences were checked for normality using the one-sample Kolmogorov-Smirnov test (Massey, 1951). The 95% confidence intervals (CI) of the mean volume difference (d) and of the limits of agreements (LOA = d ± 1.96s) were then computed as follows:
where N is the number of participants and t is the value of the t-distribution with N-1 degrees of freedom. Participants whose percent volume differences were outside the 95% CIs of the LOAs were defined as outliers.
2.4.5. Statistical analysis: control vs. stroke
We analyzed the difference in hippocampal volumes between the healthy control and stroke groups for each of the tracing methods using multi-way analysis of variance (ANOVAN, Statistics Toolbox, MATLAB Release 2015a, https://au.mathworks.com/help/stats/anovan.html Dunn and Clark). Covariates included age, body mass index (BMI), years of education, total intracranial volume (TIV) and sex. There were no significant interactions between group and any of the covariates or between the covariates themselves. Thus, the regression model included only linear terms. The MATLAB Tukey-Kramer default setting was used for multiple comparison correction and significance was tested at α = 0.05. A similar analysis was carried for comparison between the various methods, but this was done separately for each study group (control and stroke). The TIV estimated by FreeSurfer was used with the rest of the methods including manual tracing.
2.4.6. Methods ranking
This was based on the sum of performance measures. Since we found the ranking of the methods based on ICC, R, RC and W were nearly the same for both control and stroke groups, we combined the control and stroke measures for each method. The other measures were: process yield, a measure (=1 − |1 − a|) based on best-fit line slope ‘a’, a function f(p) = 1 if p < 0.05, = 0.95 otherwise; based on ‘p’ values from ANOVAN analysis of control vs. stroke, and a measure (=1 − n1/N) where n1 is the count of outliers from the BA plot for each method and N is the number of participants.
To avoid any bias in the ranking of methods, we analyzed correlations between the comparison measures and removed measures that were redundant. If a measure significantly correlated to multiple measures, that measure was removed first. If a measure significantly correlated to only one other measure, we eliminated the one which correlated higher to the other non-redundant measures. We used Spearman's rank correlation to eliminate redundant measures given there was an outlier in the W measure and f(p) values could only be discrete. In addition, all scores were not normally distributed. The overall score for each method was then computed as the sum of the non-redundant measures. Scores were also computed as percentages of the maximum score that could be achieved (equal to the count of non-redundant measures).
3. Results
Hippocampal volumes for 39 healthy and 105 stroke participants were estimated using manual tracing and eight publicly available segmentation tools including AdaBoost, ABHS, Hammers, LPBA40, Neuro, FIRST, FreeSurfer (cross-sectional algorithm) and FreeSurfer ‘Subfields’.
Hippocampal asymmetry has been reported in normal cohorts (Lucarelli et al., 2013; Middlebrooks et al., 2017) with the right hippocampus shown to be larger than the left. In this study, we also report a larger right hippocampus with a hippocampal asymmetry ranging between 1.2% and 10.6% depending on the segmentation method used (see Table 1). Except for FIRST, all methods showed that this hippocampal asymmetry is significant.
Table 1.
Left (LV) and right (RV) hippocampal volumes per segmentation method: Hippocampal asymmetry was computed as the difference in right and left volumes (RV-LV) normalized by their average.
| Method | LV | RV | 2*(RV-LV)/(RV + LV) | P |
|---|---|---|---|---|
| ABHS | 3416 | 3514 | 2.8% | < 0.001 |
| AdaBoost | 3839 | 4007 | 4.3% | < 0.001 |
| FreeSurfer | 3973 | 4115 | 3.5% | < 0.001 |
| FIRST | 3746 | 3792 | 1.2% | 0.17 |
| Hammers | 2266 | 2449 | 7.8% | < 0.001 |
| LPBA40 | 4253 | 4338 | 2.0% | 0.002 |
| Manual | 3506 | 3713 | 5.7% | < 0.001 |
| Neuro | 3054 | 3397 | 10.6% | < 0.001 |
| Subfields | 3513 | 3646 | 3.7% | < 0.001 |
It is imperative that an automated segmentation method would be comparable to manual tracing in estimating the hippocampal volume irrespective of group or hemisphere. We have found that this comparison was similar for both left and right hippocampi irrespective of method used (e.g., see correlation plots in Supplemental Fig. 1). Hence, we present our findings below based on hippocampal volumes averaged across hemispheres.
Supplemental Fig. 1.
Correlation of left and right hippocampal volumes from automated segmentations to manual tracing.
3.1. Process yield
Segmentation process yields were quite high. In the case where stroke lesions were not masked, all methods except for AdaBoost and FreeSurfer successfully segmented hippocampi for all 144 participants. AdaBoost could not obtain volumes for two participants resulting in a slightly lower yield at 98.6% and FreeSurfer yield was at 99.3% after missing out on one participant. In the case where the stroke lesions were masked, FreeSurfer lost 1.4% in yield after missing on the segmentation of two participants who had large stroke lesions across a multitude of image slices. Yields for FIRST, Hammers, LPBA40 and Neuro were not affected by stroke lesion masking.
3.2. Impact of stroke lesion on hippocampal volume estimation
We evaluated the impact of lesion removal on the estimation of hippocampal volumes. Fig. 2 shows a comparison of group mean volumes for masked and unmasked lesion segmentations. Except for LPBA40 (p = 0.01), there was no significant difference in mean volumes for FIRST (p = 0.4), FreeSurfer (p = 1), Hammers (p = 1) or Neuro (p = 0.96). More importantly, the hippocampal volumes from the unmasked lesion segmentations agreed more, and correlated better, with manually traced volumes (see Table 2). Correlation and concordance measures (Pearson's R, Lin's RC, Shrout's ICC, and Kendall's W) for the automated methods were higher (by 0–0.27) in the case of unmasked lesion segmentation, except for Lin's coefficient of concordance RC, which was higher for masked lesion segmentation with LPBA40. This was driven by a significantly lower mean volume which became closer to mean volume estimated by manual tracing. For the remainder of this analysis, we compare manual hippocampal tracing to automated segmentation without stroke lesion masking.
Fig. 2.
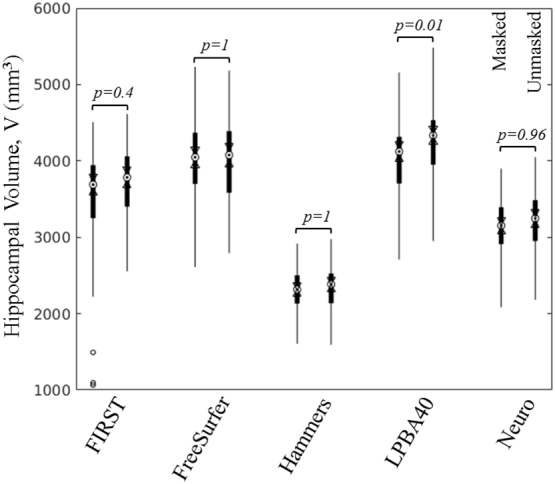
Distribution of hippocampal volumes from automated segmentation of the stroke population with lesions masked (left for each pair) and unmasked. Except for LPBA40, volume estimation was not significantly affected by lesion masking. On each filled box, the central mark (dot inside circle) indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers (±3 standard deviations). Two medians are different, with 95% confidence, if their box notches (range between arrows) do not overlap.
Table 2.
Correlation and concordance coefficients between manual tracing and automated methods: Comparison between segmentation with and without lesion masking.
| Method | Lesion | Pearson's R | Lin's RC | Shrout's ICC | Kendall's W |
|---|---|---|---|---|---|
| FIRST | Masked | 0.58 | 0.56 | 0.56 | 0.87 |
| Unmasked | 0.83 | 0.79 | 0.83 | 0.90 | |
| FreeSurfer | Masked | 0.82 | 0.60 | 0.81 | 0.90 |
| Unmasked | 0.86 | 0.63 | 0.85 | 0.92 | |
| Hammers | Masked | 0.89 | 0.12 | 0.80 | 0.94 |
| Unmasked | 0.89 | 0.13 | 0.81 | 0.95 | |
| LPBA40 | Masked | 0.83 | 0.60 | 0.82 | 0.90 |
| Unmasked | 0.86 | 0.45 | 0.85 | 0.93 | |
| Neuro | Masked | 0.86 | 0.54 | 0.85 | 0.92 |
| Unmasked | 0.88 | 0.63 | 0.87 | 0.93 |
3.3. Hippocampal volume distributions
After the elimination of participants (3 strokes and 1 control) that failed segmentation or were poorly segmented by any of the methods, a common set of 140 participants (38 controls and 102 strokes) was used for comparison between the methods. The distributions of raw (not corrected) hippocampal volumes for both controls and stroke participants are shown in Supplemental Fig. 2. In general, the figure shows close distributions among methods except for Hammers segmentation showing distinctively lower hippocampal volumes. The mean volumes for both control and stroke groups from the Hammers segmentation were shown to be significantly lower compared to all other methods, including manual tracing. For the stroke group, the LPBA40 and Neuro methods were also shown to have significantly different mean volumes compared to the other methods (see Table 3).
Supplemental Fig. 2.
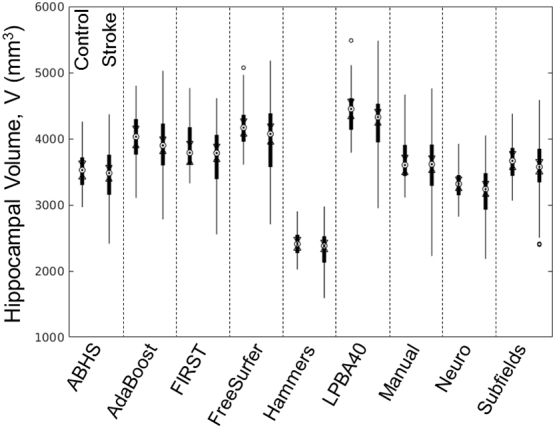
Hippocampal volume distributions (raw data): Hammers produced the smallest estimates for hippocampal volumes compared to the rest of the methods.
Table 3.
Multiple comparison test (Tukey-Kramer correction): p-values above and below diagonal are for control and stroke respectively.
| Method | ABHS | AdaBoost | FIRST | FreeSurfer | Hammers | LPBA40 | Manual | Neuro | Subfields |
|---|---|---|---|---|---|---|---|---|---|
| ABHS | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.36 | 0.21 | 0.51 | |
| AdaBoost | 0.00 | 0.77 | 0.26 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| FIRST | 0.00 | 0.20 | 0.00 | 0.00 | 0.00 | 0.17 | 0.00 | 0.10 | |
| FreeSurfer | 0.00 | 0.85 | 0.00 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 | |
| Hammers | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| LPBA40 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Manual | 0.48 | 0.00 | 0.37 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | |
| Neuro | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Subfields | 0.83 | 0.00 | 0.11 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 |
For the control group, there was no difference in mean hippocampal volume for ABHS−Manual (p = 0.36), ABHS−Neuro (p = 0.21), ABHS−Subfields (p = 0.51), AdaBoost−FIRST (p = 0.77), AdaBoost−FreeSurfer (p = 0.26), FIRST−Manual (p = 0.17), FIRST−Subfields (p = 0.10), FreeSurfer−LPBA40 (p = 0.10) and Manual−Subfields (p = 1). For the stroke group, there was no difference in mean volume for ABHS−Manual (p = 0.48), ABHS−Subfields (p = 0.83), AdaBoost−FIRST (p = 0.20), AdaBoost−FreeSurfer (p = 0.85), FIRST−Manual (p = 0.37), FIRST−Subfields (p = 0.11), and Manual−Subfields (p = 1).
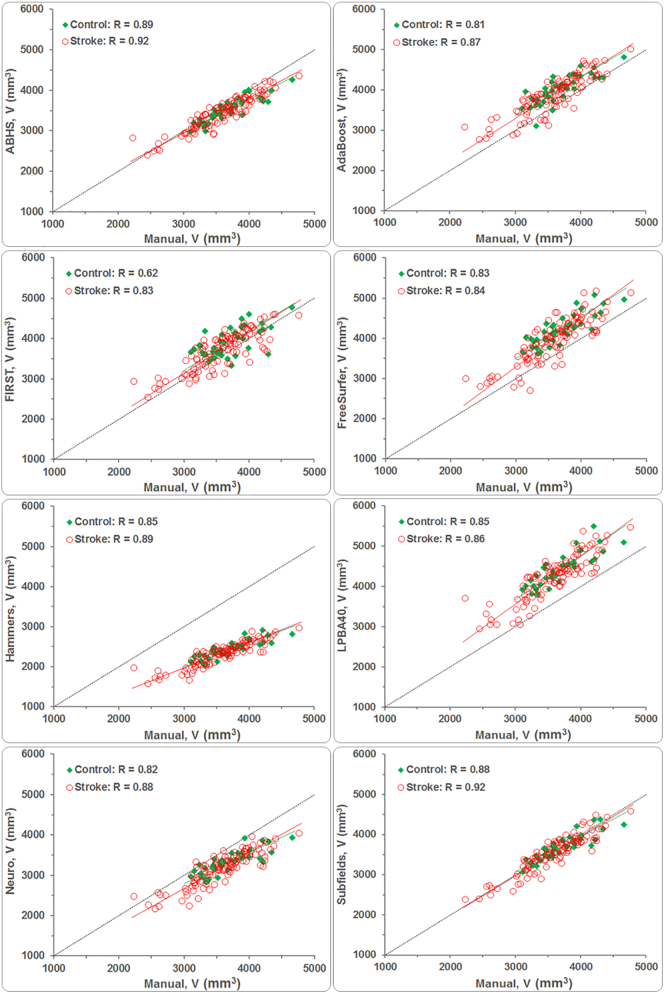
3.4. Correlation and concordance of estimated volumes
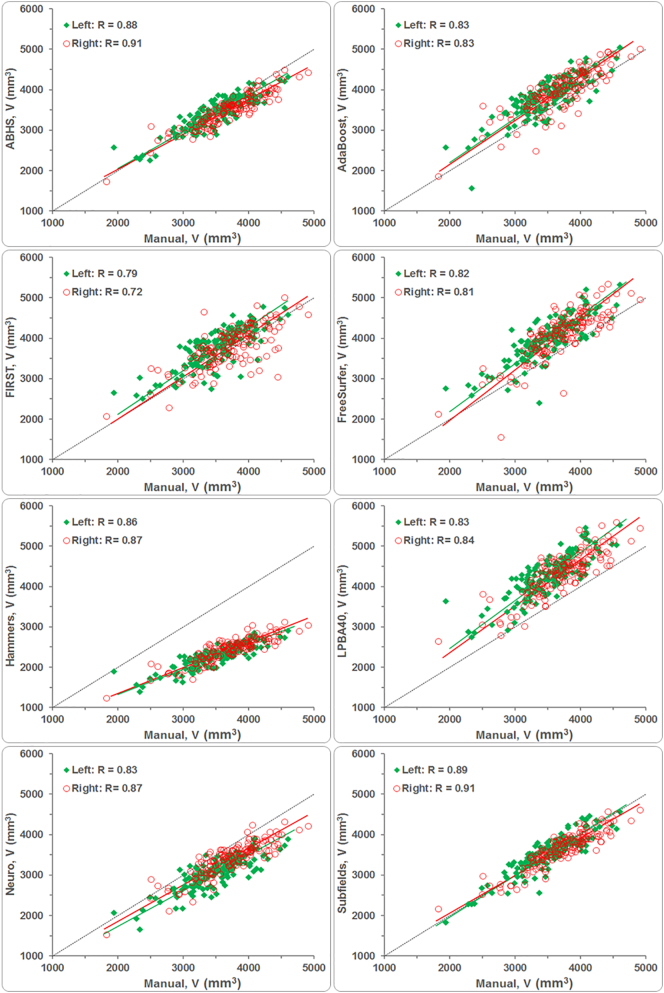
The measures of agreement are summarized in Table 4. For both control and stroke groups, ABHS and ‘Subfields’ correlated the most to manual tracing, RABHS(Control, Stroke) = (0.89, 0.92) and RSubfields(Control, Stroke) = (0.88, 0.92). FIRST correlated the least with manual tracing; R (Control, Stroke) = (0.62, 0.83). AdaBoost and the CAT methods generally correlated higher than FreeSurfer for both study groups. Hammers had the highest Pearson's correlation coefficients R (Control, Stroke) = (0.85, 0.89) among the CAT methods, but also underestimated hippocampal volumes the most; while LPBA40 overestimated them (see Fig. 3). FreeSurfer and AdaBoost also returned higher hippocampal volumes; shown mostly above the dotted ‘perfect-fit’ line in Fig. 3.
Table 4.
Agreement measures between manual tracing and automated segmentation methods.
| Method → Measure ↓ | ABHS | AdaBoost | FIRST | FreeSurfer | Hammers | LPBA40 | Neuro | Subfields | |
|---|---|---|---|---|---|---|---|---|---|
| Control (N = 38) | |||||||||
| Pearson's | R | 0.89 | 0.81 | 0.62 | 0.83 | 0.85 | 0.85 | 0.82 | 0.88 |
| Lin's | RC | 0.77 | 0.57 | 0.54 | 0.42 | 0.07 | 0.29 | 0.48 | 0.87 |
| Shrout's | ICC | 0.88 | 0.81 | 0.62 | 0.83 | 0.75 | 0.85 | 0.79 | 0.88 |
| Kendall's | W | 0.96 | 0.92 | 0.76 | 0.91 | 0.94 | 0.94 | 0.92 | 0.96 |
| Stroke (N = 102) | |||||||||
| Pearson's | R | 0.92 | 0.87 | 0.83 | 0.84 | 0.89 | 0.86 | 0.88 | 0.92 |
| Lin's | RC | 0.88 | 0.71 | 0.79 | 0.63 | 0.13 | 0.45 | 0.63 | 0.92 |
| Shrout's | ICC | 0.92 | 0.87 | 0.83 | 0.85 | 0.81 | 0.85 | 0.87 | 0.92 |
| Kendall's | W | 0.96 | 0.94 | 0.90 | 0.92 | 0.95 | 0.93 | 0.93 | 0.96 |
Fig. 3.
Correlation of average hippocampal volumes from automated segmentation to manual tracing.
The large proportional bias in hippocampal volumes by Hammers have caused this method to score poorly on Lin's concordance measure, with RC < 0.4 for both control and stroke groups as shown in Table 4. LPBA40, which had a positive bias in the mean, also scored poorly with the control group (RC = 0.29), but higher with the stroke group (RC = 0.45). Due to their relatively lower bias in the mean, FreeSurfer and Neuro scored fairly with the control (RC = 0.42 and 0.48) and stroke groups (RC = 0.63). The Lin's scores for ABHS and ‘Subfields’, with volume means being the closest to that of manual tracing, were higher for both study groups compared to other methods. RC scores for FIRST and AdaBoost were relatively similar with control, but FIRST (RC = 0.79) scored higher with the stroke group than did AdaBoost (RC = 0.71). In terms of Shrout's ICC measure, FIRST scored the lowest with the control group (ICC = 0.62) and Hammers scored the lowest with the stroke group (ICC = 0.81). ICC scores for the rest of the methods ranged between 0.75 and 0.88 for control and between 0.83 and 0.92 for stroke. Finally, the Kendall's test indicated a significant agreement for all methods and for both groups (W > 0.7) (Schleder et al., 2013).
The automated methods correlated to manual tracing similarly for both groups: similar slopes for best-fit lines as shown in Fig. 3. For easier visualization, best-fit lines were computed for control and stroke data combined, then they were normalized by mean hippocampal volume from manual tracing (see Fig. 4). AdaBoost, FIRST, and ‘Subfields’ had slopes being the closest to 1 (slope of ‘perfect-fit’ line). FreeSurfer (a = 1.18) and Hammers (a = 0.63) showed respectively the highest and lowest sensitivity to hippocampal volume variation compared to the rest of the methods.
Fig. 4.
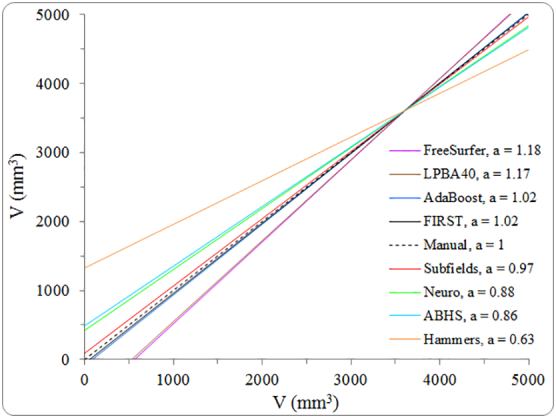
Slopes ‘a’ of best-fit lines for automatically estimated volumes plotted as function of manual volumes after removing fixed bias from all methods.
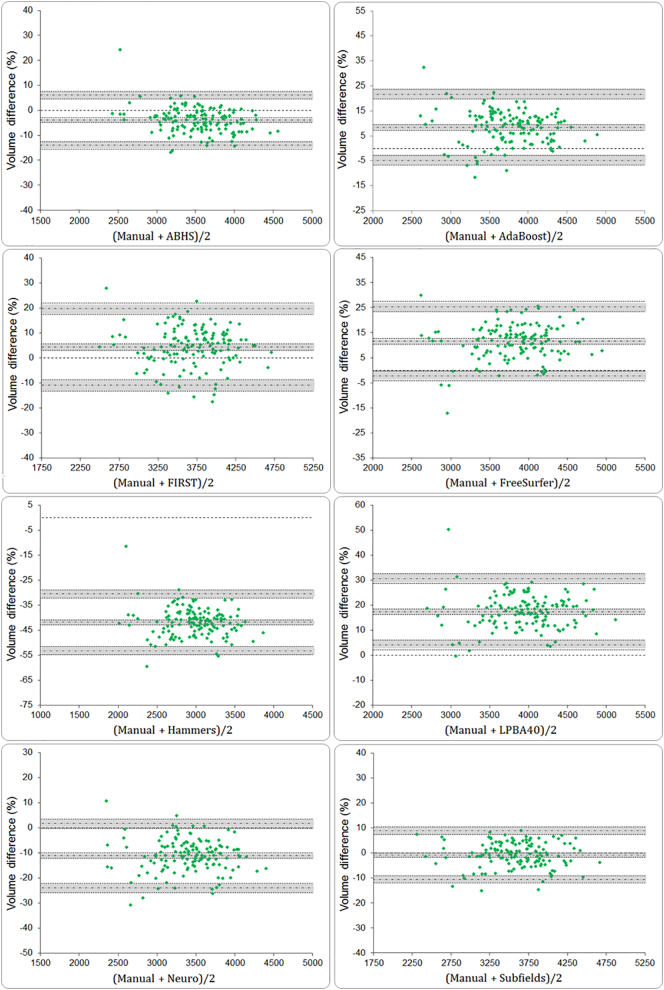
3.5. Bland-Altman plots
The BA plots are shown in Fig. 5. They confirm that FreeSurfer and LPBA40 systematically generated larger volumes compared to manual tracing while ABHS, Hammers and Neuro generated smaller volumes. But for ‘Subfields’, the BA plots suggest that this method is interchangeable with manual tracing; given that the line of equality (volume difference of zero) lies within the 95% CI of mean volume difference (d). Other than ‘Subfields’, ABHS and FIRST had mean volume difference (d) being the closest to the line of equality.
Fig. 5.
Bland–Altman plots for percent volume differences between manual and automated segmentation methods. 95% confidence intervals for mean volume difference (d) and for limits of agreements are shown (gray bands).
The 95% CIs for mean (d) and for the LOAs are determined by variability (s) in the distribution of volume differences and by sample size (N). The larger the sample size and the smaller the volume differences between the methods, the narrower are the confidence intervals. Given a wider distribution of volume differences, FIRST had the widest CIs for both (d) and LOAs.
We examined the outliers in each BA plot. Across all plots in Fig. 5, there were 32 data points showing extreme overestimation or underestimation. The distribution of outliers between methods is presented in supplemental Table 1, showing FIRST with highest outliers' percentage. Furthermore, we evaluated the correlation between the outliers' volumes, corrected by TIV, and the count (n) of segmentation methods affected. Supplemental Fig. 3 shows a significant correlation between these two variables suggesting that participants with smaller hippocampal volumes – perhaps compromised by degeneration – were estimated with less accuracy.
Supplemental Fig. 3.
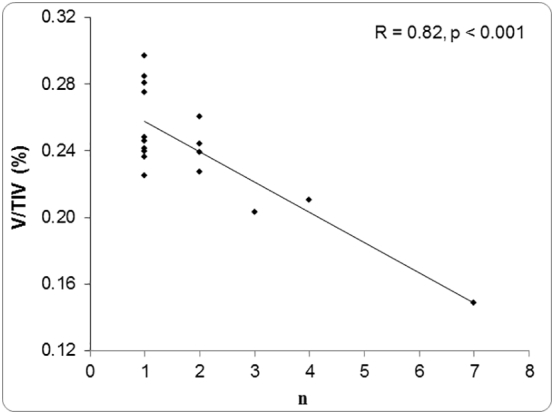
Correlation between hippocampal volume (corrected by TIV) and number of methods (n) producing volumes outside the 95% CI of LOA in BA plots (Fig. 5, V: hippocampal volume, TIV: total intracranial volume).
3.6. Control vs. stroke at three months
Between the two groups, there was no significant difference in age (p = 0.39), nor in sex (p = 0.43), but there was a significant difference in the number of years of education (p < 0.001). The mean for years of education was 12.7 ∓ 3.6 years for stroke patients and 15.5 ∓ 4.5 years for controls.
Mean hippocampal volumes − corrected for age, BMI, years of education, sex and TIV − are presented in Table 5. None of the methods showed a significant interaction between the covariates. All methods systematically showed a lower mean hippocampal volume for the stroke group (see Fig. 6). FIRST (p = 0.014), FreeSurfer (p = 0.007), manual (p = 0.049), and Neuro p = 0.017 showed that the hippocampal volume difference between the two groups was significant. On the other hand, ABHS (p = 0.061), Hammers (p = 0.058), and LPBA40 (p = 0.054) showed that this difference approached but did not reach significance.
Table 5.
Mean hippocampal volumes, V (mm3): FIRST, FreeSurfer, manual tracing, and Neuro showed significant difference between control and stroke groups.
| Method | Control |
Stroke |
P | ||||
|---|---|---|---|---|---|---|---|
| Mean | SD | V/VManual | Mean | SD | V/VManual | ||
| Manual | 3734 | 464 | 3578 | 607 | 0.049 | ||
| ABHS | 3560 | 382 | 0.95 | 3438 | 499 | 0.96 | 0.061 |
| AdaBoost | 4044 | 519 | 1.08 | 3948 | 678 | 1.10 | 0.275 |
| FIRST | 3934 | 527 | 1.05 | 3711 | 688 | 1.04 | 0.014 |
| FreeSurfer | 4267 | 532 | 1.14 | 4019 | 695 | 1.12 | 0.007 |
| Hammers | 2422 | 257 | 0.65 | 2338 | 336 | 0.65 | 0.058 |
| LPBA40 | 4408 | 505 | 1.18 | 4241 | 659 | 1.19 | 0.054 |
| Neuro | 3346 | 363 | 0.90 | 3197 | 475 | 0.89 | 0.017 |
| Subfields | 3694 | 436 | 0.99 | 3573 | 570 | 1.00 | 0.104 |
Fig. 6.
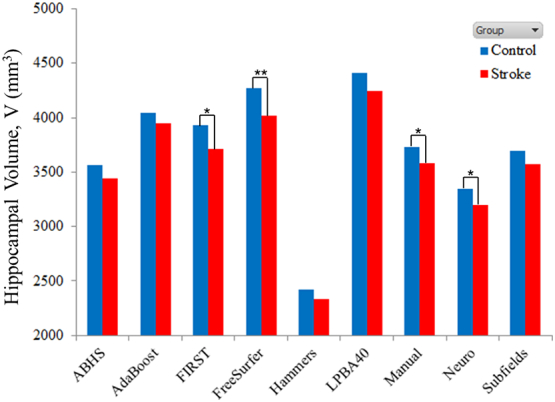
Mean hippocampal volumes at 3 months: Control vs. stroke (p < .05 *, p < .01 **).
3.7. Overall score
Calculated probability values (p-values) for Spearman's rank correlations between measures used for comparing hippocampal segmentation methods are shown in Table 6. The first redundant measures to be dropped were Pearson's R and Kendall's W, because each significantly correlated to two measures. Then, we had to decide between Lin's RC and the measure ‘1-|1-a|’ which were significantly correlated (p = 0.029). Since the correlation between Lin's RC and Shrout's ICC (p = 0.066) approached significance, while the correlation between ‘1-|1-a|’ and ICC did not approach significance (p = 0.35), we eliminated the RC measure. Furthermore, RC correlated higher to the sum of the remaining non-redundant measures compared to the measure ‘1-|1-a|’. This finding also supported the elimination of RC.
Table 6.
Probability values (p-values) of Spearman's rank correlations.
| Measure | Yield | R | RC | ICC | W | f(p) | 1 - |1-a| |
|---|---|---|---|---|---|---|---|
| R | 0.21 | ||||||
| RC | 0.43 | 0.21 | |||||
| ICC | 0.43 | 0.023 | 0.066 | ||||
| W | 0.32 | 0.001 | 0.23 | 0.042 | |||
| f(p) | 0.89 | 0.96 | 0.61 | 0.88 | 1.00 | ||
| 1 - |1-a| | 0.57 | 0.69 | 0.029 | 0.35 | 0.69 | 0.39 | |
| 1 - n1/N | 0.46 | 0.15 | 0.77 | 0.30 | 0.064 | 1.00 | 0.93 |
The remaining non-correlated measures were used to compute the overall scores for the automated segmentation methods, which ranged between 87.3% and 96.3% (see Table 7). Subfields and Hammers scored highest and lowest, respectively. For the most frequently used methods, FIRST scored better than FreeSurfer v5.3.
Table 7.
Overall score for the automated segmentation methods (percentage was computed based on maximum achievable score of 5).
| Method | Yield | ICC | f(p) | 1 - |1-a| | 1 - n1/N | Score (%) |
|---|---|---|---|---|---|---|
| Subfields | 1.00 | 0.91 | 0.95 | 0.97 | 0.98 | 4.82 (96.3) |
| AdaBoost | 0.99 | 0.86 | 0.95 | 0.98 | 0.97 | 4.75 (95.0) |
| FIRST | 1.00 | 0.79 | 1.00 | 0.98 | 0.96 | 4.73 (94.7) |
| Neuro | 1.00 | 0.86 | 1.00 | 0.88 | 0.96 | 4.71 (94.2) |
| ABHS | 1.00 | 0.91 | 0.95 | 0.86 | 0.97 | 4.69 (93.9) |
| FreeSurfer | 0.99 | 0.83 | 1.00 | 0.82 | 0.97 | 4.61 (92.2) |
| LPBA40 | 1.00 | 0.85 | 0.95 | 0.83 | 0.98 | 4.61 (92.1) |
| Hammers | 1.00 | 0.80 | 0.95 | 0.63 | 0.98 | 4.36 (87.3) |
4. Discussion
In this study, we estimated the hippocampal volumes for healthy and ischemic stroke participants using several automatic segmentation methods and compared them to volumes obtained through manual tracing based on the EADC-ADNI harmonized protocol for hippocampal segmentation.
Subfields performed best for hippocampal volume estimation; closely trailed by Adaboost and FIRST. Other studies using, mostly, older version of FreeSurfer (Cover et al., 2016; Maglietta et al., 2016; Morey et al., 2009; Mulder et al., 2014; Rana et al., 2017) have proposed this tool as a viable alternative to manual tracing for quantifying hippocampal volume in AD. We also found a substantial agreement between the newer version 5.3 of FreeSurfer and manual tracing for the estimation of hippocampal volume in healthy controls and ischemic stroke patients. Furthermore, our findings suggest that Subfields (in FreeSurfer version 6.0), AdaBoost and FIRST are better alternatives than the standard pipeline of FreeSurfer. This is based on the overall ranking of methods based on a multitude of measures. In addition to estimating overall hippocampal volume, Subfields allows for the quantification of volumes for various parts of the hippocampus. This allows one to characterize atrophy in hippocampal sub-regions, potentially providing more sensitivity in differentiating between study groups.
We note that our findings were based on method comparison relative to data acquired at the three-month timepoint. This is still relevant if one is interested in absolute hippocampal volumes at any single timepoint. For quantifying relative hippocampal volume change over time, the ranking of evaluated methods may change. For instance, we used the cross-sectional segmentation pipeline of FreeSurfer in this study in line with the rest of the methods, but for a longitudinal study, the longitudinal processing stream of FreeSurfer (Reuter et al., 2012) is preferred.
Referring to Fig. 3, the bias in hippocampal volumes shown by the various segmentation methods may be attributed to differences in datasets used for training and fine-tuning, as well as the segmentation approaches taken by these methods (atlas-based vs. model-based). In a longitudinal study, this bias can be tolerated so long it is fixed (i.e., not proportional to hippocampal size). The behaviour of the best-fit line (slope ‘a’ in Fig. 4) could predict the accuracy of the compared methods in assessing the amount of hippocampal volume change over time. Methods with slopes a > 1 will tend to overestimate hippocampal volume change over time. On the contrary, methods with slopes a < 1 will tend to underestimate volume change. AdaBoost, FIRST, and Subfields are posed to estimate a hippocampal volume change being the closest to manual estimation.
The ischemic infarcts for all stroke participants occurred away from the hippocampus except for one stroke participant who had a prior infarct involving the right hippocampus. The automated methods did not have difficulty segmenting this case, and similar to manual tracing, they all estimated a relatively smaller volume for this hippocampus. However, lesion masking was found to negatively impact on agreement measures of methods tested. The masking of the lesions could have negatively affected the quality of image registration and modified the image intensity distribution such that less accurate volumes were produced. The results here suggest that, at least for the estimation of hippocampal volume, it is more effective not to mask the stroke lesions. This recommendation may or may not apply to volumetric estimations of other structures. There are indications, though, that lesion masking may not be needed even for the estimation of brain structures other than the hippocampus. In (Werden et al., 2017), the stroke lesions were not masked, yet most FreeSurfer-reconstructed images did not require manual corrections. Additionally, the effect of lesions in multiple sclerosis was recently evaluated in (Gonzalez-Villa et al., 2017). There, it was concluded that the presence of lesions did not affect the segmentation systematically. The lesions either made the segmentation underperform or ‘surprisingly augmented’ its accuracy (Gonzalez-Villa et al., 2017).
The different comparison measures adopted in this study helped to uncover weaknesses and strengths of the evaluated methods. This was not going to be otherwise achieved if only one or two measures were used. Other than Subfields, which was shown to be interchangeable with manual tracing, the use of BA plots interestingly revealed that FIRST was the closest to be interchangeable with manual tracing despite lower scores achieved on other measures. Also, the use of BA plots helped us examine the nature of outliers. The 32 outliers examined represented data from various participants (15 strokes and 2 controls) and by different methods. None of the participants was considered an outlier for all segmentation methods at once. This confirms that none of the images had an anatomical abnormality or quality issue to the extent that it could have systematically influenced the outcome for all methods.
The evaluated methods achieved excellent overall scores (between 87.3% and 96.3%) and thus choosing one method over the other for hippocampal segmentation may prove difficult; given the tight distribution of these overall scores and the trade-offs between measures for some of the methods. The study questions and the comparison outcome objectives may eventually dictate the kind of segmentation preferred. In this study, we did not have a priori preference to what method we ought to use, but rather we wanted a method that can replicate findings from manual tracing as close as possible. Our ranking of the automated segmentation methods using an overall score based on non-redundant measures was an attempt in that direction. However, there were limitations to the design of overall ranking score. Firstly, the list of measures included in the overall score was by no means exhaustive. This list only included measures we deemed appropriate and necessary for our study objectives and did not include for instance processing times which were quantitively different between methods. Secondly, the list included a measure (RC) which seeks absolute agreement and does not tolerate bias in the data; even when this bias is not proportional. And thirdly, there were influential factors that could not be objectively quantified nor included in the overall score such as ease of implementation and amount and/or type of segmentation output data.
Despite being located remotely, stroke injuries seem to have influenced hippocampal volumes even at this early stage, as indicated by most methods (see Table 5). There are indications, though, that hippocampal volume differences between healthy participants and stroke patients early after stroke onset may be the result of an ongoing vascular burden (Werden et al., 2017). The two effects may prove challenging to untangle.
All methods showed that, at three months, the average hippocampal volume for control was larger than that in stroke; with manual showing a borderline significance (p = 0.049). The ratio V/VManual in Table 5 specifically shows the amount by which each method over or underestimated hippocampal volume for each group. The differentiation between the two groups depended on the way the methods modulated hippocampal volume (over or underestimation in same or opposite directions). For most methods, there was only about 1% difference in the amount of modulation of control volumes as opposed to stroke volumes. Consequently, the p-values for these methods were “practically” similar to that of manual. On the other hand, AdaBoost and FreeSurfer respectively overestimated control volumes by about 2% lower and higher than their overestimates of stroke volumes. Consequently, AdaBoost (p = 0.28) and FreeSurfer (p = 0.007) seem to have respectively underestimated and overestimate the difference between the two study groups.
Finally, the accurate delimitation of the hippocampus from surrounding structures proves to be particularly challenging (Maglietta et al., 2016) and is influenced by many factors, including the choice of MR scanner, image acquisition protocol, post-processing of images, the number, thickness and orientation of slices, segmentation approach, atlases, and the defined anatomical boundaries of the hippocampus itself. Even though the automated segmentation methods were assessed on the same dataset, their performance as compared to manual tracing remains relative to the aforementioned factors, especially to the contrast and resolution of the MRI dataset which were bounded by the acquisition hardware. We also remind the reader that the comparison we made between the automated segmentation methods and manual tracing was based solely on hippocampal volumes. We did not compare the anatomical maps produced by the automated segmentation methods in order to quantify their accuracy against manual hippocampal maps.
5. Conclusion
The Subfields algorithm was found superior for segmenting and quantifying absolute hippocampal volume after objectively ranking eight methods based on a number of non-correlated measures. Moreover, masking of the stroke lesion negatively affected the automatic segmentation of the hippocampus, and thus, is not recommended. Manual tracing, and a number of the automated methods, revealed a significantly compromised hippocampal state for the stroke group compared to the age-matched healthy cohort three months after stroke onset.
The following are the supplementary data related to this article.
Supplementary Tables
Acknowledgments
This work was supported by the National Health and Medical Research Council project grant (APP1020526), the Brain Foundation, Wicking Trust, Collie Trust, and Sidney and Fiona Myer Family Foundation.
The Florey Institute of Neuroscience and Mental Health acknowledges the strong support from the Victorian Government and in particular the funding from the Operational Infrastructure Support Grant. The authors acknowledge the facilities, and the scientific and technical assistance of the National Imaging Facility at the Florey Node.
The authors would like to thank Mr. Oliver Martinez and Mr. Noel Smith from the IDeA laboratory, UC Davis School of Medicine, for their contributions to the ABHS tool used in this manuscript. The authors would also like to thank the Victorian Life Sciences Computation Initiative in the University of Melbourne (http://www.vlsci.org.au/) for support of data supercomputing in SGI Altix XE Cluster.
References
- Aerts H., Fias W., Caeyenberghs K., Marinazzo D. Brain networks under attack: robustness properties and the impact of lesions. Brain. 2016;139:3063–3083. doi: 10.1093/brain/aww194. [DOI] [PubMed] [Google Scholar]
- Aljabar P., Heckemann R.A., Hammers A., Hajnal J.V., Rueckert D. Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. NeuroImage. 2009;46:726–738. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Apfel Brigitte A., Ross Jessica, Hlavin Jennifer, Meyerhoff Dieter J., Metzler Thomas J., Marmar Charles R., Weiner Michael W., Schuff Norbert, Neylan Thomas C. Hippocampal volume differences in Gulf War Veterans with Current Versus Lifetime Posttraumatic stress Disorder Symptoms. Biol. Psychiatry. 2011;69:541–548. doi: 10.1016/j.biopsych.2010.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J., Friston K.J. Unified segmentation. NeuroImage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bland J.M., Altman D.G. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 1999;8:135–160. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- Boccardi M., Bocchetta M., Apostolova L.G., Barnes J., Bartzokis G., Corbetta G., Decarli C., Detoledo-Morrell L., Firbank M., Ganzola R., Gerritsen L., Henneman W., Killiany R.J., Malykhin N., Pasqualetti P., Pruessner J.C., Redolfi A., Robitaille N., Soininen H., Tolomeo D., Wang L., Watson C., Wolf H., Duvernoy H., Duchesne S., Jack C.R., Jr., Frisoni G.B. Delphi definition of the EADC-ADNI Harmonized Protocol for hippocampal segmentation on magnetic resonance. Alzheimers Dement. 2015;11:126–138. doi: 10.1016/j.jalz.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodtmann Amy, Werden Emilio, Pardoe Heath, Li Qi, Jackson Graeme, Donnan Geoffrey, Cowie Tiffany, Bradshaw Jennifer, Darby David, Cumming Toby. Charting cognitive and volumetric trajectories after stroke: protocol for the Cognition and Neocortical volume after Stroke (CANVAS) study. Int. J. Stroke. 2014;9:824–828. doi: 10.1111/ijs.12301. [DOI] [PubMed] [Google Scholar]
- Caviness V.S., Jr., Lange N.T., Makris N., Herbert M.R., Kennedy D.N. MRI-based brain volumetrics: emergence of a developmental brain science. Brain and Development. 1999;21:289–295. doi: 10.1016/s0387-7604(99)00022-4. [DOI] [PubMed] [Google Scholar]
- Colon-Perez L.M., Triplett W., Bohsali A., Corti M., Nguyen P.T., Patten C., Mareci T.H., Price C.C. A majority rule approach for region-of-interest-guided streamline fiber tractography. Brain Imaging Behav. 2016;10:1137–1147. doi: 10.1007/s11682-015-9474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover K.S., van Schijndel R.A., Versteeg A., Leung K.K., Mulder E.R., Jong R.A., Visser P.J., Redolfi A., Revillard J., Grenier B., Manset D., Damangir S., Bosco P., Vrenken H., van Dijk B.W., Frisoni G.B., Barkhof F. Reproducibility of hippocampal atrophy rates measured with manual, FreeSurfer, AdaBoost, FSL/FIRST and the MAPS-HBSI methods in Alzheimer's disease. Psychiatry Res. 2016;252:26–35. doi: 10.1016/j.pscychresns.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Dale A.M., Fischl B., Sereno M.I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. NeuroImage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dunn O.J., Clark V.A. Wiley; New York: 1974. Applied Statistics: Analysis of Variance and Regression. [Google Scholar]
- Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J.C., Pujol S., Bauer C., Jennings D., Fennessy F., Sonka M., Buatti J., Aylward S., Miller J.V., Pieper S., Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fein G., Di Sclafani V., Tanabe J., Cardenas V., Weiner M.W., Jagust W.J., Reed B.R., Norman D., Schuff N., Kusdra L., Greenfield T., Chui H. Hippocampal and cortical atrophy predict dementia in subcortical ischemic vascular disease. Neurology. 2000;55:1626–1635. doi: 10.1212/wnl.55.11.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl Bruce, Dale Anders M. Proceedings of the National Academy of Sciences of the United States of America. Vol. 97. 2000. Measuring the thickness of the human cerebral cortex from magnetic resonance images; pp. 11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Liu A., Dale A.M. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans. Med. Imaging. 2001;20:70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Fischl B., Salat D.H., Busa E., Albert M., Dieterich M., Haselgrove C., van der Kouwe A., Killiany R., Kennedy D., Klaveness S., Montillo A., Makris N., Rosen B., Dale A.M. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B., Salat D.H., van der Kouwe A.J., Makris N., Segonne F., Quinn B.T., Dale A.M. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004;23(Suppl. 1):S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Fischl B., van der Kouwe A., Destrieux C., Halgren E., Segonne F., Salat D.H., Busa E., Seidman L.J., Goldstein J., Kennedy D., Caviness V., Makris N., Rosen B., Dale A.M. Automatically parcellating the human cerebral cortex. Cereb. Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Frisoni G.B., Jack C.R., Jr., Bocchetta M., Bauer C., Frederiksen K.S., Liu Y., Preboske G., Swihart T., Blair M., Cavedo E., Grothe M.J., Lanfredi M., Martinez O., Nishikawa M., Portegies M., Stoub T., Ward C., Apostolova L.G., Ganzola R., Wolf D., Barkhof F., Bartzokis G., Decarli C., Csernansky J.G., Detoledo-Morrell L., Geerlings M.I., Kaye J., Killiany R.J., Lehericy S., Matsuda H., O'Brien J., Silbert P., Scheltens H., Soininen S., Teipel G., Waldemar A., Fellgiebel J., Barnes M., Firbank L., Gerritsen W., Henneman N., Malykhin J.C., Pruessner L., Wang C., Watson H., Wolf M., Deleon J., Pantel C., Ferrari P., Bosco P., Pasqualetti S., Duchesne H. Duvernoy, Boccardi M. The EADC-ADNI Harmonized Protocol for manual hippocampal segmentation on magnetic resonance: evidence of validity. Alzheimers Dement. 2015;11:111–125. doi: 10.1016/j.jalz.2014.05.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodl T., Schaub A., Banac S., Charypar M., Jager M., Kummler P., Bottlender R., Zetzsche T., Born C., Leinsinger G., Reiser M., Moller H.J., Meisenzahl E.M. Reduced hippocampal volume correlates with executive dysfunctioning in major depression. J. Psychiatry Neurosci. 2006;31:316–323. [PMC free article] [PubMed] [Google Scholar]
- Gaser C., Dahnke R. 2016. CAT - a Computational Anatomy Toolbox for the Analysis of Structural MRI Data. (In HBM) [Google Scholar]
- Gemmell E., Bosomworth H., Allan L., Hall R., Khundakar A., Oakley A.E., Deramecourt V., Polvikoski T.M., O'Brien J.T., Kalaria R.N. Hippocampal neuronal atrophy and cognitive function in delayed poststroke and aging-related dementias. Stroke. 2012;43:808–814. doi: 10.1161/STROKEAHA.111.636498. [DOI] [PubMed] [Google Scholar]
- Geuze E., Vermetten E., Bremner J.D. MR-based in vivo hippocampal volumetrics: 2. Findings in neuropsychiatric disorders. Mol. Psychiatry. 2005;10:160–184. doi: 10.1038/sj.mp.4001579. [DOI] [PubMed] [Google Scholar]
- Giavarina D. Understanding Bland Altman analysis. Biochem Med (Zagreb) 2015;25:141–151. doi: 10.11613/BM.2015.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Villa S., Valverde S., Cabezas M., Pareto D., Vilanova J.C., Ramio-Torrenta L., Rovira A., Oliver A., Llado X. Evaluating the effect of multiple sclerosis lesions on automatic brain structure segmentation. Neuroimage Clin. 2017;15:228–238. doi: 10.1016/j.nicl.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousias I.S., Rueckert D., Heckemann R.A., Dyet L.E., Boardman J.P., Edwards A.D., Hammers A. Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. NeuroImage. 2008;40:672–684. doi: 10.1016/j.neuroimage.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Han X., Jovicich J., Salat D., van der Kouwe A., Quinn B., Czanner S., Busa E., Pacheco J., Albert M., Killiany R., Maguire P., Rosas D., Makris N., Dale A., Dickerson B., Fischl B. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. NeuroImage. 2006;32:180–194. doi: 10.1016/j.neuroimage.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Heckemann Rolf A., Hajnal Joseph V., Aljabar Paul, Rueckert Daniel, Hammers Alexander. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Hénon H., Pasquier F., Leys D. Poststroke Dementia. Cerebrovasc. Dis. 2006;22:61–70. doi: 10.1159/000092923. [DOI] [PubMed] [Google Scholar]
- Iglesias J.E., Augustinack J.C., Nguyen K., Player C.M., Player A., Wright M., Roy N., Frosch M.P., McKee A.C., Wald L.L., Fischl B., Van Leemput K. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. NeuroImage. 2015;115:117–137. doi: 10.1016/j.neuroimage.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Smith S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Kendall M.G., Smith B. Babington. 1939. The Problem of $m$ Rankings; pp. 275–287. [Google Scholar]
- Kim H., Chupin M., Colliot O., Bernhardt B.C., Bernasconi N., Bernasconi A. Automatic hippocampal segmentation in temporal lobe epilepsy: impact of developmental abnormalities. NeuroImage. 2012;59:3178–3186. doi: 10.1016/j.neuroimage.2011.11.040. [DOI] [PubMed] [Google Scholar]
- Klein Arno, Mensh Brett, Ghosh Satrajit, Tourville Jason, Hirsch Joy. Mindboggle: Automated brain labeling with multiple atlases. BMC Med. Imaging. 2005;5:7. doi: 10.1186/1471-2342-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliper E., Bashat D.B., Bornstein N.M., Shenhar-Tsarfaty S., Hallevi H., Auriel E., Shopin L., Bloch S., Berliner S., Giladi N., Goldbourt U., Shapira I., Korczyn A.D., Assayag E.B. Cognitive decline after stroke: relation to inflammatory biomarkers and hippocampal volume. Stroke. 2013;44:1433–1435. doi: 10.1161/STROKEAHA.111.000536. [DOI] [PubMed] [Google Scholar]
- Lin L.I. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- Lucarelli R.T., Peshock R.M., McColl R., Hulsey K., Ayers C., Whittemore A.R., King K.S. MR Imaging of Hippocampal Asymmetry at 3T in a Multiethnic, Population-based Sample: results from the Dallas Heart Study. Am. J. Neuroradiol. 2013;34:752–757. doi: 10.3174/ajnr.A3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglietta Rosalia, Amoroso Nicola, Boccardi Marina, Bruno Stefania, Chincarini Andrea, Frisoni Giovanni B., Inglese Paolo, Redolfi Alberto, Tangaro Sabina, Tateo Andrea, Bellotti Roberto. Automated hippocampal segmentation in 3D MRI using random undersampling with boosting algorithm. Pattern. Anal. Applic. 2016;19:579–591. doi: 10.1007/s10044-015-0492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier O., Schroder C., Forkert N.D., Martinetz T., Handels H. Classifiers for Ischemic Stroke Lesion Segmentation: a Comparison Study. PLoS One. 2015;10 doi: 10.1371/journal.pone.0145118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey Frank J. The Kolmogorov-Smirnov test for goodness of fit. J. Am. Stat. Assoc. 1951;46:68–78. [Google Scholar]
- M Meisenzahl, Eva, Doerthe Seifert, Ronald Bottlender, Stefan Teipel, Thomas Zetzsche, Markus Jäger, Nikolaos Koutsouleris, Gisela Schmitt, Johanna Scheuerecker, Bernhard Burgermeister, Harald Hampel, Tobias Rupprecht, Christine Born, Maximilian Reiser, Hans-Jürgen Möller, and Thomas Frodl. 2009. Differences in hippocampal volume between major depression and schizophrenia: A comparative neuroimaging study. [DOI] [PubMed]
- Middlebrooks Erik H., Quisling Ronald G., King Michael A., Carney Paul R., Roper Steven, Colon-Perez Luis M., Mareci Thomas H. The hippocampus: detailed assessment of normative two-dimensional measurements, signal intensity, and subfield conspicuity on routine 3T T2-weighted sequences. Surg. Radiol. Anat. 2017;39:1149–1159. doi: 10.1007/s00276-017-1843-x. [DOI] [PubMed] [Google Scholar]
- Mok Vincent C.T., Lam Bonnie Y.K., Wong Adrian, Ko Ho, Markus Hugh S., Wong Lawrence K.S. Early-onset and delayed-onset poststroke dementia [mdash] revisiting the mechanisms. Nat. Rev. Neurol. 2017;13:148–159. doi: 10.1038/nrneurol.2017.16. [DOI] [PubMed] [Google Scholar]
- Morey R.A., Petty C.M., Xu Y., Hayes J.P., Wagner H.R., 2nd D.V. Lewis, Labar K.S., Styner M., McCarthy G. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. NeuroImage. 2009;45:855–866. doi: 10.1016/j.neuroimage.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A.E., Avedissian C., Madsen S.K., Parikshak N., Hua X., Toga A.W., Jack C.R., Jr., Weiner M.W., Thompson P.M. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease mild cognitive impairment, and elderly controls. NeuroImage. 2008;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A.E., Toga A.W., Thompson P.M. Comparison of AdaBoost and support vector machines for detecting Alzheimer's disease through automated hippocampal segmentation. IEEE Trans. Med. Imaging. 2010;29:30–43. doi: 10.1109/TMI.2009.2021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E.R., de Jong R.A., Knol D.L., van Schijndel R.A., Cover K.S., Visser P.J., Barkhof F., Vrenken H. Hippocampal volume change measurement: quantitative assessment of the reproducibility of expert manual outlining and the automated methods FreeSurfer and FIRST. NeuroImage. 2014;92:169–181. doi: 10.1016/j.neuroimage.2014.01.058. [DOI] [PubMed] [Google Scholar]
- Pantel J., O'Leary D.S., Cretsinger K., Bockholt H.J., Keefe H., Magnotta V.A., Andreasen N.C. A new method for the in vivo volumetric measurement of the human hippocampus with high neuroanatomical accuracy. Hippocampus. 2000;10:752–758. doi: 10.1002/1098-1063(2000)10:6<752::AID-HIPO1012>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Pardoe H.R., Pell G.S., Abbott D.F., Jackson G.D. Hippocampal volume assessment in temporal lobe epilepsy: how good is automated segmentation? Epilepsia. 2009;50:2586–2592. doi: 10.1111/j.1528-1167.2009.02243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude B., Smith S.M., Kennedy D.N., Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse J.C., Giedd J.N., Rapoport J.L. Statistical approach to segmentation of single-channel cerebral MR images. IEEE Trans. Med. Imaging. 1997;16:176–186. doi: 10.1109/42.563663. [DOI] [PubMed] [Google Scholar]
- Rana A.K., Sandu A.L., Robertson K.L., McNeil C.J., Whalley L.J., R. T. Staff, Murray A.D. A comparison of measurement methods of hippocampal atrophy rate for predicting Alzheimer's dementia in the Aberdeen Birth Cohort of 1936. Alzheimers Dement (Amst) 2017;6:31–39. doi: 10.1016/j.dadm.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redolfi Alberto, Bosco Paolo, Manset David, Giovanni B., Frisoni, and Grid consortium neu Brain investigation and brain conceptualization. Funct. Neurol. 2013;28:175–190. [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Rosas H.D., Fischl B. Highly accurate inverse consistent registration: a robust approach. NeuroImage. 2010;53:1181–1196. doi: 10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Schmansky N.J., Rosas H.D., Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. NeuroImage. 2012;61:1402–1418. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltens P., Leys D., Barkhof F., Huglo D., Weinstein H.C., Vermersch P., Kuiper M., Steinling M., Wolters E.C., Valk J. Atrophy of medial temporal lobes on MRI in “probable”: Alzheimer's disease and normal ageing: diagnostic value and neuropsychological correlates. J. Neurol. Neurosurg. Psychiatry. 1992;55:967–972. doi: 10.1136/jnnp.55.10.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleder S., Pawlik M., Wiggermann P., Ott C., Fichtner-Feigl S., Muller-Wille R., Stroszczynski C., Schreyer A.G. Interobserver Agreement in MR enterography for diagnostic assessment in patients with Crohn's Disease. Rofo. 2013;184:992–997. doi: 10.1055/s-0033-1335445. [DOI] [PubMed] [Google Scholar]
- Segonne F., Dale A.M., Busa E., Glessner M., Salat D., Hahn H.K., Fischl B. A hybrid approach to the skull stripping problem in MRI. NeuroImage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Segonne F., Pacheco J., Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans. Med. Imaging. 2007;26:518–529. doi: 10.1109/TMI.2006.887364. [DOI] [PubMed] [Google Scholar]
- Seshadri S., Wolf P.A. Lifetime risk of stroke and dementia: current concepts, and estimates from the Framingham Study. Lancet Neurol. 2007;6:1106–1114. doi: 10.1016/S1474-4422(07)70291-0. [DOI] [PubMed] [Google Scholar]
- Shattuck D.W., Mirza M., Adisetiyo V., Hojatkashani C., Salamon G., Narr K.L., Poldrack R.A., Bilder R.M., Toga A.W. Construction of a 3D probabilistic atlas of human cortical structures. NeuroImage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout P.E., Fleiss J.L. Intraclass correlations: uses in assessing rater reliability. Psychol. Bull. 1979;86:420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Sled J.G., Zijdenbos A.P., Evans A.C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Strong K., Mathers C., Bonita R. Preventing stroke: saving lives around the world. Lancet Neurol. 2007;6:182–187. doi: 10.1016/S1474-4422(07)70031-5. [DOI] [PubMed] [Google Scholar]
- Tohka J., Zijdenbos A., Evans A. Fast and robust parameter estimation for statistical partial volume models in brain MRI. NeuroImage. 2004;23:84–97. doi: 10.1016/j.neuroimage.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Wang H., Suh J.W., Das S.R., Pluta J.B., Craige C., Yushkevich P.A. Multi-Atlas Segmentation with Joint Label Fusion. IEEE Trans. Pattern Anal. Mach. Intell. 2013;35:611–623. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werden E., Cumming T., Li Q., Bird L., Veldsman M., Pardoe H.R., Jackson G., Donnan G.A., Brodtmann A. Structural MRI markers of brain aging early after ischemic stroke. Neurology. 2017;89:116–124. doi: 10.1212/WNL.0000000000004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woon Fu Lye, Sood Shabnam, Hedges Dawson W. Hippocampal volume deficits associated with exposure to psychological trauma and posttraumatic stress disorder in adults: a meta-analysis. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2010;34:1181–1188. doi: 10.1016/j.pnpbp.2010.06.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables