Abstract
The third-generation EGFR inhibitor, osimertinib (AZD9291), selectively and irreversibly inhibits EGFR activating and T790 M mutants while sparing wild-type EGFR. Osimertinib is now an approved drug for non-small cell lung cancer (NSCLC) patients with activating EGFR mutations (first-line) or those who have become resistant to 1st generation EGFR inhibitors through the T790 M mutation (second-line). Unfortunately, all patients eventually relapse and develop resistance to osimertinib. Hence, it is essential to fully understand the biology underlying the development of resistance to osimertinib in order to improve its therapeutic efficacy and overcome resistance. Cellular FLICE-inhibitory protein (c-FLIP) is a truncated form of caspase-8 and functions as a key inhibitor of the extrinsic apoptotic pathway. The current study has demonstrated that osimertinib reduces c-FLIP levels via facilitating its degradation and enhances apoptosis induced by tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) primarily in NSCLC with activating EGFR mutations. Moreover, modulation of c-FLIP expression levels, to some degree, also alters the sensitivities of EGFR mutant NSCLC cells to undergo osimertinib-induced apoptosis, suggesting that c-FLIP suppression is an important event contributing to the antitumor activity of osimertinib against EGFR mutant NSCLC.
Introduction
The discovery of epidermal growth factor receptor (EGFR) activating mutations as an effective therapeutic target represented a paradigm shift in the treatment of NSCLC. Targeting EGFR activating mutations, 90% of which present as an exon 19 deletion (Del19) or exon 21 point mutation (L858R), with first and second generation EGFR tyrosine kinase inhibitors (EGFR-TKIs; e.g., erlotinib, gefitinib and afatinib) and the T790M resistance mutation with third-generation EGFR-TKIs (e.g., AZD9291; osimertinib) has provided significant clinical benefit in patients with NSCLC harboring these mutations, representing a successful example for targeted therapy against lung cancer [1], [2]. A recently completed clinical study showing that AZD9291 also achieved remarkably positive outcomes in the first-line treatment of EGFR mutation-positive advanced NSCLC, with median progression-free survival (PFS) time of 20.5 months [3], resulted in the approval of AZD9291 for the first-line treatment of EGFR mutant NSCLC. However, tumors eventually develop resistance in the clinic, resulting in disease progression; this limits the long-term efficacy of these agents either as a second-line or first-line treatment option [3]. Hence, fully understanding the mechanisms of both action of and resistance to osimertinib is highly desirable and urgently needed in the clinic in order to enhance osimertinib-based therapy and to develop effective strategies to overcome osimertinib resistance.
Cellular FLICE-inhibitory protein (c-FLIP) is a truncated form of caspase-8 that lacks enzymatic activity. It suppresses extrinsic apoptosis by blocking caspase-8 activation through competing with caspase-8 for binding to FADD in the death-inducing signaling complex (DISC) [4]. Hence, c-FLIP acts as a key inhibitor of the extrinsic apoptotic pathway induced by death receptor activation such as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)/death receptor ligation. There are multiple isoforms of c-FLIP, among which only two forms, short form (FLIPS) and long form (FLIPL), have been well characterized at the protein level in human cells [4], [5]. Both FLIPL and FLIPS are unstable proteins regulated by ubiquitination/proteasome-mediated degradation [6], [7], [8]. Elevated levels of c-FLIP have been reported in a number of different cancer types and are often correlated with poor prognosis [5], [9].
Furthermore, c-FLIP has been linked to activation of NF-κB [10], [11], a major survival signaling molecule. It was reported that silencing c-FLIP sensitized EGFR mutant NSCLCs to the first generation EGFR-TKI, erlotinib, whereas overexpression of c-FLIP rescued EGFR-mutant lung cancer cells from erlotinib treatment, presumably through modulation of NF-κB activity [12]. This study suggests that c-FLIP may play a role in regulating the response of EGFR mutant NSCLC cells to erlotinib. However, it is unknown whether erlotinib and other EGFR-TKIs modulate c-FLIP levels in NSCLC cells with activating EGFR mutations.
In this study, we assessed whether osimertinib as well as other EGFR-TKIs modulate c-FLIP levels in EGFR mutant NSCLC cells and determined the underlying mechanisms. Moreover, we studied the effect of osimertinib on TRAIL-induced apoptosis and the impact of c-FLIP modulation on cell response to osimertinib. Our results clearly show that osimertinib decreases c-FLIP levels through enhancing its protein degradation and augments TRAIL-induced apoptosis in some EGFR mutant NSCLC cell lines.
Materials and Methods
Reagents
The sources and preparation of osimertinib, CO1686, erlotinib, MG132, actinomycin D (Act D), and cycloheximide (CHX) were the same as described previously [13], [14]. Soluble recombinant human TRAIL was purchased from PeproTech, Inc. (Rocky Hill, NJ). Afatinib was obtained from the Pharmacy of the Winship Cancer Institute. EGF816 was purchased from Selleckchem (Houston, TX). Pelitinib was ordered from AdooQ Bioscience (Irvine, CA). c-FLIP mouse monoclonal antibody (7F10) was purchased from ENZO Life Sciences, Inc. (Farmingdale, NY). Other antibodies were the same as described in our previous studies [13], [14], [15], [16].
Cell Lines and Cell Culture
All cell lines used in this study and culture conditions were the same as described previously [13], [14]. PC-9 cells expressing ectopic FLIPL (PC-9/FLIPL), FLIPS (PC-P/FLIPS) and empty vector (PC-9/V) were established by infecting PC-9 cells with lentiviruses carrying FLIPL, FLIPS and vector, respectively, followed with puromycin selection as described previously [17]. Pooled cell populations were used.
Cell Survival and Apoptosis Assays
Cells were seeded in 96-well cell culture plates and treated the next day with the given agents. Viable cell numbers were determined using sulforhodamine B (SRB) assay as described previously [18]. Combinational index (CI) for drug interaction (e.g., synergy) was calculated using the CompuSyn software (ComboSyn, Inc.; Paramus, NJ). Apoptosis was evaluated with an annexin V/7-AAD apoptosis detection kit (BD Biosciences; San Jose, CA) according to the manufacturer's instructions. Caspase and PARP cleavage were also detected by Western blot analysis as additional indicators of apoptosis.
Western Blot Analysis
Preparation of whole-cell protein lysates and Western blot analysis were described previously [13], [14].
Quantitative Real-Time PCR (qRT-PCR)
Preparation of total cellular RNA, reverse transcription of cDNA and quantitative PCR reaction were performed as described previously [19] with the same primers for c-FLIP and GAPDH used in our previous studies [7], [19].
Protein Stability Assay
c-FLIP protein stability was determined with CHX chase assay as described previously [7], [20].
Gene Knockdown with Small Interfering RNA (siRNA)
Control and c-FLIP siRNAs and transfection of these siRNAs were the same as described previously [21], [22].
Results
Osimertinib and Other EGFR-TKIs Decrease c-FLIP Levels in NSCLC Cell Lines with Activating EGFR Mutations
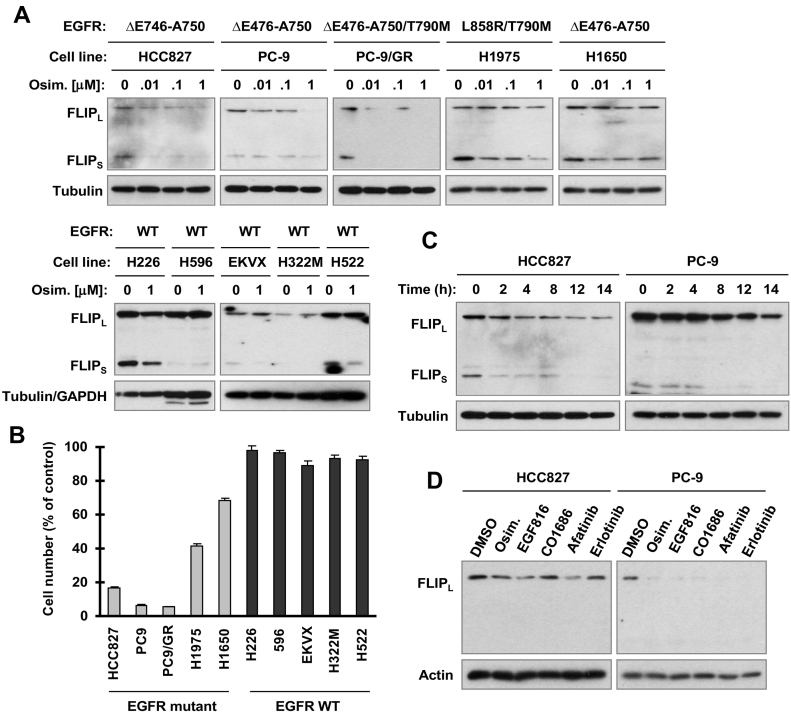
We first determined whether osimertinib alters c-FLIP levels in NSCLC cell lines with and without EGFR mutations. As presented in Figure 1A, osimertinib at concentrations ranging from 0.01 μM to 1 μM decreased the levels of c-FLIP including both FLIPL and FLIPS in several NSCLC cell lines with EGFR mutations including those with T790M mutation (PC-9/GR and H1975). Osimertinib at 1 μM high concentration decreased c-FLIP levels to some degree in H226 cells with wild-type (WT) EGFR but did not do so in other EGFR WT cell lines including H596 EKVX, H322M and H522. These EGFR WT NSCLC cell lines were all insensitive to osimertinib, particularly at concentrations of <0.1 μM (Figure 1B). c-FLIP reduction was observed at 2 h in HCC827 and at 4 h in PC-9 cells post osimertinib treatment (Figure 1C), indicating an early event. Other EGFR-TKIs including erlotinib (1st generation), afatinib (2nd generation), EGF816 and CO1686 (3rd generation) also decreased c-FLIP levels in both PC-9 and HCC827 cells, similar to osimertinib (Figure 1D).
Figure 1.
Osimertinib (A-C) and other EGFR-TKIs (D) decrease c-FLIP levels primarily in the sensitive EGFR mutant NSCLC cell lines (A-D), but not or minimally in the insensitive EGFR WT NSCLC cell lines (A and B). A, C and D, The indicated cell lines were exposed to different concentrations of osimertinib for 8 h (A), to 100 nM osimertinib for different times (C), or to 200 nM different EGFR-TKIs for 12 h (D). The given proteins were analyzed by Western blotting. B, The indicated cell lines were exposed to 10 nM osimertinib for 3 days. Cell numbers were estimated with the SRB assay. Data are means ± SDs of four replicate determinations.
Osimertinib Decreases c-FLIP Levels Through Facilitating Proteasome-Mediated Protein Degradation
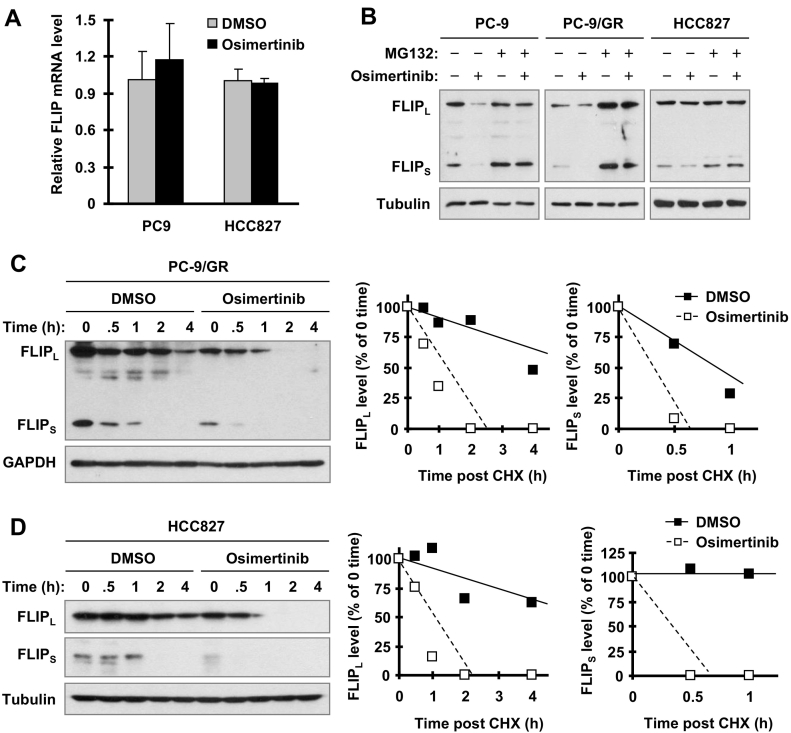
c-FLIP, including both FLIPL and FLIPS, are rapidly turned over proteins regulated by proteasome-mediated degradation [6], [7], [8]. In this study, we failed to detect c-FLIP mRNA reduction in cells treated with osimertinib (Figure 2A). Therefore we further investigated whether osimertinib facilitates c-FLIP degradation, resulting in c-FLIP reduction in EGFR mutant NSCLC cells. In three tested cell lines, PC-9. PC-9/GR and HCC827, osimertinib decreased c-FLIP levels as we demonstrated above in the absence of MG132, a widely used proteasome inhibitor, but failed to do so in the presence of MG132 (Figure 2B), suggesting that osimertinib induces proteasomal degradation of c-FLIP. Moreover, chase assays conducted in both HCC827 and PC-9/GR cell lines showed that both FLIPL and FLIPS were degraded much faster in osimertinib-treated cells than in control cells exposed to DMSO (Figure 2, C and D), further supporting our notion that osimertinib decreases c-FLIP levels through enhancing its proteasomal degradation.
Figure 2.
Osimertinib does not decrease c-FLIP mRNA levels (A), but facilitates proteasomal degradation of c-FLIP (B-D). A, The indicated cell lines were treated with 100 nM osimertinib for 8 h and then harvested for extraction of total cellular RNA and subsequent qRT-PCR. B, The given cell lines were pretreated with 10 μg/mL MG132 for 30 minutes and then co-treated with 100 nM osimertinib for an additional 4 h. The cells were then harvested for Western blotting. C and D, The indicated cell lines were exposed to 100 nM osimertinib for 8 h followed with the addition of 10 μg/ml CHX. The cells were then harvested at the indicated times for Western blotting to detect the indicated proteins. Band intensities were quantified by NIH image J software and FLIPL and FLIPS levels were presented as a percentage of levels at 0 time post CHX treatment.
Osimertinib Cooperates with TRAIL to Augment Induction of Apoptosis in Some EGFR Mutant NSCLC Cell Lines
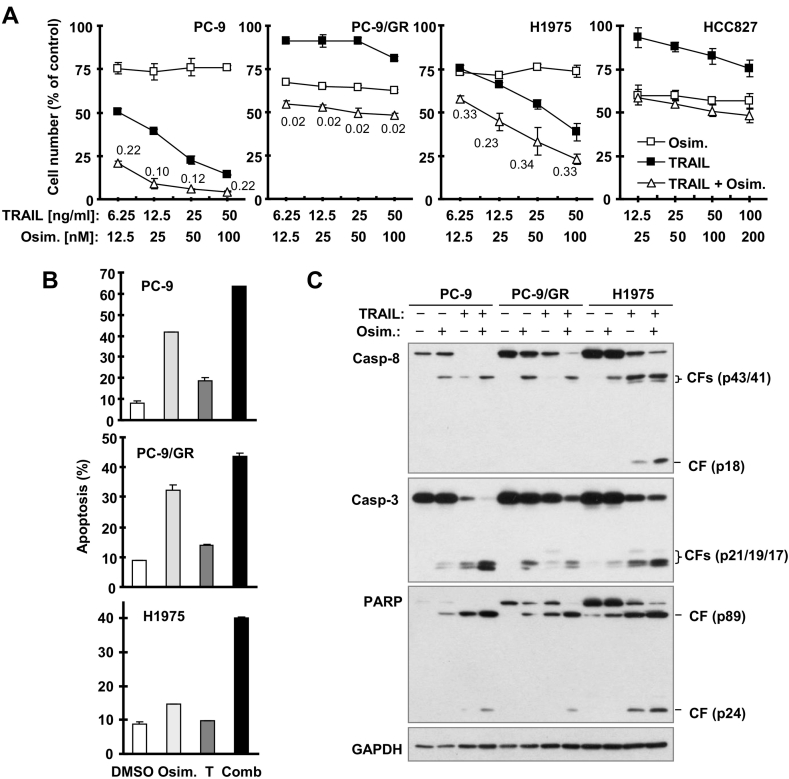
Given that c-FLIP is the sole inhibitor of TRAIL/death receptor-induced apoptosis [4], [23], we assumed that c-FLIP reduction will sensitize cancer cells to TRAIL, a well-known death ligand with cancer therapeutic potential [24], [25]. Hence we examined the effects of osimertinib in combination with TRAIL on cell survival and apoptosis in several NSCLC cell lines with EGFR mutations. The combination of osimertinib and TRAIL was clearly more potent than either agent alone in decreasing cell survival (CI< 1) in PC-9, PC-9/GR and H1975 cells, although this effect was minimal in HCC827 cells (Figure 3A). In agreement, the combination of osimertinib and TRAIL was more effective than each agent alone in inducing apoptosis (Figure 3B) including cleavage of caspase-8, caspase-3 and PARP (Figure 3C) in these cell lines. Hence, osimertinib cooperates with TRAIL to augment the induction of apoptosis in some EGFR mutant NSCLC cell lines.
Figure 3.
Osimertinib cooperates with TRAIL to decrease survival (A) and to augment induction of apoptosis in some EGFR mutant NSCLC cell lines (B and C). A, The indicated NSCLC cell lines seeded in 96-well plates were treated with different concentrations of osimertinib alone, TRAIL alone or their respective combinations as indicated for 24 h. Cell numbers were then estimated with the SRB assay. Data are means ± SDs of four replicate determinations. Combination indices (CIs) for different combinations are labeled in the graphs. B, The indicated cell lines were exposed to 100 nM osimertinib, 25 ng/ml TRAIL (T) or their combination for 24 h and then harvested for annexin V staining followed with flow cytometric analysis to detect apoptosis. Each column is mean ± SD of duplicate determinations. C. The indicated cell lines were exposed to 100 nM osimertinib, 25 ng/ml TRAIL or their combination for 16 h and then harvested for Western blotting to detect protein cleavage. CF, cleaved form.
Enforced Expression of Ectopic c-FLIP Attenuates Apoptosis Induced by Osimertinib, but Abolishes Augmented Induction of Apoptosis by Osimertinib and TRAIL Combination
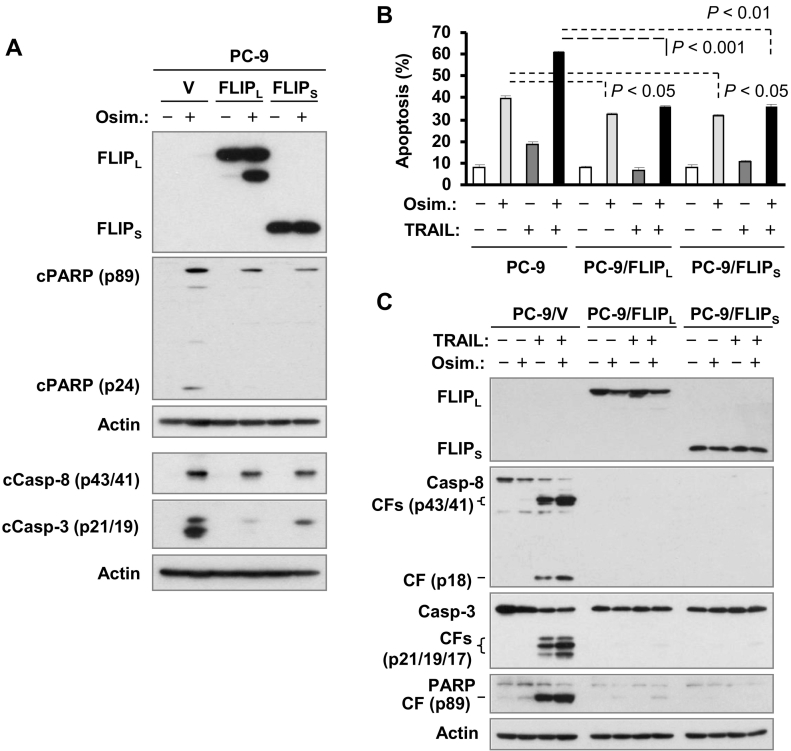
Finally, we examined the effects of ectopic c-FLIP expression on apoptosis induction by osimertinib alone or in combination with TRAIL. As presented in Figure 4A, both ectopic FLIPL and FLIPS were well detected, indicating successful expression of these tested proteins. Upon treatment with osimertinib, reduced amounts of cleaved PARP, caspase-8 and caspase-3 were detected in PC-9/FLIPL and PC-9/FLIPS cells compared with those in PC-9/V control cells (Figure 4A). Accordingly, significantly fewer annexin-V positive cells were detected in PC-9/FLIPL and PC-9/FLIPS cells than in PC-9/V cells exposed to osimertinib alone albeit with limited degrees (Figure 4B). Hence, the enforced expression of c-FLIP in part protects EGFR mutant NSCLC cells from osimertinib-induced apoptosis.
Figure 4.
Enforced expression of c-FLIP in part attenuates osimertinib-induced apoptosis (A and B) and abolishes the cooperative enhancement of apoptosis by osimertinib and TRAIL combination (B and C). A, the indicated cell lines were treated with 100 nM osimertinib for 24 h. B and C, The indicated cell lines were exposed to 100 nM osimertinib alone, 10 ng/ml TRAIL alone or their combination for 24 h (B) or 8 h (C). After the aforementioned treatments, the cells were harvested for Western blot analysis (A and C) or annexin V staining followed with flow cytometric analysis to detect apoptosis (B). Data in B are means ± SDs of duplicate determinations. CF, cleaved form; c, cleaved.
In contrast, the combination of osimertinib and TRAIL augmented the induction of apoptosis in PC-9/V cells, but not in PC-9/FLIPL and PC-9/FLIPS cells as assayed by detection of both annexin-V positive cells and caspase and PARP cleavage (Figure 4, B and C). It is clear that enforced expression of either FLIPL or FLIPS abolishes the augmented induction of apoptosis induced by the osimertinib and TRAIL combination.
Osimertinib Loses its Ability to Decrease c-FLIP Levels and to Enhance TRAIL-Induced Apoptosis in EGFR Mutant Cell Lines with Acquired Resistance to Osimertinib
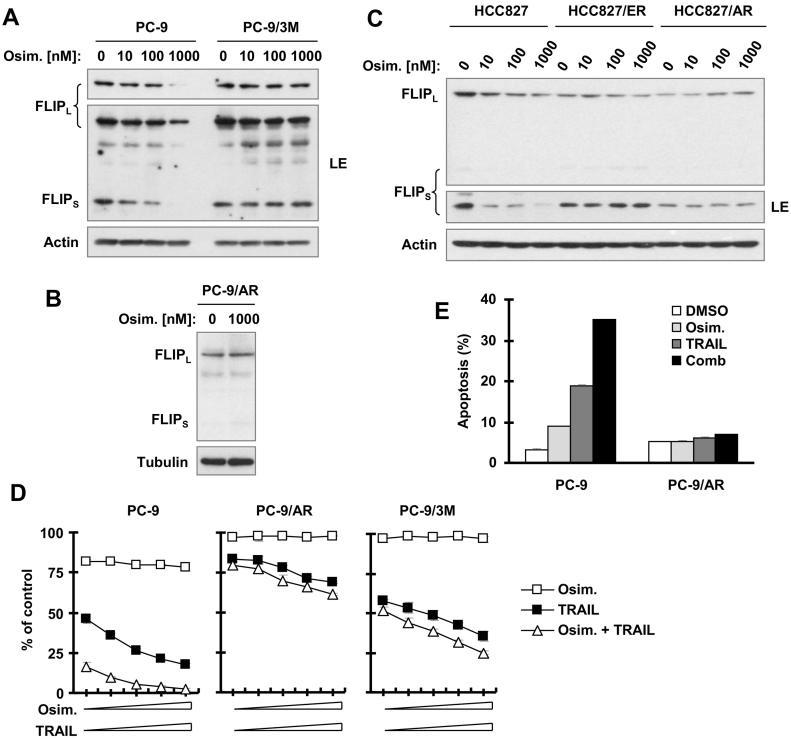
To determine whether c-FLIP modulation plays a role in the development of acquired resistance to osimertinib, we assessed the effects of osimertinib on c-FLIP modulation in several osimertinib-resistant cell lines including PC-9/AR, HCC827/ER, HCC827/AR and PC-9/3M established in our previous studies [13], [14]. As shown in Figure 5, A–C, osimertinib failed to decrease the levels of c-FLIP in these resistant cell lines even at high concentrations up to 1 μM, although it reduced c-FLIP levels even at 10 nM in their corresponding parent cell lines (PC-9 and HCC827), indicating that osimertinib loses its ability to decrease c-FLIP levels in cells resistant to AZD9291.
Figure 5.
Osimertinib loses its ability to decrease c-FLIP levels (A-C) and to augment TRAIL-induced killing (D) including apoptosis (E) in EGFR mutant NSCLC cell lines with acquired resistance to osimertinib. A-C, The given cell lines were exposed to different concentrations of osimertinib as indicated for 8 h and then harvested for Western blotting. D, The indicated cell lines seeded in 96-well plates were treated with different concentrations of osimertinib (6.25, 12.5, 15, 50 and 100 nM) alone, TRAIL (6.25, 12.5, 15, 50 and 100 ng/ml) alone or their respective combination for 24 h. Cell numbers were then estimated with the SRB assay. Data are means ± SDs of four replicate determinations. E, The indicated cell lines were treated with 100 nM osimertinib, 10 ng/ml TRAIL or osimertinib plus TRAIL for 24 h and then harvested for annexin V staining followed with flow cytometric analysis to detect apoptosis. Each column is mean ± SD of duplicate determinations.
PC-9 cells were very sensitive to TRAIL and could be further sensitized to TRAIL-dependent killing or apoptosis by inclusion of osimertinib (Figure 5, D and E). However, both PC-9/AR and PC-9/3 M cell lines became less sensitive to TRAIL and could not be further sensitized even in the presence of osimertinib (Figure 5, D and E). Therefore, cell lines with acquired resistance to osimertinib show some degree of cross-resistance to TRAIL, which cannot be overcome by the osimertinib and TRAIL combination.
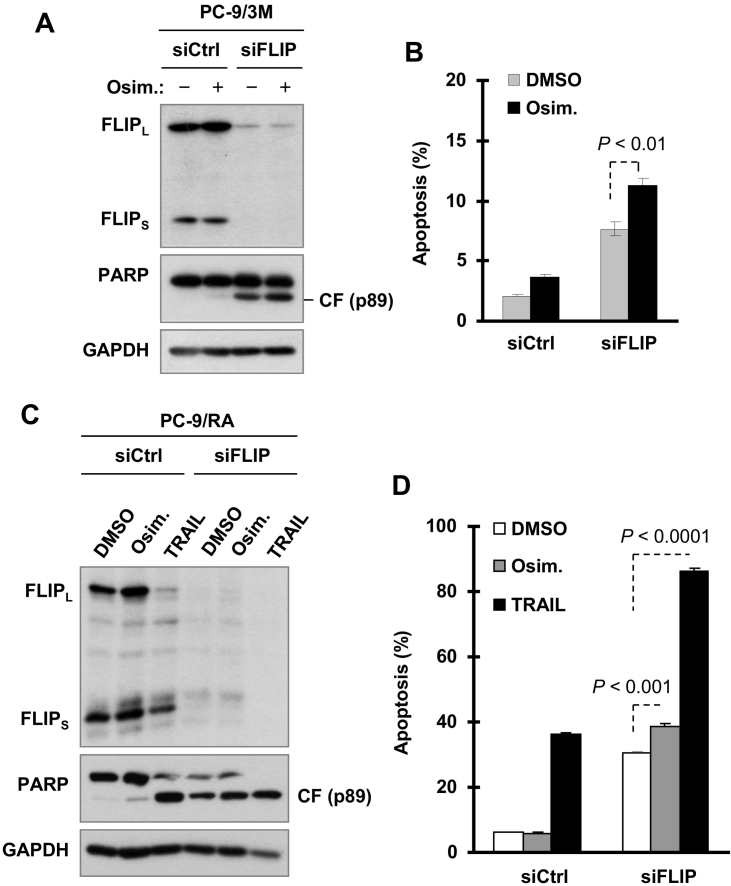
Enforced c-FLIP Suppression Through Gene Knockdown Sensitizes Osimertinib-Resistant Cells to Undergo Apoptosis
To further demonstrate the impact of c-FLIP modulation on osimertinib-induced apoptosis and the development of resistance to osimertinib, we used c-FLIP siRNA to knock down c-FLIP in PC-9/3M cells and then analyzed its impact on osimertinib-induced apoptosis. c-FLIP knockdown per se induced a degree of apoptosis in PC-9/3M cells. Treatment of these cells with osimertinib significantly enhanced apoptosis as evaluated by detection of PARP cleavage (Figure 6A) and by assaying annexin V-positive cells (Figure 6B). In PC-9/AR cells, we generated similar results albeit with a limited degree (Figure 6, C and D). As a control, c-FLIP knockdown in PC-9/AR cells substantially sensitized the cells to undergo apoptosis upon TRAIL treatment, as evidenced by the disappearance of total PARP (Figure 6C) and increase in annexin V-positive cell populations (Figure 6D). Therefore, the enforced suppression of c-FLIP in osimertinib-resistant cells, to some degree, sensitizes the cells to undergo osimertinib-induced apoptosis.
Figure 6.
c-FLIP knockdown in osimertinib-resistant cells significantly sensitizes the cells to undergo osimertinib-induced apoptosis. PC-9/3M (A and B) or PC-9/AR (C and D) cells were transfected with the given siRNAs for 24 h and then treated with DMSO, 500 nM osimertinib or 20 ng/ml TRAIL for an additional 40 h (A and B) or 48 h (C and D). c-FLIP knockdown efficiency and PARP cleavage were evaluated with Western blotting (A and C) and apoptosis was detected with annexin V/flow cytometry (B and D). Each column represents the mean ± SD of triplicate determinations.
Discussion
Although the modulation of c-FLIP expression through genetic manipulation (e.g., overexpression and gene knockdown) alters the responses of EGFR mutant NSCLC cells to erlotinib treatment [12], there are no reports documenting the downregulation of c-FLIP by EGFR-TKIs. The current study has clearly shown that osimertinib and other EGFR-TKIs decrease the levels of c-FLIP (both FLIPL and FLIPS forms) primarily in the sensitive EGFR mutant NSCLC cell lines (Figure 1). This finding hence represents the first evidence that EGFR-TKIs modulate (i.e., decrease) the levels of endogenous c-FLIP in NSCLC cell lines with activating EGFR mutations, suggesting the possible involvement of c-FLIP modulation in mediating therapeutic efficacy of osimertinib as well as other EGFR-TKIs against EGFR mutant NSCLCs. This notion is supported by the following findings: 1) c-FLIP levels were substantially reduced by osimertinib primarily in osimertinib-sensitive EGFR mutant NSCLC cell lines, but not or minimally in osimertinib-insensitive EGFR WT NSCLC cell lines (Figure 1, A and B); 2) Enforced overexpression of c-FLIP in part protected the sensitive EGFR mutant NSCLC cells from undergoing apoptosis induced by osimertinib (Figure 4); 3) Several osimertinib-resistant NSCLC cell lines lost response to osimertinib-induced c-FLIP downregulation (Figure 5); and 4) Enforced c-FLIP downregulation by siRNA-mediated gene knockdown significantly enhanced the sensitivity of osimertinib-resistant cells to osimertinib-induced apoptosis, albeit to limited degrees (Figure 6).
The development of acquired resistance to EGFR-TKIs, including osimertinib, remains the major obstacle in the clinic that prevents patients from experiencing long-term remission, resulting in eventual treatment failure [26], [27]. Therefore, strategies to effectively overcome acquired resistance are urgently needed in the clinic. Our current findings together with the previous study suggest that suppression of c-FLIP may be an effective strategy to enhance the therapeutic efficacy of osimertinib against NSCLCs with acquired resistance to this agent. Further study in this direction is warranted.
c-FLIP, including both FLIPL and FLIPS, are known to be unstable proteins subject to regulation through ubiquitination/proteasome-mediated protein degradation [6], [7], [8]. Many agents decrease c-FLIP levels through enhancing protein degradation [6], [7], [8]. In this study, osimertinib facilitated c-FLIP degradation rates as demonstrated by the CHX chase assay (Figure 2, C and D). Moreover, the presence of the proteasome inhibitor, MG132, rescued c-FLIP reduction induced by osimertinib (Figure 2B). In agreement, osimertinib did not decrease c-FLIP mRNA levels (Figure 2A). Thus, osimertinib decreases c-FLIP levels in EGFR mutant NSCLC cells through enhancing c-FLIP proteasomal degradation.
TRAIL (e.g., recombinant TRAIL) possesses cancer therapeutic potential based on its characteristic as a potent apoptosis-inducer in sensitive cancer cells [24], [28]. Importantly, endogenous TRAIL, which is primarily secreted or produced by immune cells such as cytotoxic T cells, plays a key role in eliminating cancer cells by our immune defense mechanism through TRAIL/death receptor-induced apoptosis [29], [30]. As a critical inhibitor of this extrinsic apoptosis pathway, enhanced c-FLIP downregulation or degradation is often associated with enhancement of TRAIL-induced apoptosis [5]. In this study, osimertinib cooperated with TRAIL to augment the induction of apoptosis in several EGFR mutant NSCLC cell lines (Figure 3). Once these cells became resistant to osimertinib, they exhibited some degree of cross resistance to TRAIL (Figure 5). Moreover, osimertinib failed to decrease c-FLIP levels and to augment TRAIL-induced apoptosis in these resistant cell lines (Figure 5). Therefore it is plausible to speculate that osimertinib, beyond its direct effect on inducing apoptosis of EGFR mutant NSCLC cells as we recently documented [13], may exert its anticancer activity via an indirect effect on activating the endogenous immune defense mechanism that eliminates cancer cells through TRAIL/death receptor-mediated apoptosis. Accordingly, escape from endogenous immune clearance due to resistance to TRAIL-induced apoptosis may be a critical mechanism accounting for the development of acquired resistance to osimertinib. Future research in this regard is also warranted.
Acknowledgement
We are grateful to Drs. P. A. Jänne, R. Lotan and A. N. Hata for providing some cell lines. We also thank Dr. A. Hammond in our department for editing the manuscript.
SSR and SYS are Georgia Research Alliance Distinguished Cancer Scientists.
This work was supported by the NIH/NCI R01 CA223220 (to SYS), Winship lung cancer pilot award (to SYS) and National Natural Science Foundation of China (No. 81802291; to PS).
References
- 1.Govindan R. Overcoming resistance to targeted therapy for lung cancer. N Engl J Med. 2015;372:1760–1761. doi: 10.1056/NEJMe1500181. [DOI] [PubMed] [Google Scholar]
- 2.Barnes TA, O'Kane GM, Vincent MD, Leighl NB. Third-Generation Tyrosine Kinase Inhibitors Targeting Epidermal Growth Factor Receptor Mutations in Non-Small Cell Lung Cancer. Front Oncol. 2017;7:113. doi: 10.3389/fonc.2017.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramalingam SS, Yang JC, Lee CK, Kurata T, Kim DW, John T, Nogami N, Ohe Y, Mann H, Rukazenkov Y. Osimertinib as First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2018;36:841–849. doi: 10.1200/JCO.2017.74.7576. [JCO2017747576] [DOI] [PubMed] [Google Scholar]
- 4.Kataoka T. The caspase-8 modulator c-FLIP. Crit Rev Immunol. 2005;25:31–58. doi: 10.1615/critrevimmunol.v25.i1.30. [DOI] [PubMed] [Google Scholar]
- 5.Shirley S, Micheau O. Targeting c-FLIP in cancer. Cancer Lett. 2013;332:141–150. doi: 10.1016/j.canlet.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y, Suh N, Sporn M, Reed JC. An inducible pathway for degradation of FLIP protein sensitizes tumor cells to TRAIL-induced apoptosis. J Biol Chem. 2002;277:22320–22329. doi: 10.1074/jbc.M202458200. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Cao W, Yue P, Hao C, Khuri FR, Sun SY. Celecoxib promotes c-FLIP degradation through Akt-independent inhibition of GSK3. Cancer Res. 2011;71:6270–6281. doi: 10.1158/0008-5472.CAN-11-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poukkula M, Kaunisto A, Hietakangas V, Denessiouk K, Katajamaki T, Johnson MS, Sistonen L, Eriksson JE. Rapid turnover of c-FLIPshort is determined by its unique C-terminal tail. J Biol Chem. 2005;280:27345–27,355. doi: 10.1074/jbc.M504019200. [DOI] [PubMed] [Google Scholar]
- 9.Bagnoli M, Canevari S, Mezzanzanica D. Cellular FLICE-inhibitory protein (c-FLIP) signaling: a key regulator of receptor-mediated apoptosis in physiologic context and in cancer. Int J Biochem Cell Biol. 2010;42:210–213. doi: 10.1016/j.biocel.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Kataoka T, Tschopp J. N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol Cell Biol. 2004;24:2627–2636. doi: 10.1128/MCB.24.7.2627-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golks A, Brenner D, Krammer PH, Lavrik IN. The c-FLIP-NH2 terminus (p22-FLIP) induces NF-kappaB activation. J Exp Med. 2006;203:1295–1305. doi: 10.1084/jem.20051556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, Moonsamy P, Dahlman K, Miller VA, Costa C. FAS and NF-kappaB signaling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi P, Oh YT, Deng L, Zhang G, Qian G, Zhang S, Ren H, Wu G, Legendre B, Jr., Anderson E. Overcoming Acquired Resistance to AZD9291, A Third-Generation EGFR Inhibitor, through Modulation of MEK/ERK-Dependent Bim and Mcl-1 Degradation. Clin Cancer Res. 2017;23:6567–6579. doi: 10.1158/1078-0432.CCR-17-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi P, Oh YT, Zhang G, Yao W, Yue P, Li Y, Kanteti R, Riehm J, Salgia R, Owonikoko TK. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett. 2016;380:494–504. doi: 10.1016/j.canlet.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 15.Yao W, Oh YT, Deng J, Yue P, Deng L, Huang H, Zhou W, Sun SY. Expression of death receptor 4 is positively regulated by MEK/ERK/AP-1 signaling and suppressed upon MEK inhibition. J Biol Chem. 2016;291:21694–21702. doi: 10.1074/jbc.M116.738302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh YT, Liu X, Yue P, Kang S, Chen J, Taunton J, Khuri FR, Sun SY. ERK/ribosomal S6 kinase (RSK) signaling positively regulates death receptor 5 expression through co-activation of CHOP and Elk1. J Biol Chem. 2010;285:41310–41,319. doi: 10.1074/jbc.M110.153775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Yue P, Schonthal AH, Khuri FR, Sun SY. Cellular FLICE-inhibitory protein down-regulation contributes to celecoxib-induced apoptosis in human lung cancer cells. Cancer Res. 2006;66:11115–11119. doi: 10.1158/0008-5472.CAN-06-2471. [DOI] [PubMed] [Google Scholar]
- 18.Sun SY, Yue P, Dawson MI, Shroot B, Michel S, Lamph WW, Heyman RA, Teng M, Chandraratna RA, Shudo K. Differential effects of synthetic nuclear retinoid receptor-selective retinoids on the growth of human non-small cell lung carcinoma cells. Cancer Res. 1997;57:4931–4939. [PubMed] [Google Scholar]
- 19.Qian G, Yao W, Zhang S, Bajpai R, Hall WD, Shanmugam M, Lonial S, Sun SY. Co-inhibition of BET and proteasome enhances ER stress and Bim-dependent apoptosis with augmented cancer therapeutic efficacy. Cancer Lett. 2018;435:44–54. doi: 10.1016/j.canlet.2018.07.033. [DOI] [PubMed] [Google Scholar]
- 20.Zhao L, Yue P, Khuri FR, Sun SY. mTOR complex 2 is involved in regulation of Cbl-dependent c-FLIP degradation and sensitivity of TRAIL-induced apoptosis. Cancer Res. 2013;73:1946–1957. doi: 10.1158/0008-5472.CAN-12-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Yue P, Chen S, Hu L, Lonial S, Khuri FR, Sun SY. The proteasome inhibitor PS-341 (bortezomib) up-regulates DR5 expression leading to induction of apoptosis and enhancement of TRAIL-induced apoptosis despite up-regulation of c-FLIP and survivin expression in human NSCLC cells. Cancer Res. 2007;67:4981–4988. doi: 10.1158/0008-5472.CAN-06-4274. [DOI] [PubMed] [Google Scholar]
- 22.Lin YD, Chen S, Yue P, Zou W, Benbrook DM, Liu S, Le TC, Berlin KD, Khuri FR, Sun SY. CAAT/enhancer binding protein homologous protein-dependent death receptor 5 induction is a major component of SHetA2-induced apoptosis in lung cancer cells. Cancer Res. 2008;68:5335–5344. doi: 10.1158/0008-5472.CAN-07-6209. [DOI] [PubMed] [Google Scholar]
- 23.Safa AR. Roles of c-FLIP in Apoptosis, Necroptosis, and Autophagy. J Carcinog Mutagen. 2013;Suppl. 6 doi: 10.4172/2157-2518.S6-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemke J, von Karstedt S, Zinngrebe J, Walczak H. Getting TRAIL back on track for cancer therapy. Cell Death Differ. 2014;21:1350–1364. doi: 10.1038/cdd.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim B, Allen JE, Prabhu VV, Talekar MK, Finnberg NK, El-Deiry WS. Targeting TRAIL in the treatment of cancer: new developments. Expert Opin Ther Targets. 2015;19:1171–1185. doi: 10.1517/14728222.2015.1049838. [DOI] [PubMed] [Google Scholar]
- 26.Juchum M, Gunther M, Laufer SA. Fighting cancer drug resistance: Opportunities and challenges for mutation-specific EGFR inhibitors, Drug resistance updates: reviews and commentaries in antimicrobial and anticancer. Chemotherapy. 2015;20:10–28. doi: 10.1016/j.drup.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Skoulidis F, Papadimitrakopoulou VA. Targeting the Gatekeeper: Osimertinib in EGFR T790 M Mutation-Positive Non-Small Cell Lung Cancer. Clin Cancer Res. 2017;23:618–622. doi: 10.1158/1078-0432.CCR-15-2815. [DOI] [PubMed] [Google Scholar]
- 28.Amarante-Mendes GP, Griffith TS. Therapeutic applications of TRAIL receptor agonists in cancer and beyond. Pharmacol Ther. 2015;155:117–131. doi: 10.1016/j.pharmthera.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth MJ, Takeda K, Hayakawa Y, Peschon JJ, van den Brink MR, Yagita H. Nature's TRAIL—on a path to cancer immunotherapy. Immunity. 2003;18:1–6. doi: 10.1016/s1074-7613(02)00502-2. [DOI] [PubMed] [Google Scholar]
- 30.Falschlehner C, Schaefer U, Walczak H. Following TRAIL's path in the immune system. Immunology. 2009;127:145–154. doi: 10.1111/j.1365-2567.2009.03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]