Abstract
X-ray structure of methyl-CpG binding domain (MBD) of MeCP2, an intrinsically disordered protein (IDP) involved in Rett syndrome, offers a rational basis for defining the spatial distribution for most of the sites where mutations responsible of Rett syndrome, RTT, occur. We have ascribed pathogenicity for mutations of amino acids bearing positively charged side chains, all located at the protein-DNA interface, as positive charge removal cause reduction of the MeCP2-DNA adduct lifetime. Pathogenicity of the frequent proline replacements, outside the DNA contact moiety of MBD, can be attributed to the role of this amino acid for maintaining both unfolded states for unbound MeCP2 and, at the same time, to favor some higher conformational order for stabilizing structural determinants required by protein activity. These hypotheses can be extended to transcription repressor domain, TRD, the other MeCP2-DNA interaction site and, in general, to all the IDP that interact with nucleic acids.
Keywords: DNA binding domains, MeCP2, Mutation distribution, Protein structure, Rett syndrome
Introduction
In girls under the age of 12, Rett syndrome, RTT, occurrence is estimated at 1/9000. This pathology has been associated to mutations in the X-linked MECP2 gene,1 as patients bearing this neurological disorder always exhibit modified sequences of methyl-CpG-binding protein 2, MeCP2. Along the 486 amino acid MeCP2 sequence, there are unambiguous signals confirming the role of the protein in the interaction with DNA. Indeed, AT-hook domains, critical for altering chromatin structure, have been delineated in MeCP2 transcription repressor domains, TRD,2, 3, 4 together with a methyl-CpG-binding domain, MBD.
Trying to correlate structural aspects of amino acid changes arising from missense MecP2 mutations with RTT is not straightforward, as the protein belongs to the class of intrinsically disordered proteins, IDPs,5 see Fig. 1. In this respect, it is interesting to note how many other IDPs have been found involved in neurodegenerative diseases.6
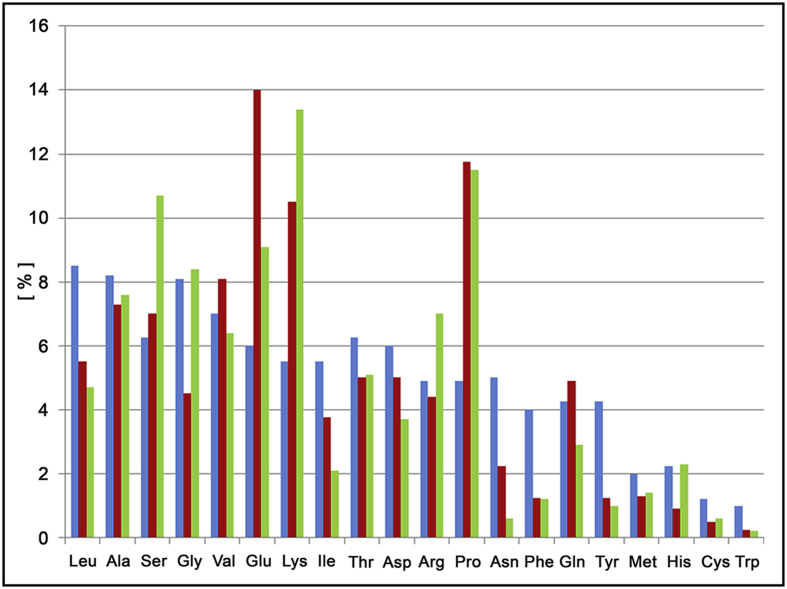
Figure 1.
Percent average amino acid occurrences in globular proteins, intrinsically disordered proteins adapted from ref (X) and in MeCP2 respectively coloured in blue, red and green (adapted from Uversky 2009).
Methods and materials
MECP2 Variation Database (RettBASE),7 a curated database of all the reported MaCP2 mutations, has been used as a reference for our bioinformatic survey. The X ray resolved structure of MaCP2 fragment bound to a 20 bp DNA has been retrieved from the Protein Data Bank with the PDB ID code 5BT2.8 The structural features of the MeCP2-DNA adduct has been analyzed with Open source PyMOL v. 1.7.1.0.9
Results and discussion
Among the 4738 records reported in the RettBASE site7 for all the observed MeCP2 variations, 3377 are related to missense mutations that are unevenly distributed along the protein sequence, as they are found mainly in 78–162 MBD, 207–255 TRD1 and 271–310 TRD2 fragments of MeCP2 sequence, see Fig. 2. However, as only a small fraction of the latter sequence changes has been unequivocally associated to RTT, we have confined our analysis to the 19 MeCP2 missense mutation sites verified by accurate clinical investigations and considered by UniProtKB10 database (https://www.uniprot.org/uniprot/P51608). Thus, from the latter database 11, 2 and 3 mutation sites are identified respectively in 78–162 MBD, 207–255 TRD1 and 271–310 TRD2 fragments of MeCP2 sequence, suggesting that DNA interacting moieties of the protein are primarily relevant for inducing RTT, see Table 1. It is interesting to note that, among the 19 UniprotKb defined mutation sites, Pro and Arg substitutions occur, by far, as the most frequent ones, as the latter amino acids are replaced five times each. This finding is consistent with the critical role of Pro for maintaining unfolded states in IDP5 and with the high relevance of Arg in establishing suitable lifetimes for protein-DNA adducts.11
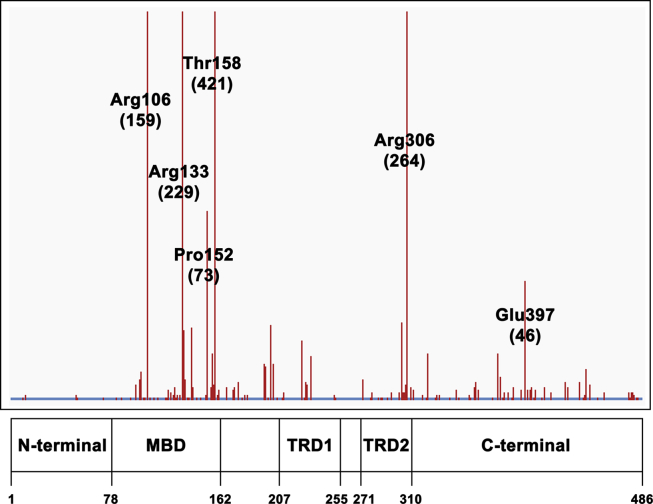
Figure 2.
Distribution of missense mutation sites along MeCP2 sequence (adapted from RettDB). Labels refer to the most frequent mutations; the corresponding occurrences are shown in parenthesis.
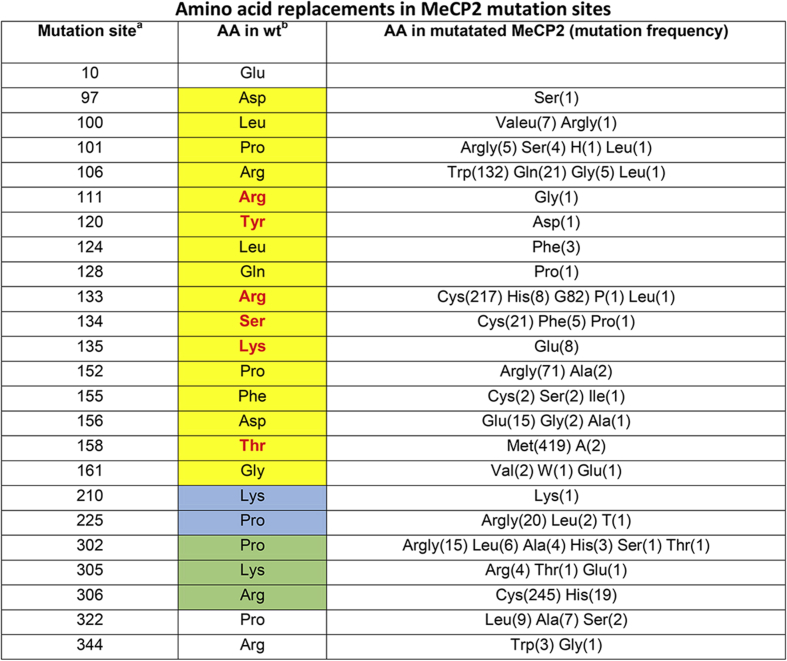
Table 1.
a) Mutation sites according to UniProtKB. b) Amino acids located in wild-type MeCP2. Yellow, blue and green boxes refer to MBD, TRD1 and TRD2 respectively. Amino acids highlighted in bold red characters are the ones that are present in the protein-DNA interface. c) Amino acid replacements with their frequency in parenthesis as retrieved from RettBASE.
Recently, a MBD-DNA adduct has been structurally characterized by X-ray crystallography12 and the corresponding structure, deposited in the Protein Data Bank8 with the ID code 5BT2, yields a powerful tool for deciphering the roles of the observed mutations in relation to RTT appearance. Thus, by using PISA database13 amino acids that are located at MeCP2-DNA interface can be identified. The presence of two Arg and one Lys at the MeCP2-DNA interface confirms how critical is the removal of positively charged side chains in this protein moiety, see Table 1. Table 1 shows also that replacements of Ser and Thr with amino acids whose side chains are not bearing hydroxyl groups, interfere with interface hydration. Interfacial water molecules, indeed, determining the dynamics of protein-DNA approach,11 strongly modulate the latter interaction.
Fig. 3 summarizes the structural distribution of pathogenic mutations in MBD, underlining the role of positively charged amino acids, particularly of Arg, in the binding to DNA. Pro, the other very frequently mutated amino acid in RTT, is outside the MBD-DNA contact region to the drive the local folding that is due to trigger the physiological intermolecular interaction. These structural features can be used for interpreting the role of Arg, Lys and Proin TRD1 and TRD2 mutation sites, see Table 1. Thus, Arg210, Lys305 and Arg306 replacements most likely affect MeCP2 interaction with DNA. Similarly, Pro225 and Pro302 mutations could inhibit the transient folding that is required for MeCP2 function.
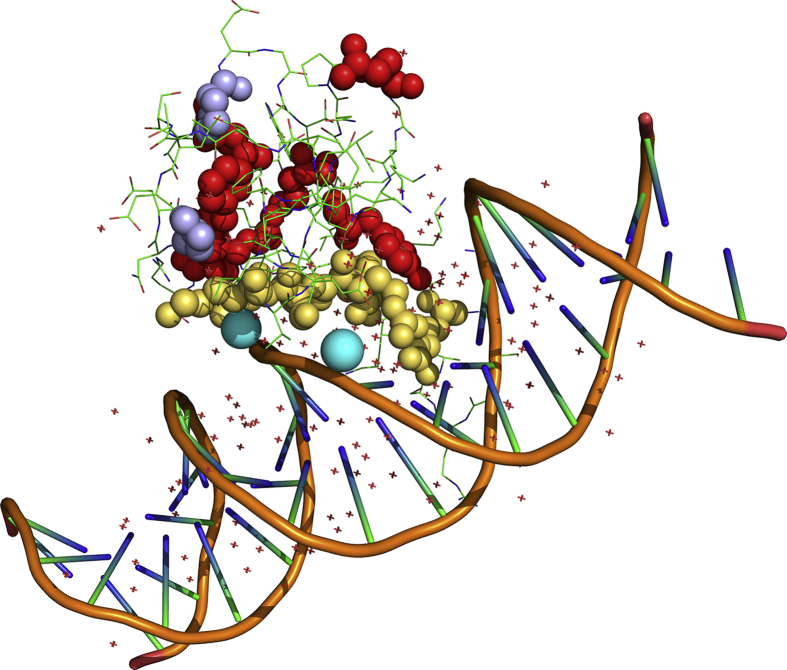
Figure 3.
Pymol representation of MBD structure (PDB ID code 5BT2). Amino acids that are involved in pathogenic mutations are shown with spheres; yellow atoms refer to interfacial amino acids; prolines are highlighted in violet. Water molecules bridging S134 and T158 to DNA chains are shown in cyan.
For alleviating RTT effects, among all the so far proposed therapeutic strategies,14 only few efforts exhibit some effectiveness.15 Possibly, CRISPR-Cas9 will provide in a near future more reliable answers to RTT damages. Thus, in future genome-editing procedures16 our present observations suggest that in all neuropathologies involving DNA binding IDPs, particular care to protect Arg, Lys and Pro from mutations should be taken.
Statement of author contribution
OS: analyzed structural data.
SG: directed the investigation.
NR: preliminary survey of Rett genetics.
VC: data bank selection and analysis.
NN: structural discussion and manuscript correction.
AS: wrote the manuscript.
FP: protein structures analysis.
Conflicts of interest
None declared.
Acknowledgements
Thanks are due to Adele Caciolli for her insightful contribution to this work.
Footnotes
In memory of Dr. Duccio Calamandrei
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Ottavia Spiga, Email: ottavia.spiga@unisi.it.
Simone Gardini, Email: simone.gardini@genomeup.com.
Nicole Rossi, Email: nicole.rossi@student.unisi.it.
Vittoria Cicaloni, Email: cicaloni@student.unisi.it.
Francesco Pettini, Email: pettini2@student.unisi.it.
Neri Niccolai, Email: neri.niccolai@unisi.it.
Annalisa Santucci, Email: annalisa.santucci@unisi.it.
References
- 1.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 2.Baker S.A., Chen L., Wilkins A.D., Yu P., Lichtarge O., Zoghbi H.Y. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013;152(5):984–996. doi: 10.1016/j.cell.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martínez de Paz A., Ausió J. MeCP2. A modulator of neuronal chromatin organization involved in Rett syndrome. Adv Exp Med Biol. 2017;978:3–21. doi: 10.1007/978-3-319-53889-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Mushtaq A.U., Lee Y., Hwang E. Biophysical characterization of the basic cluster in the transcription repression domain of human MeCP2 with AT-rich DNA. Biochem Biophys Res Commun. 2018;495(1):145–150. doi: 10.1016/j.bbrc.2017.10.169. [DOI] [PubMed] [Google Scholar]
- 5.Theillet F.X., Kalmar L., Tompa P. The alphabet of intrinsic disorder: I. Act like a Pro: on the abundance and roles of proline residues in intrinsically disordered proteins. Intrinsically Disord Proteins. 2013;1(1) doi: 10.4161/idp.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uversky V.N. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci (Landmark Ed). 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 7.Krishnaraj R., Ho G., Christodoulou J. RettBASE: Rett syndrome database update. Hum Mutat. 2017;38(8):922–931. doi: 10.1002/humu.23263. [DOI] [PubMed] [Google Scholar]
- 8.Berman H.M., Westbrook J., Feng Z. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLano W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr. 2002;40:82–92. [Google Scholar]
- 10.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardini S., Furini S., Santucci A., Niccolai N. A structural bioinformatics investigation on protein-DNA complexes delineates their modes of interaction. Mol Biosyst. 2017;13(5):1010–1017. doi: 10.1039/c7mb00071e. [DOI] [PubMed] [Google Scholar]
- 12.Chia J.Y., Tan W.S., Ng C.L., Hu N.J., Foo H.L., Ho K.L. A/T run geometry of B-form DNA is independent of bound methyl-CpG binding domain, cytosine methylation and flanking sequence. Sci Rep. 2016;6:31210. doi: 10.1038/srep31210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Chapleau C.A., Lane J., Larimore J., Li W., Pozzo-Miller L., Percy A.K. Recent progress in Rett syndrome and MeCP2 dysfunction: assessment of potential treatment options. Future Neurol. 2013;8(1) doi: 10.2217/fnl.12.79. 10.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khwaja O.S., Ho E., Barnes K.V. Safety, pharmacokinetics, and preliminary assessment of efficacy of mecasermin (recombinant human IGF-1) for the treatment of Rett syndrome. Proc Natl Acad Sci U S A. 2014;111(12):4596–4601. doi: 10.1073/pnas.1311141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ormond K.E., Mortlock D.P., Scholes D.T. Human germline genome editing. Am J Hum Genet. 2017;101(2):167–176. doi: 10.1016/j.ajhg.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]