Abstract
SKA2 (spindle and KT associated 2), also referred to as FAM33A (family with sequence similarity 33, member A), is a recently identified gene involved in cell cycle regulation, and growing evidence is implicating its roles in tumorigenesis and psychiatric disorders. It has been demonstrated that SKA2, along with its coworkers SKA1 and SKA3, constitutes the SKA complex which plays a critical role in the maintenance of the metaphase plate and/or spindle checkpoint silencing during mitosis. SKA2 is over-expressed both in cancer cell lines and clinical samples including small cell lung cancer and breast cancer, whereas downregulation of SKA2 is associated with depression and suicidal ideation. The expression of SKA2 is regulated by transcription factors including NF-κΒ and CREB, miRNAs as well as DNA methylation. In this review, we provide an overview of studies that reveal SKA2 gene and protein characteristics as well as physiological function, with a special focus on its transcription regulatory mechanisms, and also provide a summary regarding the translational opportunity of the SKA2 gene as a clinical biomarker for cancers and psychiatric disorders.
Keywords: SKA2, FAM33A, Cell cycle, Tumor, Gene expression regulation, DNA methylation
Introduction
For decades, biomedical research has tremendously improved the understanding of complicated diseases, such as cancer. Aberrant proliferation and cell cycle signals that are triggered by genetic disorder is one of the major cancer hallmarks.1 Many of the well-characterized cell cycle regulators were found to be dysregulated in cancer cells and participated in tumor progression,2 such as p53, E2F, Chk1, and Rb.3, 4, 5, 6, 7 Numerous compounds designed to target mitosis in malignant cells have been developed. Inhibitors that target CDK4/6 have been approved for breast cancer treatment.8, 9 It is important to further identify and understand genes that are required for cell cycle regulation. SKA2 (spindle and kinetochore associated complex subunit 2), also referred to as FAM33A (family with sequence similarity 33, member A), is a novel cell cycle related gene, with emerging evidence implicating its value as a biomarker and therapeutic target in both cancers and psychiatric disorders. This review provides an overview of SKA2 with respect to its gene transcription, physiological functions, and implications in cancers and psychiatric disorders.
Discovery of and characterization of SKA2 gene and protein
Identification of SKA2
SKA2 protein is a part of the spindle and kinetochore associated (SKA) complex, a heterotrimeric complex formed from SKA1, SKA2, and SKA3. SKA1 was initially discovered by Hanisch A et al as a spindle and kinetochore (KT)-associated protein. The same group then discovered SKA2 as a SKA1 binding protein.10 Not long after, SKA3 was also reported by multiple laboratories.11, 12, 13 It is now known that the heterotrimeric SKA complex accumulates on spindle microtubules and at kinetochores after nuclear envelope breakdown, and is most enriched on kinetochores at metaphase, with its primarily function as a checkpoint to ensure the timely onset of anaphase.10, 13
Gene and protein structure of SKA2
Human SKA2 gene is located in chromosome 17q22, with a full length of 45,493 bp (NC_000017.10) including four exons and three introns (Fig. 1). Two RNA splicing variants were reported (NM_182620.3 and NM_001100595.1). The longer variant NM_182620.3 is approximately 2990 bp which contains all four extrons; the open reading frame (ORF) is between 277 and 642bp, whereas the shorter NM_001100595.1 lacks the third extron and its ORF is between 357 and 584bp. Interestingly, the intron of this gene also harbors two functional miRNAs (miR-301 and miR-454).14
Figure 1.
Structure of SKA2 gene. Two splicing variants of SKA2 mRNA and isoforms of protein are shown, introns and exons are represented by straight lines and rectangles, respectively. Dash lines indicates common splicing whereas solid like represents different splicing between two variants.
Two mRNA splicing variants produce different SKA2 protein isoforms. NM_182620.3 is translated into a 121 amino acid isoform with a molecular weight of 14,188 Da containing mainly three helices, whereas shorter splicer NM_001100595.1 is translated into a 75 amino acid isoform with a molecular weight of around 8.2k Da. The two isoforms are also different at the C- terminal due to an ORF frame shift. Functional studies have been mainly focused on the longer one. Jeyaprakash et al studied the crystal structure of the SKA complex.13 This complex comprises a W shape dimer, mainly composed of 1–99 aa of Ska1, full length SKA2, and 1–101 aa of SKA3. The helix of SKA2 N- terminal (1-32aa) interacts with SKA1 (4–31aa) and SKA3 (2–30 aa) to form a short bundle. The 90Å long bundle of the SKA complex is formed by SKA2 (46–93aa) helix, SKA1 (33–87aa), and SKA3 (35–98aa). The SKA2 C- terminal (102–113aa) helix also interacts with the long bundle. Two bundles are connected by a ring formed by 15 amino acids from SKA2, and two and five amino acids from SKA1 and SKA3, respectively. To conclude, this structure suggests that SKA2 acts as a scaffold that stabilizes the SKA complex structure.
Biological function - SKA2 plays essential roles in cell cycle progress
Studies of Hanisch and colleagues demonstrated that the primary cellular localization of SKA complex is the kinetochore-microtubule interface.10 It is essential for the timely onset of anaphase during mitosis by assisting chromosomal segregation and improving movement of microspheres along a microtubule in a depolymerisation-coupled manner. During mitosis, spindle apparatus formed by rearranged microtubules provides the movement force of chromosomal segregation. The key of this step is that spindle microtubules need to stably attach to the kinetochore. The importance of stable attachment is related to two functions: the first is to ensure that chromosomes are aligned on the equatorial plate and separated into chromatids, while the second is to provide a checkpoint in spindle assembly that is essential for chromosomal alignment and segregation.
As one of the main components of the SKA complex, SKA2 physically interacts with KMN (named for the Knl1, Mis12, and Ndc80 complexes), which is a part of the protein architecture within kinetochores that links centromeric DNA to the plus ends of spindle microtubules (MTs).12 This interaction guarantees that the spindle apparatus attaches to the kinetochore. Affinity for microtubules is synergistically enhanced in the presence of the Ndc80 complex and may allow this complex to track depolymerizing microtubules.15 Suppression of SKA2 expression by RNAi resulted in the blocking of cell cycle progress during metaphase, and although cells eventually finished mitosis, the duration became significantly longer. Research has also shown that reduced expression of SKA2 dampened the stability of kinetochore fibers, and therefore provided the opportunity for some chromosomes to detach from the equatorial plate, eventually leading to the failure of mitosis, as well as the failure of spindle assembly checkpoint exit. Notably, knockdown of SKA1 or SKA3 resulted in the similar phenotype, indicating that each component of the SKA complex is essential to its functionality.10
Consistent with previous studies, recent research in live HeLa cells has shown that the SKA complex is required during depolymerization-coupled pulling (DCP), and the progressive loading of the SKA complex seemed to be indispensable for the maturing of bi-oriented kinetochores, through preventing force-dependent detachment driven by microtubule depolymerization at the poleward-moving kinetochore. Additionally, spindle assembly checkpoint involves the recruitment of Bub and Mad proteins to the kinetochore, with the SKA complex promoting the loss of Bub1 from the kinetochore, a possible mechanism of the spindle assembly checkpoint silencing.16 Another study also suggested that the SKA complex recruits Protein Phosphatase 1(PP1) which initiates the positive feedback loop to promote the recruitment of more SKA complex to the kinetochore.17 In contrast to mitosis, the SKA complex only localizes on the spindle microtubule from Pro-MI to MII stages in mouse oocyte meiosis and is primarily involved in meiotic spindle migration and anaphase spindle stability.18
To conclude, SKA2 together with SKA1 and SKA3 forms the SKA heterotrimeric complex. This complex primarily promotes stability of the equatorial plate and silences the spindle assembly checkpoint, therefore ensuring the timely progress of metaphase and entrance of anaphase during mitosis.
SKA2 and human diseases
Dysregulated expression of SKA2 is associated with aberrant cell proliferation and tumorigenesis
In normal cells, the cell cycle is tightly regulated by numerous molecules and signals, whereas aberrant cell cycle-related gene expression eventually leads to a defective cell cycle checkpoint, a major hallmark of tumorigenesis. Apart from directly regulating the cell cycle, SKA2 was also found co-localized with the glucocorticoid receptor (GR), and promoting the proliferative response to IGF-I exposure, presumably through aiding in the chaperoning of the receptor in the nucleus.19 This result suggests that the mechanism of SKA2 directly regulates cell proliferation, particularly in cancer cells, and also implicates potential functionality in the central nervous system. Dysregulation of SKA2 has been found in cancers as well as in mental disorders and elucidating the underlying mechanisms of SKA2 gene transcription may be of help to identify novel therapeutic targets and diagnostic markers for those diseases.
Rice et al compared SKA2 protein expression in normal tissues to multiple lung and breast human cancer cells line and found it to be significantly upregulated in cancer cells. Additionally, SKA2 is located in the cytoplasm of breast cancer cells, whereas in normal mammary gland cells it is primarily localized in the nucleus.19 The study from our laboratory also showed that in lung cancer patients, SKA2 mRNA expression levels are overexpressed, and differ amongst histological types and differentiation stages. Importantly, SKA2 mRNA expression was negatively correlated with prognosis.20 Similar results were found in lymph node negative (LNN) invasive ductal breast cancer specimens.21 SKA2 expression can be suppressed in a dose-dependent manner by chemotherapy drugs that act on rapid proliferation such as dexamethasone (Dex), staurosporine, phorbol ester, and trichostatin A (TSA). In vitro RNAi-mediated knockdown of SKA2 in breast cancer cell lines repressed cell cycle and proliferation.14, 19, 21 Those results strongly suggest that aberrant expression of SKA2 is involved in tumorigenesis.
SKA2 gene expression regulated by transcription factors
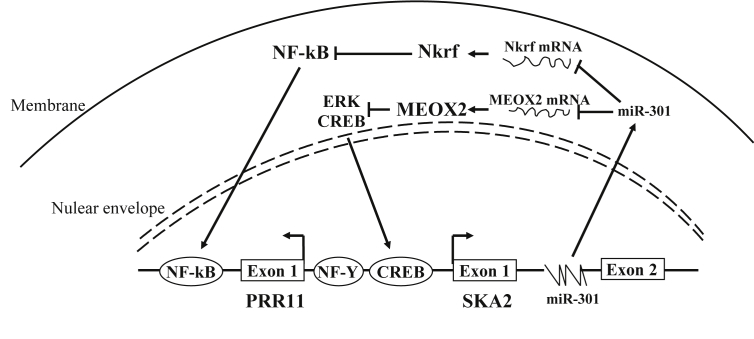
Our laboratory firstly identified the promoter of human SKA2 gene and investigated its transcriptional regulation mechanism. Bioinformatics analysis revealed the SKA2 promoter region contains a classic GC box and binding sites of transcriptional factors such as Sp1, E2F, and GATA-1, which are known to be involved in cell cycle and proliferation gene regulation. Moreover, the SKA2 gene shares the same promoter region with another novel gene PRR11 (Proline Rich 11), which has also been found to be implicated in cell cycle regulation. Use of the luciferase reporter assay demonstrated the presence of a minimal 80-bp intergenic region that functions as a core bidirectional promoter to drive transcription in both PRR11 and SKA2 orientations. EMSA and ChIP assays showed that NF-Y (Nuclear transcription factor Y) binds to and directly activates the PRR11–SKA2 bidirectional promoter, suggesting that abnormally expressed PRR11 and SKA2 may corporately disrupt the cell cycle and contribute to tumorigenesis. Indeed, PRR11 and SKA2 expression were highly correlated with each other in human lung cancer specimens and associated with poor prognosis.20
In the follow-up study, we elucidated that wildtype but not mutant p53 represses the bi-directional promotor activity and endogenous expression of PRR11 and SKA2. Furthermore, overexpression of wildtype p53 abrogated NF-Y mediated upregulation of PRR11 and SKA2. Disruption of the NF-Y binding sites decreased the p53-mediated repression of the PRR11-SKA2 bidirectional promoter. The ChIP assay showed that p53 represses PRR11-SKA2 transcription by reducing the amount of NF-Y binding to the PRR11-SKA2 promoter region. The ability of p53 to downregulate PRR11-SKA2 transcription was significantly attenuated upon siRNA-mediated depletion of nuclear factor Y subunit beta (NF-YB). Overall, our study suggested a novel mechanism of tumorigenesis after p53 loss, probably due to the inability to repress the PRR11-SKA2 bidirectional transcription unit (Fig. 2).22 More work is required to reveal the mechanism of how wildtype p53 regulates NF-Y binding affinity to the promoter.
Figure 2.
Regulatory mechanism of SKA2 gene expression. Figure represents a model of bidirectional transcription unit of SKA2 and PRR11 gene. NFY and CREB bind to a promotor region that initiates both SKA2 and PRR11 mRNA transcription. miR-301 regulates NF-κB and ERK/CREB by directly targeting Nkrf and MEOX2, respectively. NF-κB directly regulates transcription of miR-301 and SKA2. CREB directly regulates transcription of SKA2.
Additionally, in renal cell carcinoma (RCC), cyclic AMP responsive element-binding protein (CREB) was also found to target SKA2 through binding to its CRE sequence. Upregulated SKA2 expression by CREB enhanced proliferation of RCC cell lines and promoted tumorigenesis both in vitro and in vivo.23
SKA2 gene expression regulated by micro-RNAs
The SKA2 gene contains four introns, and multiple micro-RNA transcripts were found to reside in SKA2 introns. miR-301 and miR-454 are the first two that were identified hosting in SKA2 and participating in SKA2 transcriptional regulation. miR-301 regulates the ERK/CREB pathway by targeting MEOX2 (Mesenchyme Homeobox 2), therefore inducing SKA2 expression in a positive feedback manner. Suppressing miR-301 decreased the mitosis index and the clone formation ability of A549 cells.24 Overexpression of miR-301a significantly enhanced proliferation and migration in lung cancer cell lines.25 Lu et al found that both SKA2 and miR-301 can be induced by TNFα stimulation; they validated the p50-RelA binding site in the SKA2 promoter region and proposed a TNFα-NFκB-Ska2/miR-301 positive feedback regulatory model in pancreatic cancer (Fig. 2).14 SKA2 expression may be also regulated by miR-141, which directly targets 3′-UTR of SKA2 in glioma cells. Mechanism studies revealed that a glioma-related lncRNA located on chromosome 12q13.13 named Hox transcript antisense intergenic RNA (HOTAIR) may act as an endogenous “sponge” of miR-141 (a competing endogenous RNA (ceRNA) for the target SKA2 through binding miR-141), thereby retaining expression of SKA2 in glioma.26
Aberrant epigenetic modifications of SKA2 gene and implications in psychiatric function
Epigenetic modification is a gene expression regulatory mechanism which often reflects interactions between genome and environment. DNA methylation and histone modification alter DNA accessibility and chromatin structure, thereby regulating patterns of gene expression. DNA methylation typically acts to repress gene transcription. In contrast to cancers, downregulated SKA2 expression mediated by epigenetic regulation, particularly methylation, has been shown implicated in psychiatric disorders that lead to suicidal behaviors.
Guintivano et al performed genome-wide DNA methylation profiling on neuronal and glial nuclei from postmortem brain specimens, and the results showed that methylation of probe cg13989295 in the 3ʹ- UTR of SKA2 in prefrontal neurons was associated with suicide. With the same probe, SKA2 DNA methylation analysis for a collection of blood and saliva samples followed by a dexamethasone suppression test (DST) suggested that SKA2 methylation may mediate the suppression of cortisol.27 This result is consistent with another study that implicated SKA2 in glucocorticoid receptor transactivation, supporting its hypothesized role in hypothalamic pituitary adrenal (HPA) axis abnormalities.19 Importantly, SKA2 gene expression was significantly lower in surrounding tissue in suicide decedents.28 DNA methylation of the SKA2 gene and the single-nucleotide polymorphism rs7208505 genotype may have effects on suicidal behavior according to a linear model suggested by the authors. The genotype rs7208505 contains a single nucleotide polymorphism (SNP) containing a cytosine (C) variant allele instead of thymine (T) present in the common allele. This SNP allows the dinucleotide repeat (CpG) elements to occur providing a gene segment for methylation. N Sadeh et al analyzed the rs7208505 genotype, cg13989295 methylation, and methylation adjusted for genotype in 200 white non-Hispanic veterans, and they found that SKA2 methylation is associated with decreased prefrontal cortical thickness and increased PTSD severity among trauma-exposed veterans, in another collection of 466 white non-Hispanic blood sample of veterans, SKA2 methylation (at CpG locus cg13989295) was found to be associated with lifetime suicidal risk, although the association between PTSD and SKA2 were not significant.29, 30
A follow-up study of blood samples from trauma-exposed veterans and neuroimaging revealed that DNA methylation was associated with bilateral reductions of cortical thickness in frontal pole and superior frontal gyrus, and similar effects were found in the right orbitofrontal cortex and right inferior frontal gyrus. PTSD symptom severity was positively correlated with SKA2 DNA methylation and negatively correlated with cortical thickness in these regions.31 Those studies suggest its potential value as a biomarker of stress exposure and susceptibility, this hypothesis is supported by a study in 85 healthy individuals exposed to the Trier Social Stress Test, with increased SKA2 methylation being found to be associated with lower cortisol stress reactivity.32 Based on the previous findings, a recent independent study found decreased expression of SKA2 in blood from violent suicide completers compared with non-suicidal controls, establishing SKA2 as one of the proxy diagnostic probes.33
Clinical significance and translational implications
As discussed above, studies of SKA2 gene expression and epigenetic modification suggest the potential of this gene being used as biomarker for cancers and suicidal behavior prediction. Our laboratory first studied the prognosis value of SKA2 in human lung cancer patients.22 In a microarray dataset of 117 patient samples (GEO database: GSE13213), SKA2 highly expressed patients showed worse overall survival rate. Importantly, p53 wildtype and SKA2 low expressed patients exhibit best prognosis of approximately 80% 5-year survival rate, whereas the p53 mutant and SKA2 highly expressed patient group had the worst prognosis of less than 30% 5-year survival. This result also indicates that targeting of SKA2 in p53 loss non-small lung cancer patients may be of clinical therapeutic benefit, although more studies are required to evaluate this potential.
On the other hand, the SKA2 gene first received attention from the psychiatrist community due to a study showing that analysis of three independent data sets of post-mortem brains revealed signs of increased DNA methylation particularly in SKA2. This finding was extended to peripheral blood samples from other cohorts of prospectively followed individuals. In that study, SKA2 methylation in blood was found correlated to suicidal ideation when controlling for genetic variation was held, suggesting that methylation is a more reliable predictor of suicide risk than genotype.28 In another study, the authors also reported that SKA2 methylation was associated with an increased likelihood of attempted suicide among individuals with high levels of childhood trauma.27 In two subsequent studies, Niculescu and colleagues found decreased SKA2 expression levels in blood in suicide decedents compared to controls and SKA2 expression levels prospectively predicted suicidal ideation in psychiatric patients.34, 35
Those studies provide strong evidence that epigenetic alterations at the SKA2 gene are associated with multiple types of suicidal behaviors. More importantly, the detection of SKA2 methylation can be done in peripheral blood samples which largely enhanced the translational value of SKA2 as a biomarker. However, given the novelty of these findings, additional association studies in diverse samples are needed to assess the replicability and generalizability of the observed effects. On the other hand, whether this potential biomarker is able to out-compete other well-established and easily assessed risk factors has not been evaluated. Nonetheless, SKA2 explains meaningful variance which would strengthen its potential clinical utility by showing that it provides unique information about suicide risk not conveyed by clinical interviews.
Conclusions and perspectives
To conclude, SKA2 is indispensable for the cell cycle and proliferation, as well as the integrity of other signaling pathways that may be impacted by the micro-RNAs which reside in the SKA2 gene. Loss of regulation in those functions will potentially drive the cells to malignancy and promote the progression of cancers. Importantly, dysregulation of SKA2 expression might be one of the serial molecular events in tumor progression since it is regulated by p53 through NF-Y. Because many molecular mechanisms are impaired after loss of p53 including DNA damage repair, cell cycle checkpoints, and initiation of apoptosis,36 SKA2 here might be valued as a potential therapeutic target. Furthermore, whether mutant p53 binds to NF-Y in a different manner compared to the wildtype p53 remains reclusive and requires further investigation. More works need to be done to further elucidate the roles of SKA2 in diseases such as cancer, not only the longer variant/isoform but also the function and transcriptional regulation of the shorter one. On the other hand, whether SKA2 and the entire SKA complex have a role during other progression steps such as metastasis is unknown. Another important value of SKA2 is that its gene expression and DNA methylation as a potential diagnostic marker for psychiatric disorders such as PTSD and suicidal behaviors has been assessed by multiple studies using different patient cohorts. Our studies have also indicated SKA2 gene expression as a lung cancer prognosis marker. Nevertheless, more analyses with large scale clinical samples are required to evaluate this potential.
Conflicts of interest
The authors have declared no conflicts of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81672301 to Youquan Bu) and the Basic Sciences and Advanced Technology Key Project of CQ CSTC (No. cstc2017jcyjBX0069 to Youquan Bu).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2018.11.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Hartwell L.H., Kastan M.B. Cell cycle control and cancer. Science. 1994;266(5192):1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 3.Williams G.H., Stoeber K. The cell cycle and cancer. J Pathol. 2012;226(2):352–364. doi: 10.1002/path.3022. [DOI] [PubMed] [Google Scholar]
- 4.Ren B., Cam H., Takahashi Y. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16(2):245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozaki T., Nakagawara A. Role of p53 in cell death and human cancers. Cancers (Basel) 2011;3(1):994–1013. doi: 10.3390/cancers3010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malumbres M., Carnero A. Cell cycle deregulation: a common motif in cancer. Prog Cell Cycle Res. 2003;5:5–18. [PubMed] [Google Scholar]
- 7.Liu Q., Guntuku S., Cui X.S. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14(12):1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 8.Finn R.S., Crown J.P., Lang I. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 9.Dean J.L., Thangavel C., McClendon A.K., Reed C.A., Knudsen E.S. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29(28):4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 10.Hanisch A., Sillje H.H., Nigg E.A. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006;25(23):5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daum J.R., Wren J.D., Daniel J.J. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr Biol. 2009;19(17):1467–1472. doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaitanos T.N., Santamaria A., Jeyaprakash A.A., Wang B., Conti E., Nigg E.A. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 2009;28(10):1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeyaprakash A.A., Santamaria A., Jayachandran U. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol Cell. 2012;46(3):274–286. doi: 10.1016/j.molcel.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Lu Z., Li Y., Takwi A. miR-301a as an NF-kappaB activator in pancreatic cancer cells. EMBO J. 2011;30(1):57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt J.C., Arthanari H., Boeszoermenyi A. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23(5):968–980. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auckland P., Clarke N.I., Royle S.J., McAinsh A.D. Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment. J Cell Biol. 2017;216(6):1623–1639. doi: 10.1083/jcb.201607096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivakumar S., Gorbsky G.J. Phosphatase-regulated recruitment of the spindle- and kinetochore-associated (Ska) complex to kinetochores. Biol Open. 2017;6(11):1672–1679. doi: 10.1242/bio.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q.H., Qi S.T., Wang Z.B. Localization and function of the Ska complex during mouse oocyte meiotic maturation. Cell Cycle. 2012;11(5):909–916. doi: 10.4161/cc.11.5.19384. [DOI] [PubMed] [Google Scholar]
- 19.Rice L., Waters C.E., Eccles J. Identification and functional analysis of SKA2 interaction with the glucocorticoid receptor. J Endocrinol. 2008;198(3):499–509. doi: 10.1677/JOE-08-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Zhang Y., Zhang C. The gene pair PRR11 and SKA2 shares a NF-Y-regulated bidirectional promoter and contributes to lung cancer development. Biochim Biophys Acta. 2015;1849(9):1133–1144. doi: 10.1016/j.bbagrm.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Shi W., Gerster K., Alajez N.M. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71(8):2926–2937. doi: 10.1158/0008-5472.CAN-10-3369. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Weng H., Zhang Y. The PRR11-SKA2 bidirectional transcription unit is negatively regulated by p53 through NF-Y in lung cancer cells. Int J Mol Sci. 2017;18(3):534. doi: 10.3390/ijms18030534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhuang H., Meng X., Li Y. Cyclic AMP responsive element-binding protein promotes renal cell carcinoma proliferation probably via the expression of spindle and kinetochore-associated protein 2. Oncotarget. 2016;7(13):16325–16337. doi: 10.18632/oncotarget.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao G., Huang B., Liu Z. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res Commun. 2010;396(4):978–982. doi: 10.1016/j.bbrc.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 25.Chen Q., Li Y., Zhang C. MicroRNA-301a promotes growth and migration by repressing TGFBR2 in non-small cell lung cancer. Int J Clin Exp Pathol. 2017;10(2):957–971. [Google Scholar]
- 26.Bian E.B., Ma C.C., He X.J. Epigenetic modification of miR-141 regulates SKA2 by an endogenous 'sponge' HOTAIR in glioma. Oncotarget. 2016;7(21):30610–30625. doi: 10.18632/oncotarget.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaminsky Z., Wilcox H.C., Eaton W.W. Epigenetic and genetic variation at SKA2 predict suicidal behavior and post-traumatic stress disorder. Transl Psychiatry. 2015;5:e627. doi: 10.1038/tp.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guintivano J., Brown T., Newcomer A. Identification and replication of a combined epigenetic and genetic biomarker predicting suicide and suicidal behaviors. Am J Psychiatry. 2014;171(12):1287–1296. doi: 10.1176/appi.ajp.2014.14010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadeh N., Spielberg J.M., Logue M.W. SKA2 methylation is associated with decreased prefrontal cortical thickness and greater PTSD severity among trauma-exposed veterans. Mol Psychiatr. 2016;21(3):357–363. doi: 10.1038/mp.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadeh N., Wolf E.J., Logue M.W. Epigenetic variation at Ska2 predicts suicide phenotypes and internalizing psychopathology. Depress Anxiety. 2016;33(4):308–315. doi: 10.1002/da.22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadeh N., Spielberg J.M., Logue M.W. SKA2 methylation predicts reduced cortical thickness in prefrontal cortex. Mol Psychiatr. 2016;21(3):299. doi: 10.1038/mp.2016.10. [DOI] [PubMed] [Google Scholar]
- 32.Boks M.P., Rutten B.P., Geuze E. SKA2 methylation is involved in cortisol stress reactivity and predicts the development of post-traumatic stress disorder (PTSD) after military deployment. Neuropsychopharmacology. 2016;41(5):1350–1356. doi: 10.1038/npp.2015.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melhem N.M., Munroe S., Marsland A. Blunted HPA axis activity prior to suicide attempt and increased inflammation in attempters. Psychoneuroendocrinology. 2017;77:284–294. doi: 10.1016/j.psyneuen.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niculescu A.B., Levey D., Le-Niculescu H., Niculescu E., Kurian S.M., Salomon D. Psychiatric blood biomarkers: avoiding jumping to premature negative or positive conclusions. Mol Psychiatr. 2015;20(3):286–288. doi: 10.1038/mp.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niculescu A.B., Levey D.F., Phalen P.L. Understanding and predicting suicidality using a combined genomic and clinical risk assessment approach. Mol Psychiatr. 2015;20(11):1266–1285. doi: 10.1038/mp.2015.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.