Abstract
The important role of insulin-like growth factor-1 receptor (IGF-1R) in tumorigenesis has been well established. The classical model involves IGF-1R binding to IGF-1/2, the following activation of PI3K-Akt-signaling cascades, driving cell proliferation and apoptosis inhibition. Here we report a new signal transduction pathway of IGF-1R in the intestinal epithelium. Using heterozygous knockout mice (Igf1r+/−), we analyzed the expressions of viral RNA sensors MDA5 and RIG-I in the intestinal epithelium. Igf1r+/− mice exhibited higher MDA5 and RIG-I than wild-type (WT) mice, indicating that knockdown of IGF-1R could trigger MDA5 and RIG-I. IGF-1R knockdown-triggered MDA5 and RIG-I were further investigated in human colonic cancer cells. Increased MDA5 and RIG-I were clearly seen in the cytoplasm in cancer cells as well as normal human colonic cells with silenced IGF-1R. Notably, the upregulations of MDA5 and RIG-I was not affected by blockage of the PI3K-Akt pathway with LY294002. These results suggested a new signal transduction pathway of IGF-1R. Importantly, IGF-1R knockdown-triggered MDA5 and RIG-I resulted in colorectal cancer apoptosis through activation of the mitochondrial pathway. These in vitro observations were evidenced in the azoxymethane (AOM)-dextran sulfate sodium (DSS) colorectal cancer model of mice. In conclusion, knockdown of IGF-1R triggers viral RNA sensor MDA5- and RIG-I-mediated mitochondrial apoptosis in cancer cells.
Keywords: IGF-1R, viral RNA sensors, MDA5, RIG-I, mitochondrial apoptosis
Introduction
Insulin-like growth factor receptor (IGF-1R) is a key point of convergence for major signaling pathways implicated in human physiology and pathophysiology. Clinical evidence has indicated that IGF-1R has potent antiapoptotic and transforming activities and that increased IGF-1R is associated with cell survival and proliferation, cancer progression, metastasis, treatment resistance, and poor prognosis.1, 2 Preclinical studies have demonstrated that activated IGF-1R could mediate tumorigenesis, mainly through two major downstream signaling pathways: phosphatidylinositol-3-kinase (PI3K)-Akt and Ras-mitogen-activated protein kinase (Ras-MAPK) pathways.3, 4, 5
Rationally, targeting IGF-1R could serve as a promising approach in anticancer therapeutics.6, 7 To date, over 10 IGF-1R-targeted drugs, including small-molecule drugs and anti-IGF-1R antibodies, have been approved for clinical trials.8, 9 Other strategies targeting IGF-1R by small nucleotides complementary to IGF-1R have also been tested in experimental therapeutics.10 However, despite some initial promising preclinical results, the clinical disclosures subsequently have been less encouraging. Moreover, recent studies revealed that IGF-1R participates in a dynamic and complex signaling network, interacting with additional targets and pathways thereof through various crosstalk and compensatory signaling mechanisms.11 Further understanding of such mechanisms around IGF-1R, especially its interactions with other signaling pathways, could help us to shed some light on the decreased effectiveness of selective IGF-1R-targeted therapies. We anticipate that targeting these bypass signaling pathways of IGF-1R might produce a more effective therapeutic response in cancers.
Melanoma differentiation-associated gene 5 (MDA5) helicases and retinoic acid-inducible gene-I (RIG-I), two so-called RIG-I-like receptor (RLR) family proteins, have been known as sensors of double-stranded RNA (dsRNA) responsible for the induction of type I interferon (IFN).12, 13 In infected cells, MDA5 and RIG-I have distinct biological roles in the specificity of virus recognition as well as RNA-binding specificity: RIG-I recognizes short dsRNA and 5′triphosphate single-stranded (ss)RNA, while MDA5 senses long dsRNA.14 LGP2 (laboratory of genetics and physiology 2) is the third RNA helicase comprising the RIG-I-like helicase (RLH) family of receptors for detecting RNA viruses.15, 16 Polyinosinic-polycytidylic acid (poly(I:C)), a synthetic mimic of long dsRNA, has been identified as a strong activator of MDA5.17 Viral RNA mimetic synthetic 5ʹ-triphosphate RNA (pppRNA) recognizes RIG-I.18 Upon RNA viruses, RIG-I and MDA5 recognize viral nucleic acids and proteins, and they bind to the adaptor protein IFN-β promoter stimulator 1 (IPS-1), which is tethered to the outer mitochondrial membrane.19 This reciprocal interaction triggers a complex signal transduction cascade culminating in the activation of transcription factors (particularly nuclear factor κB [NF-κB] and IFN regulatory factor 3 [IRF-3]) and the production of cytokines, including type I IFN.20 Interestingly, Besch et al.21 uncovered an IFN-independent antiviral signaling pathway initiated by RIG-I and MDA5 that could activate proapoptotic signaling in human melanoma cells through the introduction of pppRNA or poly(I:C), a synthetic mimic of long dsRNA. The RIG-I- and MDA5-initiated proapoptotic pathway is more active in melanoma cells than nonmalignant cells, and thereby RIG-I- and MDA5-mediated apoptosis results in preferential tumor cell death. Due to their immunostimulatory and proapoptotic activities, RIG-I and MDA5 ligands through intracellular delivery of viral RNA mimetics might be an optimal anticancer therapy.21
Surprisingly, an unanticipated observation is that knockdown of IGF-1R could trigger cytoplasmic RNA sensors MDA5 and RIG-I as delivering poly(I:C) and pppRNA had. In our group, we had performed high-throughput sequencing (RNA sequencing) in human colonic cancer cells with knockdown of IGF-1R. The RNA sequencing data indicated a surprising result that there was a significant link between knockdown of IGF-1R and upregulations of viral RNA sensors MDA5 and RIG-I (all of these data are at GEO: GSE122534, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122534). Our further in vivo and in vitro results revealed that the knockdown of IGF-1R triggered MDA5 and RIG-I expressions in human cancer cells as well as human nonmalignant cells. Using human colonic cancer cell lines and the azoxymethane (AOM)-dextran sulfate sodium (DSS) colorectal cancer model, we confirmed that knockdown of IGF-1R triggered mitochondrial apoptosis through the upregulations of MDA5 and RIG-I. Further, we showed that increased MDA5 and RIG-I mediated the mitochondrial apoptosis through initiating the proapoptotic BH3-only proteins Bim in cancer cells. Due to normal cells being less sensitive to this endogenous proapoptotic signaling than cancer cells,21 IGF-1R knockdown-triggered MDA5- and RIG-I-mediated apoptosis could lead to preferential tumor cell death. These findings suggest that targeting IGF-1R to trigger MDA5 and RIG-I might have therapeutic potential for cancer treatment. In addition, IGF-1R knockdown also triggers MDA5 and RIG-I in human normal colonic epithelial cells. This finding provides us some clues in antivirus research that targeting IGF-1R might play roles in infected cells against the virus through triggering MDA5 and RIG-I.
Results
Heterozygous Knockout Insulin-like Growth Factor-1 Receptor Mice Demonstrate Higher Viral RNA Sensors MDA5 and RIG-I Than Their Wild-Type Littermates
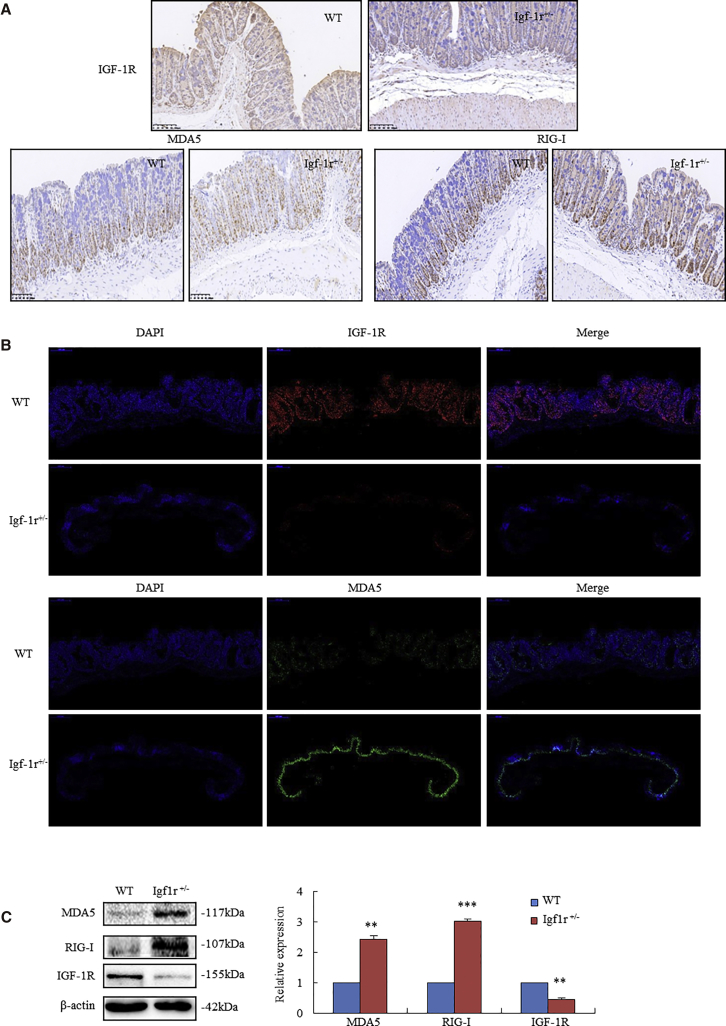
Based on the RNA sequencing data (NovelBioinformatics), we further analyzed the expressions of MDA5 and RIG-I in heterozygous knockout insulin-like growth factor-1 receptor (Igf1r+/−) mice. Due to the knockdown of IGF-1R,22 Igf1r+/− mice were determined to have a lower level of IGF-1R than their wild-type (WT) littermates (Figure 1A, first row). In Igf-1r+/− mice, immunohistochemistry (IHC) analysis showed a strong diffuse staining of MDA5 (Figure 1A, second row, left) and RIG-I (Figure 1A, second row, right), whereas only scattered positivity of MDA5 and RIG-I was in the basal cells of colonic crypt in WT mice. Immunofluorescent staining assay also showed an increased MDA5 in the intestinal epithelium of Igf-1r+/− mice (Figure 1B). In contrast to WT mice (Figure 1B, first row), Igf-1r+/− mice had a lower level of IGF-1R (Figure 1B, second row), while Igf-1r+/− mice exhibited a strong increase of MDA5 in the same intestinal epithelium (Figure 1B, fourth row). Further evidence provided by western blotting assay showed that WT mice had the basal levels of MDA5, RIG-I, and IGF-1R, while Igf1r+/− mice exhibited stronger increases in MDA5 and RIG-I in the intestinal epithelium than WT mice (Figure 1C).
Figure 1.
Knockdown IGF-1R Triggered MDA5 and RIG-I in Igf1r+/− Mice
(A) IHC analyzed the expressions of IGF-1R, MDA5, and RIG-I in Igf1r+/− mice and their WT littermates. Blue staining identified the nuclei of colonic epithelium by DAPI. WT mice exhibited a stronger diffuse staining of IGF-1R (brown) than Igf1r+/− mice. Igf1r+/− mice had a significant increase in MDA5 (brown) and RIG-I (brown) diffused membranous and cytoplasmic staining, whereas WT mice had only scattered positivity in the basal cells of colonic crypt. (B) Immunofluorescent staining of IGF-1R and MDA5 in colorectal tissues of WT and Igf1r+/− mice. First frame: DAPI stained the nuclei of colorectal epithelium (first column). Red fluorescent staining showed IGF-1R expression in colorectal epithelium. In contrast to WT mice (first row), Igf1r+/− mice demonstrated a decreased IGF-1R expression (second row). Second frame: Igf1r+/− mice showed higher MDA5 (second row, green fluorescent staining) than WT mice (first row, very weakly green fluorescent staining). (C) Western blotting analysis showed a lower level of IGF-1R and higher levels of MDA5 and RIG-I in the colorectal epithelium of Igf1r+/− mice than WT mice. (n = 6). **p < 0.01, ***p < 0.001 between Igf1r+/− mice and their WT littermates.
Knockdown of IGF-1R Triggers MDA5 and RIG-I in Human Colonic Cancer Cells
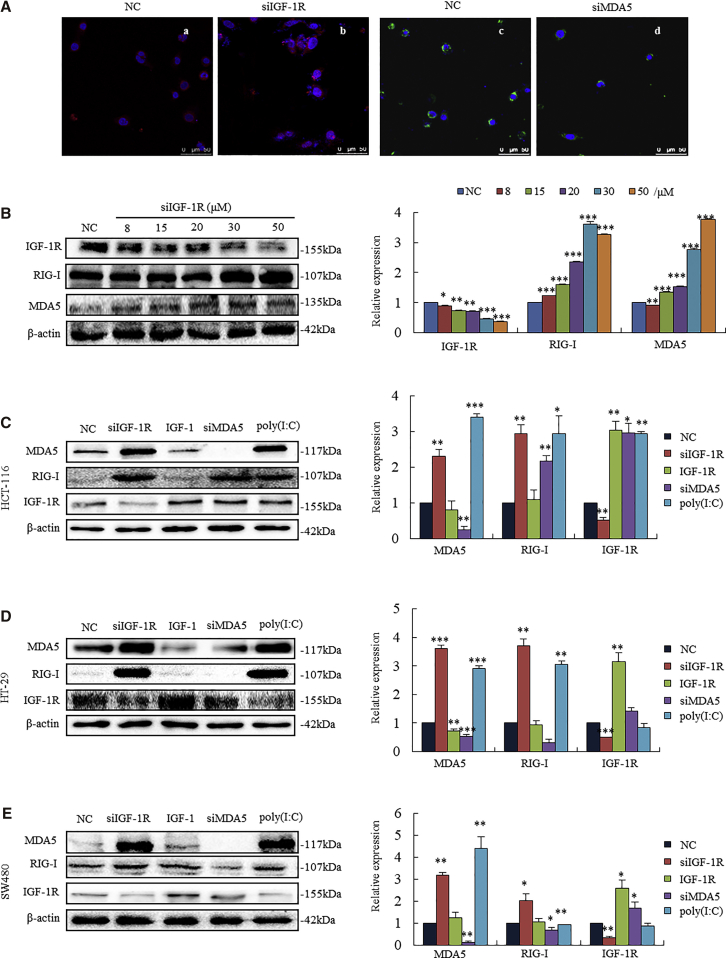
We tested the expressions of MDA5 and RIG-I in human colonic cancer cells. Immunofluorescent staining assay showed that HT-29 cells with silenced IGF-1R demonstrated an increased level of MDA5 in the cytoplasm (Figure 2A, b, red) as compared to NC (negative control) cells (Figure 2A, a), whereas silencing MDA5 did not affect the expression of IGF-1R (Figure 2A, d). These results indicate that the knockdown of IGF-1R triggers the RNA helicases in cancer cells. IGF-1R knockdown-triggered RNA helicases were further evidenced by the following assays. We used an increasing concentration of small interfering RNA of IGF-1R (siIGF-1R) to treat HT-29 cells for 48 h. Both MDA5 and RIG-I were accordingly upregulated with the decreasing IGF-1R expression (Figure 2B). Silencing IGF-1R strongly triggered MDA5 and RIG-I expressions in colonic cancer cell lines HCT-116 (Figure 2C, second lane), HT-29 (Figure 2D, second lane), and SW480 (Figure 2E, second lane). On the other hand, when IGF-1R was activated by the addition of IGF-1 (1 μM), both MDA5 and RIG-I changed irregularly in these cell lines (Figures 2C–2E, third lanes). The levels of upregulated MDA5 and RIG-I by the knockdown of IGF-1R were similar to those of poly(I:C) in the same cell lines.
Figure 2.
Knockdown of IGF-1R Triggers MDA5 and RIG-I in Human Colonic Cancer Cells
(A) Immunofluorescent staining analysis showed increased MDA5 in the cytoplasm (b, red staining) in HT-29 cells with silenced IGF-1R, whereas silenced MDA5 did not impact IGF-1R expression (d). (B) HT-29 cells showed a dose-dependent decrease in IGF-1R expression upon an increasing concentration of siIGF-1R. Both MDA5 and RIG-I were accordingly increased with a decrease in IGF-1R expression. (C–E) Expressions of MDA5, RIG-I, and IGF-1R in HCT-116 (C), HT-29 (D), and SW480 (E) cell lines. Cells transfected with siIGF-1R strongly triggered MDA5 and RIG-I (second lane). An increase in IGF-1R by IGF-1 (1 μM) resulted in the downregulation of MDA5 expressions (third lane) in HCT-116 and HT-29. Silenced MDA5 did not significantly impact IGF-1R expression (fourth lane). Cells transfected with poly(I:C) resulted in strong increases in MDA5 and RIG-I expressions (last lane). *p < 0.05, **p < 0.01, ***p < 0.001 versus NC (negative control) cells.
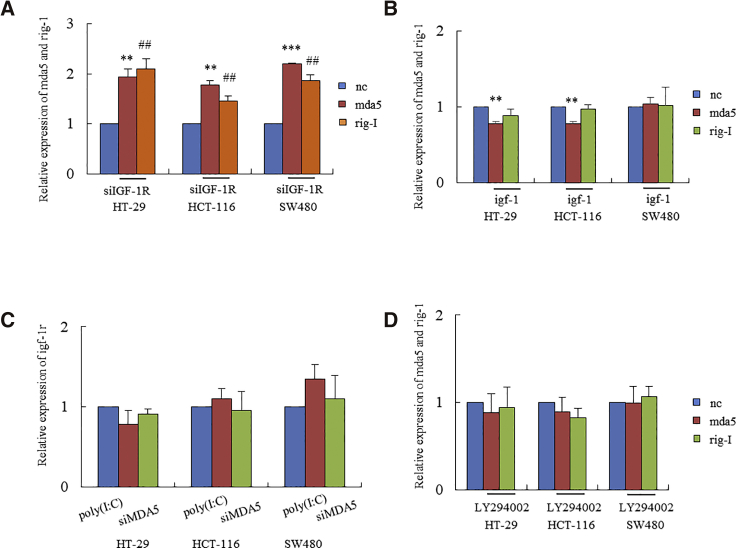
The upregulations of MDA5 and RIG-I by the knockdown of IGF-1R occurred on the mRNA level. The qRT-PCR assay showed significant increases in mda5 and rig-I in HT-29, HCT-116, and SW480 cell lines transfected with siIGF-1R (Figure 3A). On the other hand, activation of IGF-1R by the addition of IGF-1 significantly downregulated the expressions of mda5 in HT-29 and HCT-116 cells (Figure 3B). Neither increased MDA5 by poly(I:C) nor silenced MDA5 by transfection with siRNA of MDA5 (siMDA5) affected the expression of igf-1r in these cell lines (Figure 3C). We thus suggest that the knockdown of IGF-1R might unidirectionally upregulate MDA5 and RIG-I expressions in cancer cells. Further, blockage of the PI3K-Akt pathway with LY294002 did not significantly impact the expressions of MDA5 and RIG-I (Figure 3D). These results suggest a PI3K-Akt-independent pathway of IGF-1R in tumorigenesis.
Figure 3.
IGF-1R Knockdown-Triggered MDA5 and RIG-I Occurred on the mRNA Level
(A) Colonic cancer cell lines HT-29, HCT-116, and SW480 showed significant increases in mda5 (**p < 0.01, ***p < 0.001 versus NC) and rig-I (##p < 0.01 versus NC) after transfection with siIGF-1R. (B) Cell lines treated with IGF-1 reduced the levels of mda5 and rig-I, except SW480. (C) Neither increased MDA5 by poly(I:C) nor silenced MDA5 by siMDA5 impacted the expression of igf-1r. (D) LY294002-treated cells did not impact MDA5 and RIG-I expressions in cancer cells.
IGF-1R Knockdown Also Triggers MDA5 and RIG-I in Human Normal Colonic Epithelial Cells
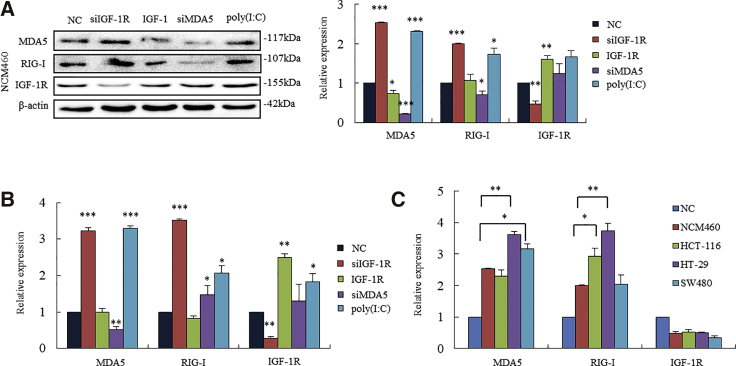
Human colonic epithelial (NCM460) cells demonstrated profiles of MDA5 and RIG-I expressions similar to those in colonic cancer cells. NCM460 cells transfected with siIGF-1R produced increases in MDA5 and RIG-I, whereas the cells with activation of IGF-1R had slight decreases in MDA5 and RIG-I by the addition of IGF-1. Consistent with cancer cell lines, NCM460 cells transfected with siMDA5 did not affect IGF-1R expression (Figure 4A). Consistent with cancer cells, increased MDA5 and RIG-I occurred on the mRNA level of normal colonic cells (Figure 4B). These findings in human normal cells provide us some clues in antivirus research that the downregulation of IGF-1R might play roles in infected cells against the virus through triggering MDA5 and RIG-I. Furthermore, we compared the increased levels of MDA5 and RIG-I in these cancer cell lines and human normal epithelial cells. Cancer cells had higher responses to the knockdown of IGF-1R than did normal cells in triggering MDA5 and RIG-I (Figure 4C).
Figure 4.
Knockdown of IGF-1R Triggers MDA5 and RIG-I in Human Nonmalignant Colonic Epithelial Cells
(A) Human colonic epithelial (NCM460) cells transfected with siIGF-1R demonstrated increased MDA5 and RIG-I expressions (second lane). Activation of IGF-1R by IGF-1 downregulated MDA5 and RIG-I slightly (third lane). Cells transfected with siMDA5 did not impact IGF-1R expression (fourth lane). Cells transfected with poly(I:C) resulted in increased MDA5 and RIG-I (last lane). (B) Knockdown IGF-1R-triggered MDA5 and RIG-I occurred on the mRNA level. *p < 0.05, **p < 0.01, ***p < 0.001 versus NC cells. (C) The comparison of increased MDA5 and RIG-I in three colonic cancer cell lines and normal colonic epithelial cells. Cancer cells show higher responses to knockdown IGF-1R in triggering MDA5 and RIG-I than human colonic epithelial normal cells. *p < 0.05, **p < 0.01 between colonic cancer cell lines and normal epithelial cells.
Knockdown of IGF-1R Triggered MDA5- and RIG-I-Mediated Mitochondrial Apoptosis in Human Colonic Cancer Cells
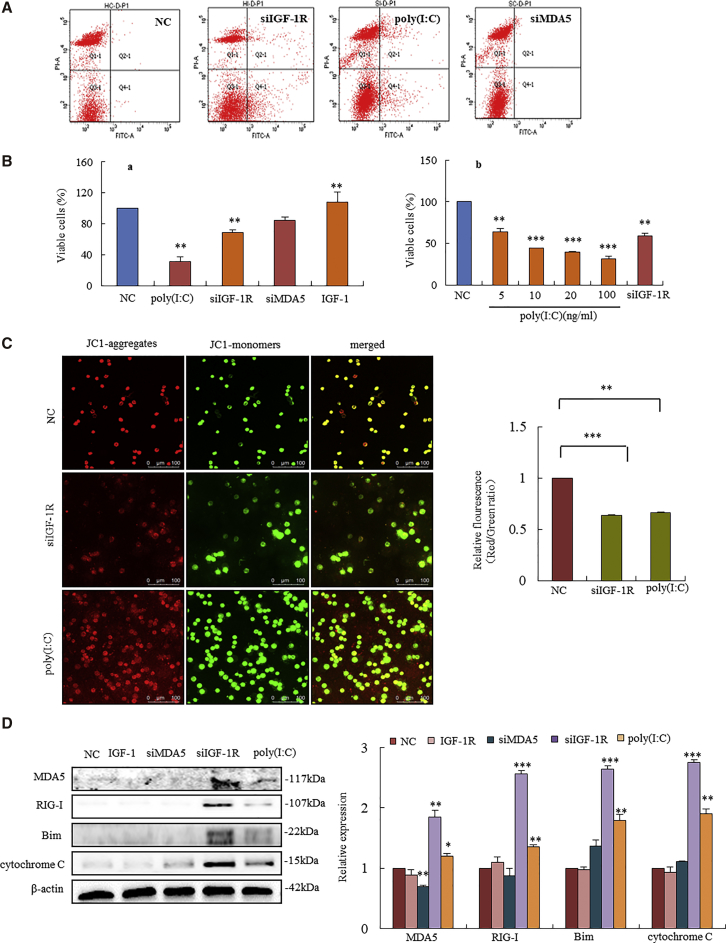
Previous study showed that poly(I:C) and pppRNA increased cytoplasmic MDA5 and RIG-I, thereby leading to mitochondrial apoptosis independently of type I IFN in human melanoma cells.21, 23 In our study, an unanticipated observation was that IGF-1R knockdown triggered cytoplasmic MDA5- and RIG-I-mediated mitochondrial apoptosis in cancer cells, as poly(I:C) had (Figure 5A). Knockdown of IGF-1R-triggered MDA5 and RIG-I consequently reduced cell survival, since IGF-1R activation by the addition of IGF-1 strongly increased cell survival (Figure 5B, a). Poly(I:C) dose-dependently decreased the survival of HT-29 cells through the activation of MDA5-mediated apoptosis. Similarly, 50 μM siIGF-1R treatment in HT-29 cells achieved the same efficacy as poly(I:C) had (Figure 5B, b).
Figure 5.
IGF-1R Knockdown-Triggered MDA5 and RIG-I Mediated Apoptosis through the Mitochondrial Pathway in Human Colonic Cancer Cells
(A) Annexin V-FITC-PI staining analysis showed increased apoptosis in HT-29 cells transfected with siIGF-1R and poly(I:C). Transfection of siMDA5 did not induce cell apoptosis. (B, a) HT-29 cell survival after treatment with poly(I:C), siIGF-1R, siMDA5, and IGF-1 for 48 h. Cell survival was strongly inhibited by the transfection of siIGF-1R and poly(I:C), but not siMDA5. IGF-1 significantly stimulated cell growth as compared to control cells. (B, b) HT-29 cell survival was dose-dependently inhibited with increasing concentrations of poly(I:C). Cells transfected with siIGF-1R (50 μM) achieved the same efficacy as poly(I:C). (C) JC-1 probe staining of HT-29 cells showed a loss of MMP in cancer cells with silenced IGF-1R, showing an increase of green fluorescence (second row), which was similar to the effect of poly(I:C) (third row). (D) Western blotting analysis showed increased Bim and cytochrome c in HT-29 cells with silenced IGF-1R (fourth lane). The efficacy of activated Bim and cytochrome c by silenced IGF-1R was higher than that by poly(I:C) (last lane). *p < 0.05, **p < 0.01, ***p < 0.001 versus NC cells.
To investigate apoptotic signaling triggered by MDA5 and RIG-I, we analyzed the levels of mitochondrial membrane potential (MMP). Loss of MMP leads to the release of cytochrome c, initiating a caspase cascade in the mitochondrial apoptosis pathway.24 Fluorescent 5,5',6,6'-tetrachloro-1,1',3,3'-tetraethyl-imidacarbocyanine iodide (JC-1) probe analysis showed a loss of MMP in HT-29 cancer cells transfected with siIGF-1R, showing a significant increase in green fluorescence (Figure 5C, second row), which was similar to the apoptotic effect that poly(I:C) had (Figure 5C, third row).
Western blotting assay showed an increase in cytochrome c and Bim in siIGF-1R-transfected cells (***p < 0.001 versus NC cells), and increased levels of these mitochondria-associated proteins were higher than those in poly(I:C)-treated cells (**p < 0.01) (Figure 5D). Neither silencing MDA5 nor activating IGF-1R by the addition of IGF-1 affected the expressions of cytochrome c and Bim. These results suggest that IGF-1R knockdown triggered MDA5- and RIG-I-mediated cancer cell apoptosis through the mitochondrial pathway.
Knockdown of IGF-1R Triggered MDA5- and RIG-I-Mediated Mitochondrial Apoptosis, thereby Leading to the Inhibition of Cancer Growth in Igf1r+/− Mice
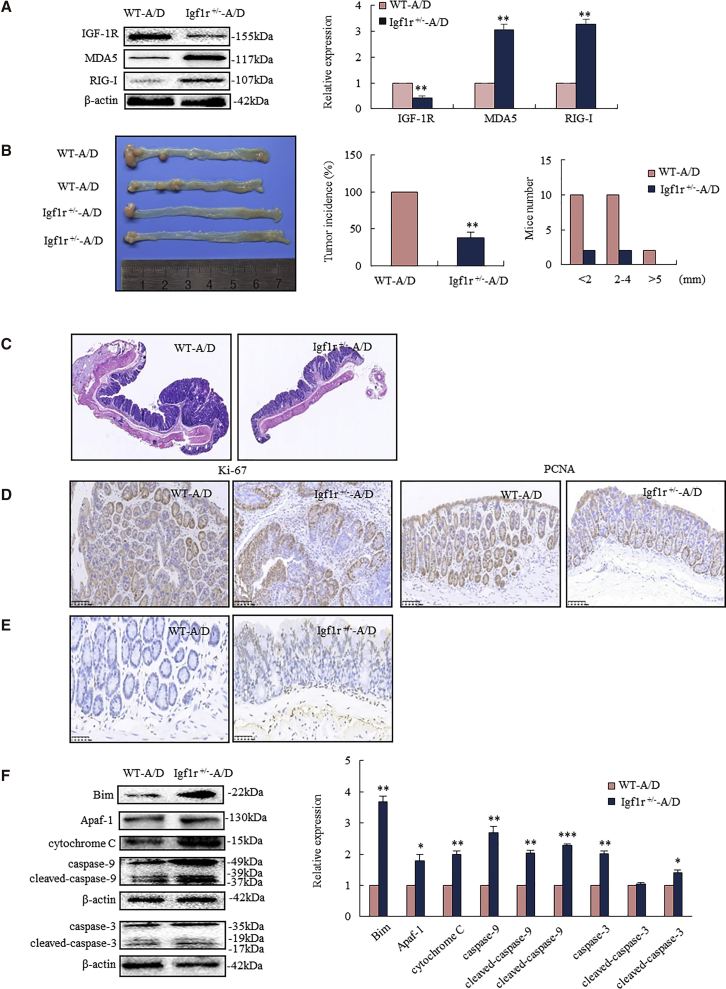
We employed the AOM-DSS colorectal cancer model to test whether knockdown IGF-1R-induced MDA5 and RIG-I could be translated in vivo.25 Igf1r+/− mice exhibited higher expressions of MDA5 and RIG-I in the intestinal epithelium than WT mice (Figure 6A). Mice were intraperitoneally injected with AOM, followed by three rounds of oral DSS. Igf1r+/− mice developed fewer and smaller-sized colorectal tumors than their WT littermates. AOM-DSS induced colorectal tumors by 100% in WT mice and only by 38% in Igf1r+/− mice (Figure 6B, middle). Analysis of the tumor size distribution showed larger tumors in WT than Igf1r+/− mice. Larger-sized tumors (>5 mm) were frequently seen in WT, but not in Igf1r+/−, mice (Figure 6B, right). Histopathological analyses showed advanced colonic adenocarcinoma in WT mice and only low-grade dysplastic mucosa in Igf1r+/− mice (Figure 6C). IHC assay showed a strong diffuse staining of Ki-67 and PCNA in cancer epithelium and high-grade dysplastic mucosa in WT mice, whereas it showed only scattered positivity in the basal cells of colorectal crypt and low-grade dysplastic mucosa in Igf1r+/− mice (Figure 6D).
Figure 6.
IGF-1R Knockdown-Triggered MDA5 and RIG-I Mediated Mitochondrial Apoptosis, Leading to Cancer Inhibition in Igf1r+/− Mice
Igf1r+/− mice and their WT littermates were exposed to AOM-DSS for inducing colorectal cancer. (A) Igf1r+/− mice had higher MDA5 and RIG-I levels in the colorectal epithelium than WT mice. (B) Igf1r+/− mice developed less colorectal tumor than WT mice (left), showing decreased incidence (middle) and size of tumor (right). (C) Histopathological analyses showed advanced adenocarcinoma in WT mice, while only low-grade dysplastic mucosa appeared in Igf1r+/− mice. (D) IHC analysis showed a strong diffuse staining of Ki-67 and PCNA in the colorectal cancer of WT mice, whereas it showed only scattered positivity in the basal cells of colorectal crypt in Igf1r+/− mice. (E) TUNEL staining analysis showed increased apoptotic epithelium in Igf1r+/− mice. (F) Western blotting analysis showed increased Bim and apoptosome (cytochrome c, apoptotic peptidase-activating factor 1 [Apaf-1], and caspase-9 and caspase-3) in colorectal epithelium of Igf1r+/− mice (n = 6). *p < 0.05, **p < 0.01, ***p < 0.001 between Igf1r+/−-A/D and WT-A/D. A/D, AOM-DSS.
Knockdown of IGF-1R induced cancer apoptosis in Igf1r+/− mice. Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) staining analysis showed an increase of apoptotic epithelium in Igf1r+/− mice (Figure 6E). Western blotting assay revealed the activation of BH3-only proteins (Bim), apoptosome (cytochrome c, apoptotic peptidase-activating factor 1 [Apaf-1], and caspase-9), and executioner caspase-3 in the intestinal epithelium of Igf1r+/− mice (Figure 6F). These results suggest that IGF-1R knockdown-triggered MDA5 and RIG-I result in cancer apoptosis through the mitochondrial pathway. IGF-1R knockdown triggering MDA5- and RIG-I-induced cancer apoptosis is sufficient to prevent AOM-DSS-driven colorectal cancer in Igf1r+/− mice.
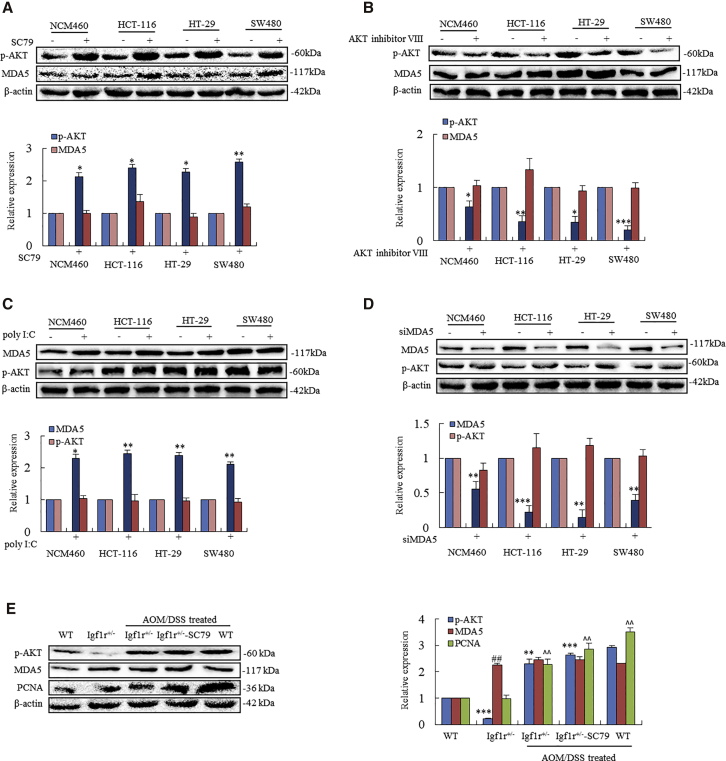
Knockdown of IGF-1R-Triggered MDA5 and RIG-I Might Be Independent of the PI3K-Akt Pathway
To investigate the association of IGF-1R knockdown-triggered MDA5 and RIG-I to the PI3K-Akt pathway, we compared the expression levels of MDA5 in the cell lines with high p-AKT induced by SC79 to the same cell lines without SC79. SC79 at 10 μM for 1.5 h significantly increased p-AKT in NCM460 cells and HCT-116, HT-29, and SW480 cell lines. At the same time, significant changes in MDA5 were not seen in both of these cell lines as compared to the cell lines without SC79 (Figure 7A). Significant changes in MDA5 were also not seen in the cell lines with knockdown AKT by 5 μM AKT inhibitor VIII (Figure 7B). On the other hand, we compared the expression levels of p-AKT in the cells with high MDA5 induced by poly(I:C) to the same cell lines without poly(I:C). There was no significant difference between the level of p-AKT in the cell lines treated with poly(I:C) and that of the cell lines without poly(I:C) treatment (Figure 7C). Downregulation of MDA5 by siMDA5 did not affect the expression of p-AKT (Figure 7D).
Figure 7.
Knockdown of IGF-1R-Triggered MDA5 and RIG-I Might Be Independent of the PI3K-Akt Pathway
(A) Human colonic epithelial (NCM460) cells and colonic cancer cells HCT-116, HT-29, and SW480 were exposed to 10 μM SC79 for 1.5 h. SC79 increased the levels of p-AKT in these cell lines (*p < 0.05, **p < 0.01 between the cells without and with SC79). Significant changes of MDA5 were not seen in these cell lines as compared to the cell lines without SC79 (p < 0.05). (B) Cells were treated with AKT inhibitor VIII (5 μM) for 1.5 h and then the levels of p-AKT and MDA5 were determined. Levels of p-AKT were significantly reduced (*p < 0.05, **p < 0.01, ***p < 0.001 between the cells without and with AKT inhibitor VIII). Significant changes of MDA5 were not seen as compared to the cells without AKT inhibitor VIII (p < 0.05). (C) Cells were treated by poly(I:C) (200 ng/mL) for 24 h and then levels of MDA5 and p-AKT were analyzed. The level of MDA5 was increased by poly(I:C) (*p < 0.05, **p < 0.01 between the cells without and with poly(I:C)). Significant changes of p-AKT were not seen between the cells without and with poly(I:C) (p < 0.05). (D) Cells were treated by siMDA5 for 24 h and then the levels of p-AKT were determined (**p < 0.01, ***p < 0.001). There was no difference between levels of p-AKT expressed in the cell lines treated by poly(I:C) and that of the cell lines without poly(I:C) (p < 0.05). (E) Igf1r+/− mice and their WT littermates were exposed to AOM-DSS for inducing colorectal cancer. Mice were then treated with SC79 (0.04 mg/g, i.p.) for 2 weeks. Igf1r+/− mice exhibited lower p-AKT (***p < 0.001) and higher MDA5 (##p < 0.01) than those in WT mice (first and second rows). During tumorigenesis, AOM-DSS induced higher p-AKT in both Igf1r+/− mice and Igf1r+/− mice exposed to SC79 (**p < 0.01, ***p < 0.001 versus without AOM-DSS) and WT mice (**p < 0.01, ***p < 0.001 versus without AOM-DSS), among which WT mice exhibited the highest level of PCNA, following Igf1r+/− mice with SC79 and Igf1r+/− mice without SC79 (ˆˆp < 0.01 versus WT). A significant difference of MDA5 was not seen in Igf1r+/− mice exposed to SC79 and without SC79 (p < 0.05).
Further, Igf1r+/− mice exposed to AOM-DSS were treated with SC79 (0.04 mg/g, intraperitoneally [i.p.], for 2 weeks) for inducing p-AKT. A significant change in MDA5 was not seen in the intestinal epithelium as compared to the Igf1r+/− mice without SC79 (Figure 7E). These results suggest that knockdown of IGF-1R-triggered MDA5 and RIG-I might be independent of the PI3K-Akt pathway in Igf1r+/− mice. Knockdown of IGF-1R-induced mitochondrial apoptosis in colonic cancer cells is mediated by the upregulations of MDA5 and RIG-I.
Discussion
IGF-1R signaling has long been recognized as a driver of tumorigenesis in multiple cancers. Classically, IGF-1R has mediated tumorigenesis through the activation of two well-established oncogenic pathways: the PI3K-Akt pathway and the Ras-MAPK pathway.26, 27, 28, 29 From then on, multiple approaches have been undertaken to modulate IGF-1R activity for the aim of anticancer therapeutics. However, most clinical disclosures have been less encouraging, probably due to the complex signaling network of IGF-1R as a key convergence in physiology. A greater understanding of cancer biology around IGF-1R has thus remained necessary for investigating an effective therapeutic response through targeting IGF-1R.
In our group, the high-throughput sequencing data in human colonic cancer cells gave us a surprising result that there is a significant link between knockdown IGF-1R and increased RNA sensors MDA5 and RIG-I. The following in vivo and in vitro experiments confirmed that knockdown IGF-1R triggers MDA5- and RIG-I-mediated mitochondrial apoptosis, leading to the inhibition of colorectal cancer. Although the proapoptotic signaling pathway is also active in nonmalignant cells, these nonmalignant cells were much less sensitive to apoptosis than cancer cells.21, 23 Further, endogenous Bcl-xL could rescue nonmalignant, but not cancer, cells from MDA5- and RIG-I-mediated mitochondrial apoptosis.23 Knockdown IGF-1R-triggered MDA5 and RIG-I might preferentially mediate apoptosis in cancer cells. Previously, Besch et al.21 showed that ligation of MDA5 and RIG-I by RNA mimetics poly(I:C) and pppRNA could trigger the mitochondrial apoptosis in human melanoma cells in an IFN-independent fashion. They suggested that tumor cell killing and immunostimulation could synergize for optimal anticancer immunochemotherapy.21 In our study, the in vitro and in vivo results showed the upregulation of MDA5 in human cancer cells as well as human normal cells through the knockdown of IGF-1R, as poly(I:C) had.
Viral RNA sensors MDA5 and RIG-I belong to the DExD/H box RNA helicase family. The classical model described in the mitochondrial antiviral signaling is that RNA virus is recognized by RIG-I and MDA5 in the cytoplasm. Due to RIG-I and MDA5 containing two N-terminal caspase recruitment domains (CARDs) for relaying signal downstream, N-terminal CARDs of RIG-I and MDA5 could trigger the intracellular signaling pathways via IPS-1, which is tethered to the outer mitochondrial membrane. This interaction then could trigger a complex signal transduction cascade, culminating in the activation of transcription factors (in particular, NF-κB and IRF-3) and the production of cytokines, including class I IFNs.21, 23, 30 Interestingly, Besch’s study21 showed that ligation of viral RNA sensors RIG-I and MDA5 by delivering poly(I:C) initiated a proapoptotic signaling pathway that was independent of type I IFNs in human melanoma cells. Delivery of viral RNA mimetic might be an optimal anticancer therapy.23
Surprisingly, our study demonstrated that the knockdown of IGF-1R could trigger viral RNA sensors MDA5 and RIG-I as poly(I:C) and pppRNA had. IGF-1R knockdown-triggered MDA5 and RIG-I also mediated the IFN-independent mitochondrial apoptosis as poly(I:C) and pppRNA had. Our results further showed that the activation of RIG-I and MDA5 by IGF-1R knockdown could dramatically increase the expression of proapoptotic BH3-only protein Bim; the apoptosomes cytochrome c, Apaf-1, and caspase-9; and the critical executioner of mitochondrial apoptosis caspase-3, thereby leading to a strong inhibition of cancer growth in Igf1r+/− mice. We thus suggest that IGF-1R knockdown-triggered MDA5 and RIG-I mediated the apoptosis of cancer cells through the mitochondrial pathway.
More surprisingly, knockdown IGF-1R also triggered MDA5 and RIG-I expressions in human nonmalignant cells. Because nonmalignant cells exhibited lower levels of MDA5 and RIG-I than cancer cells, IGF-1R knockdown-triggered MDA5- and RIG-I-mediated apoptosis might lead to preferential tumor cell death. Our further in vitro and in vivo studies revealed that IGF-1R knockdown-triggered MDA5 and RIG-I was independent of the PI3K-Akt pathway. We thus suggest a new signal transduction pathway of IGF-1R in colonic tumorigenesis. Knockdown of IGF-1R-induced mitochondrial apoptosis in colonic cancer cells is mediated by upregulations of MDA5 and RIG-I. Targeting IGF-1R to trigger MDA5 and RIG-I might have therapeutic potential for cancer treatment. In addition, the upregulations of MDA5 and RIG-I in human normal cells provide us some clues in antivirus research that targeting IGF-1R might play roles in infected cells against the virus through triggering MDA5 and RIG-I. However, the mechanisms of IGF-1R knockdown-triggered MDA5 and RIG-I still have to be uncovered. The project of investigating the signaling pathways between IGF-1R and MDA5 and RIG-I is currently underway.
In conclusion, we have identified a new pathway of IGF-1R in its signal transduction such that knockdown of IGF-1R triggers viral RNA sensors MDA5 and RIG-I in both cancer cells and normal cells. IGF-1R knockdown-triggered MDA5 and RIG-I mediate mitochondrial apoptosis in cancer cells. We anticipate that targeting IGF-1R to trigger MDA5 and RIG-I might have therapeutic potential for cancer treatment.
Materials and Methods
Igf1r+/− Mice and Care
The Committee on the Ethics of Animal Experiments of Capital Medical University approved the animal experiments and protocols (permit AEEI-2016-043). We inactivated the IGF-1R gene by homologous recombination by using the Cre-lox strategy to delete the essential exon 3 of gene, according to the reference.22 Igf1r+/− mice were generated by Cyagen Biosciences (Guangzhou, China). Igf1r+/− mice and their WT littermates were caged under a controlled room temperature, humidity, and light (12-h-light/12-h-dark cycle), and they were allowed unrestricted access to standard mouse chow, containing 52% carbohydrate, 12% fat, 23% protein, 4% fiber, 6% ash, and 3% moisture, and tap water.31
Mouse Colorectal Cancer Model
Igf1r+/− mice (both male and female, 6 weeks of age) were exposed to AOM-DSS for inducing colorectal cancer, as described.25, 32 Mice were intraperitoneally injected with 10 mg/kg AOM (Sigma), followed by three rounds of oral 1.0% DSS (MP Biomedicals). Their WT littermates were also used for all the experiments to ensure the same genetic background. At 20 weeks of age, mice were sacrificed, and colorectal cancer was analyzed by counting tumors, histopathology, and IHC assays.
Histopathology, IHC Staining, and TUNEL Analysis
Routine H&E staining of 4-μm-thick sections of colorectal tissues was used for histological, IHC, and TUNEL analyses. IHC assay was performed using antibodies against MDA5 (ab79055, Abcam), RIG-I (ab45428, Abcam), IGF-1R (ab39398, Abcam), PCNA (ab29, Abcam), and Ki-67 (ab15580, Abcam). TUNEL assay was performed by using in situ cell death detection kit (Roche, Germany).
Cell Lines, Cell Culture, and Treatment
Human colonic cancer cell lines HT-29, SW480, and HCT-116 and human colonic epithelial cell line NCM460 were purchased from China Cell Bank (authorized by ATCC). Cells were grown in RPMI 1640 medium containing 10% fetal bovine serum at 37°C in a humid atmosphere (5% CO2). Cells grown in 3.5-cm dishes were routinely transfected with siIGF-1R (5′-GCCGACACUACUACUACAATT-3′),33 siMDA5 (5′-GUAUCGUGUUAUUGGAUUA-3′),21 (Biotend, China), poly(I:C) (Amersham Biosciences), AKT activator SC79 (MedChemExpress [MCE], China), and AKT inhibitor VIII (MCE, China), respectively. The protein and cell proliferation were analyzed 48 h after transfection.
RNA Extraction and qRT-PCR
Total RNA was extracted by using TRIzol reagent in accordance with the manufacturer’s guide (Invitrogen). Reverse transcription of total miRNA was performed by using miScript reverse transcription kit (QIAGEN), according to the manufacturer’s protocol. Reverse transcribed with First Strand cDNA Synthesis kit (Toyobo, Japan). qPCR assay was performed with QuantiTect SYBR Green PCR kit (QIAGEN) in Light Cycler 480 apparatus (Roche). Real-time PCR reaction was performed in an ABI 7500 Fast Real-Time PCR System (Applied Biosystems). Primers were as follows: RIG-I-F, 5′-GACGGTCAGCAGGGTGTACT-3′; RIG-I-R, 5′-CCCGTGTCCTAACGAACAGT-3′; MDA5-F, 5′-AGAGCCCGTCCAAAGTGAAGT-3′; MDA5-R, 5′-GTTCAGCATAGTCAAAGGCAGGTA-3′;34 IGF-1R-F, 5′-TTAAAATGG CCAGAACCTGAG-3′; and IGF-1R-R, 5′-ATTATAACCAAGCCTCCCAC-3′.35
Immunofluorescence Analysis
Mouse colorectal tissues and human colonic cancer cells were routinely used to perform immunofluorescence analysis. The primary antibodies were anti-IGF-1R (ab39398, Abcam) and anti-MDA5 (ab79055, Abcam). After staining with Alexa488-labeled anti-rabbit (CA11008S, Invitrogen) or Alexa594-labeled chicken anti-goat (1387734, Invitrogen), cell nuclei were counterstained with DAPI (Sigma). Images were acquired under a confocal laser-imaging system (TCS SP8, Leica).
Annexin V-FITC-PI Staining Assay
Human colonic cancer cells were transfected with siIGF-1R, siMDA5, and poly(I:C), respectively, and their corresponding NC for 48 h. Cells were collected, and then we performed Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining assay by using a cell apoptosis detection kit (BD Biosciences). Apoptotic cells were estimated in a flow cytometer (BD Biosciences).
MMP Staining Analysis
Fluorescent probe JC-1 was used to determine MMP, which specifically to mitochondria is incorporated into the mitochondrial membrane. Human colonic cancer cells cultured in slides were stained with 5 μM JC-1 (Beyotime, China) in the dark at 37°C for 10 min and washed with PBS 3 times. Fluorescence was analyzed under a fluorescence microscope by using ImagePro Plus software.
Western Blotting Analysis
Western blotting assay was routinely performed by using antibodies against IGF-1R (ab39398, Abcam), MDA5 (ab79055, Abcam), RIG-I (ab45428, Abcam), Bim (ab32158, Abcam), cytochrome c (ab13575, Abcam), Apaf-1 (ab2000, Abcam), p-AKT (4060, Cell Signaling Technology), PCNA (13110, Cell Signaling Technology), caspase-3 (9662, Cell Signaling Technology), caspase-9 (9504, Cell Signaling Technology), and β-actin (Sigma). Immunoreactive products were visualized under a Fluorchem FC3 system by chemiluminescence (Millipore) and quantified by densitometry analyzed by using Alphaview software. Densitometric analyses of bands were normalized with β-actin functioning as a loading control.
Statistical Analysis
Data were described as mean ± SD. Statistical analysis was done with SPSS/Win19.0 software (SPSS, Chicago, IL). Comparison between Igf1r+/− and WT mice was conducted by two-tailed Student’s t test. A p value less than 0.05 was considered statistically significant.
Author Contributions
S.-Q.W. and X.-Y.Y. performed all experiments and wrote the manuscript. X.-F.Y. analyzed the data. S.-X.C. provided important intellectual contributions to the final draft. X.-J.Q. conceived and designed the study.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of China (91629303 and 81673449/81872884) and the Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission of Education (KZ201710025020 and KZ201810025033).
Contributor Information
Shu-Xiang Cui, Email: sxccui@ccmu.edu.cn.
Xian-Jun Qu, Email: quxj@ccmu.edu.cn.
References
- 1.King H., Aleksic T., Haluska P., Macaulay V.M. Can we unlock the potential of IGF-1R inhibition in cancer therapy? Cancer Treat. Rev. 2014;40:1096–1105. doi: 10.1016/j.ctrv.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasprzak A., Kwasniewski W., Adamek A., Gozdzicka-Jozefiak A. Insulin-like growth factor (IGF) axis in cancerogenesis. Mutat. Res. Rev. Mutat. Res. 2017;772:78–104. doi: 10.1016/j.mrrev.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Vigneri P.G., Tirrò E., Pennisi M.S., Massimino M., Stella S., Romano C., Manzella L. The insulin/IGF system in colorectal cancer development and resistance to therapy. Front. Oncol. 2015;5:230. doi: 10.3389/fonc.2015.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Girnita L., Worrall C., Takahashi S., Seregard S., Girnita A. Something old, something new and something borrowed: emerging paradigm of insulin-like growth factor type 1 receptor (IGF-1R) signaling regulation. Cell. Mol. Life Sci. 2014;71:2403–2427. doi: 10.1007/s00018-013-1514-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson A., Petnga W., Macaulay V.M., Weyer-Czernilofsky U., Bogenrieder T. Insulin-like growth factor (IGF) pathway targeting in cancer: role of the IGF axis and opportunities for future combination studies. Target. Oncol. 2017;12:571–597. doi: 10.1007/s11523-017-0514-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Q., Zhou Y., Ying K., Ruan W. IGFBP, a novel target of lung cancer? Clin. Chim. Acta. 2017;466:172–177. doi: 10.1016/j.cca.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Crudden C., Girnita A., Girnita L. Targeting the IGF-1R: the tale of the tortoise and the hare. Front. Endocrinol. (Lausanne) 2015;6:64. doi: 10.3389/fendo.2015.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christopoulos P.F., Corthay A., Koutsilieris M. Aiming for the Insulin-like Growth Factor-1 system in breast cancer therapeutics. Cancer Treat. Rev. 2018;63:79–95. doi: 10.1016/j.ctrv.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Chen H.X., Sharon E. IGF-1R as an anti-cancer target--trials and tribulations. Chin. J. Cancer. 2013;32:242–252. doi: 10.5732/cjc.012.10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones R.A., Campbell C.I., Wood G.A., Petrik J.J., Moorehead R.A. Reversibility and recurrence of IGF-IR-induced mammary tumors. Oncogene. 2009;28:2152–2162. doi: 10.1038/onc.2009.79. [DOI] [PubMed] [Google Scholar]
- 11.Jin M., Buck E., Mulvihill M.J. Modulation of insulin-like growth factor-1 receptor and its signaling network for the treatment of cancer: current status and future perspectives. Oncol. Rev. 2013;7:e3. doi: 10.4081/oncol.2013.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asahina Y., Izumi N., Hirayama I., Tanaka T., Sato M., Yasui Y., Komatsu N., Umeda N., Hosokawa T., Ueda K. Potential relevance of cytoplasmic viral sensors and related regulators involving innate immunity in antiviral response. Gastroenterology. 2008;134:1396–1405. doi: 10.1053/j.gastro.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Moghaddas F., Masters S.L. Monogenic autoinflammatory diseases: Cytokinopathies. Cytokine. 2015;74:237–246. doi: 10.1016/j.cyto.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barral P.M., Sarkar D., Su Z.Z., Barber G.N., DeSalle R., Racaniello V.R., Fisher P.B. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharmacol. Ther. 2009;124:219–234. doi: 10.1016/j.pharmthera.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao Y., Wan Q., Yang C., Su J. Grass carp laboratory of genetics and physiology 2 serves as a negative regulator in retinoic acid-inducible gene I- and melanoma differentiation-associated gene 5-mediated antiviral signaling in resting state and early stage of grass carp reovirus infection. Front. Immunol. 2017;8:352. doi: 10.3389/fimmu.2017.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gitlin L., Barchet W., Gilfillan S., Cella M., Beutler B., Flavell R.A., Diamond M.S., Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer C.R., Jacobson M.E., Fedorova O., Pyle A.M., Wilson J.T. Environmentally triggerable retinoic acid-inducible gene I agonists using synthetic polymer overhangs. Bioconjug. Chem. 2018;29:742–747. doi: 10.1021/acs.bioconjchem.7b00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y.M., Gale M., Jr., Akira S. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 20.Hüsser L., Alves M.P., Ruggli N., Summerfield A. Identification of the role of RIG-I, MDA-5 and TLR3 in sensing RNA viruses in porcine epithelial cells using lentivirus-driven RNA interference. Virus Res. 2011;159:9–16. doi: 10.1016/j.virusres.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Besch R., Poeck H., Hohenauer T., Senft D., Häcker G., Berking C., Hornung V., Endres S., Ruzicka T., Rothenfusser S., Hartmann G. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J. Clin. Invest. 2009;119:2399–2411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holzenberger M., Dupont J., Ducos B., Leneuve P., Géloën A., Even P.C., Cervera P., Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 23.Zitvogel L., Kroemer G. Anticancer immunochemotherapy using adjuvants with direct cytotoxic effects. J. Clin. Invest. 2009;119:2127–2130. doi: 10.1172/JCI39991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue B., Zhang Y.S., Xu H.M., Zhao C.R., Li Y.Y., Qin Y.Z., Wang R.Q., Sun D., Yuan Y., Lou H.X., Qu X.J. Riccardin D-26, a synthesized macrocyclic bisbibenzyl compound, inhibits human hepatocellular carcinoma growth through induction of apoptosis in p53-dependent way. Cancer Lett. 2013;328:104–113. doi: 10.1016/j.canlet.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Parang B., Barrett C.W., Williams C.S. AOM/DSS model of colitis-associated cancer. Methods Mol. Biol. 2016;1422:297–307. doi: 10.1007/978-1-4939-3603-8_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liefers-Visser J.A.L., Meijering R.A.M., Reyners A.K.L., van der Zee A.G.J., de Jong S. IGF system targeted therapy: Therapeutic opportunities for ovarian cancer. Cancer Treat. Rev. 2017;60:90–99. doi: 10.1016/j.ctrv.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Mutgan A.C., Besikcioglu H.E., Wang S., Friess H., Ceyhan G.O., Demir I.E. Insulin/IGF-driven cancer cell-stroma crosstalk as a novel therapeutic target in pancreatic cancer. Mol. Cancer. 2018;17:66. doi: 10.1186/s12943-018-0806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodhia K.A., Tienchaiananda P., Haluska P. Understanding the key to targeting the IGF axis in cancer: a biomarker assessment. Front. Oncol. 2015;5:142. doi: 10.3389/fonc.2015.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brouxhon S.M., Kyrkanides S., Teng X., Athar M., Ghazizadeh S., Simon M., O’Banion M.K., Ma L. Soluble E-cadherin: a critical oncogene modulating receptor tyrosine kinases, MAPK and PI3K/Akt/mTOR signaling. Oncogene. 2014;33:225–235. doi: 10.1038/onc.2012.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong X.Y., Liu W.J., Zhao M.Q., Wang J.Y., Pei J.J., Luo Y.W., Ju C.M., Chen J.D. Classical swine fever virus triggers RIG-I and MDA5-dependent signaling pathway to IRF-3 and NF-κB activation to promote secretion of interferon and inflammatory cytokines in porcine alveolar macrophages. Virol. J. 2013;10:286. doi: 10.1186/1743-422X-10-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meidenbauer J.J., Ta N., Seyfried T.N. Influence of a ketogenic diet, fish-oil, and calorie restriction on plasma metabolites and lipids in C57BL/6J mice. Nutr. Metab. (Lond.) 2014;11:23. doi: 10.1186/1743-7075-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Z.Y., Wang F., Cui S.X., Gao Z.H., Qu X.J. CXCR7/CXCR4 heterodimer-induced histone demethylation: a new mechanism of colorectal tumorigenesis. Oncogene. 2019;38:1560–1575. doi: 10.1038/s41388-018-0519-2. [DOI] [PubMed] [Google Scholar]
- 33.Kong Y.L., Shen Y., Ni J., Shao D.C., Miao N.J., Xu J.L., Zhou L., Xue H., Zhang W., Wang X.X., Lu L.M. Insulin deficiency induces rat renal mesangial cell dysfunction via activation of IGF-1/IGF-1R pathway. Acta Pharmacol. Sin. 2016;37:217–227. doi: 10.1038/aps.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen B., Hu Y., Zhang S., Zheng J., Zeng L., Zhang J., Zhu A., Wu C. Molecular characterization and expression analyses of three RIG-I-like receptor signaling pathway genes (MDA5, LGP2 and MAVS) in Larimichthys crocea. Fish Shellfish Immunol. 2016;55:535–549. doi: 10.1016/j.fsi.2016.06.032. [DOI] [PubMed] [Google Scholar]
- 35.Pfaffl M.W., Georgieva T.M., Georgiev I.P., Ontsouka E., Hageleit M., Blum J.W. Real-time RT-PCR quantification of insulin-like growth factor (IGF)-1, IGF-1 receptor, IGF-2, IGF-2 receptor, insulin receptor, growth hormone receptor, IGF-binding proteins 1, 2 and 3 in the bovine species. Domest. Anim. Endocrinol. 2002;22:91–102. doi: 10.1016/s0739-7240(01)00128-x. [DOI] [PubMed] [Google Scholar]