Abstract
CDK regulatory subunit 2 (CKS2) has a nuclear function that promotes cell division and is a candidate biomarker of chemoradioresistance in cervical cancer. The underlying mechanisms are, however, not completely understood. We investigated whether CKS2 also has a mitochondrial function that augments tumor aggressiveness. Based on global gene expression data of two cervical cancer cohorts of 150 and 135 patients, we identified a set of genes correlated with CKS2 expression. Gene set enrichment analysis showed enrichment of mitochondrial cellular compartments, and the hallmarks oxidative phosphorylation (OXPHOS) and targets of the MYC oncogene in the gene set. By in situ proximity ligation assay, we showed that CKS2 formed complex with the positively correlated MYC target, mitochondrial single-stranded DNA binding protein SSBP1, in the mitochondrion of cervix tumor samples and HeLa and SiHa cervical cancer cell lines, indicating a role in mitochondrial DNA (mtDNA) replication and thereby OXPHOS. CDK1 was found to be part of the complex. Flow cytometry analyses of HeLa cells showed cell cycle regulation of the CKS2-SSBP1 complex consistent with mtDNA replication activity. Moreover, repression of mtDNA replication and OXPHOS by acute hypoxia decreased CKS2-SSBP1 complex abundance and expression of MYC targets. By immunohistochemistry, cytoplasmic CKS2 expression was found to add to the prognostic impact of nuclear CKS2 expression in patients, suggesting that the mitochondrial function promotes tumor aggressiveness. Our study uncovers a novel link between regulation of cell division by nuclear pathways and OXPHOS in the mitochondrion that involves CKS2 and promotes chemoradioresistance of cervical cancer.
Abbreviations: CDK1, cyclin-dependent kinase 1; CKS2, CDK regulatory subunit 2; PLA, proximity ligation assay; mtDNA, mitochondrial DNA; OXPHOS, oxidative phosphorylation; SSBP1, single-stranded DNA binding protein; CCNB, cyclin B
Introduction
CDK regulatory subunit 2 (CKS2) is overexpressed and associated with aggressiveness of many human malignancies, including cancer of the uterine cervix, prostate, gastrointestinal tract, bile duct, breast, and liver [1], [2], [3], [4], [5], [6], [7], [8], [9]. In cervical cancer, high CKS2 expression has been associated with presence of lymph node metastases at diagnosis and poor survival following chemoradiotherapy [2]. Overexpression of CKS2 in cell lines from a variety of cancer types has been shown to promote cell proliferation, whereas CKS2 knockdown induced cell cycle arrest and apoptosis and inhibited cell growth, migration, and tumorigenesis in mice [1], [3], [6], [9], [10]. CKS2 has therefore been suggested as a new candidate biomarker and therapeutic target; however, a better understanding of its role in cancer pathogenesis is required before the findings can be fully exploited in the clinic [11].
Several nuclear functions of CKS2 have been suggested that might explain its role in cancer progression. The protein binds to CDKs in a complex with their activating cyclins and may facilitate transition from G2- to M-phase of the cell cycle by stimulating activation of the CDK1-cyclin B (CCNB) complex [12], [13]. CKS2 has further been shown to be chromatin bound and might be involved in transcription of the CDK1 and CCNB genes [14]. In addition, binding of CKS2 to CDK2 may allow cells to override the intra-S-phase checkpoint and continue DNA replication under conditions of replicative stress [15]. In 2010, Radulovic and coworkers [16] proposed that CKS2 also may have a mitochondrial function in complex with CDK1 and the mitochondrial single-stranded DNA-binding protein SSBP1, which is a protein required for stabilization of single-stranded DNA during mitochondrial DNA (mtDNA) replication [17]. The complex was, however, detected in a pulldown experiment on lysed lymphoma cells and in a binding assay using recombinant protein expressed in bacteria, and not demonstrated in cells in situ [16]. Although of high clinical relevance, the mitochondrial function has not been further pursued.
Global gene expression data of patient cohorts are useful for predicting cellular compartments and tumor phenotypes associated with upregulation of candidate cancer genes. To verify and add information to such large scale studies, microscopy-based techniques for visualization of protein–protein complexes in situ are essential tools [18]. In the present work, we combined gene expression analysis of two patient cohorts with protein–protein complex studies using the in situ proximity ligation assay (PLA) [19] in cell lines and tumor samples. The aim was to investigate whether CKS2 has a significant mitochondrial role in cervical cancer and how it may affect tumor phenotype. Our approach reveals a major role of CKS2 in mtDNA replication in complex with SSBP1 that represents a novel link between oxidative phosphorylation (OXPHOS) and rapid cell division in chemoradioresistant cervical cancer.
Materials and Methods
Patients and Tumor Samples
A total of 294 patients with locally advanced cervical carcinoma were included. Tumor samples were collected before the start of treatment of all patients. Samples of 285 patients (cohorts 1 and 2; Table S1) were snap frozen, stored at −80°C, and used for gene expression analysis. Samples of 50 patients, including 41 patients in cohort 1, were fixed in 4% buffered formalin and used for immunohistochemistry. All patients received radiotherapy combined with adjuvant cisplatin and followed up as described [20]. The clinical protocol was approved by The Regional Committee for Medical and Health Research Ethics in Southeast Norway (S-01129). The study was performed in accordance with the Declaration of Helsinki, and written informed consent was achieved from all patients.
Cell Cultures and Hypoxia Exposure
HeLa and SiHa cervical cancer cell lines were purchased from the American Type Culture Collection and cultivated in Dulbecco's modified Eagle medium with GlutaMAX (Life Technologies, Carlsbad, CA), as described [20]. Correct identity of the cell lines was confirmed by STR/DNA profiling using PowerPlex 21 (Promega, Madison, WI) by Eurofins Genomics (Ebersberg, Germany). Hypoxia treatment was performed in a humidified In Vivo2200 chamber (Ruskinn Technology Ltd., Brigend, UK). Exponentially growing cells were plated and incubated for 24 hours prior to hypoxia exposure (0.2% O2, 5% CO2, 95% N2) for 24 hours at 37°C. Control cells were grown under normoxia (95% air, 5% CO2) in parallel experiments.
Gene Expression Data Sets
The gene expression data of all 285 patients and normoxia- and hypoxia-treated cell lines have been published earlier [20]. In brief, gene expression profiling was performed using two different Illumina bead array versions (Illumina Inc., San Diego, CA). The patients were divided into two cohorts based on array version;: cohort 1 with 150 patients assayed by WG-6 v3 arrays and cohort 2 with 135 patients assayed by HT-12 v4 arrays (Table S1). Quantile normalized, log2-transformed data were used in the analyses. The gene expression data are available in the Gene Expression Omnibus, accession number GSE72723.
Immunofluorescence
Cells were seeded on sterilized cover slips and fixed in 10% neutral buffered formalin (Sigma Aldrich, St. Louis, MO) at room temperature and/or in ice cold methanol (−20°C) for 10 minutes. Cells were then washed with PBS, blocked, and permeabilized for 15 minutes in PBS containing 1% bovine serum albumin and 0.5% Triton X-100 (PBS-AT) and incubated for 2 hours at room temperature with the primary antibodies anti-CKS2 (mouse, 37-0300; Life Technologies), anti-CDK1 (mouse, sc-54; Santa Cruz Biotechnology, Santa Cruz, CA), anti-CDK1 (rabbit, LS-C123204; Lifespan Bioscience, Seattle, WA), or anti-SSBP1 (rabbit, HPA002866; Sigma-Aldrich) diluted in PBS-AT. The cover slips were washed with PBS and incubated with secondary antibody DyLight 549 donkey anti-mouse or Dylight 488 donkey anti-rabbit (Jackson ImmunoResearch, West Grove, PA). DNA was stained with 0.6 μg/ml Hoechst 33258. The cover slips were washed, air-dried, and mounted on slides using Prolong Gold antifade mountant (P36930; Life Technologies). The specificity of the SSBP1 antibody for mitochondria was confirmed by showing its colocalization with a mitochondria marker (Figure S1).
Microscopy was performed using an AxioImager Z1 ApoTome microscope system (Carl Zeiss, Jena, Germany) equipped with plan apochromat lenses (100×/NA1.40; 63×/NA1.40) and an AxioCam MRm camera. Images were presented as maximal projection of z-stacks using AxioVision 4.8.2 (Carl Zeiss).
Immunohistochemistry
Bright-field immunohistochemistry of CKS2, CDK1, and SSBP1 was performed on tumor sections from patients to assess cellular protein distributions (2 patients) and, for CKS2, relationship to gene expression (41 patients) and treatment outcome (47 patients; Table S1). Paraffin-embedded sections were deparaffinized and stained using the Dako EnVision+ system, as described [2], using the same antibodies as for immunofluorescence. Immunostaining of CKS2 in the cytoplasm and nucleus was scored separately and together on a three-tiered scale for both intensity (absent or weak, 1; moderate, 2; strong, 3) and extent of staining (percentage of positive tumor cells <10%, 1; 10%-50%, 2; >50%, 3) [2]. The scores of intensity and extent were multiplied to give a composite score ranging from 1 to 9 for each cellular compartment and tumor.
PLA
Duolink in situ PLA kit (Olink Bioscience, Uppsala, Sweden) was used for detection of protein–protein complexes, according to the manufacturer's description and our earlier studies [21]. The assay is based on simultaneous binding of two antibodies against each of the proteins under investigation and utilizes a proximity ligation technique coupled to an amplification reaction to detect the antibodies whenever they are less than 30 nm apart ;, i.e., when the proteins most likely are in the same complex. Because of a powerful amplification technique, each individual complex can be visualized as a distinct spot in histological sections by microscopy and quantified by image analysis or flow cytometry [19], [22].
The samples were pretreated with respect to fixation, antigen retrieval, and/or permeabilization according to the immunofluorescence protocol. All samples were blocked in Duolink blocking buffer at 37°C and incubated for 2 hours in room temperature with two primary antibodies, diluted in Duolink antibody diluent, using the same antibodies as for immunofluorescence. CDK1 rabbit antibody (LS-C123204; Lifespan Bioscience) and mouse antibody (sc-54; Santa Cruz Biotechnology) were combined with the mouse and rabbit antibodies of CKS2 and SSBP1, respectively. The samples were incubated for 1 hour at 37°C with secondary antibody PLA minus and plus probes for mouse and rabbit, and thereafter with ligation solution for 30 minutes to hybridize the two PLA probes together. An amplification solution with fluorophore labeled nucleotides was added for 100 minutes at 37°C for fluorescence detection (cell lines). For bright field microscopy (cell lines, four patient tumors), the amplification solution, containing horseradish peroxidase–labeled nucleotides and horseradish peroxidase substrate (NovaRED), was added for 120 minutes at 37°C.
For adherent cells on cover slips, Duolink mounting medium with 4′,6-diamidino-2-phenylindole nuclear stain was applied, and microscopy was performed as described for immunofluorescence. Number of PLA foci per cell was quantified in maximal projection of z-stacks images using BlobFinder software v3.2 [23] based on approximately 100 randomly selected interphase cells for each foci type (Figure S2). Suspension cells were stained for DNA with cell cycle 633 and analyzed by use of BD LSRII flow cytometer (BD Bioscience, Mountain View, CA), as described [24]. Median of the PLA histogram, as quantified in FlowJo (Tree Star Inc., Ashland, OR), was used as a measure of PLA signal. Histological sections for bright microscopy were mounted with Duolink Brightfield Mounting Medium (Olink Bioscience). Technical controls omitting primary antibodies or containing secondary antibody PLA probes combined with only one of the primary antibodies exhibited only weak or no signals (Figure S3).
Statistics
Spearman's rank test with Benjamini-Hochberg correction for multiple testing [25] was applied in correlation analyses of patient gene expression data using R version 3.1.1 [26]. Enrichment of cellular compartments and hallmarks in gene lists was searched for by using the Gene Set Enrichment Analysis (GSEA) tool with the cellular component ontology annotation and 50 hallmark gene sets from the Molecular Signatures Database [27]. Kaplan-Meier survival curves were compared with log-rank test using IBM Statistics 21 for Windows (SPSS Inc., Chicago, IL). Mann-Whitney U test or Student's t test was applied to compare data between two groups using SigmaPlot software version 12.5 (Systat software, Erkrath, Germany).
Results
Mitochondrial Compartments and Hallmarks are Associated with High CKS2 Expression in Patient Tumors
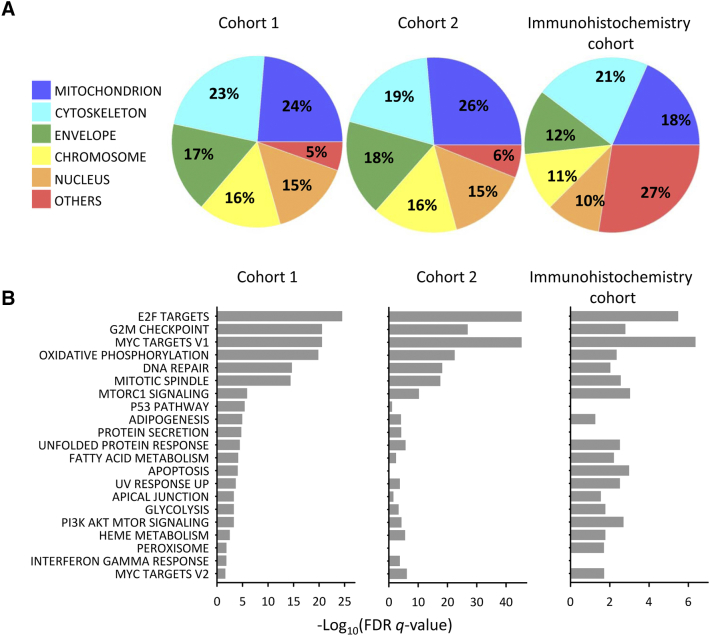
The transcriptional program associated with high CKS2 expression was determined by correlation analyses based on the global gene expression data of cohort 1 and cohort 2 patients. GSEA analysis of the 2000 most strongly correlated genes [false discovery rate (FDR) q < 0.01] was performed for the two cohorts separately to identify cellular compartments and hallmarks enriched in the program. Ontology terms representing mitochondrion, including mitochondrion, and mitochondrial part, envelope and matrix, were the most significant terms in both cohorts (Figure 1A; Table S2A). Among the other top 20 terms were nucleus, chromosome, envelope, and cytoskeleton. The genes annotated to mitochondrial terms showed only minor overlap of 4 (1.7%; cohort 1) and 10 (3.5%; cohort 2) genes with those annotated to nuclear terms (nucleolus, nucleoplasm part, nuclear body).
Figure 1.
Characterization of the transcriptional program associated with CKS2 expression in patients. Cellular compartments (A) and hallmarks (B) enriched in the transcriptional program based on 150 cohort 1 patients, 135 cohort 2 patients, and 41 patients in the immunohistochemistry cohort. (A) Percentage of unique genes annotated to the cellular compartments is presented. The cellular component ontology terms included in each compartment are defined in Table S2A.
The most significant hallmarks in both cohorts were cell cycle (E2F targets, G2M checkpoint, mitotic spindle), targets of the MYC oncogene, OXPHOS, and DNA repair (Figure 1B; Table S2B). These results were consistent with the enriched cellular compartments since cell cycle regulation and DNA repair occur to a large extent in the nucleus, OXPHOS occurs in the mitochondrion, and MYC regulates both mitochondrial and nuclear pathways. In addition, other metabolic hallmarks including fatty acid metabolism and glycolysis were significant (Figure 1B).
GSEA analysis was also performed on 2000 genes correlated with CKS2 protein expression (nominal P < .036) of 41 patients in the immunohistochemistry cohort to ensure that the above associations were valid for the protein. The score for cytoplasmic and nuclear immunostaining together was used, and this score was significantly correlated to CKS2 gene expression (rho = 0.31; P = .047). Results similar to those for CKS2 gene expression were achieved, showing mitochondrion as the second most significant cellular compartment and MYC targets and OXPHOS among the top five hallmarks (Figure 1, A and B; Table S2, A and B). All together, these results are consistent with the known nuclear functions of CKS2 and support a new, significant role in the mitochondrion.
SSBP1 Is a Strong Candidate for Connecting CKS2 to the Mitochondrial Hallmarks OXPHOS and MYC Targets
Among the cell cycle genes positively correlated with CKS2 expression in both cohorts 1 and 2 were CDK1 and CCNB1 (Table 1). The OXPHOS hallmark included positively correlated genes encoding subunits of all complexes (I-V) in the electron transfer chain (Tables 1, S3). Of relevance for OXPHOS was also a strong positive correlation of genes in mtDNA replication and transcription, like SSBP1 and mitochondrial transcription factor TFAM, and in the tricarboxylic acid cycle, like succinate-CoA ligase SUCLA2 (Table 1). MYC and several of its targets encoding mitochondrial proteins were also among the positively correlated genes (Tables 1, S3). MYC has been shown to drive proliferation of tumor cells through a positive effect on both glycolysis and OXPHOS and regulation of fatty acid metabolism [28]. Increased OXPHOS and MYC activation was therefore characteristic phenotypes of tumors with high CKS2 expression.
Table 1.
Selected CKS2 Correlating Genes⁎
| Gene | Probe ID | Description | Cohort 1 |
Cohort 2 |
||
|---|---|---|---|---|---|---|
| FDR q | rho | FDR q | rho | |||
| Nuclear and mitochondrial function | ||||||
| CDK1 | ILMN_1747911 | Cyclin-dependent kinase 1 | 3.5×10−9 | 0.54 | 1.1×10−13 | 0.64 |
| CCNB1 | ILMN_1712803 | Cyclin B1 | 0.0007 | 0.34 | 1.3×10−6 | 0.46 |
| MYC | ILMN_2110908 | MYC proto-oncogene | 0.043 | 0.23 | 1.2×10−6 | 0.47 |
| mtDNA replication and transcription | ||||||
| SSBP1 | ILMN_1809478 | Single-stranded DNA binding protein 1, MYC target | 0.0002 | 0.37 | 3.5×10−7 | 0.50 |
| TFAM | ILMN_1715661 | Transcription factor A, mitochondrial | 0.003 | 0.31 | 0.0002 | 0.37 |
| Electron transfer chain | ||||||
| NDUFB6 | ILMN_2369924 | NADH:ubiquinone oxidoreductase subunit B6, complex I | 2.0×10−5 | 0.42 | 2.6×10−13 | 0.63 |
| SDHB | ILMN_1667257 | Succinate dehydrogenase complex iron sulfur subunit B, complex II | 0.015 | 0.26 | 0.001 | 0.32 |
| UQCRC2 | ILMN_1718853 | Ubiquinol-cytochrome c reductase core protein 2, complex III | 0.036 | 0.24 | 2.5×10−3 | 0.41 |
| COX6A1 | ILMN_1783636 | Cytochrome C oxidase subunit 6A1, complex IV | 4.7×10−4 | 0.35 | 1.5×10−8 | 0.52 |
| ATP5O | ILMN_1791332 | ATP synthase peripheral stalk subunit OSCP, complex V | 0.001 | 0.34 | 2.0×10−5 | 0.41 |
| Tricarboxylic acid cycle | ||||||
| SUCLA2 | ILMN_1660787 | Succinate-CoA ligase ADP-forming beta subunit | 9.4×10−6 | 0.43 | 2.0×10−6 | 0.45 |
Correlation coefficient rho and FDR-adjusted P value (q) from Spearman's rank correlation analysis of CKS2 versus genes in the gene expression data sets of cohorts 1 and 2.
SSBP1 appeared as one of the MYC targets (Table 1) and seemed to be a reliable target, being identified with different techniques [29], [30]. Its activity is required for replication of the mitochondrial genome [17], which consists of 37 genes encoding OXPHOS proteins [31]. SSBP1 and its role in mtDNA replication could therefore connect CKS2 to the OXPHOS and MYC targets hallmarks and were a likely partner in a mitochondrial complex with CKS2.
CKS2 Forms PLA Foci with SSBP1 and CDK1 in the Mitochondria
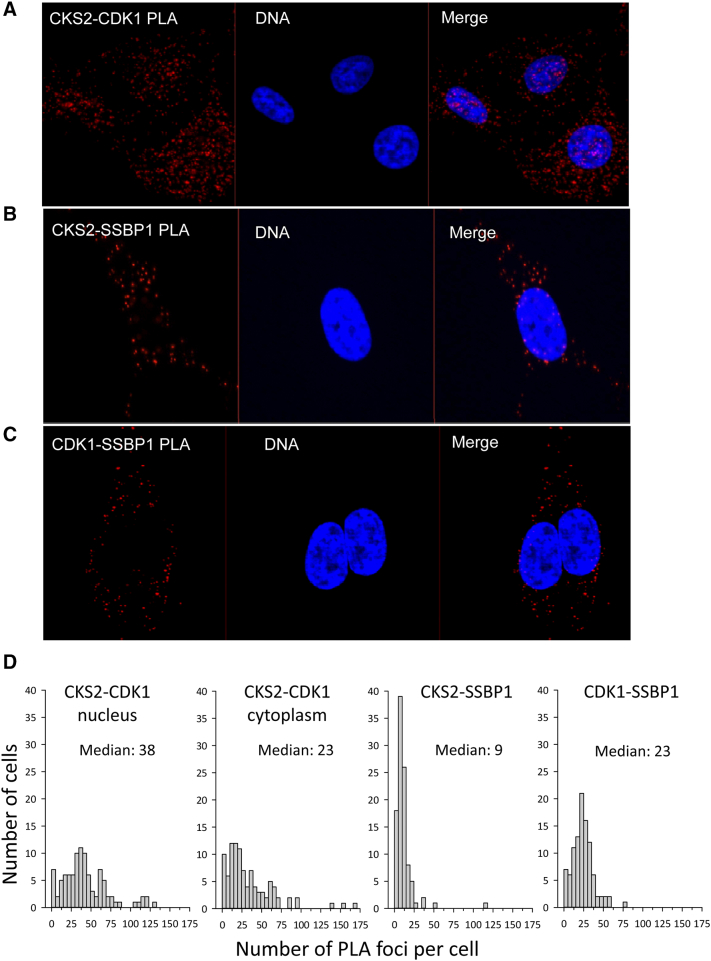
CKS2 protein was expressed in both the nucleus and cytoplasm of HeLa cells (Figure S4). CKS2 is known to bind to CDK1 in the nucleus [12], [13], [14], [32], and we first confirmed that this interaction could be detected with PLA. CDK1 showed a more diffuse expression pattern in the nucleus than CKS2 but with sparing of the nucleoli. Indeed, nuclear CKS2-CDK1 PLA foci were detected in interphase cells (Figures 2A and S5A) that disappeared from the chromatin during mitosis (Figure S5B). Quantitative image analysis showed a median number of 38 nuclear CKS2-CDK1 foci per interphase cell (Figure 2D).
Figure 2.
CKS2, CDK1, and SSBP1 protein–protein complexes in HeLa cells. Immunofluorescence images (maximal projection of z-stacks) of (A) CKS2-CDK1;, (B) CKS2-SSBP1;, and (C) CDK1-SSBP1 PLA foci in interphase cells. The PLA foci are shown in red and DNA in blue. In panel A, foci are present in the nucleus and cytoplasm. In panels B and C, foci are present in the cytoplasm only, including a few lying above the nucleus. (D) Frequency distribution showing number of PLA foci per cell for the foci types in panels A-C. A total of 100 randomly selected interphase cells in an exponentially growing cell culture were counted. For CKS2-CDK1, data for nuclear and cytoplasmic foci are shown separately.
CKS2 was further found to form PLA foci with SSBP1 in the mitochondria, with a median number of nine per interphase cell (Figure 2, B and D). CDK1-SSBP1 foci were also seen and were more abundant than the CKS2-SSBP1 foci (P < .001) with a median of 23 foci per interphase cell. In addition, CKS2 and CDK1 formed PLA foci in the same cytoplasmic regions, and the cytoplasmic CKS2-CDK1 foci were equally abundant as the CDK1-SSBP1 foci with a median of 23 foci per interphase cell (Figures 2, A and D; S5). CKS2 therefore seemed to be in a complex with SSBP1 and CDK1 in the mitochondrion. Moreover, these complexes were seen in both interphase and mitotic cells (Figure S5; data not shown).
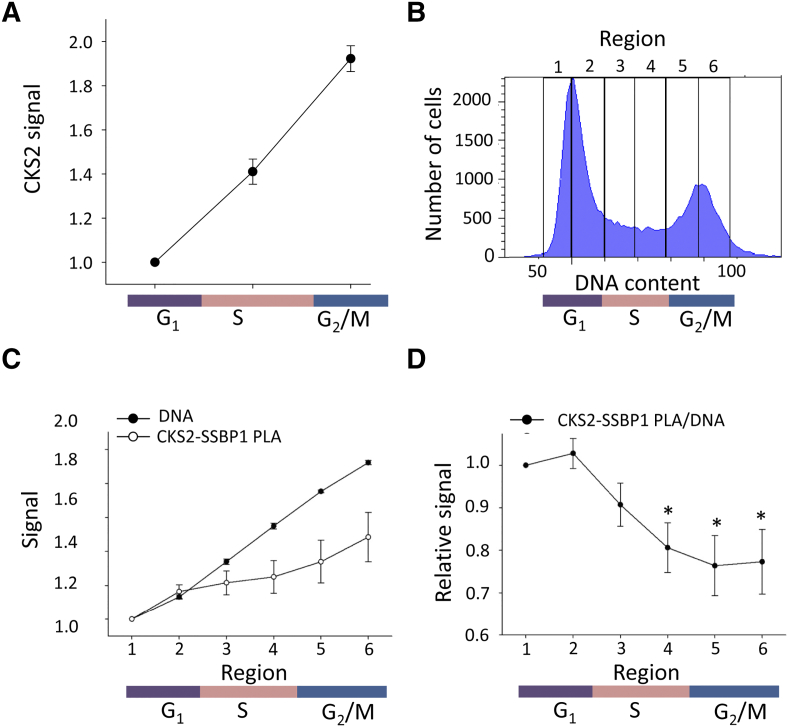
Cell Cycle Regulation of CKS2-SSBP1 Foci Abundance
In flow cytometric analysis of HeLa cells, CKS2 protein expression changed in accordance with DNA content, showing a doubling from G1- to G2-phase and therefore no cell cycle regulation (Figure 3A). MtDNA replication, however, is strictly regulated and adapted to cell cycle progression [33], [34]. To investigate whether similar regulation pattern occurred for the abundance of CKS2-SSBP1 foci, we compared the PLA signal at different times in cell cycle with total cellular DNA content. Based on the DNA histogram, the PLA signal was gated into six regions, two of each cell cycle phase, to resolve details in the regulation pattern (Figure 3B). During G1-phase (regions 1 and 2), the CKS2-SSBP1 PLA signal increased in accordance with DNA content (Figure 3C). From G1- to S-phase (regions 2-4), the signal showed only a minor increase but started to increase towards the same rate as the total DNA content in late S-phase and early G2-phase (regions 4-6; Figure 3C). Relative to total DNA, the PLA signal was highest in G1-phase, decreased significantly in S-phase, and leveled off in early G2-phase (Figure 3D). The high relative abundance of CKS2-SSBP1 foci in G1-phase and resumption in early G2 coincided with peaks of mtDNA replication [33]. The CKS2-SSBP1 foci number therefore appeared to be regulated throughout the cell cycle in a manner consistent with the known SSBP1 activity in the mitochondrion.
Figure 3.
Cell cycle regulation of CKS2 and CKS2-SSBP1 complex in HeLa cells by flow cytometry. (A) CKS2 signal in G1-, S-, and G2-phase of the cell cycle. (B) DNA histogram showing the six regions used for gating of data presented in panels C and D. (C) CKS2-SSBP1 PLA signal and DNA content in cells gated as indicated in panel B. (D) Relative CKS2-SSBP1 PLA signal in cells gated as indicated in panel B. PLA signal divided by DNA content is shown. (C, D) Data are shown relative to the first gate in G1-phase (region 1). (A, C, D) Each point represents mean value of at three (A) and five (C, D) independent experiments. The bars represent standard error of the mean. *, statistically significant difference (P < .05) compared to second region in G1-phase (region 2).
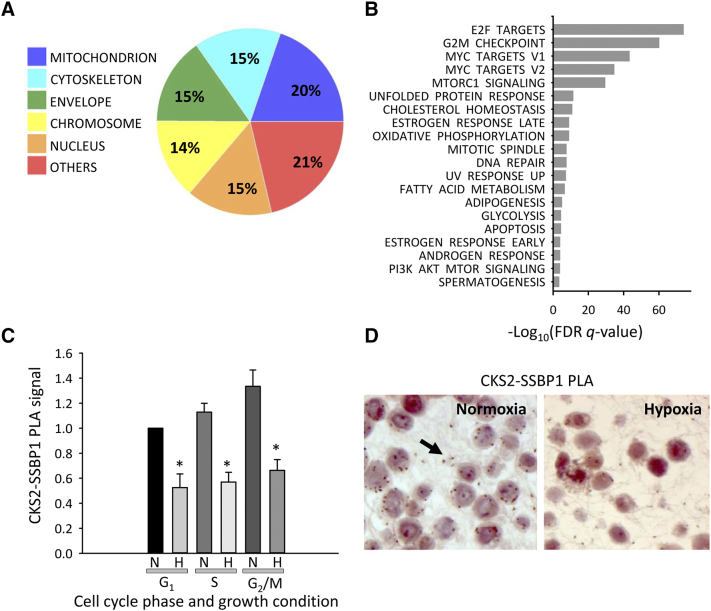
Repression of MtDNA Replication and OXPHOS
Exposure of cells to acute hypoxic stress has been shown to impair mitochondrial processes, including mtDNA replication [35]. Changes in CKS2-SSBP1 PLA signal under acute, severe hypoxia (0.2% O2, 24 hours) were measured in HeLa and SiHa cells to see whether repression of mtDNA replication had any effect on protein–protein complex abundance. In GSEA analysis of genes downregulated by a factor of 1.5 or more by hypoxia in either of the cell lines, we found significant enrichment of ontology terms representing mitochondrion and hallmarks related to mitochondrial function, including OXPHOS and MYC targets encoding mitochondrial proteins (Figure 4, A and B; Table S4, A-C), consistent with impaired mtDNA replication. Moreover, the ontology terms and hallmarks were similar to those associated with CKS2 expression in the patient data sets (Figure 1), suggesting that the same tumor phenotype was affected in the cell line experiment. A significant reduction in the CKS2-SSBP1 PLA signal by hypoxia was found in both cell lines, as demonstrated for individual cell cycle phases by flow cytometry in HeLa cells (Figure 4C) and visualized in histological sections of SiHa cells (Figure 4D), in line with a role of this complex in mtDNA replication.
Figure 4.
Gene expression and CKS2-SSBP1 complex analysis of HeLa and SiHa cells following repression of mtDNA replication by acute hypoxia. Cellular compartments (A) and hallmarks (B) enriched in a list of genes downregulated by hypoxia. (A) Percentage of unique genes annotated to the cellular compartments is presented. The cellular component ontology terms included in each compartment are defined in Table S2A. (C) CKS2-SSBP1 PLA signal in G1-, S-, and G2-phase under normoxia and hypoxia in HeLa cells measured by flow cytometry. The columns represent mean value of three independent experiments relative to the value of G1-phase normoxia. The bars represent standard error of the mean. *, statistically significant difference (P < .05) compared to normoxia. (D) CKS2-SSBP1 PLA foci (red dots) in histological sections of normoxic (left) and hypoxic (right) SiHa cells. Arrow points to PLA foci in left panel.
CKS2 in Foci with SSBP1 in Patient Tumors and in Association with Clinical Markers
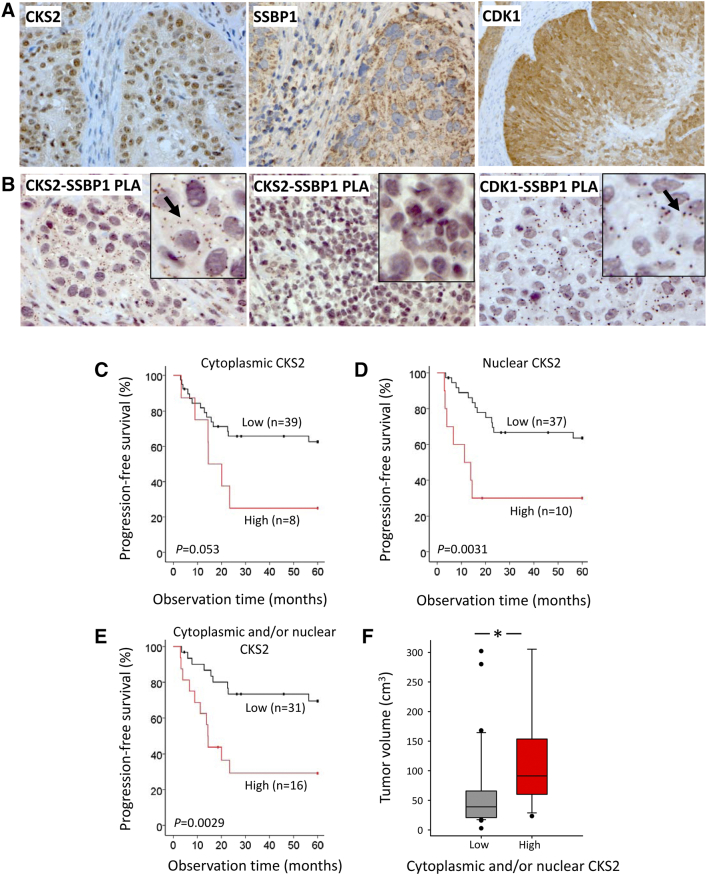
In patients, CKS2 protein was present in the nucleus and/or cytoplasm of tumor cells in about half (45%) of the 47 cases tested, SSBP1 was seen in the cytoplasm, and CDK1 was seen in the nucleus and cytoplasm (Figure 5A). Histological sections from four tumors were subjected to PLA. CKS2-SSBP1 foci were detected in the cytoplasm of tumors with high cytoplasmic CKS2 expression but not in tumors with low expression (Figure 5B). CDK1 also formed foci with SSBP1 in the cytoplasm (Figure 5B). The CKS2-SSBP1 and CDK1-SSBP1 complexes observed in cell lines therefore appeared to be present in patient tumors as well.
Figure 5.
Tumor protein–protein complexes and survival analysis in patients. (A) Cytoplasmic and nuclear CKS2 expression (left), cytoplasmic SSBP1 expression (middle), and cytoplasmic and nuclear CDK1 expression (right) by immunohistochemistry in histological sections. (B) CKS2-SSBP1 PLA foci (brown dots) in two different tumors (left, middle) and CDK1-SSBP1 PLA foci (brown dots; right). Arrow points to PLA foci in left and right inset panels. (C) Kaplan-Meier curves of progression-free survival for patients with high (composite score >0) and low (composite score of zero) cytoplasmic CKS2 expression. (D) Kaplan-Meier curves of progression-free survival for patients with high (composite score >2) and low (composite score ≤2) nuclear CKS2 expression. (E) Kaplan-Meier curves of progression-free survival for patients with high and low cytoplasmic and/or nuclear CKS2 expression. (F) Box plot of tumor volume for 31 patients with low and 16 patients with high cytoplasmic and/or nuclear CKS2 expression. *, statistically significant difference (P < .05) between the patient groups. (C-E) P value from log-rank test and number of patients are indicated.
Prognostic value of CKS2 expression was investigated for the cytoplasmic and nuclear expression separately in 47 patients. High cytoplasmic CKS2 expression was seen in eight patients. There was a clear tendency towards lower progression-free survival probability of these patients (P = .053), with a separation of 40% in survival probability between the high- and low-expression groups (Figure 5C). Comparison of patient groups based on nuclear CKS2 expression led to a highly significant (P = .0031) but somewhat smaller separation of 34% between the groups (Figure 5D). Only two patients had both high cytoplasmic and nuclear expression. The association of cytoplasmic CKS2 expression to outcome was therefore not a consequence of a correlation with nuclear expression. The strongest association to outcome (P = .0029) and a separation of 38% between the groups were seen when combining the data for the cytoplasmic and nuclear compartment (Figure 5E). Moreover, based on the combined data, patients in the high-expression group had a significantly larger tumor size than those in the low-expression group (P = .003; Figure 5F). These observations indicate that the mitochondrial function contributes to the prognostic impact of CKS2 and that the mitochondrial and nuclear function combined promotes tumor growth.
Discussion
We present evidences for a mitochondrial function of CKS2 in complex with SSPB1 in cervical cancer that involves regulation of mtDNA replication. The use of gene expression data from patient tumors led to clinically relevant predictions of cellular compartments and functions that were consistent across two independent cohorts and in analysis based on CKS2 protein expression. The global, unsupervised design enabled broad insight into the transcriptional program associated with high CKS2 expression, depicting mitochondrion as a major compartment and increased OXPHOS and MYC activation as associated tumor phenotypes. The predictions were confirmed in CKS2-SSBP1 PLA studies in cell lines, and the CKS2-SSBP1 complex was demonstrated to occur in patient tumors. The mitochondrial function adds to the known nuclear role of CKS2 in cell cycle progression. Mitochondrial activity to generate energy is known to cross talk with the cell division machinery, but the molecular connections between these processes are poorly elucidated [36]. Our results suggest that CKS2 is part of this connection, providing cancer cells with energy for rapid cell division and thereby representing a novel mechanism underlying chemoradioresistance.
PLA studies in cell lines showed that the interaction between CKS2 and SSBP1 was consistent with SSBP1 function in mtDNA replication. MtDNA replication precedes nuclear DNA synthesis in G1-phase and is reactivated towards the end of S-phase, providing energy for the G1/S and G2/M transitions [36]. By comparing the abundance of CKS2-SSBP1 foci to total DNA content, we found that the interaction was regulated throughout the cell cycle in a manner consistent with mtDNA replication activity. Moreover, OXPHOS repression decreased the abundance of CKS2-SSBP1 foci. CKS2 therefore appears to have a role in mtDNA replication in complex with SSBP1 and, thus, in regulation of key OXPHOS proteins.
Our gene expression analysis predicted MYC activation to be involved in the function of CKS2 in mtDNA replication. In particular, MYC transcriptional activity may have contributed to SSBP1 upregulation in tumors with high CKS2 expression. Moreover, OXPHOS repression in cell lines downregulated MYC targets and decreased the abundance of the CKS2-SSPB1 complex. It is therefore possible that MYC-mediated upregulation of SSBP1 promotes the formation of this complex. MYC activation has been shown to be associated with aggressive cervical cancer in previous work by us and others [37], [38], and it is tempting to speculate that the CKS2-SSBP1 complex plays a role in the MYC-related aggressive phenotype.
It is likely that CDK1 is part of the CKS2-SSBP1 complex since CDK1-SSPB1 and CKS2-CDK1 foci occurred in the same cytoplasmic regions as the CKS2-SSBP1 foci. The data of PLA foci number per cell may suggest a model of a complex consisting of the three proteins. Proteins in PLA foci can be up to 30 nm apart [18]. A higher number of CDK1-SSBP1 than CKS2-SSBP1 foci therefore points to CDK1 as a direct binding partner to SSBP1, whereas CKS2 probably interact indirectly with SSBP1 through a binding to CDK1. This is consistent with the cytoplasmic CKS2-CDK1 foci, which were more abundant than the CKS2-SSBP1 foci and equally abundant as the CDK1-SSBP1 foci. In this model, due to mobility of the proteins, CKS2 and SSBP1 will occasionally, but not always, be close enough to form PLA foci. Our model is also in accordance with the study by Radulovic and coworkers [16] showing that the CKS2-SSBP1 interaction depended on CKS2 being bound to CDK1. Regardless of the validity of this model, our data suggest that CDK1 participates in the function of CKS2 in mtDNA replication.
The function of CKS2 in mtDNA replication was suggested to be of clinical importance by visualizing CKS2-SSBP1 PLA foci in patient tumors. Moreover, high cytoplasmic CKS2 expression was shown to contribute to the prognostic impact of CKS2 in survival analysis and the relationship of CKS2 to tumor volume. Although we cannot exclude that cytoplasmic expression may reflect other roles of CKS2, this indicates that the CKS2-SSBP1 complex is important for tumor aggressiveness. Moreover, whether transient OXPHOS repression by acute hypoxia followed by reoxygenation may influence the intratumor distribution of CKS2-SSBP1 complexes and possibly the existence of aggressive subpopulations may be of high clinical relevance and requires further investigation. Our findings support the use of CKS2 as biomarker of chemoradioresistance in cervical cancer and encourage development of CKS2 targeted therapy. Mitochondrial processes, including mtDNA replication and OXPHOS, have been proposed as promising therapeutic targets [39]. In this context, CKS2 targeting might be a particularly powerful approach, interfering both directly with the cell division machinery and indirectly with the supply of energy for rapid cell proliferation.
Footnotes
Funding: The work was supported by the South-East Norway Regional Health Authority (grant no. 2015020) and the Norwegian Cancer Society (grant no. 107438-PR-2007-0179).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.01.002.
Appendix A. Supplementary data
Supplementary figures
Supplementary tables
References
- 1.Kita Y, Nishizono Y, Okumura H, Uchikado Y, Sasaki K, Matsumoto M, Setoyama T, Tanoue K, Omoto I, Mori S. Clinical and biological impact of cyclin-dependent kinase subunit 2 in esophageal squamous cell carcinoma. Oncol Rep. 2014;31:1986–1992. doi: 10.3892/or.2014.3062. [DOI] [PubMed] [Google Scholar]
- 2.Lyng H, Brovig RS, Svendsrud DH, Holm R, Kaalhus O, Knutstad K, Oksefjell H, Sundfor K, Kristensen GB, Stokke T. Gene expressions and copy numbers associated with metastatic phenotypes of uterine cervical cancer. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen DY, Zhan YH, Wang QM, Rui G, Zhang ZM. Oncogenic potential of cyclin kinase subunit-2 in cholangiocarcinoma. Liver Int. 2013;33:137–148. doi: 10.1111/liv.12014. [DOI] [PubMed] [Google Scholar]
- 4.Shen DY, Fang ZX, You P, Liu PG, Wang F, Huang CL, Yao XB, Chen ZX, Zhang ZY. Clinical significance and expression of cyclin kinase subunits 1 and 2 in hepatocellular carcinoma. Liver Int. 2010;30:119–125. doi: 10.1111/j.1478-3231.2009.02106.x. [DOI] [PubMed] [Google Scholar]
- 5.Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, Febbo PG, Balk SP. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka F, Matsuzaki S, Mimori K, Kita Y, Inoue H, Mori M. Clinicopathological and biological significance of CDC28 protein kinase regulatory subunit 2 overexpression in human gastric cancer. Int J Oncol. 2011;39:361–372. doi: 10.3892/ijo.2011.1056. [DOI] [PubMed] [Google Scholar]
- 7.Wang JI, Xu LH, Liu Y, Chen JN, Jiang H, Yang SJ, Tan H. Expression of cyclin kinase subunit 2 in human breast cancer and its prognostic significance. Int J Clin Exp Pathol. 2014;7:8593–8601. [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JJ, Fang ZX, Ye HM, You P, Cai MJ, Duan HB, Wang F, Zhang ZY. Clinical significance of overexpressed cyclin-dependent kinase subunits 1 and 2 in esophageal carcinoma. Dis Esophagus. 2013;26:729–736. doi: 10.1111/dote.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu MH, Luo Y, Qin SL, Wang ZS, Mu YF, Zhong M. Up-regulated CKS2 promotes tumor progression and predicts a poor prognosis in human colorectal cancer. Am J Cancer Res. 2015;5:2708–2718. [PMC free article] [PubMed] [Google Scholar]
- 10.Lan YS, Zhang YY, Wang JH, Lin CH, Ittmann MM, Wang F. Aberrant expression of Cks1 and Cks2 contributes to prostate tumorigenesis by promoting proliferation and inhibiting programmed cell death. Int J Cancer. 2008;123:543–551. doi: 10.1002/ijc.23548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You H, Lin H, Zhang Z. CKS2 in human cancers: clinical roles and current perspectives. Mol Clin Oncol. 2015;3:459–463. doi: 10.3892/mco.2015.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patra D, Wang SX, Kumagai A, Dunphy WG. The Xenopus Suc1/Cks protein promotes the phosphorylation of G(2)/M regulators. J Biol Chem. 1999;274:36839–36842. doi: 10.1074/jbc.274.52.36839. [DOI] [PubMed] [Google Scholar]
- 13.van Zon W, Ogink J, ter Riet B, Medema RH, Riele HT, Wolthuis RMF. The APC/C recruits cyclin B1-Cdk1-Cks in prometaphase before D box recognition to control mitotic exit. J Cell Biol. 2010;190:587–602. doi: 10.1083/jcb.200912084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinsson-Ahlzen HS, Liberal V, Grunenfelder B, Chaves SR, Spruck CH, Reed SI. Cyclin-dependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol Cell Biol. 2008;28:5698–5709. doi: 10.1128/MCB.01833-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberal V, Martinsson-Ahlzen HS, Liberal J, Spruck CH, Widschwendter M, McGowan CH, Reed SI. Cyclin-dependent kinase subunit (Cks) 1 or Cks2 overexpression overrides the DNA damage response barrier triggered by activated oncoproteins. Proc Natl Acad Sci U S A. 2012;109:2754–2759. doi: 10.1073/pnas.1102434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radulovic M, Crane E, Crawford M, Godovac-Zimmermann J, Yu VPCC. CKS proteins protect mitochondrial genome integrity by interacting with mitochondrial single-stranded DNA-binding protein. Cellular Proteomics;Mol Cell Proteomics. 2010;9:145–152. doi: 10.1074/mcp.M900078-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young MJ, Copeland WC. Human mitochondrial DNA replication machinery and disease. Curr Opin Genet Dev. 2016;38:52–62. doi: 10.1016/j.gde.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raykova D, Koos B, Asplund A, Gelleri M, Ivarsson Y, Danielson UH, Soderberg O. Let there be light! Proteomes. 2016;4 doi: 10.3390/proteomes4040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 20.Fjeldbo CS, Julin CH, Lando M, Forsberg MF, Aarnes EK, Alsner J, Kristensen GB, Malinen E, Lyng H. Integrative analysis of DCE-MRI and gene expression profiles in construction of a gene classifier for assessment of hypoxia-related risk of chemoradiotherapy failure in cervical cancer. Clin Cancer Res. 2016;22:4067–4076. doi: 10.1158/1078-0432.CCR-15-2322. [DOI] [PubMed] [Google Scholar]
- 21.Halle C, Lando M, Sundfor K, Kristensen GB, Holm R, Lyng H. Phosphorylation of EGFR measured with in situ proximity ligation assay: Relationship to EGFR protein level and gene dosage in cervical cancer. Radiother Oncol. 2011;101:152–157. doi: 10.1016/j.radonc.2011.05.052. [DOI] [PubMed] [Google Scholar]
- 22.Leuchowius KJ, Weibrecht I, Landegren U, Gedda L, Soderberg O. Flow cytometric in situ proximity ligation analyses of protein interactions and post-translational modification of the epidermal growth factor receptor family. Cytometry A. 2009;75a:833–839. doi: 10.1002/cyto.a.20771. [DOI] [PubMed] [Google Scholar]
- 23.Allalou A, Wahlby C. BlobFinder, a tool for fluorescence microscopy image cytometry. Comput Methods Programs Biomed. 2009;94:58–65. doi: 10.1016/j.cmpb.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson M, Ragnum HB, Julin CH, Yeramian A, Clancy T, Frikstad KAM, Seierstad T, Stokke T, Matias-Guiu X, Ree AH. Hypoxia-independent gene expression signature associated with radiosensitisation of prostate cancer cell lines by histone deacetylase inhibition. Br J Cancer. 2016;115:929–939. doi: 10.1038/bjc.2016.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate — a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 1995;57:289–300. [Google Scholar]
- 26.R Development Core Team Vienna, Austria: R Foundation for Statistical Computing; 2008. R: a language and environment for statistical computing. http://www.R-project.org URL. [2015]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goetzman ES, Prochownik EV. The role for Myc in coordinating glycolysis, oxidative phosphorylation, glutaminolysis, and fatty acid metabolism in normal and neoplastic Tissues. Front Endocrinol. 2018;9 doi: 10.3389/fendo.2018.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal P, Yu KB, Salomon AR, Sedivy JM. Proteomic profiling of Myc-associated proteins. Cell Cycle. 2010;9:4908–4921. doi: 10.4161/cc.9.24.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolthuis R, Clay-Farrace L, van Zon W, Yekezare M, Koop L, Ogink J, Medema R, Pines J. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 33.Chatre L, Ricchetti M. Prevalent coordination of mitochondrial DNA transcription and initiation of replication with the cell cycle. Nucleic Acids Res. 2013;41:3068–3078. doi: 10.1093/nar/gkt015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schieke SM, Mccoy JP, Finkel T. Coordination of mitochondrial bioenergetics with G(1) phase cell cycle progression. Cell Cycle. 2008;7:1782–1787. doi: 10.4161/cc.7.12.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuhrmann DC, Brune B. Mitochondrial composition and function under the control of hypoxia. Redox Biol. 2017;12:208–215. doi: 10.1016/j.redox.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salazar-Roa M, Malumbres M. Fueling the cell division cycle. Trends Cell Biol. 2017;27:69–81. doi: 10.1016/j.tcb.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 37.Halle C, Lando M, Svendsrud DH, Clancy T, Holden M, Sundfor K, Kristensen GB, Holm R, Lyng H. Membranous expression of ectodomain isoforms of the epidermal growth factor receptor predicts outcome after chemoradiotherapy of lymph node–negative cervical cancer. Clin Cancer Res. 2011;17:5501–5512. doi: 10.1158/1078-0432.CCR-11-0297. [DOI] [PubMed] [Google Scholar]
- 38.Vijayalakshmi N, Selvaluxmi G, Mahji U, Rajkumar T. c-Myc oncoprotein expression and prognosis in patients with carcinoma of the cervix: an immunohistochemical study. Eur J Gynaecol Oncol. 2002;23:135–138. [PubMed] [Google Scholar]
- 39.Dickerson T, Jauregui CE, Teng Y. Friend or foe? Mitochondria as a pharmacological target in cancer treatment. Future Med Chem. 2017;9:2197–2210. doi: 10.4155/fmc-2017-0110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary tables