Abstract
Background
Magnetic resonance imaging (MRI) is used to follow-up multiple sclerosis (MS) and evaluate disease progression and therapy response via lesion quantification. However, there is a lack of automated post-processing techniques to quantify individual MS lesion change.
Objective
The present study developed a secondary post-processing algorithm for MS lesion segmentation routine to quantify individual changes in volume over time.
Methods
An Automatic Follow-up of Individual Lesions (AFIL) algorithm was developed to process time series of pre-segmented binary lesion masks. The resulting consistently labelled lesion masks allowed for the evaluation of individual lesion volumes. Algorithm performance testing was executed in seven early MS patients with four MRI visits, and MS experienced readers verified the accuracy.
Results
AFIL distinguished 328 individual MS lesions with a 0.9% error rate to track persistent or new lesions based on expert assessment. A total of 121 new lesions evolved within the observed time period. The proportional courses of 69.1% lesions in the persistent lesion population exhibited varying volume, 16.9% exhibited stable volume, 3.4% exhibiting continuously increasing, and 0.5% exhibited continuously decreasing volume.
Conclusion
This algorithm tracked individual lesions to automatically create an individual lesion growth profile of MS patients. This approach may allow for characterization of patients based on their individual lesion progression.
Keywords: Multiple sclerosis, Therapy monitoring, Medical image analysis, Lesion tracking, Volumetric assessment, Quantitative magnetic resonance imaging
Highlights
-
•
This algorithm can be used for individual tracking of lesion volumes or can read-out parameter changes of quantitative MR images in lesions.
-
•
This method allows the characterization of patients based on their individual lesion growth profile.
-
•
In early MS most lesions had varying volume, few were stable, and very few increased or decreased continuously.
1. Introduction
Multiple sclerosis (MS) is an inflammatory demyelinating neurodegenerative disease of the human central nervous system that is characterized by the accumulation of multifocal demyelinating lesions (plaques) in white matter (WM) and grey matter (GM) of the brain and spine. These histological changes are reflected as quantifiable volumes of deviant magnetic properties in conventional magnetic resonance imaging (MRI), e.g., in T2-weighted imaging data (T2 lesions). The quantification of brain lesion load gained further importance in MS monitoring under disease-modifying therapies (DMTs). Since further (1) newly appearing or enlarging T2 lesions under DMT are accepted indicators of sub-clinical disease progression and inflammatory activity the modern concept of No Evidence of Disease Activity (NEDA) therapy monitoring has incorporated these lesion features as one of four progression markers besides (2) the brain tissue volume change as a surrogate for neurodegeneration, (3) the occurrence of clinical relapses, and, (4) the measurable clinical disability progression (Rudick et al., 2010; Stangel et al., 2015).
Several tools are available to measure lesion load change via comparison of two MRI examinations as required in NEDA, but no approaches allow for multiple time-point analyses to quantify individual lesion development and obtain a more precise perception of disease activity. Conventional MRI data post-processing analysis research recently focused on reliable, precise and reproducible automated MS lesion segmentation to quantify lesion changes and measure total brain lesion volumes. Llado et al. provided a comprehensive review on these developments (Llado et al., 2012).
Commercially available segmentation tools, such as LesionQuant™ (www.cortechslabs.com/lesionquant) and publicly available open-source software, such as the Lesion Segmentation Tool (LST; www.statistical-modelling.de/lst) detect new T2 lesions and enlarging volume lesions between two time points. However, the underlying approaches used measure the volume change in brain lesions in its entirety but do not allow for the evaluation of individual lesion development or separate analyses between multiple time points.
Further, alternative binary map-based approaches do not allow for the transfer of individual lesions to volumes-of-interest to quantify more specific MRI measures longitudinally. Newer quantitative MRI techniques that are sensitive to myelination and axon integrity revealed a distinct variability in magnetization transfer ratio (MTR) imaging (Rocca et al., 2003; Siemonsen et al., 2016), fractional anisotropy (FA) derived from diffusion tensor imaging (DTI) (Filippi et al., 2001) and myelin water fraction (MWF) (Faizy et al., 2016) within lesions. These novel in vivo insights reveal a heterogeneous MS lesion pathology (Filippi and Rocca, 2011), and the individual quantification of these changes remains a scientifically important but technically unmet need.
The aim of this work was to develop a secondary algorithm for time-series analysis for application in conjunction with map-based lesion segmentation procedures. We developed an algorithm to (1) analyse individual lesion volume development and (2) provide a read-out basis to examine individual MS lesion MRI measures and quantitative MRI properties in a multiple time-point imaging series.
2. Material and methods
The expected heterogeneous lesion longitudinal behaviour and volume development necessitated preconditions to identify different lesion courses. The algorithm considered stable, growing, or shrinking lesion volumes, as well as resolving, new, reappearing, confluent and separating lesion behavior. All algorithm developments were implemented in MATLAB® 2014b using the Image Processing Toolbox®. Required input data were pre-segmented lesions of an entire imaging time series represented as binary masks in the NIfTI format (Neuroimaging Informatics Technology Initiative). The ITK-SNAP (Yushkevich et al., 2006) (www.itksnap.org) software was used to examine MRI data and evaluate the algorithm results. The AFIL algorithm and a sample dataset is provided in the git repository (https://teahub.io/carolinekoehler/AFIL).
2.1. Algorithm development
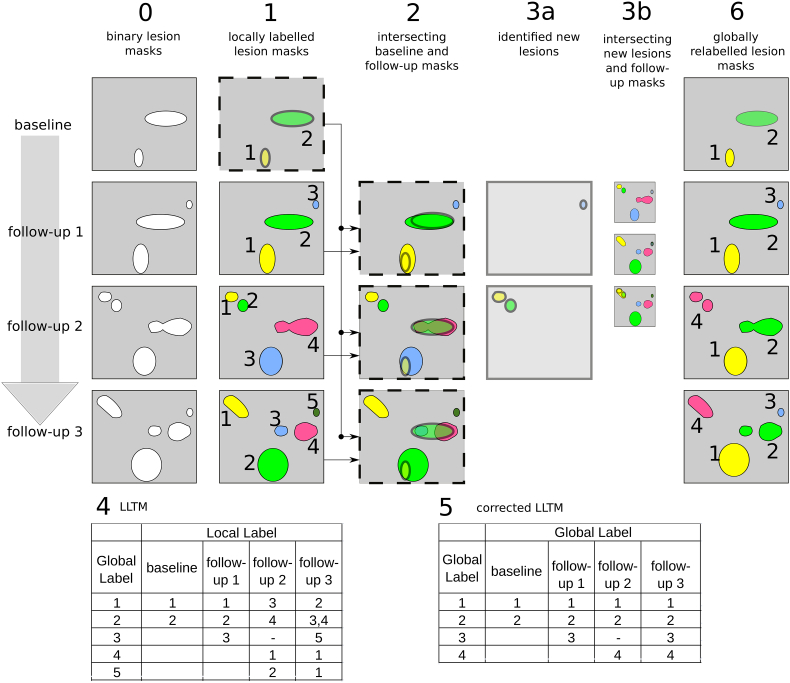
The algorithm traverses through six steps in which labels are addressed in the lesion masks of time series as illustrated in Fig. 1.
Fig. 1.
Illustrated algorithm workflow: (0) Binary lesion masks of four time points; (1) Automatic assignment of labels in longitudinal lesion masks; (2) Identification of lesion label intersections in consecutive lesion masks: Overlaid baseline mask and appropriate follow-up mask; (3) Determining new lesions in the time series: a) Note that the third labelled lesion in follow-up 1 and first and second labels in follow-up 2 do not intersect with lesions of the previous time point; b) new lesions continue in step 2 and were tested for intersection; (4) Assigning a global label to corresponding lesions in a time series: Corresponding local labels and newly identified lesions of the time series were tracked in rows of the LLTM. Note that a consecutive global label was assigned for a new lesion; (5) Determining confluent and separating lesions in the corrected LLTM. Two entries were found for global label 2, which indicates a separated lesion, and two intersections were found for local label 1 in follow-up 3 with previous time points, which indicates a confluent lesion; (6) Relabelled lesion masks: Local labels of the time series of corresponding lesions were overwritten by an appropriate global label.
2.1.1. Assigning interim labels in longitudinal lesion masks
Binary lesion masks of each time point within the series were automatically assigned an interim label using the bwlabeln function of the Image Processing Toolbox® to define connected voxel sets in a neighborhood of six connections. The local interim label of the same lesion may eventually change between time points because of new lesions.
2.1.2. Identification of lesion label intersections in consecutive lesion masks
The algorithm determined the intersections of every subsequent interim labelled lesion of the follow-up time point using locally labelled lesions of the labelled baseline time point. Two lesion volumes were considered to intersect if they overlapped by at least one voxel in baseline and follow-up.
2.1.3. Determining new lesions in the time series
The algorithm distinguished new lesions that exhibited no intersection with lesions of a previous time point in every follow-up time point (see Fig. 1, column 3a top down). The newly identified lesions were subsequently checked for intersections with lesions in follow-up time points (see Fig. 1, column 3b).
2.1.4. Assigning a global label to corresponding lesions in a time series
A Label Lesion Tracking Matrix (LLTM) was generated for the time series to preserve the local labels of intersecting lesions. An additional global label was assigned to track any individual lesion using the same label over time. New lesions were added to the LLTM for each time point, and these lesions were also assigned a consecutive global label. Therefore, the LLTM tracked each lesion in the dataset from onset to theoretically possible resolution within the time series.
2.1.5. Determining confluent and separating lesions for corrected LLTM
Confluent lesions exhibited multiple intersections with baseline lesions, and separating lesions possessed the same “mother” lesion origin. Separating lesions inherited the same global label of the “mother” lesion to facilitate the traceability, and confluent lesions were assigned the same global label in previous time points retrospectively. The LLTM was subsequently corrected with multiple rows of confluent lesions deleted from the matrix (see Fig. 1).
2.1.6. Determining confluent and separating lesions for corrected LLTM
Local labels were replaced by the appropriate persistent global label of the corrected LLTM in the final step and saved as a relabelled NIfTI lesion mask. Therefore, the output data were persistently labelled lesion masks because the global labels remained during the entire tracking period.
2.2. Accuracy evaluation
Expert MS readers verified the algorithm accuracy to track lesions using visual assessment. Relabeled lesion masks were evaluated as translucent overlays on FLAIR images of each time point. This accuracy was controlled in each individual lesion course if the global label was correctly assigned for the different cases of lesion development. An error rate was calculated as the number of lesion courses that received falsely assigned global labels within the time series divided by the total number of corrected lesion courses.
2.3. Recruitment and MRI test data acquisition
A subset of a longitudinal MRI dataset of a previously published study in clinically isolated syndrome (Kitzler et al., 2018) (CIS) was used as a test dataset (mean age 30 years (range 22–39); gender (M/F) 3/4). Within the study an age and gender adapted healthy control group of 21 subjects was scanned and seven individuals with a high number of baseline existing lesions were selected. CIS patients were recruited immediately after diagnosis and all cases used in this study converted to clinically definite MS during the study. For all patients the extended disability status scale (EDSS) was available at baseline and after 12 month. Written informed consent was obtained from all patients and healthy controls prior to data acquisition. Table 1
Table 1.
Clinically isolated syndrome and early MS patient characteristics. During the study course 5/7 patients received a treatment initiation (Copaxone).
| Baseline | 12 Months | |
|---|---|---|
| Mean EDSS (range) | 1,5 (0–2) | 1,3 (0–2) |
| Median T2 lesion load volume in mm3 (range) | 2247 (374–5384) | 4053 (1891–7911) |
| Median number of T2 lesions (range) | 21 (6–73) | 39 (14–81) |
A clinical MRI test data set was acquired using a 1.5 T MRI scanner (Siemens Magnetom Sonata, Siemens Healthineers, Erlangen, Germany) equipped with an 8-channel radiofrequency head coil within another clinical trial that was approved by the local Dresden University Hospital ethics committee. 3D-FLAIR data were acquired for each patient (FOV = 22 cm, matrix = 256 × 210, slice thickness = 3 mm, TE = 353 ms, TR = 6000 ms, TI = 2200 ms) at baseline (immediately after onset of initial disease-related symptoms) and after 3, 6 and 12 months. A T1-weighted anatomical reference scan (2D-T1 spin echo: TE =12 ms, TR = 500 ms, FOV = 22 cm, matrix = 256 × 256, slice thickness = 3 mm) and a fast low-angle shot image (FLASH: TE = 2.0 ms, TR = 5.7 ms, flip angle = 18°, FOV = 22 cm, matrix = 128 × 128, slice thickness = 1.7 mm) were acquired.
2.4. Data-processing
The FMRIB Software Library (FSL 5.0) (Jenkinson et al., 2012) involving brain extraction (BET) and affine intra-subject co-registration with the Linear Image Registration Tool (FLIRT) (Jenkinson and Smith, 2001) was used for image pre-processing. The FLASH image was used as registration target because it had a smaller slice thickness than the T1-weighted image. Registering every subsequent time point of T1 and FLAIR data to the baseline FLASH image enabled efficient longitudinal analysis. T2 lesions were identified as focal areas of elevated MRI signal intensity in the FLAIR data. A semi-automatic segmentation was applied by using z-score thresholding of z > 4 against normalized FLAIR intensity with preceding normalization by a robust maximum (98%) (Kitzler et al., 2012; 2018). The resulting binary lesion maps were manually edited. Lesions >3 voxels (8.25 mm3) were accepted in the final lesion masks. Two experienced raters manually performed lesion mask re-evaluation and editing.
3. Results
3.1. Algorithm accuracy evaluation
A total of 352 individual lesion courses (baseline existing and newly appearing) were tracked in all seven patients over four standardized time points. Correction for separating and confluent lesions (see Fig. 1; 4 LLTM) ultimately provided estimates of 328 lesion development courses. Within this set stable, growing, shrinking, but also reappearing and resolving, confluent and separating lesions were reliably labelled (see Table 2).
Table 2.
Categorized courses of all tracked lesion developments (see section 2.1). Based on AFIL algorithm results all four time points were checked with regard to their global label consistency. Performance verification results of expert visual inspection of relabelled lesion masks are provided (right/false).
| Number of tracked T2 lesions |
||
|---|---|---|
| Correct | False | |
| 1) Corresponding (growing, shrinking, stable) | 162 | 0 |
| 2) New | 121 | 1 |
| 3) Resolving | 22 | 0 |
| 4) Reappearing | 9 | 0 |
| 5) Confluent | 24 | 0 |
| 6) Separatinga | 5 | 0 |
| 7) Successively confluent and separatingb | 9 | 2 |
| Σ | 352 | 3 |
| Corrected total number of lesion courses | 328 | |
| Error rate (false/total # of lesion courses) | 0.9% | |
Result of the segmentation of initially confluent conglomerates and subsequently consolidating multiple lesion centres.
Rare lesion course complicating lesion assignment.
AFIL algorithm tracking errors evolved due to unexpected lesion development. Simultaneous coalescence and separation occurred in two cases, and lesions were falsely relabelled (case 7, Table 2). Another lesion revealed a centripetal extension and resolution of the primary lesion area, and the overlap between baseline and the final time point map was missed, which created a false persistent global label (case 2, Table 2).
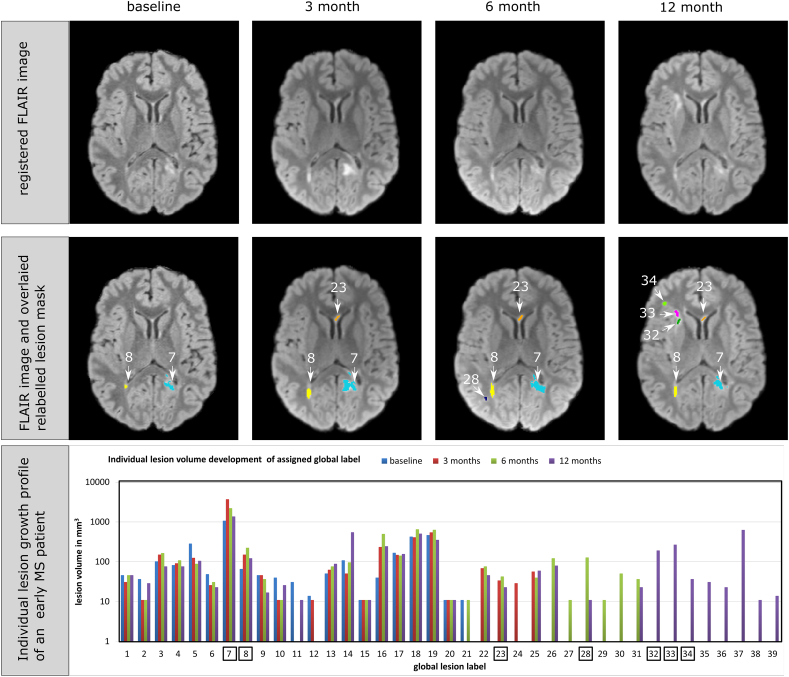
3.2. Individual application example
Fig. 2 shows an examplary application of the AFIL algorithm to a time series of segmented lesion masks to investigate the individual lesion growth profile of a MS patient. The same slice of the FLAIR image and the overlaid re-labelled lesion mask is displayed for each time point. The colour-coded bar groups illustrate the volume development of all individual lesion courses of the subject. Most lesions exhibited dynamic increases and decreases in volume over the tracked time period (varying). A minor number of lesions were stable or continuously growing.
Fig. 2.
Individual lesion growth profile of an early MS example patient: Selected slices of co-registered FLAIR images before normalization of the same subject at baseline, at 3, 6, and 12 months (first row) and the same slices superimposed with globally labelled lesions (middle row). Individual lesion volume development is represented by colour-coded bar groups of 39 lesion courses in the same subject. Encircled global labels highlight lesions in the displayed slice (bottom row). Note that the numeric labelling starts from existing lesions (left) with the volumes of all four time points and subsequently evolving lesions and volumes at their effective appearance (continuing to right). The displayed sample slice timeline shows a marked variation in lesion extent for baseline existing lesions 7 and 8 and consecutively appearing lesions 23, 28, and 32 through 34. Lesions 12, 27, 29, and 30 appeared within the study period but were not detectable in follow-up, and lesions 11 and 21 are missing an interval time point. Since this finding primarily affects very small lesions, it may arise due to segmentation failure, variation in the FLAIR signal or volume reduction below the threshold.
3.3. Cohort lesion volume expansion courses
Individual lesion volumes of the cohort were assessed over time. Therefore, lesions were characterized according to volume development into lesion expansion courses. Lesions were defined as growing in volume if they enlarge more than +8.25 mm3 (3 Voxel) and as shrinking if they decrease −8.25 mm3 in subsequent time points. If the volume varied less ± 8.25 mm3 the lesion was defined as stable.
Combined analyses of all seven patients within the 207 baseline existing lesions revealed that 35 (16.9%) lesions were stable in volume, 21 (10.1%) lesions resolved, 1 (0.5%) lesion shrank continuously and 7 (3.4%) lesions exhibited an ongoing volume increase. As many as 143 (69.1%) lesions revealed consecutive volume increase and decrease (varying) within the tracked period. Notably, only 74 (52%) of the baseline existing varying lesions exhibited an overall larger volume after 12 months. The amount of resolved and shrinking lesions was higher in new lesions.
4. Discussion
New and/or enlarging MS lesions are clinically of the utmost importance, which is reflected by their incorporation as progression surrogates in the modern therapy monitoring strategy NEDA (Rudick et al., 2010; Stangel et al., 2015). However, the individual dynamic characteristics of lesion progression in MS may not be fully recognized and understood because in vivo visualization techniques are inadequate with regard to their separate volume development.
High disease activity with many new and enlarging lesions was identified for all patients of this study using the NEDA criteria. Notably, the multiple time point analyses performed with AFIL demonstrated that most lesions did not continuously grow and even subsequently shrank during the tracked period. Consequently, shrunken lesions that subsequently enlarged were detected as growing lesions when comparing two time points even if the volume comparison to baseline did not change. Therefore, the conventional comparison of only two time points may overestimate the interpretation of disease activity. We demonstrated the feasibility of a novel algorithm to automatically create an individual lesion growth profile of MS patients. The main developed technical feature was a persistent labelling of each lesion object within a time series of lesion masks that were applied in conjunction with adapted conventions of presumed lesion development.
The possible annulment of antidromic individual lesion volume changes in a collective lesion analysis was noted previously as a limit of existing tools (Llado et al., 2012). These tools are primarily based on subtraction or deformation field-based approaches (Llado et al., 2012; Moraal et al., 2009; Rey et al., 2002) in which volume change reflects the entire lesion compartment. The ordinary cumulative lesion growth obtained as the intersection of differences of all lesions between two different lesion maps does not reflect the complete characteristics, as shown in our application. The presented analysis algorithm AFIL overcomes this restriction.
Most lesions underwent enlargement and shrinkage within the study period of the analysed sample data set. The net effect of currently assessable short-term lesion growth may represent a different feature than that of continuous growing revealed by our approach. The algorithm also measured shrinking and resolving lesions, which provided a more general overview of disease progression and possible tissue recovery. Based on radiological decisions to distinguish adjacent lesion a connectivity of 6 was chosen because otherwise nearby lesions might detect as one lesion. The chosen connectivity has an impact on the number of tracked lesions as well as categorization as confluent or separating lesions. This matter may provide further exploration and research on optimal tracking result in future.
The AFIL algorithm operates independently of the lesion segmentation routine used. Thus, individual lesion tracking can be performed if lesions have previously been segmented using any segmentation technique that creates binary masks. However, AFIL requires accurate input data, i.e., it depends on good quality pre-segmentation results. A precise image registration of longitudinal data was also crucial for the quantification accuracy of lesion volume changes. AFIL does not improve the prior lesion segmentation, but errors within lesion segmentation may strongly affect the results of its lesion tracking.
A threshold minimum (>3 voxel equal to a volume of 8.25 mm3) was used to accept a pre-segmented lesion as an individual object within lesion masks to limit image registration errors and partial volume effects at lesion borders. The applied threshold caused a higher number of resolved and reappearance results in small lesions (see example in Fig. 2). In case of registration mismatches and larger single lesions (at the size of a multitude of the threshold minimum) the algorithm will still detect the longitudinal corresponding lesion, if at least the labelled lesion masks areas will consistently feature an overlap. In contrast if two lesions are closely located to each other registration mismatch might cause misclassification when lesions are composited by the algorithm due to artificial overlap erroneously as confluent lesions. The volume change is thereby only affected in the latter case. In summary, the higher the mismatch the higher the tracking failure will be. In any case to reduce that sufficient motion correction and registration should be applied before the AFIL algorithm run to reduce mismatch as much as possible. However, the observed AFIL error rate for repeatedly labelled lesions in subsequent maps was minor, with 3 errors in 328 observations (0.9%).
The intended read-out of MR-derived tissue characteristics (e.g., MTR, FA, MWF, T1, and T2) using the labelled lesions as volumes of interest and its relevance for the classification of MS lesions will be examined in further clinical applications. Individual longitudinal volume quantification may also be an appropriate approach to recognize slowly growing chronic active lesions in late and secondary progressive MS.
AFIL uses binary segmentations, and it may be further used to compare lesions of different imaging modalities to monitor volumetric or parametric changes in other diseases, such as brain tumors, cerebral microangiopathies or extracranial lesions.
5. Conclusion
The introduced novel algorithm reliably allowed individual tracking of MS lesions in sets of pre-segmented binary lesion maps and can be used in conjunction with any segmentation routine to track individual objects. The automated individual quantification of MS lesion volume changes provides a new in vivo insight into the dynamic nature of these lesions and the possibility to observe meaningful patterns for disease monitoring. AFIL fully operates in a 3D-resolved manner of the persistently labelled lesions, and it may be further transferred into an individual lesion region-of-interest. This characteristic prospectively allows for its application to the read-out of single lesion values over time of any registered quantitative MRI metric in further studies.
Acknowledgments
Acknowledgment
We thank all patients and their families for their agreement to the MRI study. We further thank Janka Russig and Kerstin Liesk for their assistance in acquiring the imaging data.
Funding
This work was supported by a NOVARTIS research grant MFTY720A_FVTW028. We thank NOVARTIS Pharma GmbH, Germany for encouragement and generous sponsorship. The Author(s) declare(s) that there is no conflict of interest.
References
- Faizy T.D., Thaler C., Kumar D. Heterogeneity of multiple sclerosis lesions in multislice myelin water imaging. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0151496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi M., Rocca M.A. MR imaging of multiple sclerosis. Radiology. 2011;259:659–681. doi: 10.1148/radiol.11101362. [DOI] [PubMed] [Google Scholar]
- Filippi M., Cercignani M., Inglese M., Horsfield M.A., Comi G. Diffusion tensor magnetic resonance imaging in multiple sclerosis. Neurology. 2001;56:304–311. doi: 10.1212/wnl.56.3.304. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Smith S.A. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C.F., Behrens T.E., Woolrich M.W., Smith S.M. Fsl. NeuroImage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Kitzler H.H., Su J., Zeineh M. Deficient MWF mapping in multiple sclerosis using 3D whole-brain multi-component relaxation MRI. NeuroImage. 2012;59:2670–2677. doi: 10.1016/j.neuroimage.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzler H.H., Wahl H., Eisele J.C. Multi-component relaxation in clinically isolated syndrome: lesion myelination may predict multiple sclerosis conversion. NeuroImage. 2018;20:61–70. doi: 10.1016/j.nicl.2018.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llado X., Ganiler O., Oliver A. Automated detection of multiple sclerosis lesions in serial brain MRI. Neuroradiology. 2012;54:787–807. doi: 10.1007/s00234-011-0992-6. [DOI] [PubMed] [Google Scholar]
- Moraal B., Meier D.S., Poppe P.A. Subtraction MR images in a multiple sclerosis multicenter clinical trial setting. Radiology. 2009;250:506–514. doi: 10.1148/radiol.2501080480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey D., Subsol G., Delingette H., Ayache N. Automatic detection and segmentation of evolving processes in 3D medical images: application to multiple sclerosis. Med. Image Anal. 2002;6:163–179. doi: 10.1016/s1361-8415(02)00056-7. [DOI] [PubMed] [Google Scholar]
- Rocca M.A., Iannucci G., Rovaris M., Comi G., Filippi M. Occult tissue damage in patients with primary progressive multiple sclerosis is independent of T2-visible lesions—a diffusion tensor MR study. J. Neurol. 2003;250:456–460. doi: 10.1007/s00415-003-1024-1. [DOI] [PubMed] [Google Scholar]
- Rudick R.A., Lee J.C., Cutter G.R. Disability progression in a clinical trial of relapsing-remitting multiple sclerosis: eight-year follow-up. Arch. Neurol. 2010;67:1329–1335. doi: 10.1001/archneurol.2010.150. [DOI] [PubMed] [Google Scholar]
- Siemonsen S., Young K.L., Bester M. Chronic T2 lesions in multiple sclerosis are heterogeneous regarding phase MR imaging. Clin. Neuroradiol. 2016;26(4):457–464. doi: 10.1007/s00062-015-0389-8. https://www.ncbi.nlm.nih.gov/pubmed/?term=Siemonsen+Chronic+T2+lesions+in+multiple+sclerosis (Epub 2015 Apr 18) [DOI] [PubMed] [Google Scholar]
- Stangel M., Penner I.K., Kallmann B.A., Lukas C., Kieseier B.C. Towards the implementation of 'no evidence of disease activity' in multiple sclerosis treatment: the multiple sclerosis decision model. Ther. Adv. Neurol. Disord. 2015;8:3–13. doi: 10.1177/1756285614560733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P.A., Piven J., Hazlett H.C. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]