Abstract
Accumulating evidence indicates that long non-coding RNAs (lncRNAs) can play a pivotal role in regulation of diverse cellular processes. In particular, lncRNAs can serve as master gene regulators at transcriptional and posttranscriptional levels, leading to tumorigenesis. In this review, we discuss latest developments in lncRNA-meditated gene expression at the post-transcriptional level, including gene splicing, mRNA stability, protein stability and nuclear trafficking.
Keywords: Alternative splicing, LncRNA, Posttranscriptional regulation, Protein stability, RNA binding proteins, RNA stability
Introduction
The advances in functional genomics have revealed that although large numbers of RNAs are transcribed from the human genome, protein coding genes account for a very small fraction of total transcripts.1, 2 The rest of transcripts are non-coding RNAs, including microRNAs and long non-coding RNAs (lncRNAs). These non-coding transcripts were initially considered as transcriptional noises,3 however it has become increasingly apparent that they may play a critical role in diverse cellular processes from normal development to disease processes.4, 5, 6, 7 There are two major classes of non-coding RNAs based on their size, 1) small ncRNAs less than 200 nt in length, represented by microRNAs and 2) long non-coding RNAs (lncRNAs) larger than 200 nt in length. MicroRNA are well characterized and are known to induce mRNA degradation or block mRNA translation via the RNA interference pathway.8 On the other hand, lncRNAs are less well characterized. As a matter of fact, little is known for their functions for the vast majority of lncRNAs. To date, the non-code database lists over 96,000 human lncRNAs (http://www.noncode.org), whereas more than 56,000 human lncRNAs have been catalogued on LNCipedia (http://www.lncipedia.org), and the number of lncRNAs continues to grow.9, 10, 11
Multiple studies have shown that a significant number of lncRNAs are emerging as key players in various layers of cellular processes.12, 13 Although we are just beginning to understand the function of lncRNAs, increasing evidence suggests that lncRNAs may positively or negatively regulate gene expression at the transcriptional and/or post-transcriptional level.14, 15 The focus of this review is on post-transcriptional regulation of gene expression by lncRNAs.
LncRNA-mediated gene expression
Due to their role in regulation of gene expression, lncRNAs have been implicated in control of a variety of cellular functions and disease processes such as stem cell maintenance and cancer metastasis.16, 17, 18, 19, 20, 21 This may have to do with their ability to interact with DNA, RNA or protein. For example, lncRNAs may serve as 1) signals for transcription; 2) decoys to titrate transcription factors; 3) guides for chromatin-modifying enzymes to be recruited to target genes and 4) scaffolds to bring together multiple proteins to form functional ribonucleoprotein complexes.12, 22, 23, 24 Apparently, the mechanism of lncRNA-mediated gene regulation is much more sophisticated than what we had previously anticipated.
LncRNAs function in many cases as transcriptional regulators. At the transcriptional level, lncRNAs may directly act on transcriptional complexes or serve as a scaffold to recruit various components including RNA polymerase II or alter chromatin structure. LncRNAs can also function as critical players in controlling the remolding of chromatin structure. A good example is lncRNA HOTAIR that mediates the transcriptional silencing of HOXD locus via recruitment of the polycomb chromatin remodeling complex,12, 25 through which lncRNAs can regulate large numbers of genes.26
At the post-transcriptional level, lncRNAs may interact with a variety of RNA binding proteins (RBPs), leading to alternations of mRNA stability, splicing, protein stability and subcellular localization. In addition, lncRNAs may bind to complementary RNA sequences of target genes as post-transcriptional regulators. Thus, lncRNAs can impact almost every aspect of gene expression. Here, we will provide a few of examples to illustrate how lncRNAs can regulate gene expression at the post-transcriptional level.
LncRNA-associated ribonucleoprotein (RNP) complexes
Although lncRNAs are very powerful as master gene regulators, they do not act alone. Instead, they often work through various partnerships. The most common partnerships are the ribonucleoprotein (RNP) complexes consisting of lncRNAs and RNA binding proteins (RBPs). Interaction of lncRNAs with these proteins to form RNP complexes is critical for lncRNAs to exert their function as gene regulators. There are numerous examples of such interactions. Among them is the interaction of lncRNAs with RBPs such as chromatin remodeling enzymes such as EZH2 and PRC2.16, 20 As a histone methyltransferase, EZH2 is a phosphorylated protein when it is active; and the phosphorylation at threonine residue 345 has been shown to be critical to its interaction with HOTAIR.27 Similarly, linc-HOXA1 RNA represses Hoxa1 by interaction with the protein PURB as a transcriptional cofactor.28 LincRNA-p21 has been shown to be a p53 transcriptional target.29 When lincRNA-p21 binds to hnRNP K to form a RNP complex, this complex mediates global gene repression and apoptosis in the p53 pathway. On the other hand, PANDA (P21 associated ncRNA DNA damage activated) is able to block apoptosis through interaction with the transcription factor NF-YA to limit expression of pro-apoptotic genes. Linc-RoR interacts with hnRNP I to suppress p53 in response to DNA damage.30 Moreover, hnRNP I can also interact with other lncRNAs such as UCA1 to suppress p27 expression.31 Finally, lncRNA CTBP1-AS can interact with PSF to cause the global androgen-mediated gene repression.32 These examples highlight the importance of lncRNA-ribonucleoprotein complexes in gene regulation.
Post-transcriptional regulation of gene expression
The post-transcriptional regulation involves the stability and distribution of the different transcripts, such as alternative splicing, nuclear degradation (exosome), processing, and nuclear export, where RBPs often play an important role. This may also include protein modifications and the protein subcellular localization. Alterations of these events have been implicated in tumorigenesis.
In eukaryotes, after a gene is transcribed, an initial product of transcription is pre-mRNA, which is processed into mature mRNA by removing introns in most cases. This process is also called gene splicing. In addition, RNA processing also includes other events such as mRNA export, localization, translation and stability, which often involves multiple protein factors, such as RBPs. RBPs achieve these events through an RNA recognition motif (RRM) that binds a specific sequence or secondary structure of the transcripts, including the 5′ and 3′-UTR (untranslated region) of the transcript. In addition to transcripts, proteins can also be subject to post-translational modifications such as phosphorylation, acetylation and ubiquitination. Through these modifications, proteins may change their activity, stability or subcellular localization.
Thus, RBPs participate in both RNA processing and protein modifications. Especially for hnRNP proteins, they are very important to RNA processing events such as pre-mRNA splicing, mRNA export, localization, translation and stability.33 These proteins are often abundantly present in the cells and most of them are resided in the nucleus. Emerging evidence suggests that lncRNAs can directly or indirectly participate in these processes by the formation of RNP complexes. Therefore, we will discuss how lncRNAs regulate mRNA splicing, mRNA stability, protein stability and protein subcellular localization through RBPs.
Regulation of mRNA splicing
Higher eukaryotes utilize alternative splicing of pre-mRNA to achieve increased transcriptome and proteomic complexity. Alternative pre-mRNA splicing is not only a mechanism to generate protein diversity, but it is often tissue or cell type specific. In particular, alternative splicing patterns are often associated with clinical features.34 For example, while normal cells express a predominant form of pyruvate kinase (PKM1), a variety of tumors express a tumor specific isoform, PKM2.35
There are several ways that lncRNAs can be involved in alternative splicing. In most cases lncRNAs regulate gene splicing through interaction with splicing factors. The core complex for gene splicing is the spliceosome, during splicing additional splicing factors are required to determine the splice sites. Thus, splicing factors are critical to gene splicing, and they are often abundantly expressed. There are a large number of splicing factors reported in literature. Based on SpliceAid 2 databases (www.introni.it/spliceaid.html),36 there are 72 known splicing factors including two well-known protein families, serine/arginine (SR)-rich proteins37 and hnRNP proteins.38 For example, SR proteins carry one or two RNA recognition motifs (RRMs) at the N-terminus, and a characteristic arginine/serine-rich (RS) domain at the C-terminus. The RS domain is a target for extensive phosphorylation, and the phosphorylation status of the RS domain is important for the activity of SR proteins in splicing.
However, not all of these splicing factors are required for splicing of each gene. In other words, these splicing factors are selectively used for splicing of a given gene. So a question is how splicing factors are specifically selected for gene splicing. Although studies suggest that the relative amount of splicing factors in the nucleus may be an important factor, given a broad spectrum of substrates for any splicing factor in general, splicing factors lack the specificity. Thus, involvement of lncRNAs in these events would provide another layer of control, in particular for the specific splicing patterns in a given cellular context.
A relatively well-established lncRNA associated with pre-mRNA splicing is MALAT1. Tripathi et al use computer prediction and RNA immunoprecipitation assays combined with knockdown experiments and identify that SRSF1 can interact with MALAT1 via its RRM domain.39 For example, MALAT1 is required for proper localization of SRSF1 as well as several other splicing factors to nuclear speckles. Depletion of MALAT1 leads to changes in alternative splicing of a subset of transcripts.39, 40 Importantly, this type of interaction SR proteins with MALAT1 in the nuclear speckles alters their phosphorylation status and thus, leading to alterations in alternative splicing patterns. Therefore, MALAT1 could act as a ‘‘molecular sponge’’ by interacting with SR proteins in the nuclear speckles, and thereby modulate the concentration of splicing competent SR proteins in cells. Through this type of RNP complex formation, lncRNAs such as MALAT1 can add another layer of regulation for alternative splicing. In addtion, suppression of MALAT1 decreases expression of the splicing factor RBFOX2, which in turn promotes alternative processing of the pro-apoptotic tumor suppressor gene KIF1B in ovarian cancer.41 Thus, MALAT1 facilitates a pro-metastatic phenotype by promoting alternative RNA processing and differential expression of anti-apoptosis and epithelial to mesenchymal transition (EMT)-related genes.41
Chromatin-mediated splicing control is another mechanism that lncRNAs can regulate alternative splicing. For example, a nuclear antisense lncRNA derived from FGFR2 locus promotes epithelial-specific alternative splicing of FGFR2. In this case, the lncRNA acts through recruitment of Polycomb-group proteins and the histone demethylase KDM2a to create a chromatin environment that impairs binding of a repressive chromatin-splicing adaptor complex important for mesenchymal-specific splicing.42
Other examples of lncRNA-splicing factor interaction that regulates alternative splicing are those interacting with hnRNP protein family. Apparently, some of hnRNP proteins can serve as a splicing silencer binding to silencing domain, whereas others can bind to enhancer regions of pre-mRNA. By affecting the binding of the factors with pre-mRNA, lncRNAs may specifically regulate splicing for a set of genes in a given cellular context or environmental conditions. There are a large number of such examples. In this scenario, a recent report suggests that lncRNA PNCTR contains hundreds of short tandem repeats which can serve as PTBP1 (hnRNP I) binding sites. Apparently, this feature enables to sequester a substantial fraction of PTBP1 in a nuclear body.43 PNUTS is a protein coding gene, however, it can also be transcribed into a lncRNA, called lncRNA-PNUTS through alternative splicing. PNUTS and lncRNA-PNUTS appear to have distinct biological functions. While PNUTS is ubiquitously expressed, expression of lncRNA-PNUTS depends on the status of hnRNP E1 and tumor context.44 Together, they regulate EMT and tumor progression.
In addition to hnRNP proteins, lncRNAs can also interact with other types of splicing factors. For example, lnc-Spry1 associates with U2AF65 splicing factor, suggesting a role in alternative splicing. Depletion of lnc-Spry1 induces, as TGF-β, isoform switching of fibroblast growth factor receptors, resulting in FGF-2-sensitive cells.45 Thus, lnc-Spry1 regulates the expression of TGF-β-regulated gene targets. RNA-binding motif protein 4 (RBM4) is a multiple functional RBP, involved in cellular processes like alternative splicing of pre-mRNA and translation regulation by modulating alternative 5′-splice site and exon selection. For example, when lncRNA TPM1-AS interacts with RBM4, their interaction prevents binding of RBM4 to TPM1 pre-mRNA and promotes endogenous exon 2a inclusion of TPM1.46
Our own study suggests that BC200 regulates alternative splicing of Bcl-x. Bcl-x is a member of the well-known Bcl-2 family that play key roles in apoptosis.47 Alternative splicing of Bcl-x can lead to expression of Bcl-xL or Bcl-xS with an opposite effect on cell apoptosis. While Bcl-xL has an anti-apoptotic function and is often upregulated in several cancers, Bcl-xS is a pro-apoptotic protein that antagonizes the survival functions of Bcl-xL.48 In this case, BC200 overexpression promotes Bcl-xL, whereas BC200 KO promotes Bcl-xS. The mechanism involves the interaction between BC200 and splicing factor hnRNP A2/B1. Of interest, hnRNP A2/B1 has been implicated in splicing of many genes, however, many of them such as RON, CASP9, IRF-3 and A-Raf are not affected by BC200. Therefore, it is possible that BC200 facilitates binding of hnRNP A2/B1 to Bxl-x pre-mRNA through the complementary sequences.31
PCGEM1 is a prostate-specific transcript. Our study suggests that PCGEM1 plays a role in castration resistance. Specifically, PCGEM1 promotes expression of androgen receptor (AR) splice variants such as AR3 in prostate cancer. AR3 is a predominant and clinically important form that has been shown to play a significant role in castration resistance.49 Of significance, PCGEM1 interacts with splicing factors hnRNP A1 (silencer) and U2AF65 (enhancer). Moreover, androgen deprivation induces PCGEM1 and causes its accumulation in nuclear speckles. This androgen deprivation induced PCGEM1 can regulate the competition between hnRNP A1 and U2AF65 for AR pre-mRNA. While the interaction of PCGEM1 with hnRNP A1 suppresses AR3 by exon skipping, its interaction with U2AF65 promotes AR3 by exonization.50
Other factors may also be involved in regulation of alternative splicing. For example, RNA methylation may also play role in alternative splicing because RNA methylation can impact the accessibility of hnRNPs to pre-mRNAs. For instance, N6-methyladenosine (M6A) can serve as a switch to regulate gene expression and RNA maturation.51 Finally, lncRNAs can also be subject to alternative splicing, as protein coding genes. In this case it involves MBNL3 which is member 3 of the muscle blind-like family of proteins. As a splicing factor, MBNL3 has been implicated in regulation of alternative splicing and in the pathophysiology of myotonic dystrophy. In addition, MBNL3 also plays an oncogenic role because knockdown of MBNL3 abolishes hepatocellular carcinoma tumorigenesis.52 Furthermore, MBNL3 induces lncRNA-PXN-AS1 exon 4 inclusion which is important for its interaction with PXN 3′-UTR. This intermolecular RNA–RNA interactions between PXN-AS1 exon 4 and the PXN 3′UTR is critical to PXN-AS1-mediated upregulation of the PXN protein level.52
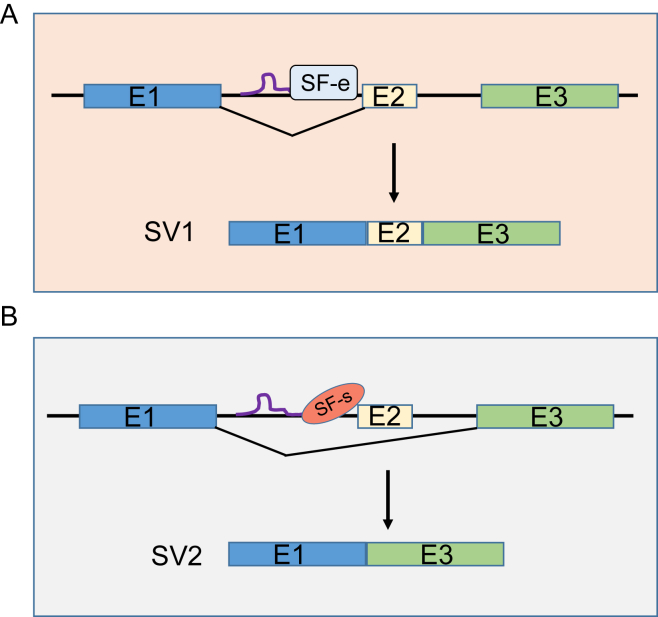
Therefore, although regulation of alternative splicing is a complex process, how lncRNAs are involved in this process can be described in a simplified model as shown in Fig. 1. In this case, a lncRNA binds to pre-mRNA to help recruit an enhancer splicing factor (SF-e) so that exon 1 (E1) and exon 2 (E2) are connected, forming splice variant 1 (SV-1) (Fig. 1, top). By contrast, when the bound lncRNA helps recruit a silencer splicing factor (SF-s), E2 is excluded such that E1 and E3 are connected, forming SV-2 (Fig. 1, bottom). It is known that splicing factors select splice sites often in a concentration-dependent manner and thus, the relative expression of these factors may decide a particular splice pattern.53 On the other hand, the involvement of lncRNAs in this process may provide more flexibility and/or specificity. In this way the cell can better respond to different environmental cues.
Fig. 1.
A simplified model for lncRNA-mediated alternative splicing. A, LncRNA promotes inclusion of Exon2 (E2) by interaction with enhancing splicing factor (SF-e) to form splicing variant 1 (SV-1). B, LncRNA promotes skipping of Exon2 (E2) by interaction with silencing splicing factor (SF-s) to form splicing variant 2 (SV-2).
Regulation of mRNA stability
Many genes such as c-Myc have a relative short mRNA half-life, which could be a consequence of the interaction of their 3′-UTR with RBPs. Given the ability of lncRNAs to interact with RBPs, it is anticipated that this type of interaction between lncRNAs and RBPs will impact not only the amount of RBPs in the pool, but also the function of those lncRNAs that share the same binding sites with other genes including coding and noncoding genes. In this way, the mRNA molecules are either stabilized or destabilized. Various types of RBPs could play a role in determining the mRNA stability and, thus mRNA level.
LncRNA OCC-1 can interact with HuR and recruit ubiquitin E3 ligase β-TrCP1 to HuR, such that HuR is downregulated by destabilization.54 Since HuR serves as a stabilizing factor for a large number of mRNAs, it ultimately causes downregulation of these HuR-targeted mRNAs. Our own study suggests that Linc-RoR interacts with hnRNP I (stabilizing factor) and AUF1 (destabilizing factor), respectively, with an opposite consequence to their interaction with c-Myc mRNA.55 In particular, interaction of Linc-RoR with AUF1 inhibits AUF1 to bind to c-Myc mRNA.55
As for alternative splicing, hnRNPs can also play an important role in this aspect. For instance, FIRRE physically interacts with hnRNP U, regulating the stability of mRNAs of selected inflammatory genes through targeting the AU-rich elements of their mRNAs in cells following LPS stimulation.56 As a NF-κB regulated lncRNA, FIRRE plays a role in immune response to LPS because FIRRE regulates the RNA stabilization of VCAM1 and IL12p40 through targeting their AREs along with hnRNPU.56
In addition, other kinds of RBPs can also regulate mRNA stability through interactions with lncRNAs. For instance, LIN28 may function as a positive RBP where lncRNA MACC1-AS1 can stabilize MACC1 mRNA and thus, post-transcriptionally regulates MACC1 expression.57 Cyclin D1 (CCND1) is an oncogene, and acts as a critical regulator of cell cycle progression. A recent study shows that lncRNA LAST can stabilize CCND1 mRNA. In this case, CNBP (CCHC-type zinc finger nucleic acid binding protein) serves as RBP to bind to the 5′-UTR of CCND1 mRNA to protect against possible nuclease targeting. LAST promotes this interaction between CNBP and the 5′-UTR of CCND1 mRNA, resulting in increased expression of CCND1.58 Similarly, IGF2BP1 is a well-known RBP. LIN28B-AS1 is able to regulate mRNA stability of LIN28B by directly interacting with IGF2BP1 but not with LIN28B.59 In some cases, lncRNAs can interact directly with the target genes to regulate their expression. For instance, lncRNA uc.173 interacts with the primary transcript of miR-195, leading to its degradation.60
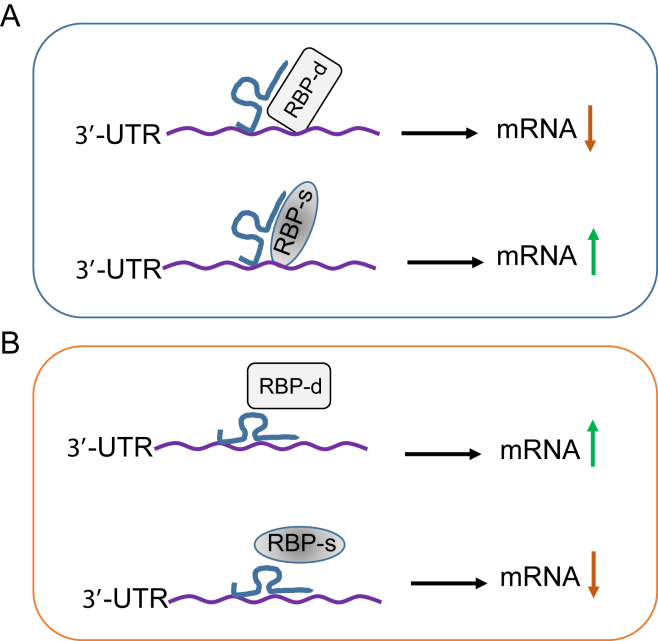
We present a diagram (Fig. 2) to summarize what we have just discussed regarding lncRNA-mediated mRNA stability. Since a number of RBPs control mRNA stability, alteration of expression of these RBP will also impact the mRNA stability. Thus, they are important post-transcriptional gene regulators. It is known that many mRNA species carry AU-rich elements (AREs) at the 3′-UTR. AREs are one of the most common determinants of RNA stability in mammalian cells by various RBPs including stabilizing and destabilizing factors. In this case, HuR serves as a stabilizing factor and, AUF1 and TTP are destabilizing factors. Depending on which factors are interacted with a given lncRNA, a consequence could be an increase or decrease for targeted mRNA (Fig. 2). One scenario would be the binding of lncRNA to the UTR, which would help to recruit RBP. In the case of stabilizing RBP, this binding would increase the mRNA stability, and thus, the mRNA level is increased. If a destabilizing RBP is recruited to this region, then the mRNA stability is decreased, leading to a decreased level of mRNA. On the other hand, if the binding of lncRNA to the UTR prevents the binding of RPB to the UTR, the opposite consequence to the first scenario would occur, i.e., binding of lncRNA to the UTR would cause a decreased level of mRNA for stabilizing RBP whereas increased level of mRNA for destabilizing RBP. Of cause, there exist many other possibilities. For example, it is possible that RBP first interacts with lncRNA and then this interaction may be able to determine whether they come to the targeted UTR. Another possibility would be that two or more lncRNAs can compete for the same RBP and the outcome of this competition would determine the fate of lncRNA-RBP complex.
Fig. 2.
LncRNAs serve as positive or negative regulator through interaction with RNA binding proteins (RBPs). A, Binding of lncRNA to 3-UTR facilitates recruiting RBPs. B, Binding of lncRNA to 3-UTR prevents RBP interaction with 3′-UTR. RBP-d, destabilizing RBP; RBP-s, stabilizing RBP.
Regulation of protein stability
The protein stability is important because it can also impact relative amount for a given protein. In particular, proteins like p53 have a relatively short half-life under normal physiological conditions. However, DNA damage can greatly increase p53 stability by suppression of the major p53 inhibitor MDM2. Apparently, MDM2 is not the only factor controlling its stability. Several studies indicate that lncRNAs can also regulate this type of protein stability, which may involve protein modifications such as ubiquitination or phosphorylation.
For example, lncRNA DINO (Damage Induced Noncoding) regulates p53 stability in a p53 dependent manner.61 DINO contributes to the p53-mediatd phenotypes, including cell cycle arrest and apoptosis in response to DNA damage. Importantly, DINO interacts with p53, and stabilize p53 so that the induced p53 is able to positively regulate the downstream targets including DINO itself. Thus, the ability of DINO to stabilize p53 protein makes DINO a critical player in DNA damage-p53 regulatory network. Another example is lncRNA GUARDIN which is also a p53-responsive lncRNA.62 GUARDIN is important for BRCA1 stability because GUARDIN serves as an RNA scaffold to facilitate the heterodimerization of BRCA1 and BARD1. This may explain why GUARDIN is essential for genomic stability.
Several other proteins are also implicated in DNA damage repair pathway through regulation of protein stability. One example is chromodomain-helicase-DNA-binding protein 7 (CHD7) which is often dysregulated in cancers such as pancreatic cancer. A low level of CHD7 is associated with increased recurrence-free survival (RFS) and overall survival (OS) in response to adjuvant gemcitabine treatment.63 In neuroblastoma, CHD7 is critical for SOX9 expression; 6p22lncRNAs are able to regulate CHD7 protein stability via modulating the cellular localization of USP36.64 Similarly, HULC is able to stabilize silent information regulator 1 (Sirt1) protein in hepatocellular carcinoma cells because HULC can upregulate ubiquitin-specific peptidase 22(USP22), and suppress ubiquitin-mediated degradation of Sirt1 protein by removing the conjugated polyubiquitin chains from Sirt1,65 leading to autophagy and chemoresistance.
FEZF1-AS1 is overexpressed in colorectal cancer and this increased expression of FEZF1-AS1 is associated with poor survival. FEZF1-AS1 could bind to the pyruvate kinase 2 (PKM2) protein and increase its stability, resulting in increased cytoplasmic and nuclear PKM2 levels. Increased cytoplasmic PKM2 promotes pyruvate kinase activity and lactate production (aerobic glycolysis), whereas FEZF1-AS1-induced nuclear PKM2 upregulation can further activate STAT3 signaling.66 LncRNA SLCO4A1-AS1 is also upregulated in colorectal cancer and its overexpression is associated with poor prognosis and tumor metastasis. This may have to do with its ability to interact with β-catenin and to enhance its stability by impairing the interaction of β-catenin with GSKβ and inhibiting its phosphorylation, leading to activation of Wnt/β-catenin signaling.67 Also in colorectal cancer, lncRNA SNHG15 is upregulated. SNHG15 maintains Slug stability by suppression of its ubiquitination and degradation through interaction with the zinc finger domain of Slug.68
In non-small cell lung cancer LSINCT5 interacts with HMGA2 and increases the stability of HMGA2 by inhibiting proteasome-mediated degradation.69 LINC00473 can interact with oncoprotein survivin and regulate its stability in hepatocellular carcinoma. Moreover, LINC00473 could recruit deubiquitinase USP9X to inhibit the ubiquitination level of survivin and then increase survivin expression.70 In cervical cancer lnc-UICC is highly expressed and it can directly interacts with the p-STAT3, and increases its protein stability by protecting it from proteasome-dependent degradation.71 LncRNA NBAT1 is able to interact with PSMD10 and promotes its degradation, such that the occupancy of PSMD10 and HSF1 in the ATG7 promoter is decreased and ATG7 transcription is suppressed.72
Finally, as discussed above, EZH2 serves as a key epigenetic regulator and EMT inducer, and it participates in metastasis in a variety of cancers. LncRNA ANCR is involved in regulation of the stability of EZH2. Specifically, ANCR promotes the interaction between CDK1 and EZH2 in such a way that EZH2 phosphorylation is increased, leading to EZH2 ubiquitination and hence degradation.73
Regulation of protein subcellular localization
In addition to the protein level, protein localization is also important for its function. For example, transcription occurs in the nucleus, and as such transcription factors must exert their function in the nucleus. Therefore, protein localization is an important mechanism for efficient functional regulation. This is especially true for proteins that have similar roles in multiple compartments with differing needs. A good example is nuclear receptors. These receptors need to be translocated into the nucleus to function in response to their corresponding ligands. For other types of proteins, their nuclear translocation is subject to protein modifications or interactions with other proteins. Accumulating evidence suggests that lncRNAs can play a role in modulating these processes.
It is well known that NF-κB is a pleiotropic transcription factor for a large number of genes. Under normal conditions, NF-κB stays in the cytoplasm as a heterotrimeric complex consisting of the subunits p50, p65, and the inhibitory subunit IκBα. In response to inducing stimuli such as cytokines, IκBα undergoes phosphorylation, ubiquitination and proteolytic degradation. The p65 subunit then undergoes phosphorylation and moves into the nucleus where it binds to specific DNA sequence and can activate the transcription of hundreds of genes.74 The phosphorylation of IκBα is catalyzed by IκBα kinase (IKK), which consists of three subunits, IKK-α, IKK-β, and IKK-γ (also called NEMO). Aberrant regulation of NF-κB and the signaling pathways that control its activity often leads to inflammation, drug/radiation resistance, and tumorigenic potential of cancer cells.75 Several lncRNAs have been shown to regulate NF-κB activity. For example, NF-κB Interacting LncRNA (NKILA) interacts with the NF-κB/IκB complex in such a way that NKILA prevents phosphorylation of IκB by IKK.76 NKILA forms a stable heterotrimeric complex. Furthermore, NKILA is upregulated by NF-κB, forming a negative feedback loop.76 Therefore, this example illustrates that lncRNAs can function as a scaffold to recruit various proteins in signaling pathways.
In the cell nucleus, there are several types of sub-nuclear structures, such as nucleoli, promyelocytic leukemia (PML) bodies and paraspeckles. These sub-nuclear structures are specific types of RNP complexes, and lack a membrane separating them from the nucleoplasm and thus they are often very dynamic. They play a role in regulating the expression of certain genes in differentiated cells by nuclear retention of RNA. Especially for nuclear paraspeckles, a large number of studies have been carried out because their abnormal formation is often associated with pathological conditions such as cancer and neurodegenerative diseases. As RNP complexes, they carry core paraspeckle proteins such as PSF/SFPQ, P54NRB/NONO, and PSPC1 where lncRNAs such as NEAT1 may act as platforms for such nuclear organization.77
Of significance, NEAT1 and paraspeckle formation are increased in cells upon exposure to a variety of environmental stresses. This may probably be due to the possibility that NEAT1 is a p53 inducible gene. For example, activation of p53, pharmacologically or by oncogene-induced replication stress, stimulates the formation of paraspeckles.78 In contrast, NEAT1 knockdown prevents paraspeckle formation, and sensitizes preneoplastic cells to DNA-damage-induced cell death and impairs skin tumorigenesis. Structural studies indicate that NEAT1 can be divided into several modular domains and the middle domain is responsible for paraspeckle assembly through interaction with NONO/SFPQ.79 Of interest, a recent study show that mitochondrial proteins are also involved in regulation of NEAT1 expression and paraspeckle formation, suggesting a cross-regulation between NEAT1-mediated formation of paraspeckles and mitochondria80
There are several examples that lncRNAs can impact protein nuclear localization. LncRNA Firre can interface with and modulate nuclear architecture across chromosomes.81 MAL is a cell differentiation protein, and functions as a coactivator of serum response factor (SRF) for transcription in responds to G-actin. Along with SRF, MAL can bind to corresponding promoters for transcription. In the other word, MAL has to get into the nucleus to function. In this regard, lncRNA CRYBG3 has been shown to interact with G-actin to inhibit its polymerization such that this interaction between CRYBG3 and G-actin blocks nuclear localization of MAL. A consequence is that SRF is kept away from the promoter region of several immediate early genes such as JUNB and Arp3.82 Finally, direct interaction of lncRNAs with target proteins can also impact their nuclear trafficking. For example, TP53TG1 can interact with YBX1, a transcription factor for growth promoting genes, and this interaction prevents its nuclear localization, leading to transcription repression.83
LncRNAs as oncogenes or tumor suppressors
We have discussed how lncRNAs regulate splicing, mRNA/protein stability and protein subcellular localization, and all of these processes ultimately impact tumorigenesis. Thus, like protein-coding genes, lncRNAs can function as oncogenes or tumor suppressor genes. There is overwhelming evidence supporting the role of lncRNAs in cancer in the literature. For example, HOTAIR is remarkably overexpressed in breast tumors and the upregulation of HOTAIR in primary breast tumors is a strong prognosis marker of patient outcomes such as metastasis and patient survival.16 Furthermore, ectopic expression of HOTAIR causes altered histone H3 Lys27 (H3K27) methylation pattern and increases invasiveness. In contrast, the depletion of HOTAIR results in the opposite cellular phenotype. In prostate cancer, a number of lncRNAs are dysregulated, including PCGEM1 and ANRIL.84, 85, 86 For example, ANRIL plays a role in repression of the tumor suppressors INK4a/p16 and INK4b/p15.86, 87 Our own work suggests that while Linc-RoR30 may function as an oncogene through suppression of p53 in response to DNA damage, loc28519488 and GAS589 may play a tumor suppressive role through the “competitive endogenous RNA” (CeRNA) mechanism.90
In particular, recent two studies provide further supporting evidence for the role of lncRNAs in cancer by analysis of large TCGA clinical specimens. For instance, epigenetic landscape analysis reveals that in contrast to the CpG island hypermethylation phenotype in cancer, over 1,000 lncRNAs are hypomethylated at their promoters. Among them is EPIC1 (epigenetically-induced lncRNA1). Overexpression of EPIC1 is associated with poor prognosis in luminal B breast cancer patients and enhances tumor growth in vitro and in vivo.91 This study suggests that most lncRNAs may play an oncogenic role. The second study shows that a number of potential oncogenic or tumor suppressive lncRNAs are dysregulated; perturbations of these lncRNAs alter proliferation of breast and gynecologic cancer cells.92 Of interest, although most lncRNAs are dysregulated in a tumor-specific manner, some, including OIP5-AS1, TUG1, NEAT1, MEG3, and TSIX, synergistically dysregulate cancer pathways in multiple tumor contexts.92 Of great interest, a recent study suggests that lncRNAs may also impact immunotherapy because a large number of tumor-specific antigens can be derived from non-coding regions.93 Together, these findings highlight the significance of lncRNAs in cancer.
However, a provocative and controversial example is MALAT1. Vast literature evidence indicates that MALAT1 plays an oncogenic role in a variety of cancers. By contrast, a recent study suggests that MALAT1 serves as metastasis-suppressing lncRNA in breast cancer. They provide ample of evidence to support this conclusion, such as targeted inactivation of the MALAT1 gene in a transgenic mouse model of breast cancer; knockout of MALAT1 in human breast cancer cells, overexpression of MALAT1 suppresses breast cancer metastasis in transgenic, xenograft, and syngeneic models. Underlying mechanism may involve regulation of YAP pathway.94 Thus, it remains to be seen whether these findings can be further verified in different laboratories.
Concluding remarks and future directions
LncRNAs have received considerable attention in the past decades and are emerging as potentially important players in regulation of biological processes. Further evidence indicates that lncRNAs can impact various aspects of cancer initiation, progression and metastasis by regulation of gene expression. Although lncRNAs have been shown to play a critical role in regulation of gene expression at transcriptional level, accumulating evidence supports the importance of lncRNA-mediated post-transcriptional regulation in cancer.
We have learned that various mechanisms are involved in lncRNA-mediated gene regulation. However, no matter what mechanism is involved, lncRNAs have to work with their partners, (DNA, RNA, protein or even small molecules). A very large group of important lncRNA partners are RBPs which were once considered “boring” proteins, and it turns out that they may be the most interesting proteins in lncRNA-gene regulation network. They participate in the post-transcription events. These RBPs may interact with 3′-UTRs of mRNA to regulate their stability, or they interact exon or intron of pre-mRNA to regulate their splicing. Importantly, lncRNAs may regulate these interactions so that lncRNA can increase or decrease mRNA/protein level or change splicing patterns or their nuclear trafficking. Therefore, identification of lncRNA-associated partners is a critical step to the understanding of lncRNA-mediated gene expression. We expect that high throughput mass spectrometry combined with other techniques such as RNA immunoprecipitation-seq (RIP-seq) would greatly speed up this line of investigation. Therefore, a better understanding the roles of lncRNA-associated RBPs in these events during tumorigenesis would open new avenues for targeted therapies.
Conflict of interest
The authors declare no confliction of interest.
Acknowledgement
This work was supported by NIH grant R01 CA154989 (YM).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Birney E., Stamatoyannopoulos J.A., Dutta A. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 3.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 5.Wilusz J.E. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859(1):128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Chen L., Chen B. Mammalian ncRNA-disease repository: a global view of ncRNA-mediated disease network. Cell Death Dis. 2013;4:e765. doi: 10.1038/cddis.2013.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 8.Hayes J., Peruzzi P.P., Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Mercer T.R., Mattick J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 10.Blythe A.J., Fox A.H., Bond C.S. The ins and outs of lncRNA structure: how, why and what comes next? Biochim Biophys Acta. 2016;1859(1):46–58. doi: 10.1016/j.bbagrm.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Koirala Pratirodh, Zou D.H., Mo Yin-Yuan. Long non-coding RNAs as key regulators of cancer metastasis. J Cancer Metastasis Treat. 2016;2(1):10. [Google Scholar]
- 12.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornienko A.E., Guenzl P.M., Barlow D.P., Pauler F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa F.F. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357(2):83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 16.Gupta R.A., Shah N., Wang K.C. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttman M., Donaghey J., Carey B.W. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung T., Wang Y., Lin M.F. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khalil A.M., Guttman M., Huarte M. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prensner J.R., Iyer M.K., Balbin O.A. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai M.C., Manor O., Wan Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 23.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long Y., Wang X., Youmans D.T., Cech T.R. How do lncRNAs regulate transcription? Sci Adv. 2017;3(9):eaao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huarte M., Rinn J.L. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko S., Li G., Son J. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24(23):2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maamar H., Cabili M.N., Rinn J., Raj A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013;27(11):1260–1271. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huarte M., Guttman M., Feldser D. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang A., Zhou N., Huang J. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23(3):340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J., Zhou N., Watabe K. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takayama K., Horie-Inoue K., Katayama S. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32(12):1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han S.P., Tang Y.H., Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430(3):379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 34.Xiong Y., Deng Y., Wang K. Profiles of alternative splicing in colorectal cancer and their clinical significance: a study based on large-scale sequencing data. EBioMedicine. 2018;36:183–195. doi: 10.1016/j.ebiom.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamada M., Suematsu M., Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin Cancer Res. 2012;18(20):5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- 36.Piva F., Giulietti M., Burini A.B., Principato G. SpliceAid 2: a database of human splicing factors expression data and RNA target motifs. Hum Mutat. 2012;33(1):81–85. doi: 10.1002/humu.21609. [DOI] [PubMed] [Google Scholar]
- 37.Shepard P.J., Hertel K.J. The SR protein family. Genome Biol. 2009;10(10):242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geuens T., Bouhy D., Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135(8):851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tripathi V., Ellis J.D., Shen Z. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernard D., Prasanth K.V., Tripathi V. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29(18):3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon M.A., Babbs B., Cochrane D.R., Bitler B.G., Richer J.K. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol Carcinog. 2019 Feb;58(2):196–205. doi: 10.1002/mc.22919. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez I., Munita R., Agirre E. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol. 2015;22(5):370–376. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yap K., Mukhina S., Zhang G., Tan J.S.C., Ong H.S., Makeyev E.V. A short tandem repeat-enriched RNA assembles a nuclear compartment to control alternative splicing and promote cell survival. Mol Cell. 2018;72(3):525–540 e513. doi: 10.1016/j.molcel.2018.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grelet S., Link L.A., Howley B. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19(9):1105–1115. doi: 10.1038/ncb3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez-Mateo C., Torres B., Gutierrez G., Pintor-Toro J.A. Downregulation of Lnc-Spry1 mediates TGF-beta-induced epithelial-mesenchymal transition by transcriptional and posttranscriptional regulatory mechanisms. Cell Death Differ. 2017;24(5):785–797. doi: 10.1038/cdd.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang G.W., Zhang Y.L., Liao L.D., Li E.M., Xu L.Y. Natural antisense transcript TPM1-AS regulates the alternative splicing of tropomyosin I through an interaction with RNA-binding motif protein 4. Int J Biochem Cell Biol. 2017;90:59–67. doi: 10.1016/j.biocel.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 47.Hetz C. BCL-2 protein family. Essential regulators of cell death. Preface. Adv Exp Med Biol. 2010;687 [vii-viii] [PubMed] [Google Scholar]
- 48.Minn A.J., Boise L.H., Thompson C.B. Bcl-x(S) anatagonizes the protective effects of Bcl-x(L) J Biol Chem. 1996;271(11):6306–6312. doi: 10.1074/jbc.271.11.6306. [DOI] [PubMed] [Google Scholar]
- 49.Guo Z., Yang X., Sun F. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69(6):2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z., Zhou N., Huang J. Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget. 2016;7(13):15481–15491. doi: 10.18632/oncotarget.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan J.H., Liu X.N., Wang T.T. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19(7):820–832. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- 53.van der Houven van Oordt W., Diaz-Meco M.T., Lozano J., Krainer A.R., Moscat J., Caceres J.F. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149(2):307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lan Y., Xiao X., He Z. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46(11):5809–5821. doi: 10.1093/nar/gky214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang J., Zhang A., Ho T.T. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 2016 Apr 20;44(7):3059–3069. doi: 10.1093/nar/gkv1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Y., Liu X., Xie M. The NF-kappaB-responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J Immunol. 2017;199(10):3571–3582. doi: 10.4049/jimmunol.1700091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y., Liu Y., Lin L. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17(1):69. doi: 10.1186/s12943-018-0820-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao L., Zhang P., Li J., Wu M. LAST, a c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote CCND1 mRNA stability in human cells. Elife. 2017;6 doi: 10.7554/eLife.30433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C., Gu Y., Zhang E. A cancer-testis non-coding RNA LIN28B-AS1 activates driver gene LIN28B by interacting with IGF2BP1 in lung adenocarcinoma. Oncogene. 2018 Oct 23 doi: 10.1038/s41388-018-0548-x. [Epub ahead of print] PubMed PMID: 30353165. [DOI] [PubMed] [Google Scholar]
- 60.Xiao L., Wu J., Wang J.Y. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of MicroRNA 195. Gastroenterology. 2018;154(3):599–611. doi: 10.1053/j.gastro.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmitt A.M., Garcia J.T., Hung T. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet. 2016;48(11):1370–1376. doi: 10.1038/ng.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu W.L., Jin L., Xu A. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. 2018;20(4):492–502. doi: 10.1038/s41556-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 63.Colbert L.E., Petrova A.V., Fisher S.B. CHD7 expression predicts survival outcomes in patients with resected pancreatic cancer. Cancer Res. 2014;74(10):2677–2687. doi: 10.1158/0008-5472.CAN-13-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mondal T., Juvvuna P.K., Kirkeby A. Sense-antisense lncRNA pair encoded by locus 6p22.3 determines neuroblastoma susceptibility via the USP36-CHD7-SOX9 regulatory axis. Cancer Cell. 2018;33(3):417–434. doi: 10.1016/j.ccell.2018.01.020. e417. [DOI] [PubMed] [Google Scholar]
- 65.Xiong H., Ni Z., He J. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36(25):3528–3540. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 66.Bian Z., Zhang J., Li M. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24(19):4808–4819. doi: 10.1158/1078-0432.CCR-17-2967. [DOI] [PubMed] [Google Scholar]
- 67.Yu J., Han Z., Sun Z., Wang Y., Zheng M., Song C. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through beta-catenin-dependent Wnt pathway. J Exp Clin Cancer Res. 2018;37(1):222. doi: 10.1186/s13046-018-0896-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang H., Li T., Qu Y. Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor slug and promotes colon cancer progression. Cancer Lett. 2018;425:78–87. doi: 10.1016/j.canlet.2018.03.038. [DOI] [PubMed] [Google Scholar]
- 69.Tian Y., Zhang N., Chen S., Ma Y., Liu Y. The long non-coding RNA LSINCT5 promotes malignancy in non-small cell lung cancer by stabilizing HMGA2. Cell Cycle. 2018;17(10):1188–1198. doi: 10.1080/15384101.2018.1467675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen H., Yang F., Li X., Gong Z.J., Wang L.W. Long noncoding RNA LNC473 inhibits the ubiquitination of survivin via association with USP9X and enhances cell proliferation and invasion in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2018;499(3):702–710. doi: 10.1016/j.bbrc.2018.03.215. [DOI] [PubMed] [Google Scholar]
- 71.Su K., Zhao Q., Bian A., Wang C., Cai Y., Zhang Y. A novel positive feedback regulation between long noncoding RNA UICC and IL-6/STAT3 signaling promotes cervical cancer progression. Am J Cancer Res. 2018;8(7):1176–1189. [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng T., Li D., He Z., Feng S., Zhao S. Long noncoding RNA NBAT1 inhibits autophagy via suppression of ATG7 in non-small cell lung cancer. Am J Cancer Res. 2018;8(9):1801–1811. [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z., Hou P., Fan D. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24(1):59–71. doi: 10.1038/cdd.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gupta S.C., Sundaram C., Reuter S., Aggarwal B.B. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta. 2010;1799(10-12):775–787. doi: 10.1016/j.bbagrm.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aggarwal B.B. Nuclear factor-kappaB: the enemy within. Cancer Cell. 2004;6(3):203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 76.Liu B., Sun L., Liu Q. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 77.Clemson C.M., Hutchinson J.N., Sara S.A. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adriaens C., Standaert L., Barra J. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22(8):861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 79.Yamazaki T., Souquere S., Chujo T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol Cell. 2018;70(6):1038–1053. doi: 10.1016/j.molcel.2018.05.019. e1037. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y., Hu S.B., Wang M.R. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat Cell Biol. 2018;20(10):1145–1158. doi: 10.1038/s41556-018-0204-2. [DOI] [PubMed] [Google Scholar]
- 81.Hacisuleyman E., Goff L.A., Trapnell C. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21(2):198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pei H., Hu W., Guo Z. Long noncoding RNA CRYBG3 blocks cytokinesis by directly binding G-Actin. Cancer Res. 2018;78(16):4563–4572. doi: 10.1158/0008-5472.CAN-18-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Diaz-Lagares A., Crujeiras A.B., Lopez-Serra P. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc Natl Acad Sci U S A. 2016;113(47):E7535–E7544. doi: 10.1073/pnas.1608585113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Srikantan V., Zou Z., Petrovics G. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 2000;97(22):12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Petrovics G., Zhang W., Makarem M. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23(2):605–611. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 86.Yap K.L., Li S., Munoz-Cabello A.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kotake Y., Nakagawa T., Kitagawa K. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Q., Huang J., Zhou N. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41(9):4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Z., Zhu Z., Watabe K. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20(11):1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Z., Yang B., Zhang M. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018 Apr 9;33(4):706–720.e9. doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chiu H.S., Somvanshi S., Patel E. Pan-cancer analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep. 2018;23(1):297–312. doi: 10.1016/j.celrep.2018.03.064. e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Laumont C.M., Vincent K., Hesnard L. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med. 2018;10(470) doi: 10.1126/scitranslmed.aau5516. [DOI] [PubMed] [Google Scholar]
- 94.Kim J., Piao H.L., Kim B.J. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018 Dec;50(12):1705–1715. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]