Abstract
Purpose of the research
Although single nucleotide polymorphisms of membrane-spanning 4A (MS4A) (rs670139) and several other susceptibility genes have shown interaction effects on the risk of Alzheimer's disease (AD), little is known about the interaction effects of apolipoprotein E (APOE) with MS4A (rs670139) on cognitive performances, and the underlying pathogenesis is unclear. The study aimed to investigate the APOE-MS4A (rs670139) interaction effects on cognitive performances, cortical volumes, and functional connectivity (FC) in brain networks.
Principal results
Cognitive performances were characterized in each genotypic group, and were compared between normal controls and patients in each genotypic group. APOE-MS4A interaction effects on memory and executive function scores, cortical volumes, and FC in brain networks were demonstrated. Significant effects of APOE-MS4A interactions on FC were observed in executive control network (ECN) (T maxima = 4.99, false discovery rate-corrected p < .001), the calculation score (F3, 87 = 6.218; p = .015), and the volume in prefrontal (F3, 87 = 4.374; p = .039) and orbitofrontal cortices (F3, 87 = 6.022; p = .016). The calculation score was correlated with each frontal volume (cc) (ρ = 0.304; p = .004) and genetic interaction-associated FC in ECN (ρ = 0.282; p = .008). Variations in genotypes affected the relationship between the calculation score and each frontal volume (cc).
Major conclusions
These findings indicate that the genetic interaction effects on FC in ECN might contribute to pathogenic mechanisms underlying the interaction effects of APOE-MS4A on calculation ability in AD.
Abbreviations: Aβ, amyloid β; AD, Alzheimer's disease; ANOVA, analysis of variance; apoE, apolipoprotein E; CDR, Clinical Dementia Rating; CVVLT, Chinese Version Verbal Learning Test; DLPFC, dorsolateral prefrontal cortex; DMN, default mode network; DSB, Digit Span Backward; ECN, executive control network; ε4+ carriers, APOE-ε4 carriers; ε4 non-carriers, APOE-ε4 non-carriers; ε4+/GG-carriers, APOE-ε4 carriers with MS4A-rs670139-GG genotype; ε4+/T-allele-carriers, APOE-ε4 carriers with MS4A-rs670139-T-allele genotype; ε4−/GG-carriers, APOE-ε4 non-carriers with MS4A-rs670139-GG genotype; ε4−/T-allele-carriers, APOE-ε4 non-carriers with MS4A-rs670139-T-allele genotype; FC, functional connectivity; FDR, false discovery rate; GM, grey matter; HWE, Hardy-Weinberg equilibrium; MMSE, Mini-Mental State Examination; MNI, Montreal Neurological Institute; MRI, magnetic resonance imaging; NFTs, neurofibrillary tangles; NCs, normal controls; OFI, orbital frontoinsular; PCC, posterior cingulate cortex; rs-fMRI, resting state-functional MRI; SPM 12, Statistic Parametric Mapping software version 12; TIV, total intracranial volume; TMB, Trail Making Test B
Keywords: APOE, Calculation, Executive control network, Genetic interaction, MS4A
Highlights
-
•
APOE-MS4A interactions affect executive control network (ECN) and calculation score.
-
•
Calculation score is correlated with functional connectivity in ECN.
-
•
Variation in ECN is associated with genetic effects on calculation performances.
-
•
The calculation score is correlated with each frontal volume.
-
•
Genotypic variation affects the relationship between calculation and frontal volume.
1. Introduction
The early symptoms in the evolution of Alzheimer's disease (AD) are memory deficits, followed by a progressive decline in visuospatial, attention, and executive function (Petersen and Edotor, 2003). Executive function consists of complex attention, planning, initiation, monitoring, inhibition, and flexibility in goal-oriented behavior (Cahn-Weiner et al., 2007). Executive impairment has been associated with loss of independence in patients with AD (Boyle et al., 2003). Executive function deficits in the early stage of AD are shown to be associated with a rapid decline in instrumental activities of daily living in later stages of dementia (Cahn-Weiner et al., 2007). In addition, symptomatic treatment of executive dysfunction at the mild dementia phase of AD has been suggested to improve the quality of life of the patients (Martyr and Clare, 2012). The pathophysiological process underlying the severity of executive deficits in AD patients may be associated with frontal atrophy (Morgen et al., 2014), decreased functional connectivity (FC) within the executive control network (ECN) (Seeley et al., 2007), and genetic factors (Morgen et al., 2014).
With regards to genetic effects, a meta-analysis demonstrates that apolipoprotein E (APOE-ε4) carriers perform significantly worse on executive function tasks than non-carriers do (Wisdom et al., 2011). The potential mechanism underlying the executive impairment is that APOE-ε4 carriers have lower prefrontal volume than non-carriers do (Wishart et al., 2006; Honea et al., 2009; Bender and Raz, 2012). However, APOE-ε4 carriers sometimes show better performance on executive function tasks than non-carriers do (Wolk and Dickerson, 2010). Increased FC between the prefrontal cortex and the anterior cingulate cortex has been suggested as a neural basis for better executive performances in APOE-ε4 carriers than in non-carriers (Machulda et al., 2011); however, a link between APOE-ε4 carrier genotype and decreased neuronal activity in prefrontal areas, as revealed by fluorodeoxyglucose-positron emission tomography has been observed in the prefrontal areas (Arenaza-Urquijo et al., 2015). These discrepancies may be due to interaction effects of APOE with age (Bender and Raz, 2012), years of education (Arenaza-Urquijo et al., 2015), or genes (Morgen et al., 2014) on the influence of APOE-ε4 carrier status on executive performances, prefrontal volumes, or prefrontal activity.
In addition to APOE-ε4 carrier status, single nucleotide polymorphisms in nine susceptibility genes-namely ABCA7 (Reitz et al., 2013), CR1, CLU, PICALM, BIN1 (Seshadri et al., 2010), CD2AP, CD33, EPHA1 (Naj et al., 2011), and membrane-spanning 4A (MS4A) (Naj et al., 2011)-have been shown to be associated with the risk of AD. These genes fall into several functional pathways, including immune response, cholesterol metabolism, and synaptic function (Karch et al., 2012; Tanzi, 2012). As ion channel adaptor proteins (Zuccolo et al., 2010), MS4A family members are involved in immune response (Antunez et al., 2011) and cellular signal transduction (Tanzi, 2012) in AD. The MS4A minor allele T (Hollingworth et al., 2011) in rs670139 has been found to be associated with increased amyloid β (Aβ) plaques and intracellular neurofibrillary tangles (NFTs) (Karch et al., 2012). This minor allele of MS4A (rs670139) may have a detrimental effect on clinical severity by affecting the NFT load (Karch et al., 2012); however, MS4A-rs670139-T-allele sometimes is shown to have a protective role in the pathogenesis of AD (Naj et al., 2011). Discrepancies between the previous studies suggest the possible effects of gene-environment or gene-gene interactions (epistasis).
Genetic interactions between APOE and PICALM have been found to be associated with structural and clinical heterogeneity of AD (Morgen et al., 2014). Evidence has revealed the interaction effects of MS4A (rs670139) with CLU (Lambert et al., 2013) or CD33 (Lambert et al., 2013; Ebbert et al., 2014) on conferring risk of AD. Even though MS4A and APOE are both involved in the control of inflammation (Antunez et al., 2011; Dorey et al., 2014), synaptic function (Herz et al., 2009; Tanzi, 2012), Aβ generation (Karch et al., 2012; Dorey et al., 2014), and NFT phosphorylation (Han et al., 2010; Karch et al., 2012), little is known about the interaction effects of MS4A with APOE on these biologic mechanisms. Nonetheless, the interaction between MS4A and APOE is likely to have phenotypic relevance, which would contribute to the heterogeneity of AD in terms of clinical symptoms, cortical volumes, and FC.
In the present study, we characterized the pattern of cognitive performances in 91 CE patients with different genotypes, and we compared the cognitive performances between these AD patients and normal controls (NCs). We investigated the possible interaction effects of APOE with MS4A (rs670139) on memory and executive function scores, hippocampal and frontal volume, and FC in default mode network (DMN) and ECN. Through these analyses, this study aimed to explore the contribution of these genetic variants to clinical and pathological heterogeneity in AD.
2. Materials and methods
2.1. Inclusion and exclusion criteria
Ninety-one patients with AD were enrolled from the Department of Neurology of Chang Gung Memorial Hospital from 2011 to 2017. They were diagnosed according to the International Working Group criteria (Dubois et al., 2010) with a clinically typical AD. The diagnostic biomarkers included the medial temporal lobe atrophy on magnetic resonance imaging (MRI), and/or reduced glucose metabolism in temporoparietal regions on fluorodeoxyglucose-positron emission tomography, and/or increased florbetapir retention on amyloid positron emission tomography. The exclusion criteria included a history of clinical stroke, a modified Hachinski ischemic score > 4 (Rosen et al., 1980), or depression. The patients were included on the basis of a consensus of panels composed of neurologists, neuroradiologists, and experts in nuclear medicine. Only mild-stage AD patients with a Clinical Dementia Rating (CDR) score of 0.5 or 1 were included in this study. All AD patients were under stable treatment with acetylcholine esterase inhibitors from the time of diagnosis. NCs were recruited from an outpatient clinic with a 1:1 age-, sex-, and education (years)-matched to AD patients. Twenty-nine included NCs had no underlying neurological or psychiatric disorder, no subjective or objective cognitive deficits, and no neuroimaging abnormalities.
2.2. Study design
The study was approved by Chang Gung Memorial Hospital's Institutional Review Committee on Human Research, and all of the participants and their authorized caregivers (when appropriate) provided written informed consent. Cognitive testing and MRI were all performed within a period of four weeks.
2.3. Genotyping
Genomic DNA was extracted from blood samples using a commercial kit (Gentra Puregene Blood Kit, Qiagen, USA), followed by genotyping for SNP rs670139 at the MS4A gene using the polymerase chain reaction-restriction fragment length polymorphism method (Elias-Sonnenschein et al., 2013). The APOE genotype was also determined. Genotyping was conducted by an operator blinded to the clinical data. Patients were classified into two genotypic groups based on the MS4A SNP: rs670139-GG carriers (GG-carriers) and rs670139-T-allele carriers (T-allele-carriers). Those with one or two APOE-ε4 alleles were defined as APOE-ε4 carriers (ε4+ carriers), and the remainder were classified as APOE-ε4 non-carriers (ε4 non-carriers). Among the 53 ɛ4 non-carriers, 52 non-carriers were homozygous for the ɛ3 allele (ɛ3/ɛ3) and one non-carrier was heterozygous (ɛ2/ɛ3). In addition, 33 ɛ4+ carriers were heterozygous (ɛ3/ɛ4), and five carriers were homozygous (ɛ4/ɛ4). The χ2 test was used to assess whether the allele frequencies agreed with expectation in Hardy-Weinberg equilibrium (HWE). Statistical significance was set at P < .05.
2.4. MRI acquisition
MRI scans were acquired on a GE 3 T Signa Excite scanner (GE Medical System, Milwaukee, WI). The scanning protocol of T1-weighted imaging included inversion-recovery-prepared, three-dimensional, spoiled, gradient-recalled acquisition in a steady-state sequence with a repetition time/inversion time of 8600 ms/450 ms, 240 × 240 mm field of view, and 1-mm slice thickness. Resting state-functional MRI (rs-fMRI) scans were performed with the patients' eyes closed using a T2*-weighted echo-planar imaging sequence (TR 2500 ms, TE 45 ms, FOV 240 × 240 mm, flip angle 10°, thickness 4 mm, and 200 scans of 32 contiguous axial slices) with a total scanning time of 10 min per subject.
2.5. Structural MRI pre-processing
The Statistic Parametric Mapping software version 12 (SPM 12) (http://www.fil.ion.ucl.ac.uk/spm/software/) was used to pre-process T1 MRI scans, including removal of non-relevant tissue, intensity and spatial normalization to the Montreal Neurological Institute (MNI) space, and tissue segmentation. Using Segmentation in SPM 12, the images were segmented into grey matter (GM) and white matter. The raw volume (cc) in hippocampus, orbitofrontal and prefrontal cortices, and total intracranial volume (TIV) (cc) were estimated with surface-based atlas maps in Computational Anatomy Toolbox 12 in SPM12 (Gaser and Dahnke, 2016). TIV-adjusted regional volume was calculated by raw volume (cc) divided by TIV (cc).
2.6. Rs-fMRI pre-processing
The first 10 volumes of rs-fMRI were discarded to reduce fluctuations in MRI signals. The steps of pre-processing rs-fMRI included slice time correction, realignment, segmentation, normalization into standard stereotactic MNI spaces, spatial smoothing using a Gaussian Kernel of 6 mm, and resampling to 2 × 2 × 2 mm3. Subsequently, images were detrended and filtered to 0.008 ∼ 0.09 Hz. Bad volumes were detected and repaired if global average blood oxygen level dependent signal from scan to scan exceeded 1% variation and/or if framewise displacement exceeded 0.25 mm/TR. White matter and cerebrospinal fluid signals were regressed out from each voxel using an anatomical component-based noise correction method as implemented in CONN toolbox (http://www.nitrc.org/projects/conn) (Behzadi et al., 2007; Whitfield-Gabrieli and Nieto-Castanon, 2012).
2.7. Neuropsychological assessments
Memory function was examined using Chinese Version Verbal Learning Test (CVVLT) (Chang et al., 2010), assessing number of items retrieved over four learning trials of a 9-word list in free recall after 10 min (CVVLT-10 min). A modified Rey–Osterrieth Complex Figure copy for visuospatial function (Boone, 2000) was also evaluated. The CDR and Mini-Mental State Examination (MMSE) (Folstein et al., 1975; Morris, 1993) were used to assess the general intellectual function. Executive function was evaluated with Digit Span Backward (DSB) and Trail Making Test B (TMB) (Weintraub et al., 2009). In addition, calculation ability was determined, and the genetic and neural basis for calculation ability was investigated in the present study. Early calculation impairment in AD manifests as difficulties with complex calculation procedures, especially with multiplication, followed by complex addition and subtraction, and lastly by simple addition and subtraction (Mantovan et al., 1999). Difficulties in multiplication in AD patients have been suggested to arise from a monitoring deficit, which is one of the executive function components (Cahn-Weiner et al., 2007). Calculation processing has been associated with increased functional activity in the prefrontal cortex, suggesting the role of ECN in the calculation ability (Menon et al., 2000). Although the calculation ability depends on previous math skills, linguistic abilities, and executive function (Jogi and Kikas, 2016), the calculation score might be associated with executive performances after controlling for educational level of each patient. Moreover, evaluating the calculation performance might play an important role in AD because it has been suggested as a tool for cognitive rehabilitation in dementia patients (Kawashima et al., 2005). Our calculation tests comprised two additions, two subtractions, and one multiplication. Each addition and each subtraction consisted of one and two items respectively, and the multiplication is composed of a multiplicand of >100 and a multiplier of <100 (e.g. 214 × 35). Patients were asked to perform the computations in a written form. Calculation performance was assessed using number of correct answers over the five calculation tests.
2.8. Statistical analysis
2.8.1. Comparison of clinical and volumetric variables
Clinical and volumetric data were expressed as mean ± standard deviation. Based on the study rationale, the patients were further classified into four genotypic groups: ε4+ carriers with MS4A-rs670139-GG genotype (ε4+/GG-carriers); ε4+ carriers with MS4A-rs670139-T-allele genotype (ε4+/T-allele-carriers); ε4 non-carriers with MS4A-rs670139-GG genotype (ε4−/GG-carriers); and ε4 non-carriers with MS4A-rs670139-T-allele genotype (ε4−/T-allele-carriers). The Mann-Whitney U test for continuous variables and χ2 test for dichotomous variables were used to compare among each genotypic group and control group. All statistical analyses for continuous variables were conducted using SPSS software (SPSS version 22 for Windows®, SPSS Inc., Chicago, IL).
2.8.2. Interaction effects on cognitive and volumetric variables
Analysis of variance (ANOVA) was used to identify significant effects of the interactions between APOE (ε4+ carriers versus ε4 non-carriers) and MS4A (GG- versus T-allele-carriers) on cognitive function scores and regional volume.
We used partial correlation to analyze the relationship between each frontal volume and each executive function score in all AD patients and AD patients in each genotypic group. We then used Fisher transformation to further analyze the differences in correlation coefficient value of ρ, which indicates the relation of each executive function score with each frontal volume between different genotype group.
2.8.3. Interaction effects on cognition and FC in brain networks
An ANOVA was used to investigate the effects of the interactions between APOE and MS4A (rs670139) on FC within DMN anchored by the posterior cingulate cortex (PCC) seed (radius 10 mm), and on FC within ECN anchored by the dorsal lateral prefrontal cortex (DLPFC) and those of orbital frontoinsular (OFI) seeds (radius 10 mm). MNI coordinates of PCC seed were x = −2, y = −36, z = 35 (Margulies et al., 2009), while those of DLPFC seed were x = 44, y = 36, z = 20 and OFI seed were x = 38, y = 26, z = −10 (Seeley et al., 2007). The ANOVA was only conducted in AD patients.
The ANOVA was computed in CONN toolbox (Whitfield-Gabrieli and Nieto-Castanon, 2012) with threshold of uncorrected p < .001 at peak-level and false discovery rate (FDR)-corrected p < .05 at cluster-level, using second-level analysis of relative functional inter-regional covariance, the FC in brain networks. The whole-brain correlation maps were converted into z-score maps using a Fisher's r-to-z transformation. After demonstrating the genetic effect on FC in the brain networks, FC between each seed and each peak cluster was further extracted (Whitfield-Gabrieli and Nieto-Castanon, 2012) to correlate with the cognitive function scores.
Because education (years) had significant correlation with each executive function score (p < .05), the correlation of executive function scores with other variables was analyzed using a partial correlation after controlling for education (years).
3. Results
3.1. Clinical and pathological characteristics
In total, 91 CE patients (32 GG-carriers and 59 T-allele-carriers) and 29 NCs completed the study. AD patients within each genotypic group had significantly lower scores in MMSE, CDR, and CVVLT-10 min than NCs (P < .05). AD patients in the genotypic groups except the ε4−/GG-carrier group had lower hippocampal volume than NCs (P < .05). Other detailed results of comparison between NCs and patients in each genotypic group are presented in Table 1. The distribution of the MS4A-rs670139-TT genotype conformed to HWE with χ2 = 0.006 and P = .939, and the distribution of the APOE-ε4 carrier genotype conformed to HWE with χ2 = 0.836 and P = .360. Allele frequencies did not violate the expectation in HWE.
Table 1.
Demographic and clinical data in the genotypic groups.
| Normal controls | ε4+/GG-carriers | ε4+/T-allele-carriers | ε4−/GG-carriers | ε4−/T-allele-carriers | P value | |
|---|---|---|---|---|---|---|
| Sample size (n) | 29 | 17 | 21 | 15 | 38 | |
| Age (years) | 69.9 ± 5.1 | 70.0 ± 6.2 | 72.2 ± 8.1 | 70.1 ± 7.4 | 72.6 ± 8.2 | 0.453 |
| Sex (woman/man) | 12/17 | 9/8 | 11/10 | 4/11 | 15/23 | 0.510 |
| Education (years) | 8.8 ± 4.3 | 8.2 ± 5.5 | 7.7 ± 5.3 | 9.7 ± 4.4 | 8.4 ± 5.0 | 0.798 |
| MMSE | 27.0 ± 2.3 | 20.3 ± 6.5* (<0.001) | 21.9 ± 4.9* (0.001) | 21.8 ± 6.6* (0.002) | 23.1 ± 5.4* (0.002) | <0.001 |
| Clinical Dementia Rating | 0.0 ± 0.0 | 0.6 ± 0.3* (<0.001) | 0.6 ± 0.3* (<0.001) | 0.6 ± 0.2* (<0.001) | 0.5 ± 0.2* (<0.001) | <0.001 |
| Memory function score | ||||||
| CVVLT-10 min | 7.0 ± 1.6 | 2.5 ± 3.4* (<0.001) | 3.2 ± 3.4* (<0.001) | 5.0 ± 2.2*†a (0.014) | 5.3 ± 2.1*†b (0.008) | <0.001 |
| Visuospatial function score | ||||||
| Modified ROCF copy | 16.7 ± 0.9 | 13.6 ± 5.4* | 14.8 ± 4.1 | 14.6 ± 4.4 | 14.8 ± 4.8 | 0.140 |
| Executive function score | ||||||
| TMB (seconds) | 72.7 ± 35.7 | 94.8 ± 37.6* (0.040) | 97.1 ± 34.8* (0.016) | 93.1 ± 29.1 (0.068) | 88.7 ± 34.8 (0.065) | 0.097 |
| Digital span backward | 4.5 ± 1.4 | 3.1 ± 1.7* (0.004) | 3.7 ± 1.6 (0.077) | 4.1 ± 1.7 (0.466) | 3.7 ± 1.3* (0.028) | 0.038 |
| Calculation | 4.5 ± 1.1 | 3.2 ± 1.9* (0.002) | 4.1 ± 1.0†c (0.358) | 4.2 ± 1.3†d (0.560) | 3.6 ± 1.4* (0.008) | 0.011 |
| Regional volume (cc) | ||||||
| Hippocampus | 3.7 ± 0.4 | 2.8 ± 0.7* (<0.001) | 3.1 ± 0.6* (<0.001) | 3.4 ± 0.6†e (0.153) | 3.4 ± 0.6*†f (0.022) | <0.001 |
| Orbitofrontal gyrus | 19.5 ± 2.2 | 19.1 ± 2.3 | 18.6 ± 1.8 | 18.4 ± 2.7 | 19. ± 2.6 | 0.540 |
| Prefrontal gyrus | 63.3 ± 6.5 | 61.5 ± 7.0 | 62.3 ± 5.4 | 60.5 ± 7.8 | 61.9 ± 8.1 | 0.784 |
Data are presented as mean ± standard deviation (P value compared with normal controls); P value denotes significant differences among groups on Mann-Whitney U test for continuous variables, and χ2 test for dichotomous variables. *P < .05, compared with normal controls. †P < .05, compared with ε4+/GG-carriers; P of †a = 0.05, †b < 0.001, †c = 0.037, †d = 0.033, †e = 0.003, and †f = 0.001. CVVLT-10 min, free recall after 10 min in Chinese version of the Verbal Learning Test; ε4+/GG-carriers: APOE-ε4 carriers (ε4+ carriers) with MS4A-rs670139-GG genotype; ε4+/T-allele-carriers: ε4+ carriers with MS4A-rs670139-T-allele genotype; ε4−/GG-carriers: APOE-ε4 non-carriers (ε4− carriers) with MS4A-rs670139-GG genotype; ε4−/T-allele-carriers: ε4− carriers with MS4A-rs670139-T-allele genotype; MMSE, Mini-Mental State Examination; ROCF, Rey-Osterrieth Complex Figure; TMB, Trail Making Test B.
Because education (years) significantly correlated with the scores in calculation (ρ = 0.417, p < .001), DSB (ρ = 0.324, p = .002), and TMB (seconds) (ρ = −0.362, p < .001), the correlations of the three cognitive scores with other variables were analyzed after controlling for education (years).
To investigate the relationship between the calculation score and executive performances, we correlated the calculation score with DSB and TMB (seconds) scores after controlling for education (years). The calculation score was significantly correlated with TMB (seconds) (ρ = −0.373, p < .001) and DSB (ρ = 0.447, p < .001) scores. The results indicated that the calculation ability was associated with executive performances in AD patients.
3.2. Effects of genotypic variation on cognitive performances and regional volumes
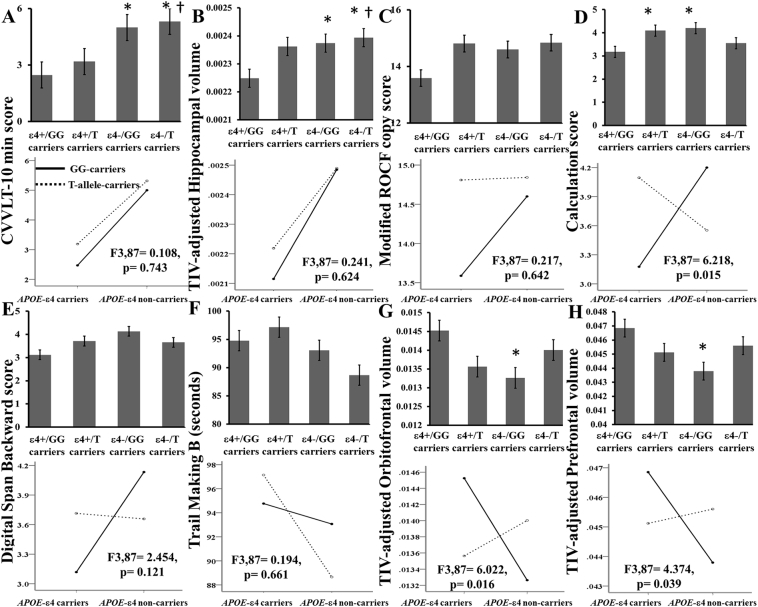
Dose-dependent gradients were observed in CVVLT-10 min score (Fig. 1A) and hippocampal volumes in the four genotypic groups (Fig. 1B), suggesting the main effect of APOE on memory performance and hippocampal volumes. Neither dose-dependent gradient nor interaction effects were observed in modified ROCF copy score (Fig. 1C).
Fig. 1.
Interaction effects of APOE with MS4A (rs670139) on cognitive function scores (A, C–F) and TIV-adjusted volume (B, G-H). *P < .05, compared to the ε4+/GG-carriers. †P < .05, compared with ε4+/T-allele-carriers. ε4+/GG-carriers: APOE-ε4 carriers (ε4+ carriers) with MS4A-rs670139-GG genotype; ε4+/T-allele-carriers: ε4+ carriers with MS4A-rs670139-T-allele genotype; ε4−/GG-carriers: APOE-ε4 non-carriers (ε4- carriers) with MS4A-rs670139-GG genotype; ε4−/T-allele-carriers: ε4- carriers with MS4A-rs670139-T-allele genotype; GG-carriers: MS4A-rs670139-GG carriers; T-allele-carriers: MS4A-rs670139-T-allele carriers. CVVLT-10 min: free recall after 10 min in Chinese version of the Verbal Learning Test; TIV: total intracranial volume.
Regarding executive function scores and frontal volumes, this study showed that there were a significant effect of APOE-MS4A (rs670139) interaction on the calculation score (p = .015; Fig. 1D, Table 2) and a borderline effect of APOE-MS4A (rs670139) interaction on the DSB score (Fig. 1E, Table 2), but the interaction effect on the TMB score (seconds) was not significant (p > .05; Fig. 1F). Moreover, there were interaction effects of APOE with MS4A (rs670139) on the volumes in orbitofrontal and prefrontal cortices (p < .05; Fig. 1G–H, Table 2).
Table 2.
Seed-to-voxel analysis reveals brain regions with significant effects of APOE-MS4A (rs670139) interactions on cognitive function scores, regional volume, and functional connectivity in brain networks.
| Seed | Cluster | MNI (x, y, z) | Cluster size | T | p-FDR of size |
|---|---|---|---|---|---|
| PCC | Left frontal lobe | −26, 4, 24 | 876 | 5.78 | <0.001 |
| DLPFC | Left supramarginal gyrus | −46, −26, 24 | 445 | 4.70 | 0.003 |
| Right medial orbitofrontal gyrus | 6, 70, −4 | 1208 | 4.22 | <0.001 | |
| Orbital Frontoinsular | Right middle orbitofrontal gyrus | 42, 54, −16 | 329 | 4.59 | 0.009 |
| Left middle frontal gyrus | −36, 18, 42 | 1309 | 4.71 | <0.001 | |
| Cognitive function score or regional volume | Main effects | F3,87 | p value |
|---|---|---|---|
| Digital Span Backward score | APOE-ε4 carrier genotype | 1.965 | 0.165 |
| MS4A (rs670139) | 0.031 | 0.860 | |
| APOE-ε4 carrier genotype × MS4A (rs670139) | 2.454 | 0.121 | |
| Calculation score | APOE-ε4 carrier genotype | 0.586 | 0.446 |
| MS4A (rs670139) | 0.187 | 0.667 | |
| APOE-ε4 carrier genotype × MS4A (rs670139) | 6.218 | 0.015 | |
| TIV-adjusted orbitofrontal volume | APOE-ε4 carrier genotype | 1.411 | 0.238 |
| MS4A (rs670139) | 0.103 | 0.749 | |
| APOE-ε4 carrier genotype × MS4A (rs670139) | 6.022 | 0.016 | |
| TIV-adjusted prefrontal volume | APOE-ε4 carrier genotype | 2.312 | 0.132 |
| MS4A (rs670139) | 0.002 | 0.964 | |
| APOE-ε4 carrier genotype × MS4A (rs670139) | 4.374 | 0.039 | |
T maxima, and contiguous voxels of cluster size are shown. The significance clusters are detected with thresholds of FDR-corrected P < .05 at the cluster-level and uncorrected P < .001 at the peak-level. DLPFC, dorsolateral prefrontal cortex; FDR, false discovery rate; MNI (x, y, z), local maxima coordinates on the Montreal Neurological Institute template brain; PCC, posterior cingulate cortex; TIV, total intracranial volume.
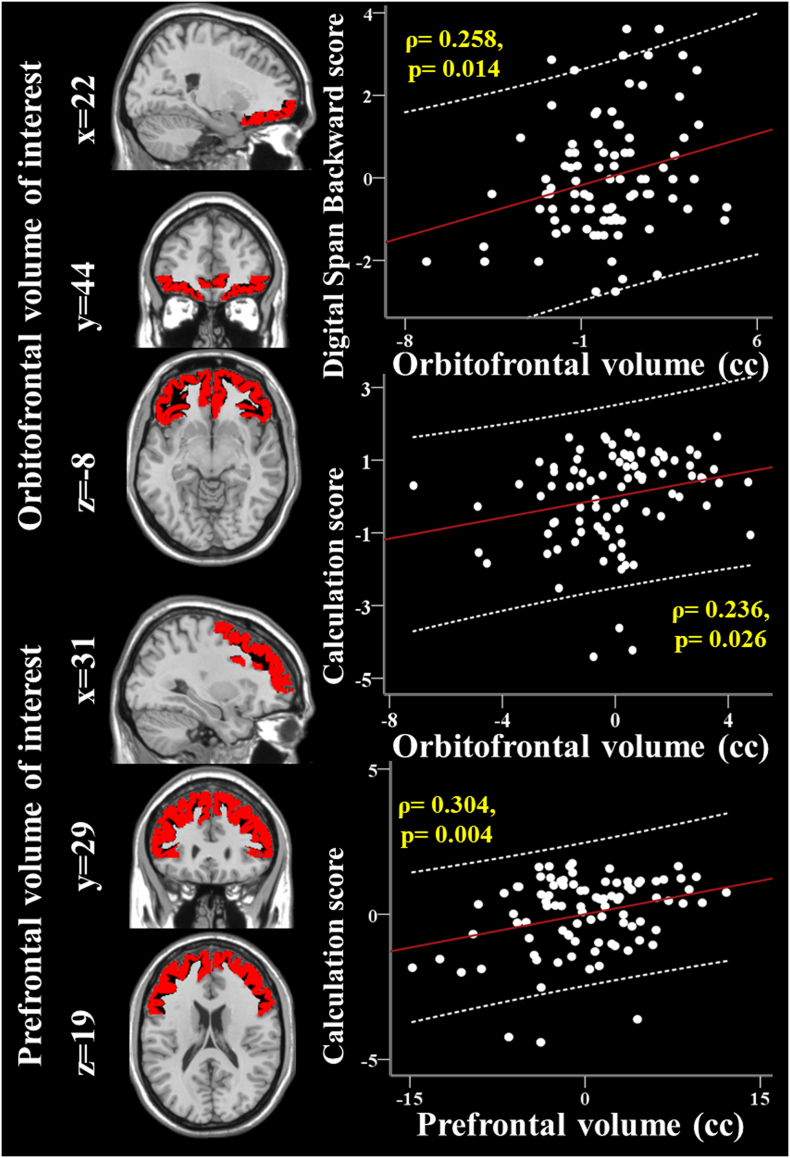
Because there were genetic interaction effects on calculation and DSB scores, as well as orbitofrontal and prefrontal volumes, we correlated the two executive function scores with each frontal volume. Each volume of interest is shown on left panel of Fig. 2. After controlling for education (years) and TIV (cc), DSB score was correlated with orbitofrontal volume (cc) (ρ = 0.258, p = .014; Fig. 2) and calculation score had correlation with both orbitofrontal (ρ = 0.236, p = .026; Fig. 2) and prefrontal (ρ = 0.304, p = .004; Fig. 2) volumes (cc).
Fig. 2.
Each volume of interest rendered on the Montreal Neurological Institute template and partial regression plots indicate the correlation of orbitofrontal and prefrontal volumes (cc) with each cognitive function score. Residuals are plotted for each variable to adjust for the effects of total intracranial volume (cc) and education (years). The 95% confidence interval is the area enclosed by the dashed curves.
3.3. Genetic interaction effects on FC in brain networks and scores in calculation and DSB
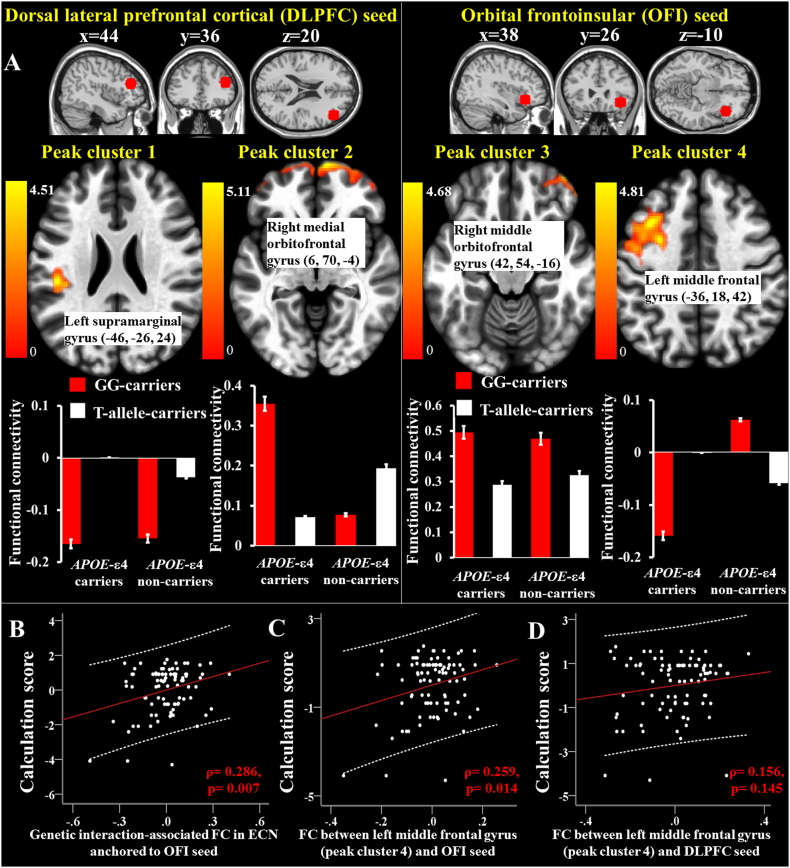
ANOVA showed interaction effects of APOE with MS4A (rs670139) on FC within DMN anchored by the PCC seed (Table 2), and within ECN anchored by the DLPFC and OFI seeds (Table 2 and Fig. 3A). DLPFC and OFI seeds are shown on top of Fig. 3. The genetic interaction-associated FC in DMN and ECN anchored by the DLPFC seed did not correlate with the calculation or DSB scores (p > .05).
Fig. 3.
Statistic maps of interaction effects of APOE with MS4A (rs670139) on functional connectivity (FC) in executive control network (ECN). ECN is anchored by the DLPFC and OFI seeds (A), and partial regression plots show correlation between calculation score and FC (B-D). Residuals are plotted for each variable to adjust for the effect of education (years). The 95% confidence interval is the area enclosed by the dashed curves. GG-carriers: MS4A-rs670139-GG carriers; T-allele-carriers: MS4A-rs670139-T-allele carriers.
In ECN anchored by the OFI seed, APOE-MS4A interaction-associated FC in the left middle frontal gyrus (x = −36, y = 18, z = 42; peak cluster 4 in Fig. 3A) had correlation with the calculation score (ρ = 0.286, p = .007, Fig. 3B), but not with the DSB score (p > .05). We further analyzed the association between the calculation score and FC of left middle frontal gyrus (peak cluster 4) with the OFI and DLPFC seeds, respectively. We demonstrated that the calculation score was correlated with FC between the left middle frontal gyrus (peak cluster 4) and the OFI seed (ρ = 0.259, p = .014, Fig. 3C), but not with FC between the left middle frontal gyrus (peak cluster 4) and the DLPFC seed (p = .145; Fig. 3D).
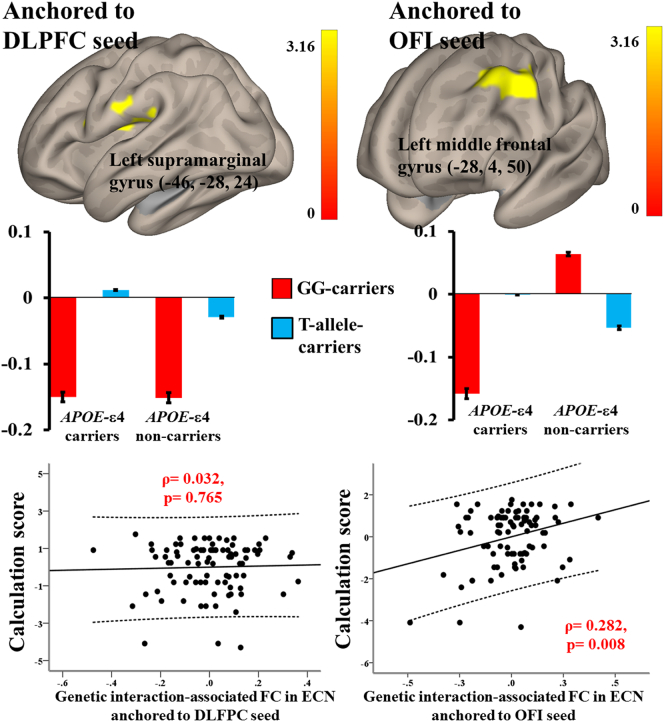
We further added GM maps as covariance of no interest to the genetic interaction effects on FC within ECN anchored by the DLPFC and OFI seeds (Fig. 4).
Fig. 4.
Statistic maps of interaction effects of APOE with MS4A (rs670139) on functional connectivity (FC) in executive control network (ECN) after adding grey matter maps as covariance of no interest. Genetic interaction affects FC between the dorsal lateral prefrontal cortical (DLPFC) seed and the left supramarginal gyrus and FC between the orbital frontoinsular (OFI) seed and the left middle frontal gyrus (upper row), and partial regression plots show correlation between calculation score and FC (lower row). Residuals are plotted for each variable to adjust for the effect of education (years). The 95% confidence interval is the area enclosed by the dashed curves. GG-carriers: MS4A-rs670139-GG carriers; T-allele-carriers: MS4A-rs670139-T-allele carriers.
In ECN anchored by the OFI seed, APOE-MS4A interaction-associated FC in the left middle frontal gyrus (x = −28, y = 4, z = 50; T maxima = 4.99; Cluster size = 1446; FDR-corrected p < .001) had correlation with calculation score (ρ = 0.282, p = .008, Fig. 4). In ECN anchored by the DLPFC seed, APOE-MS4A interaction-associated FC in the left supramarginal gyrus (x = −46, y = −28, z = 24; T maxima = 4.70; Cluster size = 527; FDR-corrected p = .001) did not correlate with calculation score (p = .765, Fig. 4).
3.4. Genetic interaction on the calculation score, FC, and frontal volume
Genetic interaction-associated FC between left middle frontal gyrus (peak cluster 4 in Fig. 3A and peak cluster in right panel of Fig. 4) and the OFI seed had a correlation with calculation score. The APOE-ε4 carrier genotype had detrimental effects on FC of left middle frontal gyrus with the OFI seed (peak cluster 4 in Fig. 3A and peak cluster in right panel of Fig. 4) and on calculation score (Fig. 1D) in GG-carriers, but not in T-allele-carriers. Collectively, these results indicated that the genetic interaction effects on FC and the calculation score were similar.
In contrast, the interaction effects between APOE and MS4A on calculation score were different from that on each frontal volume. The ε4+ carriers had lower calculation scores but higher frontal volumes than ε4 non-carriers in GG-carrier group. We further analyzed the differences in ρ value, which indicated the correlation of the calculation score with each frontal volume between different genotypic groups (Table 3).
Table 3.
Correlations of calculation score with each frontal volume in each genotypic groups after controlling for education (years) and total intracranial volume (cc).
| ε4+/GG-carriers | ε4+/T-allele-carriers | ε4−/GG-carriers | ε4−/T-allele-carriers | |
|---|---|---|---|---|
| Orbitofrontal volume (cc) | 0.163 (0.561) | 0.198 (0.415) | 0.481 (0.096) | 0.349 (0.037) |
| Prefrontal volume (cc) | 0.243 (0.383) | −0.409 (0.082) | 0.760 (0.003)* | 0.420 (0.011)* |
| APOE-ε4 carriers | APOE-ε4 non-carriers | |||
| Orbitofrontal volume (cc) | 0.128 (0.458) | 0.337 (0.016) | ||
| Prefrontal volume (cc) | −0.006 (0.972) | 0.466 (0.001)* | ||
| GG-carriers | T-allele-carriers | |||
| Orbitofrontal volume (cc) | 0.168 (0.374) | 0.260 (0.050) | ||
| Prefrontal volume (cc) | 0.368 (0.045) | 0.259 (0.051) | ||
Data are represented as ρ (p) values; bold fonts highlight p < .05. *indicates the correlation coefficient with a significant difference from that in the square; ε4+/GG-carriers: APOE-ε4 carriers (ε4+ carriers) with MS4A-rs670139-GG genotype; ε4+/T-allele-carriers: ε4+ carriers with MS4A-rs670139-T-allele genotype; ε4−/GG-carriers: APOE-ε4 non-carriers (ε4- carriers) with MS4A-rs670139-GG genotype; ε4−/T-allele-carriers: ε4- carriers with MS4A-rs670139-T-allele genotype; GG-carriers: MS4A-rs670139-GG carriers; T-allele-carriers: MS4A-rs670139-T-allele carriers.
A comparison using Fisher transformation showed significant differences in ρ value, indicating the correlation of the calculation score with prefrontal volume (cc) between ε4+ carriers and non-carriers (P = .028), between ε4+/T-allele-carriers and ε4−/GG-carriers (P = .001), and between ε4+/T-allele-carriers and ε4−/T-allele-carriers (P = .006) (Table 3). The results indicated that the genotypic variation affected the correlation of calculation score with each frontal volume (Table 3).
4. Discussion
4.1. Main findings
This study has three main findings. First, an APOE-MS4A (rs670139) interaction effect on calculation score was observed, and the calculation score was correlated with genetic interaction-associated FC in ECN anchored by the OFI seed. Second, there were APOE-MS4A (rs670139) interaction effects on orbitofrontal and prefrontal volume and on FC in ECN anchored by the DLPFC and OFI seeds. The genetic interaction effects were found on FC in ECN mainly between DLPFC seed and left supramarginal gyrus (peak cluster 1 in Fig. 3), as well as between the OFI seed and the left middle frontal gyrus (peak cluster 4 in Fig. 3). After controlling for GM maps, the genetic interaction effects on FC in ECN were still significant between DLPFC seed and left supramarginal gyrus and between the OFI seed and the left middle frontal gyrus (Fig. 4). Third, genotypic variation affected the pathological-clinical relationship between the prefrontal volume and the calculation score.
4.2. Interaction effects of APOE with MS4A (rs670139) on FC in ECN
Previous studies have found the evidence of the interaction effect of MS4A (rs670139) with CLU (Lambert et al., 2013) or CD33 (Lambert et al., 2013; Ebbert et al., 2014) on the AD risk. Although APOE-MS4A interaction effects are not associated with the increased AD risk (Hollingworth et al., 2011), we found an interaction effect between APOE and MS4A on FC in ECN, suggesting that the genetic interaction effects are involved in the pathological changes in patients with AD.
Greater FC between the prefrontal cortex and the anterior cingulate cortex has been found in APOE-ε4 carriers than in non-carriers in cognitively normal elderly individuals (Machulda et al., 2011). However, another study demonstrates that APOE-ε4 carriers have lower neuronal activity in prefrontal areas than non-carriers do (Arenaza-Urquijo et al., 2015). The present study showed that APOE-ε4 carriers had higher FC between left middle frontal gyrus and OFI seed than non-carriers did in MS4A-rs670139-T-allele carrier group, rather than in MS4A-rs670139-GG carrier group. Taken together, these results suggest that modulation effects of MS4A genotype might be potentially implicated in the pathogenic mechanisms underlying the discrepant effects of APOE-ε4 carrier status on frontal activity or FC of the frontal cortices with other cortical regions shown in previous studies.
The epistasis effect may occur because both APOE and MS4A modulate neuronal synaptic function. APOE encodes apolipoprotein E (apoE) that is involved in the neuronal integrity and regeneration process (Ignatius et al., 1986). MS4A family members are involved in cellular signaling transduction (Tanzi, 2012). The genotypic interaction effect on synaptic function might be the molecular basis of APOE-MS4A interaction effects on FC in brain networks.
4.3. Interaction effects of APOE with MS4A (rs670139) on executive performances
With regards to the effect of the APOE-ε4 carrier status on executive performances and its neural basis, the APOE-ε4 carriers sometimes have better executive performances than non-carriers do (Wolk and Dickerson, 2010), but sometimes APOE-ε4 carriers show worse executive performances than non-carriers do (Wisdom et al., 2011). Because the present study demonstrated that APOE-ε4 carriers had lower calculation scores than non-carriers in the MS4A-rs670139-GG carrier group, but had better calculation scores than non-carriers in the MS4A-rs670139-T-allele carrier group, genetic interaction effects might be the potential neural basis for the discrepant relationships of the APOE-ε4 carrier status with executive impairment.
ApoE4 encoded by APOE-ε4 carrier genotype has been shown to modulate both detrimental toll-like receptor and beneficial interleukin-4R-nuclear receptor pathways in chronic neuroinflammation (Tai et al., 2015). MS4A family members have been found to be associated with phagocytic function in microglia and immune responses in brain (Karch et al., 2012). Pro-inflammatory cytokines have been demonstrated to be associated with working memory (Yang et al., 2013). The genotypic interaction effects on neuroinflammation might contribute to the pathogenic mechanisms underlying APOE-MS4A interaction effects on the executive performances in the calculation tasks.
4.4. Relationship between executive performances and frontal volumes
The association between executive performances and activation in the prefrontal cortex has been attributed to the critical prefrontal cortex involvement in tasks that require encoding action sequences, coordinating action, and maintaining abstract sequential movement plans (Badre et al., 2009). In agreement with the critical role of the frontal lobe in the executive function status as shown in previous studies, the current study indicated that both DSB and calculation scores correlate with the frontal volume.
Moreover, the present study revealed that DSB score was positively correlated with the orbitofrontal volume, and the calculation score was positively correlated with the orbitofrontal and prefrontal volumes. In this regards, previous studies also indicate that orbitofrontal cortex is involved in the manipulation and monitoring of information in working memory (Barbey et al., 2011), and the inferior and middle prefrontal cortices are associated with the calculation process (Menon et al., 2000).
However, the clinicopathological relationships were discrepant between different genotypic groups. Significant differences were observed in the correlation coefficient of calculation scores with the prefrontal volume between APOE-ε4 non-carriers and APOE-ε4 carriers, between ε4−/GG-carriers and ε4+/T-allele-carriers, and between ε4−/T-allele-carriers and ε4+/T-allele-carriers.
Previous study has shown that APOE-ε4 allele dose-dependently modulates the relationship between executive performances and the cortical volume in cognitively normal aging individuals (Cacciaglia et al., 2018). Our results supported the observation and expanded on some of the previous findings. The present study demonstrated a positive correlation of the calculation score with each frontal volume in the APOE-ε4 non-carriers with AD, but not in APOE-ε4 carriers with AD. In addition, the calculation score was correlated with prefrontal volume in MS4A-rs670139-GG-carriers with AD, and the score was also correlated with orbitofrontal volume in MS4A-rs670139-T-allele carriers with AD. Collectively, these results suggested that both APOE-ε4 carrier status and MS4A (rs670139) genotype modulated the correlation of the calculation score with the prefrontal volume in AD patients.
4.5. Calculation scores and FC between the OFI seed and the left middle frontal gyrus in ECN
The APOE-MS4A (rs670139) interaction effects on calculation scores and each frontal volume were different; however, the interaction effects on the calculation score and FC of the OFI seed with the left middle frontal gyrus were similar. The APOE-ε4 carrier genotype had detrimental effects on the calculation score and FC between left middle frontal gyrus and the OFI seed in the GG-carrier group, but showed protective effects on these variables in the T-allele-carrier group. In addition, the genetic interaction-associated FC was correlated with the calculation score. The genetic interaction effects on FC of the OFI seed with the left middle frontal gyrus might be the neural basis for the APOE-MS4A interaction effect on the calculation performance.
The relationship between neuronal activity in the middle frontal gyrus and calculation ability has also been demonstrated in one previous study (Menon et al., 2000). Another study further demonstrates that calculation ability is associated with co-activation in the prefrontal and right anterior-insular cortices (Cowell et al., 2000). In consistent with these findings, the present study indicated that the calculation performance was associated with the FC between the OFI seed and middle frontal gyrus.
In addition, the DLPFC and inferior parietal gyrus are activated during calculation tasks, as demonstrated by fMRI (Rickard et al., 2000). The present study demonstrated that genetic effects on the calculation score and FC of the DLPFC seed with the left supramarginal gyrus paralleled each other; however, FC of the left supramarginal gyrus with the DLPFC seed did not correlate with the calculation score. Therefore, it would be difficult to determine whether the inferior parietal gyrus is involved in the calculation tasks.
4.6. Therapeutic consideration in the future
Identifying the interaction effect of MS4A (rs670139) risk factor with APOE might be useful to illustrate the pathogenesis of cognitive variation in AD and to optimize the treatment strategies for AD.
4.7. Limitations
The present study has three limitations. First, although calculation impairment was evident in AD patients, different patterns of calculation dysfunction were not characterized in this study. It is not clear how math skills, linguistic abilities, and executive function affected calculation performance separately. Therefore, to avoid the effects of math skills or linguistic abilities, this present study controlled for the educational level in each correlation analysis. Second, pathological effects of Aβ and NFTs on the genotype-associated variation in the clinical presentation were not elucidated in this study. Further investigation would be needed to determine how pathological depositions influence the genetic interaction effects on cognitive performances, FC in brain networks, and the cortical volume would be needed. Third, the genetic interaction effects on cognitive performances, FC, and cortical volume were only investigated in AD patients. The present analyses were limited to AD patients, due to the small sample size of NC group, to obtain a more genetically and cognitively homogenous study population. Further study would be needed to illustrate how genetic interaction affects pathophysiological changes in NCs.
5. Conclusions
We found interaction effects of APOE with MS4A (rs670139) on calculation performance. The genetic interaction effects on FC of the brain networks might be the neural basis for the APOE-MS4A interaction effects on the cognitive performances. Furthermore, this study showed that genotypic variation affected the relationship between the prefrontal volume and the calculation score.
Ethics approval and consent to participate
The study was approved by Chang Gung Memorial Hospital's Institutional Review Committee on Human Research, and all of the participants and their authorized caregivers provided written informed consent.
Submission declaration and verification
This work is not published previously, not under consideration for publication elsewhere, and it is approved by all authors.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by Chang Gung Memorial Hospital [CMRPG8G1521 and CMRPG8E0541] by National Science Council [106-2314-B-182A-070 and 106-2314-B-182A-169].
Authors' contributions
YTC participated in the design of the study and drafted the manuscript. YTC and MS performed the statistical analysis. CWH, JJL and WNC participated in the sequence alignment and clinical evaluation of patients. EM and CCC conceived the study, and participated in its design and coordination, and helped draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
References
- Antunez C., Boada M., Gonzalez-Perez A., Gayan J., Ramirez-Lorca R., Marin J., Hernandez I., Moreno-Rey C., Moron F.J., Lopez-Arrieta J., Mauleon A., Rosende-Roca M., Noguera-Perea F., Legaz-Garcia A., Vivancos-Moreau L., Velasco J., Carrasco J.M., Alegret M., Antequera-Torres M., Manzanares S., Romo A., Blanca I., Ruiz S., Espinosa A., Castano S., Garcia B., Martinez-Herrada B., Vinyes G., Lafuente A., Becker J.T., Galan J.J., Serrano-Rios M., Vazquez E., Tarraga L., Saez M.E., Lopez O.L., Real L.M., Ruiz A. The membrane-spanning 4-domains, subfamily A (MS4A) gene cluster contains a common variant associated with Alzheimer's disease. Genome Med. 2011;3:33. doi: 10.1186/gm249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenaza-Urquijo E.M., Gonneaud J., Fouquet M., Perrotin A., Mezenge F., Landeau B., Egret S., De la Sayette V., Desgranges B., Chetelat G. Interaction between years of education and APOE epsilon4 status on frontal and temporal metabolism. Neurology. 2015;85:1392–1399. doi: 10.1212/WNL.0000000000002034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badre D., Hoffman J., Cooney J.W., D'Esposito M. Hierarchical cognitive control deficits following damage to the human frontal lobe. Nat. Neurosci. 2009;12:515–522. doi: 10.1038/nn.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbey A.K., Koenigs M., Grafman J. Orbitofrontal contributions to human working memory. Cereb. Cortex. 2011;21:789–795. doi: 10.1093/cercor/bhq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y., Restom K., Liau J., Liu T.T. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A.R., Raz N. Age-related differences in memory and executive functions in healthy APOE varepsilon4 carriers: the contribution of individual differences in prefrontal volumes and systolic blood pressure. Neuropsychologia. 2012;50:704–714. doi: 10.1016/j.neuropsychologia.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone K.B. The Boston qualitative scoring system for the Rey-Osterrieth complex figure. J. Clin. Exp. Neuropsychol. 2000;22:430–434. doi: 10.1076/1380-3395(200006)22:3;1-V;FT430. [DOI] [PubMed] [Google Scholar]
- Boyle P.A., Malloy P.F., Salloway S., Cahn-Weiner D.A., Cohen R., Cummings J.L. Executive dysfunction and apathy predict functional impairment in Alzheimer disease. Am. J. Geriatr. Psychiatry. 2003;11:214–221. [PubMed] [Google Scholar]
- Cacciaglia R., Molinuevo J.L., Falcon C., Brugulat-Serrat A., Sanchez-Benavides G., Gramunt N., Esteller M., Moran S., Minguillon C., Fauria K., Gispert J.D. Effects of APOE-epsilon4 allele load on brain morphology in a cohort of middle-aged healthy individuals with enriched genetic risk for Alzheimer's disease. Alzheimers Dement. 2018;14:902–912. doi: 10.1016/j.jalz.2018.01.016. [DOI] [PubMed] [Google Scholar]
- Cahn-Weiner D.A., Farias S.T., Julian L., Harvey D.J., Kramer J.H., Reed B.R., Mungas D., Wetzel M., Chui H. Cognitive and neuroimaging predictors of instrumental activities of daily living. J. Int. Neuropsychol. Soc. 2007;13:747–757. doi: 10.1017/S1355617707070853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.C., Kramer J.H., Lin K.N., Chang W.N., Wang Y.L., Huang C.W., Lin Y.T., Chen C., Wang P.N. Validating the Chinese version of the Verbal Learning Test for screening Alzheimer's disease. J. Int. Neuropsychol. Soc. 2010;16:244–251. doi: 10.1017/S1355617709991184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell S.F., Egan G.F., Code C., Harasty J., Watson J.D. The functional neuroanatomy of simple calculation and number repetition: a parametric PET activation study. NeuroImage. 2000;12:565–573. doi: 10.1006/nimg.2000.0640. [DOI] [PubMed] [Google Scholar]
- Dorey E., Chang N., Liu Q.Y., Yang Z., Zhang W. Apolipoprotein E, amyloid-beta, and neuroinflammation in Alzheimer's disease. Neurosci. Bull. 2014;30:317–330. doi: 10.1007/s12264-013-1422-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B., Feldman H.H., Jacova C., Cummings J.L., Dekosky S.T., Barberger-Gateau P., Delacourte A., Frisoni G., Fox N.C., Galasko D., Gauthier S., Hampel H., Jicha G.A., Meguro K., O'Brien J., Pasquier F., Robert P., Rossor M., Salloway S., Sarazin M., de Souza L.C., Stern Y., Visser P.J., Scheltens P. Revising the definition of Alzheimer's disease: a new lexicon. Lancet Neurol. 2010;9:1118–1127. doi: 10.1016/S1474-4422(10)70223-4. [DOI] [PubMed] [Google Scholar]
- Ebbert M.T., Ridge P.G., Wilson A.R., Sharp A.R., Bailey M., Norton M.C., Tschanz J.T., Munger R.G., Corcoran C.D., Kauwe J.S. Population-based analysis of Alzheimer's disease risk alleles implicates genetic interactions. Biol. Psychiatry. 2014;75:732–737. doi: 10.1016/j.biopsych.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias-Sonnenschein L., Verhey F.R.J., Visser P.J. Effects of MS4A variants on cerebrospinal fluid beta-amyloid 1-42. Alzheimer's Dement. 2013;9:P229–P230. [Google Scholar]
- Folstein M.F., Folstein S.E., McHugh P.R. "Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Gaser, Dahnke R. 2016. CAT-A Computational Anatomy Toolbox for the Analysis of Structural MRI Data. (HBM 2016) [Google Scholar]
- Han M.R., Schellenberg G.D., Wang L.S. Genome-wide association reveals genetic effects on human Abeta42 and tau protein levels in cerebrospinal fluids: a case control study. BMC Neurol. 2010;10:90. doi: 10.1186/1471-2377-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J., Chen Y., Masiulis I., Zhou L. Expanding functions of lipoprotein receptors. J. Lipid Res. 2009;50:S287–S292. doi: 10.1194/jlr.R800077-JLR200. Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M., Abraham R., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Jones N., Stretton A., Thomas C., Richards A., Ivanov D., Widdowson C., Chapman J., Lovestone S., Powell J., Proitsi P., Lupton M.K., Brayne C., Rubinsztein D.C., Gill M., Lawlor B., Lynch A., Brown K.S., Passmore P.A., Craig D., McGuinness B., Todd S., Holmes C., Mann D., Smith A.D., Beaumont H., Warden D., Wilcock G., Love S., Kehoe P.G., Hooper N.M., Vardy E.R., Hardy J., Mead S., Fox N.C., Rossor M., Collinge J., Maier W., Jessen F., Ruther E., Schurmann B., Heun R., Kolsch H., van den Bussche H., Heuser I., Kornhuber J., Wiltfang J., Dichgans M., Frolich L., Hampel H., Gallacher J., Hull M., Rujescu D., Giegling I., Goate A.M., Kauwe J.S., Cruchaga C., Nowotny P., Morris J.C., Mayo K., Sleegers K., Bettens K., Engelborghs S., De Deyn P.P., Van Broeckhoven C., Livingston G., Bass N.J., Gurling H., McQuillin A., Gwilliam R., Deloukas P., Al-Chalabi A., Shaw C.E., Tsolaki M., Singleton A.B., Guerreiro R., Muhleisen T.W., Nothen M.M., Moebus S., Jockel K.H., Klopp N., Wichmann H.E., Pankratz V.S., Sando S.B., Aasly J.O., Barcikowska M., Wszolek Z.K., Dickson D.W., Graff-Radford N.R., Petersen R.C., van Duijn C.M., Breteler M.M., Ikram M.A., Destefano A.L., Fitzpatrick A.L., Lopez O., Launer L.J., Seshadri S., Berr C., Campion D., Epelbaum J., Dartigues J.F., Tzourio C., Alperovitch A., Lathrop M., Feulner T.M., Friedrich P., Riehle C., Krawczak M., Schreiber S., Mayhaus M., Nicolhaus S., Wagenpfeil S., Steinberg S., Stefansson H., Stefansson K., Snaedal J., Bjornsson S., Jonsson P.V., Chouraki V., Genier-Boley B., Hiltunen M., Soininen H., Combarros O., Zelenika D., Delepine M., Bullido M.J., Pasquier F., Mateo I., Frank-Garcia A., Porcellini E., Hanon O., Coto E., Alvarez V., Bosco P., Siciliano G., Mancuso M., Panza F., Solfrizzi V., Nacmias B., Sorbi S., Bossu P., Piccardi P., Arosio B., Annoni G., Seripa D., Pilotto A., Scarpini E., Galimberti D., Brice A., Hannequin D., Licastro F., Jones L., Holmans P.A., Jonsson T., Riemenschneider M., Morgan K., Younkin S.G., Owen M.J., O'Donovan M., Amouyel P., Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honea R.A., Vidoni E., Harsha A., Burns J.M. Impact of APOE on the healthy aging brain: a voxel-based MRI and DTI study. J. Alzheimers Dis. 2009;18:553–564. doi: 10.3233/JAD-2009-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius M.J., Gebicke-Harter P.J., Skene J.H., Schilling J.W., Weisgraber K.H., Mahley R.W., Shooter E.M. Expression of apolipoprotein E during nerve degeneration and regeneration. Proc. Natl. Acad. Sci. U. S. A. 1986;83:1125–1129. doi: 10.1073/pnas.83.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogi A.L., Kikas E. Calculation and word problem-solving skills in primary grades - Impact of cognitive abilities and longitudinal interrelations with task-persistent behaviour. Br. J. Educ. Psychol. 2016;86:165–181. doi: 10.1111/bjep.12096. [DOI] [PubMed] [Google Scholar]
- Karch C.M., Jeng A.T., Nowotny P., Cady J., Cruchaga C., Goate A.M. Expression of novel Alzheimer's disease risk genes in control and Alzheimer's disease brains. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima R., Okita K., Yamazaki R., Tajima N., Yoshida H., Taira M., Iwata K., Sasaki T., Maeyama K., Usui N., Sugimoto K. Reading aloud and arithmetic calculation improve frontal function of people with dementia. J. Gerontol. A Biol. Sci. Med. Sci. 2005;60:380–384. doi: 10.1093/gerona/60.3.380. [DOI] [PubMed] [Google Scholar]
- Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., Destafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., Russo G., Thorton-Wells T.A., Jones N., Smith A.V., Chouraki V., Thomas C., Ikram M.A., Zelenika D., Vardarajan B.N., Kamatani Y., Lin C.F., Gerrish A., Schmidt H., Kunkle B., Dunstan M.L., Ruiz A., Bihoreau M.T., Choi S.H., Reitz C., Pasquier F., Cruchaga C., Craig D., Amin N., Berr C., Lopez O.L., De Jager P.L., Deramecourt V., Johnston J.A., Evans D., Lovestone S., Letenneur L., Moron F.J., Rubinsztein D.C., Eiriksdottir G., Sleegers K., Goate A.M., Fievet N., Huentelman M.W., Gill M., Brown K., Kamboh M.I., Keller L., Barberger-Gateau P., McGuiness B., Larson E.B., Green R., Myers A.J., Dufouil C., Todd S., Wallon D., Love S., Rogaeva E., Gallacher J., St George-Hyslop P., Clarimon J., Lleo A., Bayer A., Tsuang D.W., Yu L., Tsolaki M., Bossu P., Spalletta G., Proitsi P., Collinge J., Sorbi S., Sanchez-Garcia F., Fox N.C., Hardy J., Deniz Naranjo M.C., Bosco P., Clarke R., Brayne C., Galimberti D., Mancuso M., Matthews F., Moebus S., Mecocci P., Del Zompo M., Maier W., Hampel H., Pilotto A., Bullido M., Panza F., Caffarra P., Nacmias B., Gilbert J.R., Mayhaus M., Lannefelt L., Hakonarson H., Pichler S., Carrasquillo M.M., Ingelsson M., Beekly D., Alvarez V., Zou F., Valladares O., Younkin S.G., Coto E., Hamilton-Nelson K.L., Gu W., Razquin C., Pastor P., Mateo I., Owen M.J., Faber K.M., Jonsson P.V., Combarros O., O'Donovan M.C., Cantwell L.B., Soininen H., Blacker D., Mead S., Mosley T.H., Jr., Bennett D.A., Harris T.B., Fratiglioni L., Holmes C., de Bruijn R.F., Passmore P., Montine T.J., Bettens K., Rotter J.I., Brice A., Morgan K., Foroud T.M., Kukull W.A., Hannequin D., Powell J.F., Nalls M.A., Ritchie K., Lunetta K.L., Kauwe J.S., Boerwinkle E., Riemenschneider M., Boada M., Hiltuenen M., Martin E.R., Schmidt R., Rujescu D., Wang L.S., Dartigues J.F., Mayeux R., Tzourio C., Hofman A., Nothen M.M., Graff C., Psaty B.M., Jones L., Haines J.L., Holmans P.A., Lathrop M., Pericak-Vance M.A., Launer L.J., Farrer L.A., van Duijn C.M., Van Broeckhoven C., Moskvina V., Seshadri S., Williams J., Schellenberg G.D., Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machulda M.M., Jones D.T., Vemuri P., McDade E., Avula R., Przybelski S., Boeve B.F., Knopman D.S., Petersen R.C., Jack C.R., Jr. Effect of APOE epsilon4 status on intrinsic network connectivity in cognitively normal elderly subjects. Arch. Neurol. 2011;68:1131–1136. doi: 10.1001/archneurol.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovan M.C., Delazer M., Ermani M., Denes G. The breakdown of calculation procedures in Alzheimer's disease. Cortex. 1999;35:21–38. doi: 10.1016/s0010-9452(08)70783-4. [DOI] [PubMed] [Google Scholar]
- Margulies D.S., Vincent J.L., Kelly C., Lohmann G., Uddin L.Q., Biswal B.B., Villringer A., Castellanos F.X., Milham M.P., Petrides M. Precuneus shares intrinsic functional architecture in humans and monkeys. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20069–20074. doi: 10.1073/pnas.0905314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyr A., Clare L. Executive function and activities of daily living in Alzheimer's disease: a correlational meta-analysis. Dement. Geriatr. Cogn. Disord. 2012;33:189–203. doi: 10.1159/000338233. [DOI] [PubMed] [Google Scholar]
- Menon V., Rivera S.M., White C.D., Glover G.H., Reiss A.L. Dissociating prefrontal and parietal cortex activation during arithmetic processing. NeuroImage. 2000;12:357–365. doi: 10.1006/nimg.2000.0613. [DOI] [PubMed] [Google Scholar]
- Morgen K., Ramirez A., Frolich L., Tost H., Plichta M.M., Kolsch H., Rakebrandt F., Rienhoff O., Jessen F., Peters O., Jahn H., Luckhaus C., Hull M., Gertz H.J., Schroder J., Hampel H., Teipel S.J., Pantel J., Heuser I., Wiltfang J., Ruther E., Kornhuber J., Maier W., Meyer-Lindenberg A. Genetic interaction of PICALM and APOE is associated with brain atrophy and cognitive impairment in Alzheimer's disease. Alzheimers Dement. 2014;10:S269–S276. doi: 10.1016/j.jalz.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Morris J.C. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., Larson E.B., Bird T.D., Boeve B.F., Graff-Radford N.R., De Jager P.L., Evans D., Schneider J.A., Carrasquillo M.M., Ertekin-Taner N., Younkin S.G., Cruchaga C., Kauwe J.S., Nowotny P., Kramer P., Hardy J., Huentelman M.J., Myers A.J., Barmada M.M., Demirci F.Y., Baldwin C.T., Green R.C., Rogaeva E., St George-Hyslop P., Arnold S.E., Barber R., Beach T., Bigio E.H., Bowen J.D., Boxer A., Burke J.R., Cairns N.J., Carlson C.S., Carney R.M., Carroll S.L., Chui H.C., Clark D.G., Corneveaux J., Cotman C.W., Cummings J.L., Decarli C., Dekosky S.T., Diaz-Arrastia R., Dick M., Dickson D.W., Ellis W.G., Faber K.M., Fallon K.B., Farlow M.R., Ferris S., Frosch M.P., Galasko D.R., Ganguli M., Gearing M., Geschwind D.H., Ghetti B., Gilbert J.R., Gilman S., Giordani B., Glass J.D., Growdon J.H., Hamilton R.L., Harrell L.E., Head E., Honig L.S., Hulette C.M., Hyman B.T., Jicha G.A., Jin L.W., Johnson N., Karlawish J., Karydas A., Kaye J.A., Kim R., Koo E.H., Kowall N.W., Lah J.J., Levey A.I., Lieberman A.P., Lopez O.L., Mack W.J., Marson D.C., Martiniuk F., Mash D.C., Masliah E., McCormick W.C., McCurry S.M., McDavid A.N., McKee A.C., Mesulam M., Miller B.L., Miller C.A., Miller J.W., Parisi J.E., Perl D.P., Peskind E., Petersen R.C., Poon W.W., Quinn J.F., Rajbhandary R.A., Raskind M., Reisberg B., Ringman J.M., Roberson E.D., Rosenberg R.N., Sano M., Schneider L.S., Seeley W., Shelanski M.L., Slifer M.A., Smith C.D., Sonnen J.A., Spina S., Stern R.A., Tanzi R.E., Trojanowski J.Q., Troncoso J.C., Van Deerlin V.M., Vinters H.V., Vonsattel J.P., Weintraub S., Welsh-Bohmer K.A., Williamson J., Woltjer R.L., Cantwell L.B., Dombroski B.A., Beekly D., Lunetta K.L., Martin E.R., Kamboh M.I., Saykin A.J., Reiman E.M., Bennett D.A., Morris J.C., Montine T.J., Goate A.M., Blacker D., Tsuang D.W., Hakonarson H., Kukull W.A., Foroud T.M., Haines J.L., Mayeux R., Pericak-Vance M.A., Farrer L.A., Schellenberg G.D. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen R., Edotor . Oxford University Press; New York, NY: 2003. Mild Cognitive Impairment: Aging to Alzheimer's Disease. [Google Scholar]
- Reitz C., Jun G., Naj A., Rajbhandary R., Vardarajan B.N., Wang L.S., Valladares O., Lin C.F., Larson E.B., Graff-Radford N.R., Evans D., De Jager P.L., Crane P.K., Buxbaum J.D., Murrell J.R., Raj T., Ertekin-Taner N., Logue M., Baldwin C.T., Green R.C., Barnes L.L., Cantwell L.B., Fallin M.D., Go R.C., Griffith P., Obisesan T.O., Manly J.J., Lunetta K.L., Kamboh M.I., Lopez O.L., Bennett D.A., Hendrie H., Hall K.S., Goate A.M., Byrd G.S., Kukull W.A., Foroud T.M., Haines J.L., Farrer L.A., Pericak-Vance M.A., Schellenberg G.D., Mayeux R. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4,and the risk of late-onset Alzheimer disease in African Americans. JAMA. 2013;309:1483–1492. doi: 10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard T.C., Romero S.G., Basso G., Wharton C., Flitman S., Grafman J. The calculating brain: an fMRI study. Neuropsychologia. 2000;38:325–335. doi: 10.1016/s0028-3932(99)00068-8. [DOI] [PubMed] [Google Scholar]
- Rosen W.G., Terry R.D., Fuld P.A., Katzman R., Peck A. Pathological verification of ischemic score in differentiation of dementias. Ann. Neurol. 1980;7:486–488. doi: 10.1002/ana.410070516. [DOI] [PubMed] [Google Scholar]
- Seeley W.W., Menon V., Schatzberg A.F., Keller J., Glover G.H., Kenna H., Reiss A.L., Greicius M.D. Dissociable intrinsic connectivity networks for salience processing and executive control. J. Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S., Fitzpatrick A.L., Ikram M.A., Destefano A.L., Gudnason V., Boada M., Bis J.C., Smith A.V., Carassquillo M.M., Lambert J.C., Harold D., Schrijvers E.M., Ramirez-Lorca R., Debette S., Longstreth W.T., Jr., Janssens A.C., Pankratz V.S., Dartigues J.F., Hollingworth P., Aspelund T., Hernandez I., Beiser A., Kuller L.H., Koudstaal P.J., Dickson D.W., Tzourio C., Abraham R., Antunez C., Du Y., Rotter J.I., Aulchenko Y.S., Harris T.B., Petersen R.C., Berr C., Owen M.J., Lopez-Arrieta J., Varadarajan B.N., Becker J.T., Rivadeneira F., Nalls M.A., Graff-Radford N.R., Campion D., Auerbach S., Rice K., Hofman A., Jonsson P.V., Schmidt H., Lathrop M., Mosley T.H., Au R., Psaty B.M., Uitterlinden A.G., Farrer L.A., Lumley T., Ruiz A., Williams J., Amouyel P., Younkin S.G., Wolf P.A., Launer L.J., Lopez O.L., van Duijn C.M., Breteler M.M. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai L.M., Ghura S., Koster K.P., Liakaite V., Maienschein-Cline M., Kanabar P., Collins N., Ben-Aissa M., Lei A.Z., Bahroos N., Green S.J., Hendrickson B., Van Eldik L.J., Ladu M.J. APOE-modulated Abeta-induced neuroinflammation in Alzheimer's disease: current landscape, novel data, and future perspective. J. Neurochem. 2015;133:465–488. doi: 10.1111/jnc.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi R.E. The genetics of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub S., Salmon D., Mercaldo N., Ferris S., Graff-Radford N.R., Chui H., Cummings J., Decarli C., Foster N.L., Galasko D., Peskind E., Dietrich W., Beekly D.L., Kukull W.A., Morris J.C. The Alzheimer's Disease Centers' Uniform Data Set (UDS): the neuropsychologic test battery. Alzheimer Dis. Assoc. Disord. 2009;23:91–101. doi: 10.1097/WAD.0b013e318191c7dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S., Nieto-Castanon A. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2012;2:125–141. doi: 10.1089/brain.2012.0073. [DOI] [PubMed] [Google Scholar]
- Wisdom N.M., Callahan J.L., Hawkins K.A. The effects of apolipoprotein E on non-impaired cognitive functioning: a meta-analysis. Neurobiol. Aging. 2011;32:63–74. doi: 10.1016/j.neurobiolaging.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Wishart H.A., Saykin A.J., McAllister T.W., Rabin L.A., McDonald B.C., Flashman L.A., Roth R.M., Mamourian A.C., Tsongalis G.J., Rhodes C.H. Regional brain atrophy in cognitively intact adults with a single APOE epsilon4 allele. Neurology. 2006;67:1221–1224. doi: 10.1212/01.wnl.0000238079.00472.3a. [DOI] [PubMed] [Google Scholar]
- Wolk D.A., Dickerson B.C. Apolipoprotein E (APOE) genotype has dissociable effects on memory and attentional-executive network function in Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10256–10261. doi: 10.1073/pnas.1001412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Cudaback E., Jorstad N.L., Hemingway J.F., Hagan C.E., Melief E.J., Li X., Yoo T., Khademi S.B., Montine K.S., Montine T.J., Keene C.D. APOE3, but not APOE4, bone marrow transplantation mitigates behavioral and pathological changes in a mouse model of Alzheimer disease. Am. J. Pathol. 2013;183:905–917. doi: 10.1016/j.ajpath.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo J., Bau J., Childs S.J., Goss G.G., Sensen C.W., Deans J.P. Phylogenetic analysis of the MS4A and TMEM176 gene families. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.