Classical studies have consistently identified some specific gene mutations in classic Hodgkin lymphoma (cHL), mainly affecting members of the NF-kappaB and JAK/STAT pathways1–4. However, this knowledge is yet to be exploited in the clinical milieu. Molecular analyses of cHL have been limited because of the paucity of Hodgkin and Reed-Sternberg (HRS) cells, which usually account for < 5% of cells as identified by standard CD30 immunostaining. Various enrichment strategies, such as laser capture microdissection (LCM), could facilitate the identification of genetic alterations5. Also, in recent years, the sensitivity and specificity of next-generation sequencing (NGS) techniques have been greatly improved by simultaneously testing selected genes, arranged in comprehensive gene panels. This is even possible with formalin-fixed, paraffin-embedded (FFPE) tissue samples.
However, very few NGS studies have described the genomic landscape of the disease. The first whole-exome sequencing analyses of primary Hodgkin and Reed-Sternberg (HRS) cells found beta-2-microglobulin (B2M) to be the most commonly altered gene6. Recently, our group have identified concurrent genetic lesions in relevant signaling pathways, such as those of JAK-STAT, NF-kappaB, and BCR, as well as in epigenetic regulators7. These findings are largely concordant with other NGS studies in cHL cell lines8. More recently, Tiacci et al9 have reported frequent mutations affecting genes of the JAK-STAT pathway, as well as mutations in GNA13, XPO1, and ITPKB. However, none of these works set out to identify variants associated with response to treatment.
The identification of specific genetic lesions and better biological characterization of the subgroup of patients with refractory disease remain major research goals. Here we analyze the genomic characteristics of 12 cHL tumors, corresponding to selected patients with diseases that are primary refractory to conventional therapy, and compare the genetic variants identified in the HRS cells from primary tumors and relapsed tumors.
Initially, formalin-fixed paraffin-embedded (FFPE) tumor samples and clinical data from 20 cHL patients were obtained from the records of the participating institutions. All patients were intentionally selected because of their refractoriness to conventional treatment: primary-progressive disease (absence of complete remission after treatment) or early relapse (less than 12 months after complete remission). All cases received ABVD therapy. We collected representative tumor blocks from both, the original pretreatment and the relapse biopsy. All the samples and data were collected through the MDACC Madrid Biobank, in accordance with the technical and ethical procedures of the Spanish National Biobank Network, including anonymization processes and obtaining written informed consent according to the Helsinki Declaration. Approval was obtained from the institutional review board (Clinical Research Ethical Committee, ref. 354/12). Clinical characteristics of the patients are summarized in Table 1. After LCM, DNA extraction and library construction, 12 cases fulfilled the minimum quality criteria and the generated libraries were sequenced and their data analyzed.
Table 1.
Clinical characteristics of the cases
| Case # | Histological subtype | Bone marrow involvement | Bulky mass | Ann Arbor stage | Age | Sex | IPS | First-line therapy | Response | Follow-up |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MC | no | yes (mediastinal) | IIB | 23 | F | 0 | ABVD (x4) + Rt | CR + R | DOD (13 months) |
| 2 | NS | no | yes (mediastinal) | IIB | 31 | F | 4 | ABVD (x6) + Rt | CR + R | Alive (90 months) |
| 3 | MC | no | yes (retroperitoneal) | IIIB | 27 | M | 1 | ABVD (x6) | CR + R | Alive (64 months) |
| 4 | NS | NA | no | IIA | 40 | M | 2 | ABVD (x4) | CR + R | Alive (84 months) |
| 5 | NS | no | no | IIB | 41 | M | 2 | ABVD (x4) | PD | DOD (36 months) |
| 6 | NS | no | yes | IIB | 32 | F | 3 | ABVD (x6) + Rt | PD | DOD (10 months) |
| 7 | NS | no | no | IIB | 59 | M | 4 | ABVD (x6) | PD | DOD (7 months) |
| 8 | NS | yes | yes (mediastinal) | IV | 23 | F | 2 | ABVD (x6) + Rt | PD | AWD (42 months) |
| 9 | NS | no | no | IIIA | 42 | M | 2 | ABVD (x6) | PD | DOD (27 months) |
| 10 | MC | yes | no | IV | 21 | M | 5 | ABVD (x6) | PD | Lost |
| 11 | MC | yes | yes (abdominal) | IV | 69 | F | 3 | ABVD (x6) + Rt | CR + R | AWD (32 months) |
| 12 | NS | yes | no | IV | 73 | M | 4 | ABVD (x6) | PD | DOD (15 months) |
MC mixed cellularity, NS nodular sclerosis, IPS International Prognostic Score, ABVD adriamycin, bleomycin, vinblastine, and dacarbazine, Rt radiotherapy, CR + R complete remission and early relapse, PD progressive disease, DOD dead of disease, AWD alive with disease
We enriched the tumor cells by LCM using a Palm MicroBeam V4 microscope (Carl Zeiss Inc., Oberkochen, Germany) equipped with a catapult system for contamination-free sample isolation. In brief, 5-μm adjacent sections from FFPE tissues were cut, deparaffinized and immunostained with anti-CD30 antibody (clone BerH2, Dako/Agilent, Madrid, Spain). Parallel sections were also stained with hematoxylin and eosin to assess morphology, content and distribution of HRS cells.
All experimental procedures were repeated in the original pretreatment biopsy and the relapse biopsy, comparing each duplicated sample and discarding non-concordant variants. From each case, 15,000–25,000 individually picked HRS cells duplicated per case were isolated from FFPE tissue. We also isolated normal lymphocytes from the reactive background (morphologically normal lymphocytes, CD30-negative, duplicated per case). This procedure was repeated in the original pretreatment biopsy and the relapse biopsy. Thus, eight different experiments were analyzed by NGS for each patient (all the workflow is depicted in Supplementary Figure 1).
After cell enrichment, we performed targeted analysis of 35 genes involved in B cell-related pathways (Supplemental Table 1), previously reported as being the most frequently mutated in cHL6,7,9, using Ion Torrent PGM (Thermo Fisher Scientific, NY, USA) technology with a modified protocol from that previously published7. DNA was extracted from duplicates of isolated HRS cells and CD30-negative fractions using the Gentra Puregene Tissue Kit (Qiagen, Germantown, MD). Libraries were constructed starting with 10 ng of genomic DNA and following the manufacturer´s protocol. Sequencing (BAM) files have been deposited in the NCBI Sequence Repository (SRA: PRJNA506444). The data were analyzed with the Torrent Suite program. All variants were examined with Integrative Genomics Viewer (IGV) software10, discarding non-concordant variants. Functional consequences of the SNVs were predicted using the publicly available PROVEAN (shift and polyphen-2) and CONDEL algorithms.
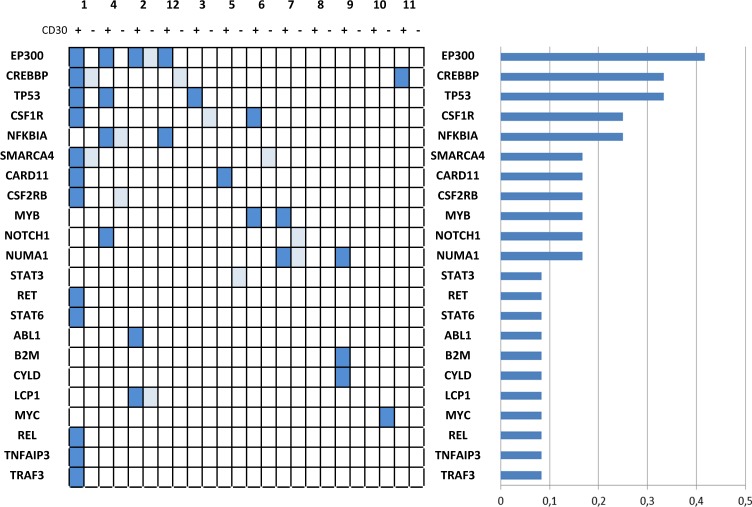
Using this protocol for targeted sequencing, and after filtering SNPs, non-concordant, and silent variants, 42 candidate somatic SNVs were identified in the CD30-positive fraction and 15 in the CD30-negative fraction, from the 238 initial candidate variants (Fig. 1 and Supplementary Table 2 summarize the results after comparing each duplicated sample and discarding non-concordant variants). These figures could be related to genetic instability11, extensive clonal diversity and mutations with very low variant allele frequencies, as described in HRS cells7 and tumors9, or could arise because some of the individual variants are sequencing errors. An astringent filtering process, and the fact that, in all cases, the analysis was done in duplicate, guarantee that the risk of false-positive results is very low, even at the cost of losing valuable information about low-frequency mutations and intratumor heterogeneity.
Fig. 1. Distribution and frequency of variants.
NGS analyses were repeated in the initial pretreatment and the relapse biopsies, comparing duplicated samples and discarding non-concordant variants. Light blue indicates variants detected in the CD30-negative fraction. The histogram on the right indicates relative frequencies
Most cases presented gene variants that had previously been described in cHL6,7,9. We confirmed the previously reported prevalent mutations affecting the NF-kappaB pathway, associated with JAK/STAT activation, B2M mutations, and mutations affecting the BCR pathway.
Interestingly, mutations affecting the TP53 gene were overrepresented in this series of refractory cases (3 out of 12 cases, 25%), with some variants that are redundantly detected in different cases, such as C238Y (cases 1 and 3) or Y234* (cases 3 and 4). These results contrast with classic studies that focused solely on p53 pathway mutations in cHL, and previous NGS studies of unselected cHL only identified rare TP53 mutations (Supplementary Table 3). It is worth noting that TP53 mutations have been more frequently described recently in refractory cHL through the use of more sensitive NGS techniques12. This is entirely unsurprising since mutations of the p53 pathway are a classic prognostic factor associated with chemoresistance in most cancers.
In addition, the most frequently mutated genes, EP300 and CREBBP, correspond to epigenetic regulators. Identical variants were detected in different cases and also in the CD30-positive and CD30-negative fractions, such as EP300 N1776H or CREBBP P1083L. EP300 encodes a transcriptional coactivator protein and functions as a histone acetyltransferase and regulates transcription via chromatin remodeling. Confirming previous results7,9, mutations in the histone acetylation domains of EP300 and CREBBP are frequent in cHL (one or both alterations are present in 5 out of 12 cases, 41.6%), similar to what is seen in DLBCL and follicular lymphoma. Furthermore, a recent report has presented evidence that histone acetyltransferase might act as a tumor suppressor that controls MHCII expression and promote tumor immune control and evasion13, and that drugs targeting histone acetyltransferase activities and chromatin remodeling, such as BET inhibitors, represent a novel therapeutic approach that have recently demonstrated efficacy in solid malignancies and lymphomas.
This study is obvious limited by the small number of cases, experimental complexity, and the impossibility of analyzing a parallel cohort of samples of “good responders” as control patients. Nevertheless, it seems clear that mutational frequencies are different from the distribution reported in previous unselected series (Supplementary Table 3). However, in a very recent study using a targeted NGS panel commercially available in archival tumor samples particularly enriched with relapsed patients, the most commonly mutated gene was also TP53, detected in 11 patients (22%). Possible comparisons are limited by the obvious methodological differences between the various studies.
In our previous work7, in which 34% of the cases were primary refractory cHL, EP300 was also one of the most frequently mutated genes (occurring in 14% of cases). There were no statistically differences between mutational patterns of responders and non-responders in the original analyses, probably due to the small sample size. Nevertheless, further Kaplan–Meier analyses of failure-free survival (FFS), using the records from the 46 patients for whom we have complete clinical information and follow-up, revealed a significant association between the presence of CREBBP mutations and FFS (Supplemental Figure 2). Although we must recognize that a formal comparison between these different studies has obvious limitations.
An important observation in our study is the discovery of several mutations in the CD30-negative fraction in most of the experiments, using an LCM protocol with a very low probability of being subject to common contamination. We can speculate that these results might indicate a clonally related population, in a stem cell or less differentiated state, that is not morphologically or phenotypically distinguishable, as other authors have previously suggested14. In any case, additional experiments are necessary to corroborate this finding and fully understand its pathogenic significance.
In conclusion, it seems that several recurrent mutational events are present in primary refractory cHL that could be used as biomarkers and eventually exploited for therapy. TP53 mutations seem to represent a relevant predictive marker also in cHL, and the highly prevalent mutations of epigenetic regulators EP300 and CREBBP suggest a rationale for alternative therapeutic strategies that need to be investigated further.
Supplementary information
Acknowledgements
This work was supported by grants from the Plan Nacional de I + D + I co-financed by the ISCIII-Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER), PI12/1832, the Spanish Association for Cancer Research (AECC), and Programas para Grupos de Investigación de la Comunidad Autónoma de Madrid (Biomedicina 2017). MSB currently holds a Miguel Servet II contract (CPII16/00024) supported by ISCIII-MINECO AES-FEDER (Plan Estatal I + D + I 2013–2016) and the Fundación de Investigación Biomédica Puerta de Hierro.
Author contributions
E.M. designed and performed the experiments and the targeted NGS panel. S.F. analyzed the sequencing data. A.A., R.F., M.G.-C., M.P., and M.E. provided clinical and diagnostic expertise and contributed to data interpretation. M.S.-B., C.M., M.A.P., and J.F.G. designed the experiments, directed the research, and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41408-019-0195-7).
References
- 1.Emmerich F, et al. Inactivating I kappa B epsilon mutations in Hodgkin/Reed-Sternberg cells. J. Pathol. 2003;201:413–420. doi: 10.1002/path.1454. [DOI] [PubMed] [Google Scholar]
- 2.Schmitz RH, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J. Exp. Med. 2009;206:981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weniger MA, et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene. 2006;25:2679–2684. doi: 10.1038/sj.onc.1209151. [DOI] [PubMed] [Google Scholar]
- 4.Liu X, et al. Mutations of NFKBIA in biopsy specimens from Hodgkin lymphoma. Cancer Genet. Cytogenet. 2010;197:152–157. doi: 10.1016/j.cancergencyto.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Tiacci E, et al. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood. 2012;120:4609–4620. doi: 10.1182/blood-2012-05-428896. [DOI] [PubMed] [Google Scholar]
- 6.Reichel J, et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood. 2015;125:1061–1072. doi: 10.1182/blood-2014-11-610436. [DOI] [PubMed] [Google Scholar]
- 7.Mata E, et al. Analysis of the mutational landscape of classic Hodgkin lymphoma identifies disease heterogeneity and potential therapeutic targets. Oncotarget. 2017;8:111386–111395. doi: 10.18632/oncotarget.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, et al. The mutational landscape of Hodgkin lymphoma cell lines determined by whole-exome sequencing. Leukemia. 2014;28:2248–2251. doi: 10.1038/leu.2014.201. [DOI] [PubMed] [Google Scholar]
- 9.Tiacci E, et al. Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood. 2018;131:2454–2465. doi: 10.1182/blood-2017-11-814913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomic Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2012;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Re D, Zander T, Diehl V, Wolf J. Genetic instability in Hodgkin’s lymphoma. Ann. Oncol. 2002;13(Suppl 1):19–22. doi: 10.1093/annonc/13.S1.19. [DOI] [PubMed] [Google Scholar]
- 12.Liang WS, et al. Comprehensive genomic profiling of hodgkin lymphoma reveals recurrently mutated genes and increased mutation burden. Oncologist. 2018;23:1–10. doi: 10.1634/theoncologist.2017-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashwah H, et al. Inactivation of CREBBP expands the germinal center B cell compartment, down-regulates MHCII expression and promotes DLBCL growth. Proc. Natl Acad. Sci. USA. 2017;114:9701–9706. doi: 10.1073/pnas.1619555114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones RJ, et al. Circulating clonotypic B cells in classic Hodgkin lymphoma. Blood. 2009;113:5920–5926. doi: 10.1182/blood-2008-11-189688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.