Abstract
In Drosophila, female development is governed by a single RNA-binding protein, Sex-lethal (Sxl), that controls the expression of key factors involved in dosage compensation, germline homeostasis and the establishment of female morphology and behaviour. Sxl expression in female flies is maintained by an auto-regulatory, positive feedback loop with Sxl controlling splicing of its own mRNA. Until now, it remained unclear how males prevent accidental triggering of the Sxl expression cascade and protect themselves against runaway protein production. Here, we identify the protein Sister-of-Sex-lethal (Ssx) as an inhibitor of Sxl auto-regulatory splicing. Sxl and Ssx have a comparable RNA-binding specificity and compete for binding to RNA regulatory elements present in the Sxl transcript. In cultured Drosophila cells, Sxl-induced changes to alternative splicing can be reverted by the expression of Ssx. Moreover, in adult male flies ablation of the ssx gene results in a low level of productive Sxl mRNA splicing and Sxl protein production in isolated, clonal cell populations. In sum, this demonstrates that Ssx safeguards male animals against Sxl protein production to reinforce a stable, male-specific gene expression pattern.
INTRODUCTION
In most species of higher eukaryotes, sexual reproduction is the favoured form of reproduction. By combining alleles from two different individuals, increased variation in the offspring can be achieved which allows rapid adaption to changing environments, as well as the cleansing of harmful mutations from a population. Species that reproduce sexually usually generate two types of individuals, males and females. Not surprisingly, the genetic programs that determine sex and control sexual differentiation need to be robust in order to ensure survival of the population.
In Drosophila the switch gene Sex-lethal (Sxl) plays a key role in sex determination and sexual differentiation. It encodes an RNA-binding protein that acts as the master regulator of female development by controlling synthesis of key factors with functions critical for dosage compensation, germline homeostasis, morphology, and behaviour (1–5). Full length Sxl protein is expressed only in female animals where it post-transcriptionally regulates gene expression on multiple levels. Its function in somatic tissues has been particularly well studied where it acts both in the nucleus and the cytoplasm to control processing, nuclear export, and translation of its RNA targets (6).
mRNAs originating from the Sxl gene are also detected in male flies. However, inclusion of a poison cassette exon (exon L3) with a premature termination codon in males results in the translation of truncated and non-functional proteins. In female animals, production of full-length Sxl protein is initiated in pre-cellular embryos by an X-chromosome counting mechanism that activates the Sxl ‘establishment’ promoter, SxlPe. Sxl protein expressed from SxlPe instructs productive splicing of Sxl transcripts originating from the ‘maintenance’ promoter (SxlPm) which is activated at a later stage in development irrespective of sexual identity. Regulation occurs by Sxl suppressing inclusion of the poison exon in its own primary transcript, thus generating an auto-regulatory, positive feedback loop for self-sustained expression (4). Hence, synthesis of functional Sxl protein acts as a molecular switch that, once activated, commits to female development.
Owing to this feedback loop, Sxl expression in females is lasting, but non-expression in males appears a rather unstable state. In males, expression of functional Sxl protein presumably occurs at a non-zero rate due to fluctuations inherent to gene expression and occasional mis-splicing. In these animals small amounts of aberrantly produced, functional Sxl protein could trigger the feedback loop, amplifying the signal and flipping the switch into the stable, female state which is characterized by sustained expression of Sxl. However, forced ubiquitous expression of Sxl in male flies is deleterious and causes lethality (7,8). Hence, one would predict additional safeguard mechanisms to operate in males, protecting against Sxl protein production snowballing out of control.
Here we identify Sister-of-Sex-lethal (Ssx) as a protein that can inhibit Sxl-mediated auto-regulatory splicing. Sxl and Ssx are paralogs originating from a gene duplication event early in Drosophilid evolution (9,10) and the two proteins share a high degree of identity within their central, RNA-binding domain. While Sxl has been well studied in the past decades, the function of Ssx remained enigmatic. Its knockout does neither result in a morphological phenotype nor does it affect viability in either sex under standard laboratory conditions, even in combination with mutations in Sxl (9). Transposon insertion into the Ssx locus however is immunocompromising and the mutant flies quickly succumb to gram-positive bacterial infection, but not to infection with gram-negative pathogens, suggesting a function in immunity (11). We have previously reported, that Ssx can associate with msl-2 mRNA to repress its translation like its paralog Sxl (12).
Here we show that in contrast to Sxl, Ssx is expressed in both sexes. The two proteins have a comparable RNA-binding activity and associate with similar uridine-rich sequences. We further demonstrate that both proteins compete for binding to the same regulatory RNA sequences present in the Sxl primary transcript. When overexpressed in cultured Drosophila cells, Ssx promotes inclusion of the Sxl poison exon L3 most probably by acting as a competitive inhibitor of Sxl auto-regulatory feedback to alternative splicing. In line with this finding, even in the absence of a morphological phenotype, adult male flies that are hemizygous mutant for ssx exhibit detectable levels of productive Sxl mRNA splicing and Sxl protein expression in isolated, clonal cell populations. This demonstrates that, through competition with Sxl for the same binding sites, Ssx reinforces a male-specific gene expression pattern by protecting against accidental triggering of the Sxl auto-regulatory, positive feedback loop.
MATERIALS AND METHODS
RT-PCR
Total RNA was prepared from flies or cultured cells using Trizol (Sigma). Reverse transcription was performed according to manufacturer's protocol using Superscript II (ThermoFisher Scientific) in combination with oligo-dT or random hexamer primers. cDNAs were subjected to 30 cycles of semi-quantitative PCR. The sequences of the primers targeting the sxl, ssx, tra and msl-2 mRNAs are listed in Supplementary Table S2.
DNA constructs
For transfection experiments in cultured insect cells, annealed oligonucleotides encoding a FLAG-3xHA sequence were introduced into a modified pCaSpeR-HS vector (13) using the EcoRI restriction site. Subsequently the full length open reading frames of Sxl and Ssx were PCR amplified and cloned in-frame with the tag using the EcoRI and XbaI restriction sites. For fly transgenesis, the ssx coding sequence (with an N-terminal FLAG-3xHA-tag) was cloned into the pUASt-attB and pUASp-attB vectors. To generate a repair template to target the 3′ end of the Sxl coding region, approx. 1,000nt long sequences derived from the ssx locus were inserted into NotI-SacII and KpnI–SpeI restriction sites of the pT-GEM(0) vector (a gift from Benjamin White, Addgene plasmid # 62891, 14) generating pT-GEM(0)-Sxl.
Recombinant protein and antibody production
Recombinant Sxl and Ssx proteins were produced as described before (12). Antibodies against Ssx were raised in rabbits injected with purified Ssx-RBD4 or a Ssx-derived peptide encompassing aa19–34 (DIEGSGDNVGRDDGTD) using 28-day immunization protocol (Eurogentec, Belgium).
Electromobility shift assays
To allow ribonucleoprotein complex formation, 10 fmol of 32P-labeled U16 RNA were incubated for 30 min at 4°C with the indicated amounts of protein in a reaction containing 10 mM Tris/Cl pH 7.4, 50 mM KCl, 1 mM EDTA, 1 mM DTT and 0.2 μg/μl yeast tRNA. Subsequently, complexes were resolved by 8% native PAGE (acrylamid/bisacrylamid 37.5:1) for 2 h at 4°C and 10 V/cm. Detection occurred using the Personal Molecular Imager System (Bio-Rad).
Tissue culture
Drosophila SL2 and Kc167 cells were propagated at 80% confluency in Express Five SFM supplemented with 10× Glutamax. Cells were transfected with Fugene HD (Promega) following the manufacturer's instructions and incubated for 48 h at 25°C before harvesting.
For knockdown of Ssx and Sxl, double-stranded RNAs were generated by run-off transcription on PCR products generated from the 5′ region of Sxl or Ssx respectively, introducing T7 promoter sequences with the primers. For efficient RNAi in SL2 cells a single treatment of 1.2 × 106 cells for three days with 30 μg of dsRNA in a six-well format was sufficient.
Western blotting
Cultured cells were harvested and resuspended in lysis buffer (20 mM Tris/Cl pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 2% SDS). For extract preparation from flies or embryos, whole animals were homogenized in lysis buffer, followed by extensive centrifugation. Protein concentration of cleared lysates was determined using the BioRad protein assay reagent. Equal amounts of total protein were subjected to western blotting using the following antibodies: rabbit anti-Ssx (1:1000, described above), mouse anti-HA (1B8, Sigma Aldrich, 1:1000), monoclonal mouse anti-Sxl (M18, developed by P. Schedl, obtained from the Developmental Studies Hybridoma Bank (DHSB), created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA52242), polyclonal rabbit anti-Sxl (15, a gift from M. Hentze), mouse anti-beta-tubulin (E7, deposited to the DSHB by M. Klymkowsky), rabbit anti-GAPDH (GeneTex, 1:1,000), HRP-coupled anti-rabbit and anti-mouse light chain-specific secondary antibodies (1:10 000, Jackson Immuno Research). Detection occurred by using Clarity Western ECL substrate and the ChemiDoc Touch Imaging System (BioRad).
Ssx individual-nucleotide cross-linking and immunoprecipitation (iCLIP)
iCLIP was performed as described (16). Briefly, a 15-cm dish of SL2 cells was washed with PBS and UV-irradiated (300 mJ/cm² at 254 nm) using a UV Stratalinker 2400 (Stratagene). Next, cell extract was prepared and subjected to RNase treatment using 36 U of RNase I (Ambion). Immunoprecipitation was performed with either polyclonal Ssx-antibody-containing serum or control serum on Dynabeads Protein G (Life Technologies) for 2 h at 4°C. After washing four times with washing buffer (50 mM Tris/Cl pH7.4, 800 mM NaCl, 0.05% Tween 20), the co-immunoprecipitated RNA was dephosphorylated, ligated with a 3′-RNA linker and 5′-radiolabeled with T4 PNK and [γ-³²P]-ATP. Samples were subjected to neutral SDS-PAGE (NuPAGE, Invitrogen) and transferred to a nitrocellulose membrane. Protein/RNA-complexes were visualized by autoradiography. Ssx/RNA-complexes were cut from the membrane, proteins were digested with Proteinase K and RNA was subjected to iCLIP library preparation as previously described (16). Sequencing occurred on a MiSeq® (Illumina, 130nt single read).
Bioinformatic analyses
For analyses of iCLIP data, the raw sequence reads were subjected to adapter trimming using Cutadapt (17) and the unique molecular identifier (UMI) introduced with the primers was extracted. Reads mapping to ribosomal RNA were removed and the remaining reads were aligned to the Drosophila dm6 genome sequence using bowtie (18). Duplicate reads (based on mapping position and UMI) were removed. The remaining reads were subjected to differential gene expression analysis using the DeSeq2 package (19) comparing iCLIP samples to controls that were generated with a non-specific antiserum. Peaks were scored using ASPeak (20) and the sequences 30nt up- and downstream of each peak were retrieved for motif analysis using MEME (21).
Raw sequencing data is accessible via Gene Expression Omnibus: GEO Series GSE98189.
Fly stocks and genetics
Flies were kept under standard conditions (25°C, 12 h/12 h LD cycle). Fly lines obtained from the Bloomington Stock Center: ssxEY14203 (BDSC_20792), da-GAL4 (BDSC_55850), tubulin-Gal80ts (BDSC_7108), UAS-Stinger (BDSC_65402), vas-Cas9 (BDSC_56552 and BDSC_51324), Cre (BDSC_1092), UAS-GFP (BDSC_6874) and FM7c (BDSC_3378).
UASt-Ssx and UASp-Ssx transgenes were generated using phiC31-mediated germline transformation into attp40 (22). For overexpression of Ssx, virgins of da-GAL4, nos-GAL4, or osk-GAL4 (the latter two lines a gift from Anne Ephrussi, BDSC_44241 and BDSC_44242) were mated to UASt-Ssx or UASp-Ssx males.
Replacement of the ssx-coding region to generate a knockout allele was essentially performed as described before (23). In brief, Cas9-mediated DNA cleavage was induced in vasa-Cas9-GFP flies at two positions flanking the Ssx CDS (compare Supplementary Figure S3, using the guide sequences CTATCAAGGCTTGACACAGA and CCCAGCCAGCCGCATCCCGT, targeting the genomic region chrX:1371303..1373219). As a repair template a red fluorescent protein (RFP) donor construct (pDSRed, a gift from Melissa Harrison & Kate O’Connor-Giles & Jill Wildonger, Addgene plasmid #51019) was provided, with homology arms targeting the sequences up- and downstream of the cleavage sites. Flies expressing dsRed were selected and back-crossed to generate a homzygous ssxΔ(3xP3-RFP) stock.
For lethality tests, in three independent experiments at least 100 embryos were visually scored for embryonic, larval (L1, L2, L3), and pupal lethality on a daily basis. Genotypes of the animals are as follows: (i) ctrl: UASt-ssx/UASt-ssx, (ii) UASt-ssx, da-GAL4, (iii) ssxEY14203/ssxEY14203 or ssxEY14203/Y and (iv) ssxΔ/ssxΔ or ssxΔ/Y. The loss of individual animals (the lethality) was calculated for each developmental stage (embryo, larva, pupa) and expressed in % relative to the total number of fertilized eggs.
The Sxl-T2A-GAL4 reporter strain was constructed in a manner analogous to the knockout allele, targeting the terminal exon of the Sxl locus in vas-Cas9 embryos with guide RNA sequences that target the genomic regions chrX:7088090-109 (CCAGAAACGAAUACAAGAUGAAA) and chrX:7087935-54 (CCAGCAAAUGUACCACCGCCGCC). For homologous recombination, the plasmid pT-GEM(0)-Sxl was provided as a repair template. After injection, adult flies were crossed to FM7c males or virgins and the offspring was screened for red fluorescent eyes. Positive transformants were again crossed to FM7c to create stable stocks. Females heterozygous for Sxl-T2A-GAL4 showed a high lethality because of GAL4 toxicity and were crossed to tubulin-Gal80ts to generate Sxl-T2A-GAL4(3xP3-RFP)/FM7c; tubulin-Gal80ts flies to reduce lethality in females. The ssxΔ(3xP3-RFP), w* chromosome was recombined with a CantonS X-chromosome to generate a ssxΔ(3xP3-RFP), w+ chromosome followed by excision of the 3xP3-RFP with Cre-Recombinase. Subsequently, the ssxΔ, w+chromosome was recombined with a Sxl-T2A-GAL4(3xP3-RFP) chromosome to obtain flies with the genotype ssxΔ, w+, sxl-T2A-GAL4(3xP3-RFP). Again, to reduce GAL4 toxicity in females,ssxΔ, w+, sxl-T2A-GAL4(3xP3-RFP); tubulin-Gal80ts stocks were created. To confirm absence of ssx, males carrying the ssxΔ, w+, sxl-T2A-GAL4(3xP3-RFP) chromosome were analyzed by PCR (using the ssx-upstream and ssx-3′UTR primers and the the ssxE3 and ssxE4 primerset, Supplementary Table S2).
Visualization of GFP expression from sxl-T2A-GAL4 reporter lines
Virgins from ssxΔ, w+, Sxl-T2A-GAL4(3xP3-RFP); tubulin-Gal80ts or Sxl-T2A-GAL4(3xP3-RFP); tubulin-Gal80ts stocks were crossed to either UAS-GFP or UAS-Stinger males and raised at 18°C. After hatching, adult males were shifted to 29°C for three days to allow GFP expression. ssxΔ, w+, Sxl-T2A-GAL4(3xP3-RFP) or Sxl-T2A-GAL4(3xP3-RFP) males expressing either GFP or Stinger were cold anesthetized, screened for GFP expression with a Leica M2 Fl III and imaged with a Zeiss Axiophot combined with a Zeiss Colibri.
RESULTS
Sister-of-Sex-lethal is expressed in both sexes
In Drosophila melanogaster, Sxl mRNA isoforms are expressed in a sex-specific fashion resulting in the production of functional, full-length protein exclusively in female animals. In contrast, in male animals splicing includes an exon that contains a premature termination codon (L3) resulting in mRNAs that encode truncated and non-functional protein. Analysis of Sxl expression by RT-PCR detects the sex-specific isoforms that lack exon L3 in females while males show inclusion of the exon (Figure 1A, bottom panel).
Figure 1.
Ssx is expressed in both sexes. (A) Ssx mRNAs can be detected in male and female flies. Shown at the top is a schematic representation of the ssx locus with exons depicted as boxes and introns as lines. The shaded area represents the open reading frame of the protein isoform PB (compare Supplementary Figure S1). Positions of the primers used for the RT-PCR are indicated by arrows. Below: RT-PCR analysis of ssx and Sxl transcripts from embryos, male, and female flies. Primers for detection of ssx are depicted in the above schematics. For Sxl primers were used that flank the male-specific exon L3 and hence yield sex-specific RT-PCR products: a male-specific longer one (+L3) and a short, female-specific one (–L3). Control reactions without reverse transcriptase are shown on the right. (B) Ssx antibodies do not cross-react with Sxl. SL2 cell lysates were subjected to Western blotting with antibodies against Ssx (top panel), Sxl, Hemagglutinin (HA), or GAPDH (lower panel). As control, 10, 20 and 40 μg of total protein from untreated SL2 cells were used (lanes 1–3, CTRL) and compared to 40 μg of protein from cells that were transfected with plasmids encoding FLAG-3xHA-tagged Sxl (+F/H-Sxl) or Ssx (+F/H-Ssx). Treatment with dsRNA targeting either Sxl (lane 4) or ssx (lane 5) was used as specificity control. Molecular weight markers are indicated on the right. (C) Ssx protein is detectable in male and female cells. Cell lysates (10, 20 and 40 μg of total protein) of ‘male’ SL2 (lanes 1–3) and ‘female’ Kc167 cells (lanes 4–6) were subjected to Western blotting with antibodies directed against Sxl (upper panel), Ssx (middle panel), and β-tublin (lower panel). Molecular weight markers are indicated on the right. (D) Ssx expression is independent of sexual identity. Lysates from adult male or female flies of different genotypes (as indicated at the top) were probed for Ssx expression by western blotting. Wildtype flies are shown in lanes 5 and 6, followed by animals that express FLAG-3xHA-tagged Ssx (F/H-ssx, lanes 7 and 8). In addition, Ssx protein expression was analysed in flies homozygous/hemizygous for insertion of a transposable element in the ssx locus (ssxEY14203, lanes 1 and 2) or in which the entire ssx open reading frame has been replaced by a dsRED cassette (ssxΔ, lanes 3 and 4). The asterisk marks an unspecific signal, demonstrating equal loading of the samples.
In contrast to Sxl, we did not detect sex-specific expression of ssx mRNA isoforms. We obtained full length open reading frames from both sexes (Figure 1A, see also Supplementary Figure S1 for isoform-specific analyses) which is in agreement with publicly available high throughput sequencing datasets (modENCODE, 24).
To verify that also the Ssx protein is expressed independent of sexual identity, we generated antibodies directed against either a peptide from the N-terminal region of Ssx, or the central RNA-binding domain that encompasses the two RRMs. By Western blotting, the antisera detect a protein of approx. 50kDa in lysates from cultured Drosophila SL2 cells (Figure 1B). RNAi against ssx, but not Sxl, strongly diminishes the signal while not affecting controls, confirming the identity of the detected protein (Figure 1B, lanes 4 and 5). Upon expression of a tagged version of Ssx, an additional signal can be detected that—due to the tag—exhibits a slightly reduced mobility (Figure 1B, lane 7). Importantly, despite the high similarity between the central domains of Sxl and Ssx (Supplementary Figure S2A), the antibodies show no detectable cross-reactivity, as demonstrated by forced expression of Sxl: overexpressed, FLAG/HA-tagged Sxl protein can be detected with both anti-Sxl and anti-HA antibodies, but not with the Ssx-specific antisera (Figure 1B, lane 6).
Using the validated antibodies, we then probed for Ssx protein in two Drosophila cell lines that are derived from opposite sexes (15). Kc167 cells exhibit female characteristics and express functional Sxl protein (Figure 1C, upper panel), resulting in repression of Msl-2 (Male-specific lethal 2) protein synthesis and female-specific splicing patterns of Sxl target mRNAs. In contrast, the SL2 cell line exhibits male characteristics and expresses Msl-2 protein instead of Sxl (15). As previously suggested by RT-PCR and sequencing, the Ssx protein can be detected by Western blotting in both cell lines with higher protein levels present in the ‘male’ SL2 cells (Figure 1C). Similarly, we detect Ssx protein in lysates from both male and female adult flies (Figure 1D, lanes 5 and 6) as well as in embryos of mixed stages (Supplementary Figure S3E). In the ‘female’ cell line Kc167, we determine an ∼2-fold molar excess of Sxl protein over Ssx, indicating that in female cells the concentration of Sxl exceeds the one of Ssx (Supplementary Figure S4).
Ssx and Sxl bind U-rich sequences in an overlapping set of RNAs
Sxl and Ssx exhibit a similar domain structure with two RNA-recognition motifs centred within the proteins. While the flanking N- and C-terminal extensions are quite distinct, the RNA-binding domain is 80% identical on the amino acid level (Supplementary Figure S2A). Sxl is a well-studied RNA-binding protein with a preference for U-rich motifs and many experimentally confirmed binding sites (8,25–34). Furthermore, detailed structural data is available that allows molecular insight into how Sxl achieves RNA-binding specificity (35,36). We could previously demonstrate that—similar to Sxl—Ssx binds to U-rich RNA elements present in the msl-2 transcript (12), a finding that was independently confirmed in another study that employed a different experimental approach (37).
To identify the RNA target sites of Ssx on a transcriptome-wide scale, we performed individual-nucleotide resolution crosslinking-immunoprecipitation (iCLIP) experiments, targeting the endogenous protein. In brief, the Ssx RNA interaction was stabilized through UV-crosslinking of cultured Drosophila SL2 cells. After stringent purification of Ssx-containing ribonucleoproteins using our validated antibodies, co-purified RNAs were subjected to high throughput sequencing. Control experiments were performed using non-specific antibodies. Analyses of the iCLIP crosslink sites revealed that, relative to control samples, 92 transcripts are bound by Ssx in SL2 cells (Supplementary Table S1).
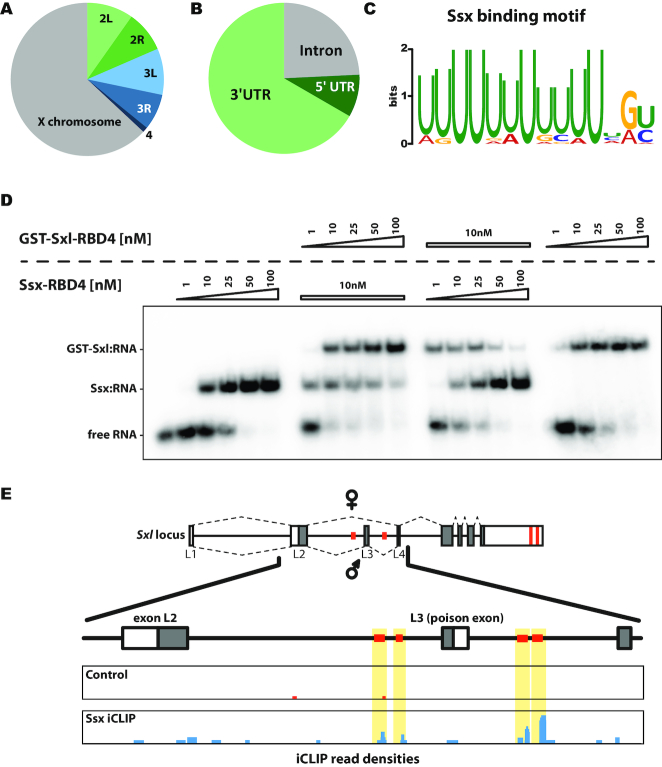
Previously, prediction of Sxl binding sites reported an increased occurrence in X chromosome-encoded transcripts (38). We also find that the majority of Ssx-bound RNAs is encoded on the X chromosome (Figure 2A), although the size and gene number of the Drosophila X chromosome is comparable to the chromosomes 2 and 3. The identified Ssx binding sites appear to be enriched in 3′ UTRs and intronic sequences (Figure 2B). The latter ones score highly in the iCLIP data but, due to processing of the RNAs, are barely detectable in total RNA isolated from cultured cells. This enrichment of iCLIP reads (over input RNA) suggests that the detected intronic binding sites are highly occupied by Ssx. In contrast, we cannot identify a single binding site within a coding sequence, potentially owing to the fact that a poly(U) sequence (the preferred binding motif of Ssx, see below) translates into a phenylalanine stretch—an amino acid sequence only rarely encountered in proteins.
Figure 2.
Identification of Ssx target mRNAs. (A) Most of the Ssx-bound RNAs (as identified by iCLIP) are encoded on the X chromosome. The chromosomal origin of transcripts that contain statistically significant clusters of iCLIP is plotted. (B) Ssx binding sites (as identified by iCLIP) are predominantly found in the 3′ UTRs of its target RNAs. (C) iCLIP strongly enriches a U-rich RNA sequence motif as putative Ssx recognition site. (D) Competitive electromobility shift assay using a short, U-rich RNA (U16) and recombinant Ssx-RBD4 (aa93–269) and GST-Sxl-RBD4 (aa122–301) proteins (see Supplementary Figure S2). Amounts of recombinant proteins are indicated for each reaction above the individual lanes, free RNA and protein:RNA complexes are indicated on the left. (E) In the absence of Sxl, Ssx associates with known Sxl binding sites in the Sxl transcript (areas highlighted in yellow, Sxl binding sites depicted as orange boxes). The Sxl gene locus is depicted schematically at the top. Exons are shown as boxes, introns as lines, the coding region is shaded in grey. The splicing pattern in females is indicated at the top (corresponding to the transcript variant RD, FBtr0331262), the male splicing pattern below (corresponding to the transcript variant RF, FBtr0331249). Individual splice events are indicated with dashed lines. Below the region spanning exons L2 to L4 is enlarged and read densities combined from replicates of Ssx iCLIP experiments (blue) and control reactions (red) are given.
Analysis of the sequences that surround the Ssx crosslink positions shows a strong enrichment for a U-rich sequence motif (Figure 2C) that closely matches the sequence motif previously determined to be recognized by Sxl. This further confirms that Sxl and Ssx recognize similar RNA sequences.
To understand if both proteins can simultaneously bind to the same U-rich RNA element, we performed competitive electromobility shift experiments using the recombinant RNA-binding domains of Sxl (denoted Sxl-RBD4) and Ssx (denoted Ssx-RBD4, Figure 2D, Supplementary Figure S2) and a well-characterized Sxl binding motif derived from msl-2 mRNA (B-site). We previously demonstrated that both proteins form RNP complexes with almost indistinguishable migration behaviour (12). In order to allow for discrimination between Sxl:RNA and Ssx:RNA complexes we used a GST-tagged version of Sxl-RBD4 (39) that produces an RNP with reduced mobility and, hence, allows resolving of the different protein-RNA complexes. When testing binding to an RNA fragment consisting of 16 Uridine residues, GST-Sxl-RBD4 and Ssx-RBD4 individually exhibit similar affinities (Figure 2D, lanes on the left and right). Upon combination of the proteins in a single reaction, we cannot observe formation of an additional complex with altered mobility (which would be indicative of a heterotrimeric complex—GST-Sxl:Ssx:RNA). Rather we observe a mixture of Sxl- and Ssx:RNA complexes. Moreover, both proteins compete for binding to the RNA and a Sxl:RNA complex can be efficiently challenged with Ssx and vice versa. Similar results are obtained when the GST tag is fused to Ssx instead of Sxl (data not shown), indicating that the tag does not affect RNP complex formation.
The iCLIP data also reveal that in SL2 cells Ssx associates with the Sxl mRNA. This RNA contains several Sxl binding sites that play an important role in post-transcriptional regulation of Sxl protein expression (Figure 2E, highlighted in orange), allowing the protein to exert control over its own synthesis. Binding sites in the 3′ UTR have been implicated in homeotic regulation with Sxl protein suppressing translation of its own transcript to prevent accumulation of excessive protein levels (40). In contrast, binding sites in the introns that flank exon 3 of the Sxl mRNA are involved in the regulation of alternative splicing and the auto-regulatory, positive feedback loop that ensures expression of functional Sxl protein in female flies (41–43). Our data indicate that, in the absence of Sxl (the ‘male’ SL2 cells do not express the protein), these binding sites are occupied by Ssx (Figure 2E).
Ssx promotes inclusion of Sxl exon L3 in cultured cells
To address if Ssx can affect alternative splicing when bound to sites flanking exon L3 of the Sxl mRNA, we analysed the effect of Sxl and Ssx overexpression in cultured Drosophila cells. We first expressed increasing amounts of FLAG-HA-tagged Ssx in the ‘female’ cell line Kc167; FLAG-HA-tagged GFP served as a control. Expression of the proteins was confirmed by Western blotting against the HA tag and with a polyclonal anti-Sxl antibody that cross-reacts with Ssx (Figure 3A). Moreover, Sxl protein levels are not affected by the forced expression of Ssx. RT-PCR analysis reveals that overexpression of Ssx but not GFP results in altered Sxl alternative splicing with low levels of inclusion of the poison exon (L3) that is typically skipped in females (Figure 3B).
Figure 3.
Ssx inhibits Sxl-mediated alternative splicing in cultured cells. (A) Overexpression of Ssx and GFP in cultured, female Drosophila cells (Kc167). Expression of the transfected constructs (FLAG-3xHA-Ssx and -GFP, as indicated above the lanes) is assessed by Western Blotting using anti-HA (lower panel) or a polyclonal anti-Sxl antibody that cross-reacts with Ssx (top panel). (B) RT-PCR analysis of alternative splicing of endogenous Sxl transcripts in transfected Kc167 cells. Splicing products that either lack or contain exon L3 are indicated on the left, molecular weight marker sizes on the right. A control reaction was performed in the absence of reverse transcriptase (panel labelled –RT control) is shown at the bottom. (C) Expression of FLAG-3xHA-tagged Sxl, Ssx and GFP in cultured, male Drosophila cells (SL2). Expression levels of the transfected proteins (as indicated above each lane) are analysed by Western Blotting against the HA tag. Sizes of the individual proteins are indicated on the left. (D) RT-PCR analysis of alternative splicing of endogenous Sxl transcripts in transfected, male SL2 cells. Transfected constructs are indicated above each lane, non-transfected cells were used as a reference (lane ctrl). Labelling as in panel B.
We then performed similar experiments in the ‘male’ SL2 cell line that does not express functional Sxl protein because splicing includes the poison exon in the mature Sxl mRNA (Figure 3D, lane 9). Upon transient expression of FLAG-HA-tagged Sxl a partial skipping of the poison exon can be observed (Figure 3C and D). In contrast, overexpression of Ssx has no detectable effect. Importantly, the Sxl-induced change to alternative splicing of the endogenous Sxl mRNA can be reverted by simultaneous overexpression of Ssx, but not by expression of GFP. Taken together our experiments suggest that Ssx can act as an inhibitor of Sxl auto-regulatory splicing through competition for the same RNA binding sites.
Loss of Ssx results in a low level of productive splicing of Sxl mRNA in male flies
To confirm these findings in living animals, we constructed several fly strains. Using the UAS/Gal4 system, we either forced ubiquitous Ssx expression by a daughterless driver (da-GAL4) or we expressed the protein in the female germline (under the control of either the oskar or nanos promoter). The progeny of females that ectopically express Ssx in the germline is viable, exhibits o visible morphological phenotypic and shows no bias towards either sex (Supplementary Figure S5).
In contrast, upon ubiquitous expression of Ssx, flies homozygous for the transgene are not viable. Even with only a moderate expression from a single transgene the animals do not survive at 25°C. However, despite marked lethality (Figure 4A), survivors can be obtained when the animals are raised at lower temperatures (18–20°C). Overexpression of Ssx in these animals is rather moderate (Figure 1D) and they exhibit no apparent phenotype or bias towards either sex. These animals provided us with an opportunity to analyse alternative splicing patterns under conditions with slightly elevated Ssx levels (see Figure 1D for expression levels in adult flies and embryos). We sorted surviving da-GAL4; UASt-ssx animals by sex and analysed by RT-PCR alternative splicing events that are known to be controlled by Sxl. This includes alternative 3′ splice site use in transformer mRNA, 5′ UTR intron retention in msl-2 mRNA, and skipping of Sxl exon L3 in female flies. The splicing patterns of the aforementioned genes are unaffected in both sexes of Ssx overexpressing flies (Figure 4B).
Figure 4.
Loss of Ssx affects Sxl alternative splicing in male flies. (A) Overexpression of Ssx in flies, but not disruption of the gene, reduces viability. Modest expression of FLAG-3xHA-tagged Ssx from a single transgene (genotype: UASt-ssx, da-GAL4) results in a significant reduction of embryonic survival and increased lethality during larval development relative to control flies that carry the transgene but not the GAL4 driver (ctrl: UASt-ssx). Survival of the ssx-mutant strains (ssxEY14203 and ssxΔ) that are homozygous/hemizygous for a defective ssx locus is comparable to wild-type animals. All flies were raised at 20°C. (B) Surviving adult flies that overexpress FLAG-3xHA-tagged Ssx under control of the daughterless promoter (lanes da:ssx) exhibit normal splicing patterns of Sxl target genes in both sexes. Sxl-mediated, sex-dependent alternative splicing of msl-2 (intron retention, top panel), tra (alternative 3′ splice site usage, third panel), and Sxl mRNA (exon skipping, bottom panel) was assessed by RT-PCR analysis. A constitutive splicing event (processing of msl-2 intron 3, second panel) is shown for comparison. Control reactions were performed in the absence of reverse transcriptase (lanes –RT control). Molecular weight marker sizes are indicated on the right of each gel, schematic representation of the alternative splicing events or indication of the exons contained in the final splicing product are shown on the left. (C, D) Ablation of Ssx expression results in low levels of skipping of Sxl exon 3 in male flies. RT-PCR analysis of alternative splicing in flies either wildtype for ssx (wt), overexpressing FLAG-3xHA-tagged Ssx under control of the daughterless promoter (da:ssx), flies harbouring an insertion of a transposable element in the ssx locus (ssxEY14203, panel C), or animals in which the Ssx open reading frame was replaced by a dsRed expression cassette (ssxΔ, panel D). While wt and Ssx-overexpressing flies exhibit the normal sex-specific splicing pattern of Sxl mRNA (compare panel B), male flies that lack functional Ssx protein produce low levels of the female-specific Sxl transcript variant that lacks exon 3 (top panels, left lanes). Splicing of the Sxl downstream targets tra and msl-2 is shown below. Labelling as in panel B.
We next turned to flies that carry a transposable element insertion in the first intron of the ssx gene (ssxEY14203, Supplementary Figure S3) resulting in a strong reduction of detectable Ssx protein in homozygous mutant females and hemizygous mutant males (Figure 1D). As previously reported, the mutation does not cause a morphological phenotype and the viability of this mutant strain remains unaffected (9). In line with this, alternative splicing of Sxl and tra mRNAs in ssxEY14203/EY14203 females follows the expected, sex-specific pattern and is comparable to wildtype controls (Figure 4C). In contrast, despite lack of a morphological phenotype, ∼5% of the ssxEY14203 hemizygous males exhibit aberrant splicing of Sxl, producing mRNAs that lack exon L3 and encode full length, functional Sxl protein. However, splicing of the Sxl downstream target tra remains unaffected in these animals and we do not detect Sxl-mediated utilization of the female-specific, downstream 3′ splice site (Figure 4C). The molecular phenotype with skipping of the exon L3 was barely detectable when the animals were raised at 25°C, but became more pronounced when lower rearing temperatures were used (18°C).
To rule out that the phenotype is caused by a yet undetected second site mutation in the ssxEY14203 strain, we employed Cas9-mediated genomic engineering to replace the entire Ssx protein-coding region by a dsRed expression cassette (Supplementary Figure S3B). The absence of Ssx protein in mutant flies is confirmed by Western blotting (Figure 1D). Again—as for the ssxEY14203 flies—neither a morphological phenotype nor a skewed sex ratio can be detected. Moreover, female knockout flies do not display changes in the alternative splicing pattern of Sxl (Figure 4D, ssxΔ). In hemizygous ssxΔ males raised at 18°C, however, we observe increased skipping of exon L3 and production of low levels of the female-specific Sxl mRNA isoform. Again, the phenotype was heterogeneous, variable in strength and affected only few individuals. In most cases, in hemizygous ssxΔ males with aberrant Sxl mRNA processing, alternative splicing of the Sxl downstream target tra remained unaffected and produced exclusively the longer mRNA isoform (Figure 4D) (we could detect small effects on tra processing only in a single sample, not shown).
Aberrant Sxl protein expression in male ssx knockout flies
Despite the presence of significant levels of the female-specific mRNA that encodes functional Sxl protein, ssxEY14203 and ssxΔ males exhibit no apparent morphological phenotype and are fully viable (9 and Figure 4A). These findings are at odds with the strong phenotypes and male-specific lethality that are associated with Sxl gain-of-function alleles which cause ectopic expression of Sxl protein in males. This raises the question if indeed significant levels of functional, full length Sxl protein are produced in male ssx mutant animals.
When multiple products are generated in a single reaction, RT-PCR often suffers from amplification biases thus precluding quantification. We therefore were concerned that we might overestimate the amount of the female-specific Sxl mRNA isoform that is generated in male ssx knockout flies. To analyse Sxl protein production directly in living animals, we turned to a genetic approach with high resolution and sensitivity that employs GFP as a reporter gene (Figure 5A). Using Cas9-mediated genomic engineering, we inserted a T2A-Gal4 cassette at the 3′ end of the Sxl coding region and in frame with the major Sxl open reading frame utilized in somatic tissues of female flies. Productive splicing of Sxl mRNA and the removal of the termination codon-containing exon L3 would therefore generate mRNAs that encode a Sxl-T2A-Gal4 fusion protein. This in turn would release Gal4 through autocatalytic cleavage to drive expression of a GFP reporter gene. Inclusion of the Sxl poison exon L3 during splicing, however, would prevent production of the full-length fusion protein and the Gal4 moiety, effectively preventing activation of the GFP reporter gene.
Figure 5.

Male ssx knockout flies aberrantly express full-length Sxl protein. (A) Schematic representation of the reporter gene strategy employed to monitor full-length Sxl protein expression in flies. A T2A-Gal4 encoding sequence was fused to the 3′ end of the Sxl open reading frame (top) to allow expression of a Sxl-T2A-Gal4 fusion protein upon productive splicing (female splicing pattern indicated by dashed lines). Auto-proteolytic cleavage of the T2A sequence in the fusion protein releases the C-terminal Gal4 moiety which activates expression of a GFP reporter gene. (B–D) GFP expression in Sxl-T2A-Gal4, UAS-GFP (panel B) and Sxl-T2A-Gal4, UAS-Stinger flies (panels C and D). Control flies with an intact ssx locus are shown on the left, ssxΔ flies in the panels on the right. Arrowheads indicate GFP-positive cells or clonal cell populations in the tibia of the metathoracic leg (panel B) and the 3rd posterior cell of the wing (panel C) or the midgut (panel D; top: ventral view of the abdomen with the GFP signal visible through the cuticula, bottom: hindgut after manual dissection of the animal). Arrows in panel C indicate GFP-positive nerve projections and neural cell bodies in the L1 and L3 veins of the wing. Scale bars: 0.5mm.
As expected, female embryos and imagines that carry the Sxl-T2A-Gal4 fusion gene exhibit ubiquitous and strong GFP expression. In contrast, GFP fluorescence is absent from most tissues of male animals, with one notable exception: the nervous system shows robust expression of the GFP reporter gene in pupae and adults. Absence of GFP fluorescence in non-neuronal tissues of male flies, however, allowed us to test for Sxl protein mis-expression outside of the nervous system upon deletion of the ssx locus. While none of the control animals that carried an X-chromosome with an intact ssx locus (sxl-T2A-GAL4(3xP3-RFP)) showed GFP fluorescence outside of neuronal tissues, ∼4% of the ssxΔ, w+, sxl-T2A-GAL4(3xP3-RFP) hemizygous males exhibited GFP expression in non-neuronal cells indicative of aberrant Sxl expression. The aberrant GFP signal was detectable in small, clonal cell populations or individual cells in different organs and anatomical structures such as the tibia of the T3 (metathoracic) leg, the 3rd posterior cell of the wing, or the midgut (Figure 5B–D). This confirms that ssx contributes to the silencing of Sxl protein expression in adult male flies to reinforce the male-specific gene expression pattern.
DISCUSSION
Regulation by feedback loops
Positive feedback loops are widely employed to amplify and convert a graded (and often weak) input signal into a binary, all-or-nothing response. This is particularly important during definition of precise borders of gene expression for pattern formation during embryonic development (44) and it conveys robustness to developmental decisions through canalization (45). Negative feedback loops also stabilize biological states by buffering against fluctuations that are inherent to gene expression or imposed upon the cell by a changing environment (46). To cope with these challenges, cells often employ signalling cascades that feed back to gene expression creating homeotic circuits (47), or miRNAs that fine-tune translational output of mRNAs to maintain proteostasis (48).
Auto-regulatory feedback control appears to be particularly frequent among RNA-binding proteins that control their own expression by various strategies, e.g. by regulated unproductive splicing and translation (RUST) (49). In most cases RBPs use negative feedback to maintain protein concentrations within a narrow physiological range. In contrast, sex determination in insects makes use of a positive feedback loop to stabilize a genetic switch, functioning as a ‘cellular memory system’ that, once triggered, maintains the female cellular state (50). However, when left unchallenged, positive feedback loops can erroneously convert a short and/or weak signal into a solid and lasting response. Hence, noise stemming from e.g. environmental fluctuations, heterozygosity, or stochastic promoter activity could result in accidental triggering of the feedback loop. It is therefore of critical importance to buffer against these fluctuations by generating a precise threshold for activation. Cellular signalling pathways employ various strategies to do so, e.g. cooperative binding of regulators or formation of opposing signalling gradients (44). Here, we report threshold formation through expression of a protein that inhibits positive feedback by direct competition with the positive regulator. This prevents accidental triggering of the feedback loop by weak input signals and stabilizes the male-specific gene expression pattern in Drosophila.
Ssx promotes inclusion of a poison exon to prevent erroneous Sxl expression in males
In Drosophila melanogaster, female development relies on an auto-regulatory, positive feedback loop that is generated via alternative splicing of Sxl. Regulatory feedback requires multiple Sxl binding sites in the RNA and involves, besides Sxl, numerous co-factors to generate a strong and robust regulation in females (6). While Sxl-mediated regulation has been well studied in somatic tissues of female flies, it has long remained unclear how male flies completely shut off Sxl protein expression and protect themselves against accidental triggering of the feedback loop. Here we report the identification of Ssx as the first inhibitor of Sxl auto-regulatory feedback to reinforce a male-specific gene expression pattern.
Sxl and Ssx bind to U-rich RNA motifs in a mutually exclusive fashion and as predicted for competitive regulation, the outcome of Sxl mRNA alternative splicing depends on the relative concentrations and RNA-binding activities of the two proteins. When both are present at similar concentrations, substantial RNA binding can be observed for Sxl (Figure 2D). Hence, in female tissues, where the concentration of Sxl exceeds the one of Ssx (Supplementary Figure S4), moderate overexpression of Ssx has no measurable impact on the female-specific and Sxl-dependent splicing patterns of tra, msl-2 and Sxl mRNAs (Figure 4B). However, if Sxl protein concentration is low, its activity in splicing regulation can be suppressed by Ssx (Figures 3 and 4). This suggests that Ssx acts as a safeguard by establishing a threshold to prevent low concentrations of Sxl from triggering the auto-regulatory, positive feedback loop in male animals. In agreement with this, loss of Ssx results in low levels of productive Sxl splicing in males and aberrant Sxl protein production (Figures 4 and 5).
Sxl expression in male flies
The aberrant expression of full-length Sxl protein in ssx mutant flies occurs in isolated cell populations at late developmental stages and does neither result in a viability defect nor a morphological phenotype. This is seemingly at odds with the strong phenotype associated with forced ubiquitous Sxl protein expression from gain-of-function alleles that results in male lethality through upsets in dosage compensation (7,8). How can this surprising discrepancy be explained? In females Sxl protein expression is primed in the pre-cellular, syncytial embryo and during later cellularization every cell receives an initial dose of the protein. This systemically triggers productive splicing of transcripts emerging from the Sxl maintenance promoter. In male animals Sxl protein production is suppressed in the pre-cellular embryo. Activation of the positive, auto-regulatory feedback loop at a later stage therefore requires a different source of functional Sxl protein, e.g. by spontaneous skipping of the poison exon during splicing. Such an event is most likely rare, presumably occurs in a rather stochastic fashion and results in production of initially only minute amounts of functional Sxl protein. Therefore, even in the absence of safeguarding mechanisms (such as the lack of Ssx), Sxl expression is likely initiated in only few cells of a male fly. This is confirmed by the finding that the Sxl-T2A-Gal4 reporter line does not show ubiquitous, low level Sxl protein production, but rather strong expression in isolated clonal populations, indicative of a rare triggering event.
The GFP reporter line also indicates Sxl protein production in neuronal tissues of male animals. The first GFP-positive cells become visible shortly before pupation and in adults GFP fluorescence is detected in the entire nervous system. In 2010, Cline et al. reported the discovery of a Sxl mRNA isoform that contains a previously unrecognized and evolutionarily conserved exon (denoted exon Z). Importantly, this RNA isoform is detectable in male flies and strongly expressed in heads (9). Moreover, exon Z-containing transcripts lack the poison exon L3 and hence they contain an open reading frame that initiates in exon Z and encodes a near full-length Sxl protein (9). Previously, low levels of antigens that cross-react with anti-Sxl antisera could be detected in male heads. These proteins exhibited a slightly increased gel mobility compared to the female-specific Sxl proteins (51), supporting the idea that they might be generated by translation of exon Z-containing transcripts. Our findings confirm that male animals express Sxl protein isoforms in neuronal tissues, however, their function remains enigmatic.
Genetic interaction of ssx and Sxl
Previously, genetic interactions between ssx and Sxl were reported (9). On the one hand, ablation of Ssx expression caused a weak fecundity defect in male flies that could be rescued by the removal of Sxl (reflecting an antagonistic behaviour as reported in this study). On the other hand, etching of distal tergites in male flies caused by the weak gain-of-function allele SxlM12 could be partially rescued by the removal of ssx, suggesting that in this context the functions of Sxl and Ssx might be related. This agrees with our previous finding that similarly to Sxl, Ssx can function as a repressor of translation (12). Taken together, these findings illustrate that Sxl and Ssx can either exhibit a (partially) redundant activity, or function as competitors, depending on the RNA target, the position and context of the binding site, and the ability to engage of additional co-factors (such as the co-repressor protein Unr in the control of msl-2 translation). Unfortunately, all alleles that express Sxl at sub-lethal doses in male animals (such as SxlM12) have been lost, precluding further genetic testing for interactions in males flies with ssx alleles (such as the newly constructed ssxΔ allele) to unravel the complex relationship of the two loci.
Based on our finding that Ssx acts as a competitive inhibitor of the Sxl auto-regulatory feedback loop, we hypothesized that Ssx protein overexpression might impact on female development which critically depends on the expression of full-length Sxl protein. Ubiquitous and strong overexpression of Ssx under the control of a daughterless driver unfortunately resulted in both male and female lethality. We therefore expressed a ssx transgene (UASp-Ssx) in female flies under the control of germ-line specific drivers (osk-Gal4 and nos-Gal4) to increase the Ssx protein dose in oocytes and early embryos and to interfere with the initiation of the Sxl auto-regulatory feedback loop to block female sexual development. However, we did not detect a skewed sex ratio in the offspring (Supplementary Figure S5). More sophisticated genetic analyses are clearly required to shed light on the antagonistic relationship of Sxl and Ssx in early female development and to understand why Ssx does not interfere with Sxl auto-regulation at embryonic stages.
Competition between RNA-binding proteins
Antagonistic function of RNA binding proteins by steric hindrance and competition for RNA regulatory elements is not unprecedented. Similar regulatory concepts are also encountered in other aspects of Sxl-dependent post-transcriptional regulation. Sxl-mediated alternative splicing of tra and msl-2 mRNAs, as well as alternative polyadenylation of enhancer of rudimentary (e(r)) transcripts is thought to involve direct competition between Sxl and processing factors. In the tra and msl-2 primary transcripts, regulation involves steric interference of Sxl with U2AF and Rox8 that are involved in the recognition of the splice sites (33,52,53). Similarly, polyadenylation of e(r) mRNA is shifted to a distal processing site by Sxl blocking interaction of CSTF with the proximal site (54).
The human Sxl homolog HuR (human antigen R) shares a regulatory relationship with the protein TTP (Tristetraprolin) reminiscent of what we describe for Ssx and Sxl. Both proteins recognize and competitively bind to AU-rich RNA elements (AREs), but elicit different downstream functions. While TTP (and its partner proteins) are mainly involved in rapid turnover of ARE-containing mRNAs, HuR stabilizes the transcripts to allow their translation (55–57). The exchange of TTP for HuR—and hence the stabilization and translational activation of ARE-containing transcripts—is facilitated by phosphorylation of TTP by p38/MK2 which lowers its affinity to the RNA (58). Conversely, ubiquitination of HuR facilitates its release by the ATPase p97 and allows binding of TTP (59). Dysregulation of the balanced activities of TTP and HuR also plays an important role in numerous diseases, including many types of cancers (60).
While our data shed light onto a function of Ssx in safeguarding against Sxl expression in adult male animals, our findings also raise numerous questions. Ssx is not only expressed in adult males, but it can be detected in all developmental stages tested irrespective of sexual identity. Its function(s) in early embryos and in females, however, remain enigmatic.
We also demonstrate that Sxl and Ssx recognize similar RNA sequence motifs and bind RNA in a competitive manner. In line with this, we and others have demonstrated that Ssx and Sxl can both associate with RNA regulatory elements present in the msl-2 mRNA which results in translational repression (12,37). Competition for RNA binding between the two proteins is most probably not restricted to the Sxl and msl-2 mRNAs since we find Ssx to be associated with numerous additional previously identified (or predicted) Sxl target RNAs. Whether Ssx also plays a role in their post-transcriptional regulation, however, remains to be determined.
We also provide additional evidence for the expression of Sxl isoforms in male animals that are most probably derived from exon Z-containing transcripts. It has been suggested that these Sxl protein isoforms serve a yet unidentified, ancestral and non-sex-specific function related to Sxl orthologs from other insect species where female sexual development is independent of Sxl (9). Further analyses of Ssx might yield insight into this enigmatic activity of Sxl and its related proteins.
DATA AVAILABILITY
Sequencing data from iCLIP experiments has been deposited at GEO under the following accession number: GSE98189.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Matthias Hentze for providing polyclonal anti-Sxl antibodies and Anne Ephrussi for fly lines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [ME4238/1-1 and SFB 960/2, B11 to J.M., KR3901/1-2 and KR3901/3-1 to M.P.K., SFB 960/2, B01 to G.M.]; Bavarian State Ministry for Education, Science and the Arts [Bavarian Research Network for Molecular Biosystems, BioSysNet, to J.M.]; German Federal Ministry of Education and Research [BMBF, 01ZX1401D to J.M.]; LOEWE program Medical RNomics [to O.R.]. Funding for open access charge: Institutional money.
Conflict of interest statement. None declared.
REFERENCES
- 1. Graindorge A., Militti C., Gebauer F.. Posttranscriptional control of X-chromosome dosage compensation. Wiley Interdiscipl. Rev. RNA. 2011; 2:534–545. [DOI] [PubMed] [Google Scholar]
- 2. Penalva L.O., Sanchez L.. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol. Mol. Biol. Rev: MMBR. 2003; 67:343–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salz H.K. Sex determination in insects: a binary decision based on alternative splicing. Curr. Opin. Genet. Dev. 2011; 21:395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Salz H.K., Erickson J.W.. Sex determination in Drosophila: the view from the top. Fly. 2010; 4:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Venables J.P., Tazi J., Juge F.. Regulated functional alternative splicing in Drosophila. Nucleic Acids Res. 2012; 40:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moschall R., Gaik M., Medenbach J.. Promiscuity in post-transcriptional control of gene expression: Drosophila Sex-lethal and its regulatory partnerships. FEBS Lett. 2017; 591:1471–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cline T.W. Two closely linked mutations in Drosophila melanogaster that are lethal to opposite sexes and interact with daughterless. Genetics. 1978; 90:683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cline T.W. Autoregulatory functioning of a Drosophila gene product that establishes and maintains the sexually determined state. Genetics. 1984; 107:231–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cline T.W., Dorsett M., Sun S., Harrison M.M., Dines J., Sefton L., Megna L.. Evolution of the Drosophila feminizing switch gene Sex-lethal. Genetics. 2010; 186:1321–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Traut W., Niimi T., Ikeo K., Sahara K.. Phylogeny of the sex-determining gene Sex-lethal in insects. Genome. 2006; 49:254–262. [DOI] [PubMed] [Google Scholar]
- 11. Ayres J.S., Freitag N., Schneider D.S.. Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics. 2008; 178:1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moschall R., Strauss D., Garcia-Beyaert M., Gebauer F., Medenbach J.. Drosophila Sister-of-Sex-lethal is a repressor of translation. RNA. 2018; 24:149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Medenbach J., Seiler M., Hentze M.W.. Translational control via protein-regulated upstream open reading frames. Cell. 2011; 145:902–913. [DOI] [PubMed] [Google Scholar]
- 14. Diao F., Ironfield H., Luan H., Diao F., Shropshire W.C., Ewer J., Marr E., Potter C.J., Landgraf M., White B.H.. Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Rep. 2015; 10:1410–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duncan K., Grskovic M., Strein C., Beckmann K., Niggeweg R., Abaza I., Gebauer F., Wilm M., Hentze M.W.. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: translational repression for dosage compensation. Genes Dev. 2006; 20:368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J.. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010; 17:909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. Journal. 2011; 17:10–12. [Google Scholar]
- 18. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kucukural A., Ozadam H., Singh G., Moore M.J., Cenik C.. ASPeak: an abundance sensitive peak detection algorithm for RIP-Seq. Bioinformatics. 2013; 29:2485–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Groth A.C., Fish M., Nusse R., Calos M.P.. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004; 166:1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Port F., Chen H.M., Lee T., Bullock S.L.. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. PNAS. 2014; 111:E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Celniker S.E., Dillon L.A., Gerstein M.B., Gunsalus K.C., Henikoff S., Karpen G.H., Kellis M., Lai E.C., Lieb J.D., MacAlpine D.M. et al.. Unlocking the secrets of the genome. Nature. 2009; 459:927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Singh R., Valcarcel J., Green M.R.. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995; 268:1173–1176. [DOI] [PubMed] [Google Scholar]
- 26. Sakashita E., Sakamoto H.. Characterization of RNA binding specificity of the Drosophila sex-lethal protein by in vitro ligand selection. Nucleic Acids Res. 1994; 22:4082–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samuels M., Deshpande G., Schedl P.. Activities of the Sex-lethal protein in RNA binding and protein:protein interactions. Nucleic Acids Res. 1998; 26:2625–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singh R., Banerjee H., Green M.R.. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor sex-lethal. RNA. 2000; 6:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bashaw G.J., Baker B.S.. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell. 1997; 89:789–798. [DOI] [PubMed] [Google Scholar]
- 30. Bell L.R., Horabin J.I., Schedl P., Cline T.W.. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991; 65:229–239. [DOI] [PubMed] [Google Scholar]
- 31. Boggs R.T., Gregor P., Idriss S., Belote J.M., McKeown M.. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell. 1987; 50:739–747. [DOI] [PubMed] [Google Scholar]
- 32. Gebauer F., Corona D.F., Preiss T., Becker P.B., Hentze M.W.. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 1999; 18:6146–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Inoue K., Hoshijima K., Sakamoto H., Shimura Y.. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature. 1990; 344:461–463. [DOI] [PubMed] [Google Scholar]
- 34. Kelley R.L., Wang J., Bell L., Kuroda M.I.. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature. 1997; 387:195–199. [DOI] [PubMed] [Google Scholar]
- 35. Handa N., Nureki O., Kurimoto K., Kim I., Sakamoto H., Shimura Y., Muto Y., Yokoyama S.. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999; 398:579–585. [DOI] [PubMed] [Google Scholar]
- 36. Hennig J., Militti C., Popowicz G.M., Wang I., Sonntag M., Geerlof A., Gabel F., Gebauer F., Sattler M.. Structural basis for the assembly of the Sxl-Unr translation regulatory complex. Nature. 2014; 515:287–290. [DOI] [PubMed] [Google Scholar]
- 37. Rogell B., Fischer B., Rettel M., Krijgsveld J., Castello A., Hentze M.W.. Specific RNP capture with antisense LNA/DNA mixmers. RNA. 2017; 23:1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kelley R.L., Solovyeva I., Lyman L.M., Richman R., Solovyev V., Kuroda M.I.. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell. 1995; 81:867–877. [DOI] [PubMed] [Google Scholar]
- 39. Grskovic M., Hentze M.W., Gebauer F.. A co-repressor assembly nucleated by Sex-lethal in the 3′UTR mediates translational control of Drosophila msl-2 mRNA. EMBO J. 2003; 22:5571–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yanowitz J.L., Deshpande G., Calhoun G., Schedl P.D.. An N-terminal truncation uncouples the sex-transforming and dosage compensation functions of sex-lethal. Mol. Cell. Biol. 1999; 19:3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horabin J.I., Schedl P.. Regulated splicing of the Drosophila sex-lethal male exon involves a blockage mechanism. Mol. Cell. Biol. 1993; 13:1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Horabin J.I., Schedl P.. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol. Cell. Biol. 1993; 13:7734–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lallena M.J., Chalmers K.J., Llamazares S., Lamond A.I., Valcarcel J.. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell. 2002; 109:285–296. [DOI] [PubMed] [Google Scholar]
- 44. Perrimon N., Pitsouli C., Shilo B.Z.. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harbor Perspect. Biol. 2012; 4:a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Waddington C. Canalization of development and the inheritance of acquired characters. Nature. 1942; 150:563–565. [Google Scholar]
- 46. Becskei A., Serrano L.. Engineering stability in gene networks by autoregulation. Nature. 2000; 405:590–593. [DOI] [PubMed] [Google Scholar]
- 47. Freeman M. Feedback control of intercellular signalling in development. Nature. 2000; 408:313–319. [DOI] [PubMed] [Google Scholar]
- 48. Ebert M.S., Sharp P.A.. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012; 149:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lareau L.F., Brooks A.N., Soergel D.A., Meng Q., Brenner S.E.. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv. Exp. Med. Biol. 2007; 623:190–211. [DOI] [PubMed] [Google Scholar]
- 50. Sawanth S.K., Gopinath G., Sambrani N., Arunkumar K.P.. The autoregulatory loop: a common mechanism of regulation of key sex determining genes in insects. J. Biosci. 2016; 41:283–294. [DOI] [PubMed] [Google Scholar]
- 51. Bopp D., Bell L.R., Cline T.W., Schedl P.. Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes Dev. 1991; 5:403–415. [DOI] [PubMed] [Google Scholar]
- 52. Sosnowski B.A., Belote J.M., McKeown M.. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell. 1989; 58:449–459. [DOI] [PubMed] [Google Scholar]
- 53. Valcarcel J., Singh R., Zamore P.D., Green M.R.. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993; 362:171–175. [DOI] [PubMed] [Google Scholar]
- 54. Gawande B., Robida M.D., Rahn A., Singh R.. Drosophila Sex-lethal protein mediates polyadenylation switching in the female germline. EMBO J. 2006; 25:1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hinman M.N., Lou H.. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci.: CMLS. 2008; 65:3168–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simone L.E., Keene J.D.. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr. Opin. Genet. Dev. 2013; 23:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. von Roretz C., Di Marco S., Mazroui R., Gallouzi I.E.. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscipl. Rev. RNA. 2011; 2:336–347. [DOI] [PubMed] [Google Scholar]
- 58. Tiedje C., Ronkina N., Tehrani M., Dhamija S., Laass K., Holtmann H., Kotlyarov A., Gaestel M.. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLos Genet. 2012; 8:e1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhou H.L., Geng C., Luo G., Lou H.. The p97-UBXD8 complex destabilizes mRNA by promoting release of ubiquitinated HuR from mRNP. Genes Dev. 2013; 27:1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang H., Ding N., Guo J., Xia J., Ruan Y.. Dysregulation of TTP and HuR plays an important role in cancers. Tumour Biol. 2016; 37:14451–14461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data from iCLIP experiments has been deposited at GEO under the following accession number: GSE98189.