Abstract
RNA can directly bind to purine-rich DNA via Hoogsteen base pairing, forming a DNA:RNA triple helical structure that anchors the RNA to specific sequences and allows guiding of transcription regulators to distinct genomic loci. To unravel the prevalence of DNA:RNA triplexes in living cells, we have established a fast and cost-effective method that allows genome-wide mapping of DNA:RNA triplex interactions. In contrast to previous approaches applied for the identification of chromatin-associated RNAs, this method uses protein-free nucleic acids isolated from chromatin. High-throughput sequencing and computational analysis of DNA-associated RNA revealed a large set of RNAs which originate from non-coding and coding loci, including super-enhancers and repeat elements. Combined analysis of DNA-associated RNA and RNA-associated DNA identified genomic DNA:RNA triplex structures. The results suggest that triplex formation is a general mechanism of RNA-mediated target-site recognition, which has major impact on biological functions.
INTRODUCTION
The recent discovery that large parts of the genome are transcribed into long non-coding (lnc)RNAs has revolutionized our understanding of how cells regulate the stability, transmission and expression of their genetic information. Yet, while the importance of lncRNAs as versatile regulators of cellular processes in health and disease is increasingly recognized, fundamental aspects regarding their regulation, structure and function remain poorly understood. LncRNAs exert their regulatory functions by establishing intermolecular interactions with proteins, DNA or other RNAs. Serving as molecular signals, guides or scaffolds, lncRNAs target chromatin-modifying enzymes or transcription regulators to specific genomic sequences either via direct interaction with nucleic acids or via specific protein partners (1–3). Improved methods of chromatin isolation, targeted RNA enrichment and high-throughput sequencing have facilitated the identification of thousands of chromatin-enriched RNAs (cheRNAs), most of them affecting transcription of neighboring genes (4,5). Previously, genome-wide methods for the identification of DNA regions that are associated with RNA have demonstrated that the lncRNAs HOTAIR, MALAT1 and NEAT1 interact with thousands of genomic loci (6,7). Various techniques have been developed to localize RNAs on crosslinked chromatin, including Chromatin Isolation by RNA Purification (ChIRP) (6), Capture Hybridization Analysis of RNA Targets (CHART) (8), RNA Antisense Purification (RAP) (9), Mapping RNA-Genome Interactions (MARGI) (10), and global RNA interactions with DNA by deep sequencing (GRID-seq) (11). These crosslink-based methods allow the global analysis of potential RNA chromatin interactions. How RNAs target specific genomic sites is still elusive.
Consistent with the concept of gene regulation by RNA discussed by Britten and Davidson almost 50 years ago (12), RNA is well adapted for regulatory roles, because base complementarity allows RNA to interact with DNA either by canonical Watson-Crick pairing with one of the DNA strands, forming R-loop structures or by direct binding to duplex DNA, forming DNA:RNA triplex structures. RNA binds in the major groove of DNA by forming Hoogsteen or reverse Hoogsteen hydrogen bonds between the purine-rich strand of duplex DNA and single-stranded RNA (13–15). Canonical rules of triplex formation comprise (i) the pyrimidine motif in which the third strand is composed of pyrimidine bases bound parallel to the purine-rich strand of DNA (forward Hoogsteen base pairing), (ii) the purine motif where the third strand is composed of purines bound antiparallel to the purine strand of DNA (reverse Hoogsteen base pairing) or (iii) the mixed motif where guanine and uracil bases bind either in parallel or antiparallel configuration with respect to the purines in duplex DNA (15,16). Although the ability of DNA to engage in triplex structures with RNA is well established, the in vivo existence and biological significance of these structures remain largely unknown.
Examples for the existence of DNA:RNA triplexes formed by lncRNA with specific DNA sequences include pRNA which represses transcription of rRNA genes by targeting DNMT3b to the rDNA promoter (17), Fendrr which regulates developmental genes by recruiting the PRC2 complex (18), PARTICLE which regulates the expression of MAT2A in response to low-dose radiation (19), MEG3 which guides PRC2 to regulatory regions of TGF-β pathway genes (20), HOTAIR which regulates adipogenic differentiation of mesenchymal stem cells (21), and PAPAS which guides CHD4/NuRD to the rDNA promoter (22). Furthermore, triple-helix formation of LINE-1 RNAs with repetitive LINE-1 elements occurs in the mouse embryo a few hours after fertilization, correlating with transcriptional activation of repetitive elements (23). Another example is KHPS1, an RNA which is synthesized in antisense orientation to the proto-oncogene SPHK1. KHPS1 activates SPHK1 transcription by binding to a homopurine stretch upstream of the transcription start site of SPHK1, which in turn facilitates recruitment of p300/CBP and E2F1, thereby establishing a transcription-permissive chromatin structure (24).
Triplex formation of RNAs with specific DNA sequences can occur both in cis and in trans, affecting the expression of neighboring and distant genes (25). Computational analyses revealed that a large population of triplex target sites is present in mammalian genomes, the majority of annotated human genes, promoters and intergenic regions containing at least one potential triplex-forming sequence (26–29). This suggests that tethering RNA to specific genomic sites might guide RNA-associated regulatory proteins to establish an epigenetic landscape that facilitates or inhibits gene expression.
However, none of the previous studies provided rigorous proof that DNA:RNA triplex structures exist in vivo and are physiologically relevant. Therefore, it is mandatory to develop methods that allow the global characterization of RNAs that guide transcriptional regulators to relevant genomic loci via direct interaction with DNA. Here, we describe a simple, fast and cost-effective method to isolate, sequence and characterize DNA:RNA triplexes in the human genome. While previous studies have investigated RNA-chromatin interactions in crosslinked chromatin, our approach does not involve crosslinking and therefore does not identify RNA that is associated with DNA via DNA-bound proteins. Applying purpose-adapted peak calling and sequence-based triplex analysis tools, we have identified a large set of RNAs originating from coding and non-coding loci with triplex-forming potential. Moreover, we have mapped genomic locations of DNA:RNA triplexes by RNA pull-down and sequencing of associated DNA. By combining different experimental approaches with bioinformatic analyses of RNA and DNA, we now have the technology to catalogue and characterize cellular DNA:RNA triplexes, which is a prerequisite for unraveling their function.
MATERIALS AND METHODS
Cell culture
HeLa S3 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS), 1% penicillin/streptomycin and 2% Na-pyruvate. U2OS cells were cultured in DMEM supplemented with 5% FBS and 1% penicillin/streptomycin.
Preparation of chromatin
Ten million cells were lysed in 10 mM Tris–HCl [pH 7.9], 100 mM KCl, 5 mM MgCl2, 0.5% NP-40, 1 mM DTT and nuclei were purified by centrifugation through 50% glycerol, 20 mM Tris–HCl [pH 7.4], 75 mM KCl, 5 mM MgCl2, 1 mM DTT. The nuclear pellet was suspended in 500 μl glycerol/urea buffer (25% glycerol, 20 mM Tris–HCl [pH 7.4], 187.5 mM KCl, 0.5 M urea, 0.5% NP-40, 7.5 mM MgCl2, 1 mM DTT), incubated on ice for 5 min and centrifuged for 3 min at 2000 × g (4). Chromatin was resuspended in 1 ml 10% glycerol, 340 mM sucrose, 10 mM Tris–HCl [pH 7.4], 10 mM KCl, 5 mM MgCl2, 1 mM DTT, centrifuged again and subjected to mild treatment (2.5 mU/μl, 5 min at 37°C) with DNase I (Sigma-Aldrich) to yield DNA fragments with an average size of >10 kb. After treatment with 0.2% Na-Sarkosyl (30), chromatin was pelleted by centrifugation through a 0.88 M sucrose cushion and resuspended in triplex buffer (10 mM Tris–HCl [pH 7.4], 50 mM KCl, 5 mM MgCl2).
Genome-wide isolation of DNA-associated RNA
To isolate chromatin-associated nucleic acids, chromatin was treated with proteinase K (0.25 μg/μl, Roche) in triplex-forming buffer containing 0.1% SDS (30 min at 37°C) followed by two rounds of phenol/chloroform extraction. After suspension in triplex buffer supplemented with 10 mM DTT, 16–20 μg of nucleic acids were incubated for 30 min at room temperature with RNase H (200 mU/μl, NEB) to digest cellular DNA–RNA heteroduplexes (R-loops). To determine the concentration of RNase H required to digest cellular R-loops, radiolabeled synthetic heteroduplexes were spiked into the samples in the presence increasing amounts of RNase H and digestion was monitored by electrophoretic mobility shift assay (EMSA) (Supplementary Figure S1A). Finally, samples were incubated for 5 min at 37°C with RNase I (3.125 mU/μl, Thermo Fisher Scientific) to yield RNA with an average size of 100–150 nucleotides. Control samples were digested with DNase I (62.5 mU/μl) together with RNase H and RNase I.
For Solid Phase Reversible Immobilization (SPRI)-based paramagnetic bead size selection, 16–20 μg of nucleic acids were treated for 30 min with 200 mU/μl RNase H and 12.5 mU/μl Shearase Plus (Zymo Research) to digest RNAs in R-loops and trim the size of genomic DNA to 5–10 kb. RNA was partially digested with RNase I (3.125 U/μl at 37°C) before size selection using 0.4 sample volumes of AMPure XP beads (Backman Coulter). After elution with triplex buffer, DNA was digested with DNase I (62.5 mU/μl, 20 min at 37°C) and associated RNA was isolated with TRI reagent.
To separate free RNA from DNA-associated RNA by immunopurification with anti-DNA antibody, 16–20 μg of nucleic acids were treated for 30 min with 200 mU/μl RNase H and 75 mU/μl Shearase Plus to digest RNAs in R-loops and trim the size of genomic DNA to 500–1000 bp. After partial digestion with RNase I (3.125 mU/μl, 37°C), 2 μg of nucleic acids were incubated with 6 μg of immobilized anti-dsDNA antibody (Abcam ab27156) in 200 μl IP buffer (10 mM Tris–HCl [pH 7.4], 50 mM KCl, 5 mM MgCl2, 0.05% Tween-20, 20 U RNasin (Promega), 40 U SUPERase In RNase inhibitor (Invitrogen)) for 2 h at room temperature. Complexes bound to Protein G-coated Dynabeads (Invitrogen) were washed twice with triplex buffer. RNA was eluted for 30 min with 2 U of TURBO DNase (Thermo Fisher Scientific), treated with 1 μg/μl proteinase K for 30 min at 37°C and recovered using TRI reagent.
To determine the size of DNA-associated RNA, RNA was 5′-end labeled using γ[32P]ATP and resolved on 10% polyacrylamide/TBE gels. For RT-qPCR, 500–1000 ng of RNA was reverse transcribed with Transcriptor Reverse Transcriptase (Roche) in the presence of random hexamer primers. cDNA was amplified using the QuantiTect SYBR Green PCR Kit (QIAGEN). Primers used in qPCR analyses are listed in Supplementary Table S1.
Genome-wide isolation of RNA-associated DNA
To fragment DNA and digest DNA–RNA heteroduplexes, 16–20 μg of chromatin-associated nucleic acids were incubated for 45 min at 37°C with 100 mU/μl dsDNA Shearase Plus and 200 mU/μl RNase H in triplex buffer containing 10 mM DTT. 3′-Biotinylated DNA oligos (11) (Supplementary Table S3) were 5′-adenylated using Mth RNA ligase (NEB), bound to MyOne Streptavidin C1 Dynabeads (Invitrogen), and ligated to cellular RNA for 2 h at room temperature using 3–3.5 μg nucleic acids from chromatin preparations in 100 μl of a buffer containing 10% PEG, 0.1% Triton X-100, 500 U T4 RNA ligase 2, truncated KQ (NEB). After washing with triplex buffer containing 0.05% Tween-20, RNA-associated DNA was eluted in 100 μl triplex buffer containing 50 ng/μl RNase A (Thermo Fisher Scientific) for 30 min at 37°C and recovered by phenol/chloroform extraction. Ligated RNA was eluted with 2U of TURBO DNase (Thermo Fisher Scientific) for 30 min at 37°C.
Electrophoretic mobility shift assays
DNA oligos were 5′-labeled with γ[32P]ATP, annealed to equimolar ratios of corresponding unlabeled oligonucleotides in 10 mM Tris-acetate [pH 7.4], 5 mM Mg-acetate, 50 mM NaCl for 10 min at 70°C and cooled down to 20°C. For triplex formation, RNA was incubated with 0.25 pmol of radiolabeled duplex oligos for 1 h at 37°C in either Triplex-buffer A (40 mM Tris-acetate [pH 7.4], 30 mM NaCl, 20 mM KCl, 5 mM Mg-acetate, 10% glycerol, PhosSTOP EASYpack (Roche), 20 U of RNasin) or Triplex-buffer B (10 mM Tris–HCl [pH 7.4], 50 mM KCl, 5 mM MgCl2, 10 μg salmon sperm DNA (Thermo Fisher Scientific), PhosSTOP EASYpack, 20U of RNasin). Triplex formation was monitored by electrophoresis on 12% polyacrylamide gels at 120 V. Sequences of DNA and RNA oligos are shown in Supplementary Table S2.
Capture of DNA by biotinylated NEAT1 RNA oligonucleotides
RNA-free genomic DNA was sheared with Bioruptor Pico (Diagenode) to an average size of 200–300 bp and 10 μg of fragmented DNA were incubated with 20 pmol of biotinylated RNA oligos (Supplementary Table S3) for 1 h at 37°C in 100 μl triplex buffer plus 40 U of RNasin. DNA–RNA oligo complexes were bound to MyOne Streptavidin C1 Dynabeads and washed three times with 500 μl triplex buffer containing 0.05% Tween-20. To elute RNA-associated DNA, beads were incubated with 25 ng/μl RNase A and 5 mU/μl of RNase I for 30 min at 37°C.
ASO-based capture of NEAT1-associated DNA
To trim the size of genomic DNA to about 200–300 bp, 16–20 μg of chromatin-associated nucleic acids were incubated for 45 min at 37°C with 100 mU/μl dsDNA Shearase Plus. A total of 3.5 μg of nucleic acids were hybridized for 4 h at 37°C with 20 pmol of biotinylated NEAT1-specific capture oligos (7) (Supplementary Table S3) in 100 μl buffer containing 50 mM Tris–HCl [pH 7.4], 50 mM NaCl, 10 mM MgCl2, 36% formamide, 40 U RNasin. After incubation with MyOne Streptavidin C1 Dynabeads for 40 min at room temperature, beads were washed three times with triplex buffer containing 0.05% Tween-20. NEAT1-associated DNA was eluted with 50 ng/μl RNase A (37°C, 30 min) and recovered by phenol/chloroform extraction. RNA was eluted in 100 μl triplex buffer containing 2 units TURBO DNase.
Preparation of libraries
RNA samples were treated with 2 U of DNase I for 20 min at room temperature, purified with TRI reagent, and rRNA was depleted with the NEBNext rRNA depletion kit (NEB). RNA from DNA-IP was not subjected to rDNA depletion. Libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit and NEBNext Multiplex Oligos for Illumina (NEB). Chromatin-associated and nuclear RNA was fragmented according the manufacturer's instructions. Libraries from U2OS cells were prepared from two independent experiments monitoring DNA-associated RNA (DNA-IP) and nuclear RNA. Libraries from HeLa S3 cells were from independent experiments monitoring DNA-associated RNA from DNA-IP (N=3) and SPRI-size selection (N=4) as well as chromatin-associated RNA (N=4) and nuclear RNA (N=2). As practically no DNA was detectable in the IgG controls (Supplementary Figure S1B), the IgG samples were not subjected to library preparation and sequencing. DNA libraries were prepared from at least three independent experiments from HeLa S3 cells using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB) and NEBNext Multiplex Oligos for Illumina (NEB). Single-end sequencing was performed on Illumina NextSeq 500 platform.
Bioinformatic analyses
Adapter sequences were removed from the sequence reads by Trim galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore) with default parameters. RNA sequence reads were aligned to human genome version hg38 using STAR with strand information and stringent parameters (31), allowing maximal one mismatch (Supplementary Table S4). Duplicate reads were filtered out. As DNA-IP and SPRI-size selection enrich triplex-forming regions in RNA, usual bioinformatic methods for RNA quantification and de-novo discovery of full transcripts cannot be used. We therefore adapted the differential peak caller THOR (32) to detect regions within transcripts enriched in particular fractions, taking into account strand information and using the bin size of 50 bp. Estimation of fragment sizes was based on Agilent Bioanalyzer profiles of libraries (Supplementary Table S4). Differential peak calling was performed by contrasting particular pairs of RNA fractions (Supplementary Figure S2B; adjusted P-value <10−500 and fold change (FC) >1 in log2; Negative Binomial test). Peaks enriched in DNA-IP (versus nuclear RNA) and SPRI-size selection (versus nuclear RNA) are defined as DNA-associated RNA or TriplexRNA regions. Chromatin-associated and nuclear RNA are defined as peaks that are enriched in these fractions as compared to RNAs isolated by DNA-IP (Supplementary Table S4). To compare TriplexRNA with chromatin-associated RNA and nuclear RNA, a gene-centric analysis similar to the strategy in Werner et al. (5) was used, which quantifies reads that overlap genes. Counts were normalized by RPKM (Figure 2A and Supplementary Figure S2E).
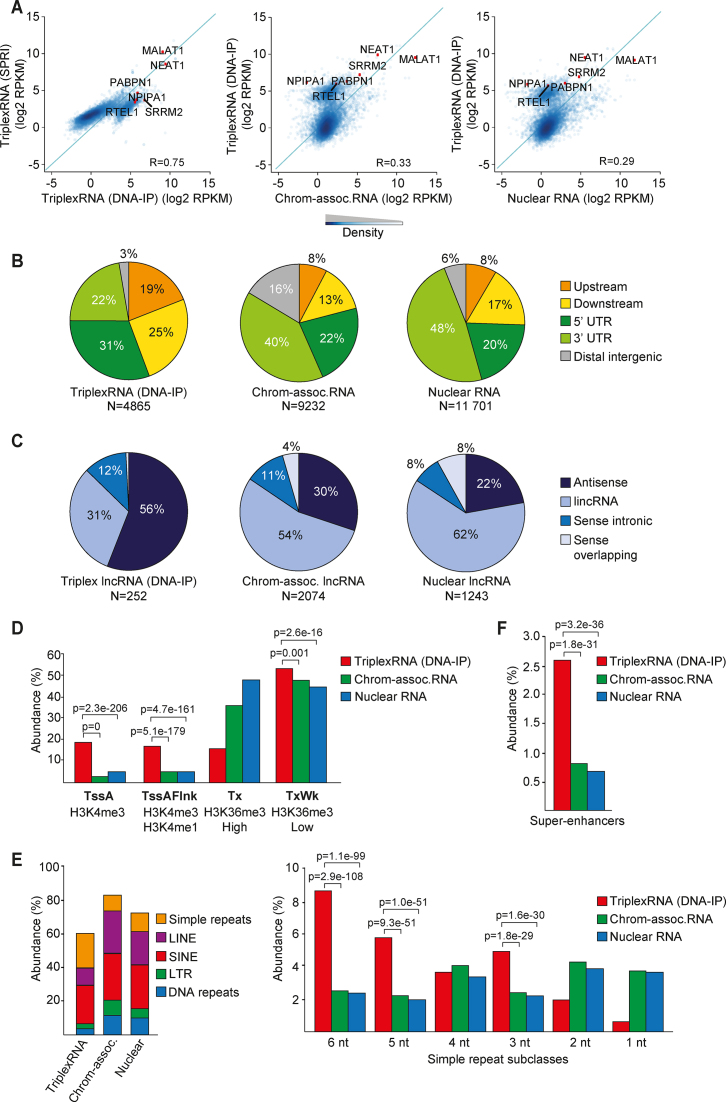
Figure 2.
Identification and global characterization of DNA-associated RNA. (A) Scatter plots showing the correlation between TriplexRNA isolated by DNA-IP and SPRI selection (left) and TriplexRNA (DNA-IP) and control RNA (middle, right). Pearson correlation coefficient (R) across 7148 genes overlapping peaks is shown. Green diagonal line x = y. Some representative genes that overlap TriplexRNAs and control RNAs are highlighted. (B) Pie charts depicting the genomic distribution of TriplexRNA (DNA-IP) compared to chromatin-associated and nuclear RNA peaks, excluding intronic and exonic gene regions. Upstream and downstream regions are defined within 2.5 kb proximity of the closest gene. (C) Pie chart showing classification of long noncoding RNAs that overlap TriplexRNA (DNA-IP), chromatin-associated and nuclear RNA. (D) Association of TriplexRNA (DNA-IP) and control RNA with ChromHMM promoter states and transcribed states. Active transcription start site (TssA), flanking active TSS (TssAFlnk), strong (Tx) and weak (TxWk) transcription regions are shown. (E) Left: Overlap of TriplexRNA (DNA-IP), chromatin-associated RNA and nuclear RNA with different classes of repeat elements. Right: Abundance of simple repeat subclasses. (F) Abundance of TriplexRNA (DNA-IP) and control RNAs overlapping super-enhancers in HeLa S3 cells. Data are from HeLa S3 cells. Adjusted P-values <0.05 in panels (D–F) were obtained from one-tailed Fisher's exact test using chromatin-associated and nuclear RNA as control.
For DNA sequencing, reads were aligned with bwa (33) allowing one mismatch (MAPQ > 1) (Supplementary Tables S5–S7). The differential peak caller THOR was used (default parameters) to find regions enriched in TriplexDNA-seq (RNA-associated DNAs) versus controls (Control DNA) each with quadruplicates (adjusted P-value <10−3, Negative Binomial test; Supplementary Table S5). A similar strategy was used to detect NEAT1-associated DNAs by comparing data from NEAT1-TFR capture experiments and from ASO-mediated capture assays with respective controls in triplicates (adjusted P-value < 10−2 and FC >1.5 and P-value < 10−10; Negative Binomial test; Supplementary Tables S6 and S7). MEME-ChIP suite (34) was applied for de novo motif analysis of top 500 peaks ranked based on P-value or randomly selected 500 peaks. The motifs with the lowest e-value are shown.
Transcript annotation was based on Gencode V24 (https://www.gencodegenes.org), the biotype field being used to define the genomic localization and features of identified RNAs (35). Repeatmasker version 4.0.7 was used to define repeat elements (http://www.repeatmasker.org/). Genomic coordinates for super-enhancers were from the Super-Enhancer Archive (http://sea.edbc.org/) (36). We used ChromHMM annotation of HeLa S3 cells for 15 chromatin states provided by ENCODE (37,38). For genomic characteristics of the peaks, we used the regulatory genomics toolbox (www.regulatory-genomics.org). The overlap was determined with Fisher's exact test.
In silico prediction of DNA:RNA triplexes
Triplex-forming potential of enriched RNA sequences was analyzed with Triplex Domain Finder (TDF), a software developed to identify triplex-forming regions (TFRs) in RNA and predict their potential to associate with specific DNA binding sites (DBS) (www.regulatory-genomics.org/tdf) (21). TDF finds RNA and DNA sequences with a minimal size of 15 bp where at least 80% follows Hoogsteen base pairing rules. We used ChromHMM to define regions with active promoter histone signatures (State 1-TssA), chromatin state annotation in HeLa S3 cells provided by ENCODE (accession ENCSR497SKR). Triplex-forming properties of top 1000 enriched regions (ranked by peak P-value) were compared with Mann–Whitney U test.
Statistical analyses
Data are reported as mean values from at least three biological replicates, with error bars representing standard deviation (SD) or standard error of the mean (SEM). For high-throughput data analyses, statistical tests are referred to in the text or figure legends. All P-values were corrected for multiple testing using Benjamini-Hochberg procedure (39).
Code availability
All software used in this manuscript is publicly available. The softwares for prediction of triple helices (Triple Helix Domain Finder) and for detection of enriched DNA and RNA regions (THOR) are part of the RGT package (version 0.11.3). The code and manuals are deposited at https://github.com/CostaLab/reg-gen. Computational analysis was based on the short read aligners STAR (version 2.5.4b— https://github.com/alexdobin/STAR/) and bwa (version 0.7.15-r1140—http://bio-bwa.sourceforge.net/), the read trimming tool trim-galore (version 0.4.5—https://www.bioinformatics.babraham.ac.uk/projects/trim_gal-ore/) and the motif analysis tools MEME-ChIP (version 4.12—http://meme-suite.org/). All statistical analysis and graphical displays were produced with R (version 3.4.3).
RESULTS
Isolation of DNA-associated RNA
To enrich and characterize RNAs that are physically associated with specific DNA sequences, we sought to establish an unbiased method that allows the isolation of RNAs that are bound to genomic DNA via Hoogsteen base pairing. The principle of our approach was to separate DNA with associated RNAs from free RNA. In brief, nuclei from U2OS or HeLa S3 cells were extracted with a buffer containing 0.5 M urea and 0.5% NP-40 to remove nucleoplasmic RNA. Isolated chromatin was treated with DNase I to fragment genomic DNA to an average size of 10 kb, washed with Sarkosyl and purified by centrifugation through a sucrose cushion (Figure 1A and B). RNAs that are associated with chromatin via DNA-bound proteins were released by proteinase K treatment and phenol/chloroform extraction. RNAs that are associated via DNA-RNA heteroduplexes (R-loops) were digested with RNase H (Supplementary Figure S1A). Finally, the samples were subjected to partial digestion with RNase I to trim the size of RNA to ∼100 nucleotides.
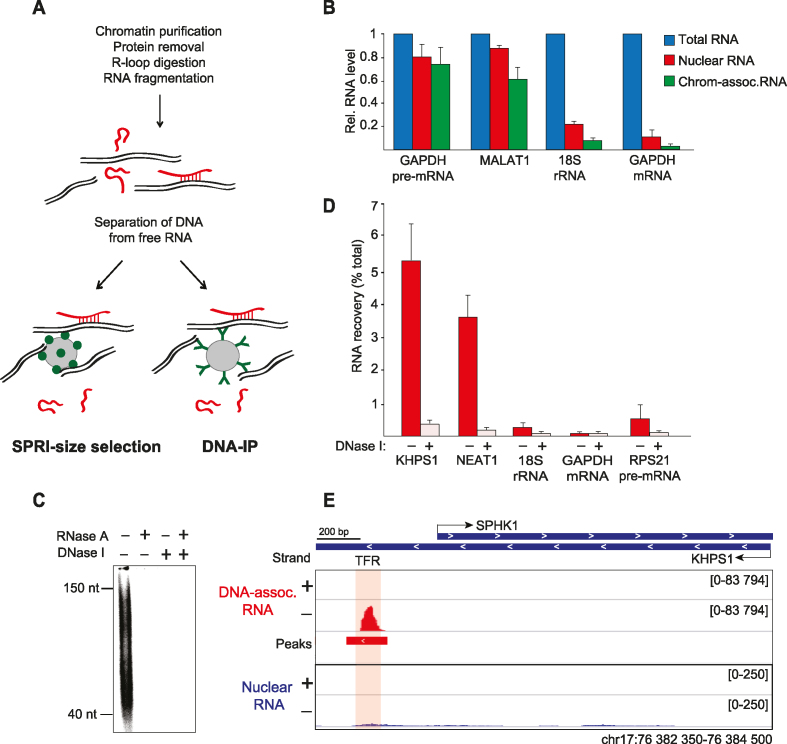
Figure 1.
Enrichment of triplex-forming RNAs. (A) Schematic overview of the method to enrich DNA-associated RNA. (B) RT-qPCR monitoring the indicated RNAs recovered from HeLa S3 cells, nuclei and purified chromatin. Values are normalized to cellular RNA (±SEM, N = 3). (C) Polyacrylamide gel electrophoresis of 5′-labeled RNA enriched by SPRI-size selection. Control samples were treated with DNase I before size selection or with RNase A before gel loading. (D) RT-qPCR analysis of DNA-associated RNA from HeLa S3 cells isolated by SPRI-size selection. Values are normalized to cellular RNA. Control samples were treated with DNase I before size selection (±SEM, N = 3). (E) RNA-seq profiles for KHPS1 in DNA-associated RNAs (DNA-IP) and nuclear RNA from U2OS cells. The overlap with the TFR of KHPS1 is shaded. Minus (–) and plus (+) strands are shown.
To enrich DNA-associated RNA, we separated free RNA from RNA that is bound to genomic DNA by Solid Phase Reversible Immobilization (SPRI)-based paramagnetic bead size selection. Alternatively, we isolated DNA-associated RNA by immunopurification with an anti-DNA antibody (DNA-IP) (Supplementary Figure S1B and S1C). With both approaches, DNA was efficiently separated from free RNA (Supplementary Figure S1D).
To ascertain that the applied methods enrich RNA that is associated with DNA, DNA was digested with DNase I, and associated RNA was analyzed by gel electrophoresis after radiolabeling with T4 polynucleotide kinase. The isolated RNA was 40–150 nucleotides in length (Figure 1C). Treatment with RNase A abolished the labeled signal, demonstrating that the RNA was free of DNA. No RNA was detected if the nucleic acids were treated with DNase I prior to SPRI-bead selection, confirming that DNA-associated RNA was recovered.
Previous studies have established that the lncRNA KHPS1 activates transcription of SPHK1 mRNA by direct binding to a stretch of purines upstream of the SPHK1 promoter. Tethering KHPS1 to this triplex-forming region (TFR) via Hoogsteen base pairing is essential for guiding KHPS1-associated epigenetic regulators to the SPHK1 promoter and for activation of SPHK1 expression (24). To validate that the established method enriched RNAs that are engaged in DNA:RNA triplex structures, we monitored the presence of lncRNA KHPS1 by RT-qPCR. Indeed, KHPS1 was enriched in DNA-associated RNA (Figure 1D). RNA recovery was abolished if the samples were treated with DNase I prior to size-separation, confirming that KHPS1 is physically associated with DNA. NEAT1 was also enriched, whereas other abundant cellular RNAs, such as 18S rRNA and GAPDH mRNA, were efficiently depleted. The specific enrichment and depletion of different RNA moieties reinforces that the fractionation procedure selectively enriched DNA-associated RNA. Sequencing of DNA-associated RNA from U2OS cells revealed an RNA peak that precisely overlapped the TFR of KHPS1 (Figure 1E). In accord with the low cellular abundance of KHPS1 (24), this lncRNA was hardly detectable in nuclear RNA. In contrast, RNAs that were easily detected in nuclear RNA, e.g. GAPDH and RPS21 mRNAs, were depleted in DNA-associated RNA, validating the fidelity of the crosslink-free fractionation procedure (Supplementary Figure S1E).
Global characterization of RNA engaged in triplex structures
To comprehensively catalogue DNA-associated transcripts, RNA-seq libraries were prepared from nuclear, chromatin- and DNA-associated RNA, which were isolated from HeLa S3 cells by either SPRI-size selection or by DNA-IP. After strand-specific high-throughput sequencing, reads were mapped to the human reference genome using stringent thresholds and standard RNA-seq pipelines (Supplementary Table S4). Biological replicates correlated well, indicating that the applied approaches yield robust and reproducible data (Supplementary Figure S2A).
To identify regions that are enriched in nuclear RNA, chromatin-associated RNA and DNA-associated RNA, we used a stringent and strand-specific adaptation of the differential peak caller THOR comparing read distributions in a pairwise fashion (Supplementary Figure S2B). Comparison of DNA-associated RNA to nuclear RNA revealed 7189 peaks in samples from DNA-IP and 3282 in SPRI-fractionated RNA (P-value < 10−500; Negative Binomial test, Supplementary Table S4). These peaks represent RNA regions that are associated with DNA via DNA:RNA triplexes and are thereafter designated ‘TriplexRNA’.
Approximately 60% of TriplexRNA in SPRI samples overlapped with RNA identified by DNA-IP. Samples derived from DNA-IP exhibited higher resolution and stronger signals compared to SPRI-size selection (Supplementary Figure S2C and S2D). Scatter plot analysis revealed a strong correlation between TriplexRNA isolated by DNA-IP and SPRI-size selection (R = 0.75), whereas their correlation with chromatin-associated and nuclear RNA was much lower (Figure 2A and Supplementary Figure S2E). This demonstrates that both triplex isolation approaches enriched RNA regions that are distinct from chromatin-associated and nuclear RNA. TriplexRNA isolated by DNA-IP was more dissimilar to chromatin-associated and nuclear RNA than RNA isolated by SPRI-size selection (R = 0.33 and 0.29 versus R = 0.62 and 0.53), indicating that DNA-IP is more specific than SPRI fractionation. To evaluate the results in an unbiased way, we analyzed the data sets from both methods.
To assess whether a fraction of recovered RNA is associated with DNA via R-loops, we compared DNA-associated RNA with published R-loop regions identified by DRIPc-seq (40). This analysis revealed that 20% of TriplexRNA peaks overlapped R-loop regions (Supplementary Figure S2F), indicating that the majority of enriched RNAs are bound to DNA via DNA:RNA triplexes rather than by R-loops. As we cannot rule out that the overlapping regions may represent cellular R-loops which were not completely digested by RNase H or constitute RNAs transcribed from R-loop-prone loci that form triplexes at distant sites, we included these sequences in the data analysis.
The peak distribution profiles of TriplexRNAs revealed the prevalence of regions originating from gene bodies and gene-proximal regions. Similar to chromatin-associated and nuclear RNA, a large fraction of TriplexRNA mapped to intronic and exonic regions of protein-coding genes (Supplementary Figure S2G). Significantly, RNAs mapping to 5′UTRs, sequences upstream and downstream of genes and antisense RNAs were enriched in TriplexRNA compared to chromatin-associated and nuclear RNA (P-value < 0.05, one-tailed Fisher's exact test) (Figure 2B and C; Supplementary Figure S2H and S2I). Among the most highly enriched lncRNAs was NEAT1, which has been shown to bind to hundreds of active genomic sites (7).
To characterize the chromatin signatures of genomic loci from which TriplexRNAs originate, we overlapped peaks with ChromHMM chromatin states which are defined by the combinatorial presence and absence of epigenetic marks in HeLa S3 cells. This analysis revealed that a significant fraction of TriplexRNA peaks were associated with transcription start sites (TSS) of active genes and their flanking regions marked by H3K4 methylation (Figure 2D and Supplementary Figure S2J). Notably, the majority of TriplexRNAs originated from weakly transcribed regions with low H3K36me3 marks, implying that low abundant regulatory RNAs are preferentially engaged in triplex structures.
Similar to nuclear and chromatin-associated RNA, a large fraction of TriplexRNA peaks overlapped with repeat elements (Figure 2E, left and Supplementary Figure S2K, left). Simple repeats were specifically enriched in TriplexRNA (P-value < 8.7e–87; one-tailed Fisher's exact test), 5 and 6 nt repeat units with diverse sequence compositions being most prominent (Figure 2E, right and Supplementary Figure S2K, right). This result is in accord with previous studies suggesting that repeat-derived sequences may represent functional domains that target regulatory RNAs to distant genomic regions (41–43). Interestingly, the fraction of RNAs originating from super-enhancers was also larger in TriplexRNA as compared to control RNA (Figure 2F and Supplementary Figure S2L), suggesting that triplex-forming RNAs serve a role in enhancer function.
Identification of DNA engaged in triplex structures
The next challenge was to experimentally identify the genomic target sites of RNAs engaged in triplex structures. To enrich RNA-associated DNA by an unbiased approach, we ligated a biotinylated linker oligonucleotide to the 3′-end of RNA and captured associated DNA on streptavidin beads. This approach is similar to MARGI-seq (10) and GRID-seq (11) except that we used native conditions to pull-down RNA from deproteinized, RNase H-treated chromatin (Figure 3A). Almost 50% of chromatin-associated RNA was bound to streptavidin beads, while no RNA was retrieved if linker ligation was omitted (Supplementary Figure S3A). After elution with RNase A, captured DNA from biological quadruplicates was subjected to deep sequencing, samples without linker ligation serving as negative control. Differential peak calling identified 2547 regions that were enriched in RNA-associated DNA, termed ‘TriplexDNA’, compared to control samples (P-value<10−3; Negative Binomial test) (Supplementary Figure S3B and Supplementary Table S5). DNA in unligated samples, which is not captured by associated RNA, will thereafter be designated ‘control DNA’. Although enrichment over background was lower in TriplexDNA-seq compared to TriplexRNA-seq, replicates exhibited similar signals in identified regions which were distinct from control DNA (Supplementary Figure S3B and S3C). This indicates that the capture approach reproducibly retrieved RNA-associated DNA. Comparison of TriplexDNA-seq with published DRIPc-seq data (40) revealed 13% overlap of RNA-associated DNA regions with R-loops (Supplementary Figure S3D). Given that R-loops form at genomic regions with strong G-clustering (44), a fraction of TriplexDNA may form both R-loops and DNA:RNA triplexes.
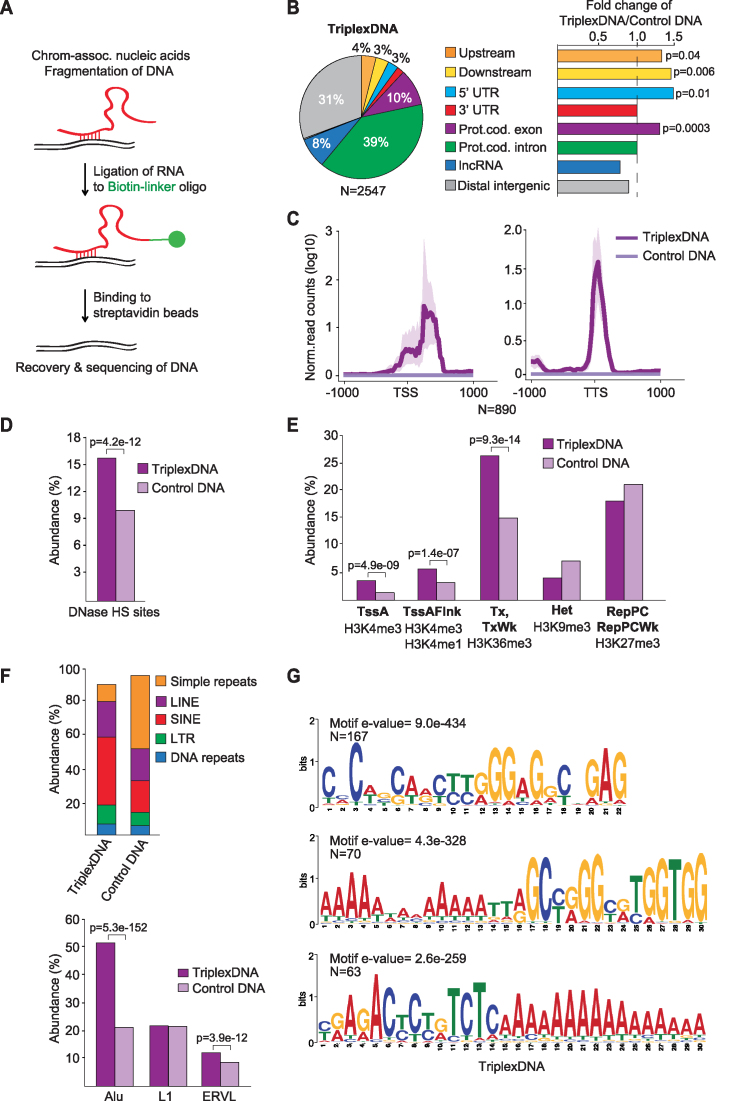
Figure 3.
Isolation and identification of RNA-associated DNAs. (A) Scheme illustrating the method used to isolate RNA-associated DNA (TriplexDNA). (B) Pie chart depicting the genomic distribution of TriplexDNA peaks. Upstream and downstream regions are defined within 2.5 kb proximity of the closest gene. The bar diagrams at the right display the fold change in the distribution of the respective regions in TriplexDNA compared to control DNA. (C) Line plots depicting the mean values of TriplexDNA-seq signals over TSS and TTS of 890 genes that overlap RNA-associated DNA peaks. Interval defined by maximum and minimum values is shaded. (D) TriplexDNA-seq regions overlapping DNase Hypersensitive Sites (DNase HS) in HeLa S3 cells provided by ENCODE. (E) Abundance of TriplexDNA regions associated with the indicated ChromHMM states. Active transcription start site (TssA), flanking active TSS (TssAFlnk), strong and weak (Tx, TxWk) transcription, heterochromatin (Het) and Polycomb-repressed (RepPC) regions are shown. (F) Top: Overlap of TriplexDNA and control DNA with different classes of repeat elements. Bottom: Abundance of predominating repeat subclasses in TriplexDNA. (G) MEME motif analysis identifying purine-rich consensus motifs in randomly selected 500 TriplexDNA peaks. Data are from HeLa S3 cells. Adjusted P-values <0.05 reported in panels (B,D-F) were obtained from one-tailed Fisher's exact test.
Peak distribution analysis showed that TriplexDNA originate both from introns (39%) and exons (10%) of protein-coding genes as well as intergenic regions (31%), lncRNAs (8%), UTRs (5%), and regions upstream or downstream of gene bodies (7%) (Figure 3B and Supplementary Figure S3E). Similar to the RNA-seq data, sequences mapping upstream and downstream of gene bodies and 5′UTRs were enriched in TriplexDNA compared to control DNA. Analysis of the read distribution along gene bodies revealed a positional preference for sequences downstream of transcription start sites (TSSs) and transcription termination sites (TTSs), suggesting that these regions preferentially engage in DNA:RNA triplex structures (Figure 3C). Enrichment of DNA regions around TSSs is in accord with previous in silico studies predicting high triplex-forming potential of promoter regions (26–28). Comparison of TriplexDNA-seq data with DNase I hypersensitive sites from ENCODE revealed that Triplex DNA is more sensitive to DNase I than control DNA (Figure 3D). Consistently, ChromHMM analysis revealed enrichment of TriplexDNA at active TSSs (H3K4me3), at TSS flanking regions (H3K4me1&3) and at transcribed loci (H3K36me3) (Figure 3E). Control DNA, on the other hand, exhibited higher prevalence of heterochromatic (H3K9me3) and Polycomb-repressed regions (H3K27me3). Together, these results reveal that triplex formation preferentially occurs at open chromatin regions.
Notably, TriplexDNA had a propensity to harbor significantly more SINE and LTR elements than control DNA (P-values <1,7e-16; one-tailed Fisher's exact test) (Figure 3F, top). Alu and ERVL subclasses were predominant (Figure 3F, bottom), supporting that these repetitive DNA sequences might serve an important function in tethering regulatory RNAs to specific genomic regions. In contrast to TriplexRNA, super-enhancers were not enriched, supporting that RNAs originating from super-enhancers form triplexes at distant regions (Supplementary Figure S3F). Moreover, MEME motif analysis identified purine-rich consensus motifs in 300 out of 500 randomly selected TriplexDNA regions, substantiating that these sequences are capable to form DNA:RNA triplexes (Figure 3G and Supplementary Figure S3G).
Validation of triplex-forming RNA and DNA regions
To confirm that the identified DNA-associated RNAs have the potential to form triplexes, we performed in silico analysis of candidate RNAs using TDF (Triple Helix Domain Finder), a software that predicts the triplex-forming potential of RNAs with DNA targets based on Hoogsteen base pairing rules (21). RNA sequences that are more likely to associate with specific DNA regions than with random DNA are termed triplex-forming regions (TFR) and their DNA binding sites DBS. TDF calculates enrichments and ranks the triplex-forming potential of RNAs by taking into account both their number of TFRs and putative DBSs.
Given that lncRNAs form triplexes at promoters (25) and TriplexDNA regions correlate with active TSSs, we used TDF to assess the triplex-forming potential of TriplexRNAs at active promoters identified by ChromHMM. This analysis revealed that the majority of TriplexRNA peaks have the potential to form triplexes with active promoters (Supplementary Figure S4A). The number of predicted TFRs and promoter-associated DBSs were significantly higher in TriplexRNA than in control RNA (Figure 4A and Supplementary Figure S4B). About half of TriplexRNA (51%) comprised purine (A,G) or mixed motifs (U,G) which bind to DNA in antiparallel orientation. The other half (49%) comprised pyrimidine (C,U) or mixed motifs (U,G) which bind to the purine-rich strand of DNA in parallel orientation (Figure 4B and Supplementary Figure S4C).
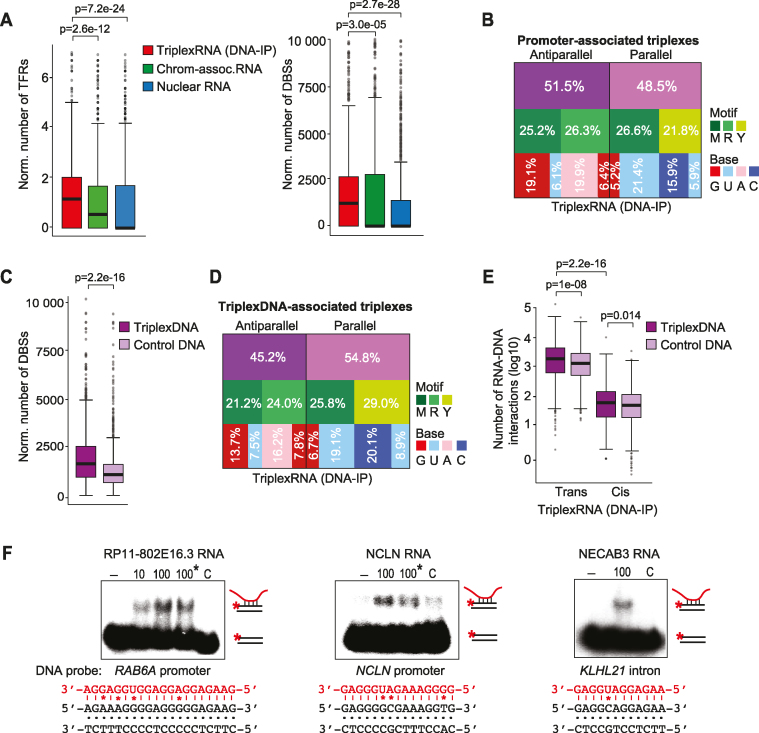
Figure 4.
Validation of triplex-forming RNA and DNAs. (A) TDF analysis predicting the potential of top 1000 enriched TriplexRNA (DNA-IP) regions (ranked by peak P-value) to bind to active promoters defined by ChromHMM. Number of TFRs in RNA (per kilobase of RNA, left) and the number of putative DBSs at promoters (per kilobase of RNA, right) are shown. Boxplot borders are defined by the 1st and 3rd quantiles of the distributions, the middle line corresponds to the median value. The top whisker denotes the maximum value within the third quartile plus 1.5 times the interquartile range (bottom whisker is defined analogously). Dark gray dots represent outliers with values higher or lower than whiskers. Further box plots are based on the same definitions. (B) Motif analysis of triplexes formed between TriplexRNA (DNA-IP) and active promoters. The diagram depicts the fraction of antiparallel and parallel triplexes with the respective motif and nucleotide composition of TFRs in TriplexRNA. (C) TDF analysis comparing the triplex-forming potential of top 2000 TriplexDNA-seq regions with top 1000 TriplexRNA (DNA-IP) (ranked by peak P-value). The number of putative DBSs (per kilobase of RNA) is shown. (D) Motif analysis of predicted triplexes formed between TriplexRNAs (DNA-IP) and TriplexDNA. The diagram depicts the fraction of antiparallel and parallel triplexes, with the respective motif and nucleotide composition of TFRs in TriplexRNA. (E) Box plot classifying triplex interactions between TriplexRNAs (DNA-IP) and TriplexDNA-seq regions as cis (>10 kb in the same chromosome) and trans (at different chromosomes) interactions, excluding underrepresented local interactions (within 10 kb distance). (F) EMSAs using 10 or 100 pmol of synthetic TriplexRNAs and 0.25 pmol of double–stranded 32P-labeled oligonucleotides comprising target regions from TriplexDNA (Supplementary Table S2). Reactions marked with an asterisk (*) were treated with 0.5 U RNase H. As a control (C), RNA without a putative TFR was used. Potential Hoogsteen base pairing between motifs and respective TFR sequences are shown; mismatches are marked (*). TriplexRNA-seq and TriplexDNA-seq data are from HeLa S3 cells. Adjusted P-values <0.05 in panels (A, C, E) are based on one-tailed Mann–Whitney test.
TDF-based analysis of the potential of RNAs identified by TriplexRNA-seq to associate with TriplexDNA revealed that the triplex-forming potential of RNA was significantly higher at RNA-associated DNA than at control DNA (Figure 4C; Supplementary Figure S4D and S4E). The length of merged DBSs in the target regions ranged from 15 to 75 bp, indicating that both short and long purine-rich sequences can engage in triplexes (Supplementary Figure S4F). Mixed motifs (U,G) were overrepresented in TFRs and there was a slight preference of TriplexRNA to bind to TriplexDNA in parallel orientation (Figure 4D and Supplementary Figure S4G).
RNAs can affect expression of neighboring genes in cis or at distant genes in trans (25). To examine whether there is a tendency for in cis or in trans DNA:RNA interactions, we classified predicted triplexes between TriplexDNA and TriplexRNA peaks in local regions (within 10 kb distance), in cis (more than 10 kb distance on the same chromosome) and in trans (at different chromosomes). The majority of RNA was engaged in trans interactions with DNA, while local interactions were underrepresented (Figure 4E and Supplementary Figure S4H–S4J).
To validate that the regions predicted by TDF are capable to engage in DNA:RNA triplexes, we performed electrophoretic mobility shift assays (EMSAs) with TriplexRNAs and putative targets identified by TriplexDNA-seq. To this end, 5′-labeled DNA oligonucleotides were annealed with complementary unlabeled DNA oligos, the duplexes were incubated with synthetic RNAs identified by TriplexRNA-seq, and triplex formation was analyzed by gel electrophoresis. Significantly, the predicted RNAs formed a low mobility DNA–RNA complex with their radiolabeled DNA targets, whereas no complex was formed with a control RNA that does not contain a potential triplex-forming sequence (Figure 4F and Supplementary Figure S4K). Treatment with RNase H did not affect the mobility of the complexes, excluding the possibility that the RNAs interact with DNA by forming DNA–RNA heteroduplexes. Altogether, these in vitro data validate the potential of TriplexRNA and TriplexDNA sequences to form DNA:RNA triplex structures.
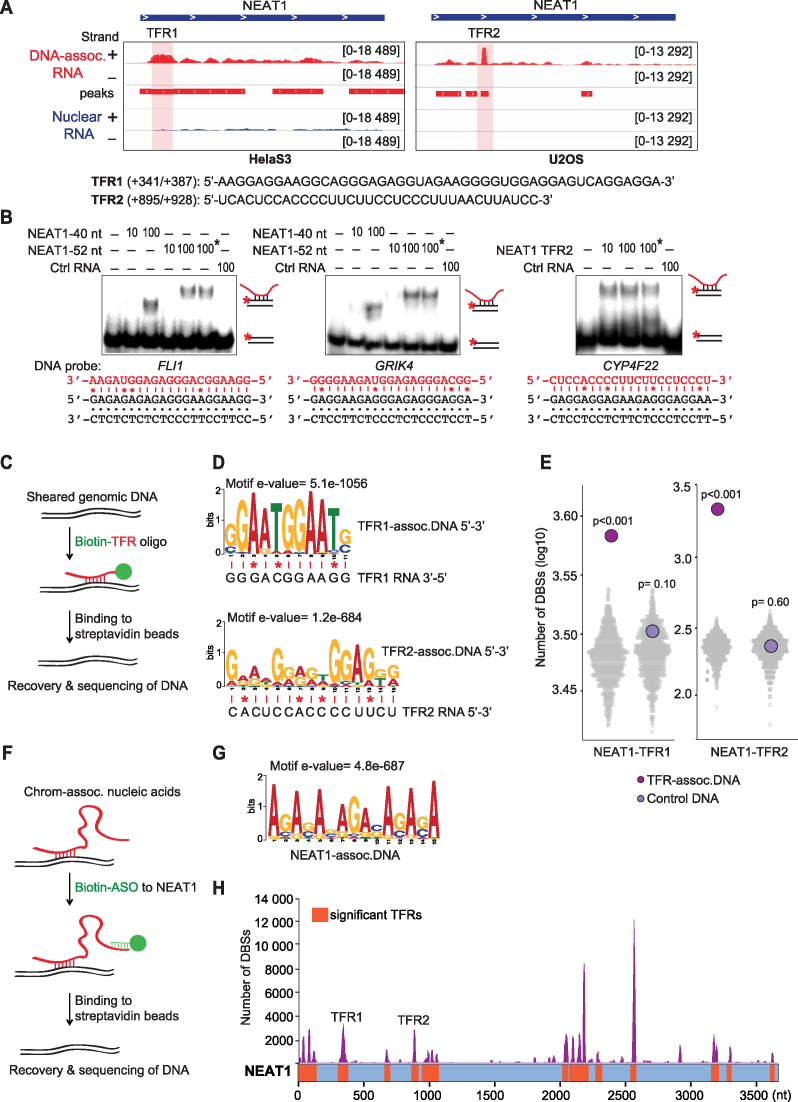
Identification of genomic sites associated with NEAT1
To validate the triplex-forming potential of a candidate RNA, we focussed on NEAT1, a prominent lncRNA identified by TriplexRNA-seq (Figure 2A). A region of NEAT1 comprising nucleotides +341/+387 (NEAT1-TFR1) was enriched in TriplexRNA-seq from HeLa S3 cells, whereas a more downstream region comprising nucleotides +895/+928 (NEAT1-TFR2) was more pronounced in U2OS cells (Figure 5A). This suggests that different TFR-containing regions of a given RNA may target different genes in a cell type-specific manner. Although NEAT1 has never been shown to associate directly with DNA, TDF analysis predicted several DBSs in CHART-seq data (7) that might be targeted by NEAT1 (Supplementary Figure S5A). To demonstrate the potential of NEAT1 to bind to the identified target regions, we performed EMSAs using radiolabeled double-stranded oligonucleotides containing predicted target gene sequences. After incubation with synthetic RNAs comprising NEAT1-TFR1 and NEAT-TFR2 sequences, the electrophoretic mobility of the double-stranded DNAs was decreased (Figure 5B and Supplementary Figure S5B). Treatment with RNase H did not affect the mobility of the complex, excluding the possibility that retardation of electrophoretic mobility was due to DNA–RNA heteroduplex formation. A control RNA that does not contain any TFR sequence did not affect the mobility of the DNA fragments, supporting sequence-specific triplex formation by the NEAT1-TFRs. Thus, the EMSA approach validates the TDF-based prediction of target DNA sequences that are bound by the TFRs of NEAT1 and confirms the ability of RNA regions identified by TriplexRNA-seq to directly associate with DNA.
Figure 5.
NEAT1 forms triplexes at numerous genomic sites. (A) NEAT1 profiles in TriplexRNA-seq (DNA-IP) (red) and nuclear RNA (blue) from HeLa S3 and U2OS cells with shaded TFR1 and TFR2. Minus (-) and plus (+) strands are shown. The position and sequence of NEAT1-TFR1 and -TFR2 are shown below. (B) EMSAs using 10 or 100 pmol of synthetic NEAT1 versions comprising TFR1 (40 or 52 nt) or TFR2 incubated with 0.25 pmol of double–stranded 32P-labeled oligonucleotides which harbor sequences of NEAT1 target genes predicted from CHART-seq (Supplementary Table S2). Reactions marked with an asterisk (*) were treated with 0.5 U RNase H. As a control, RNA without a putative TFR was used. Potential Hoogsteen base pairing between motifs and respective TFR sequences are shown; mismatches are marked (*). (C) Schematic depiction of the TFR-based capture assay. Biotinylated RNA oligos covering NEAT1-TFR1 and NEAT1-TFR2 were used to capture genomic DNA. (D) MEME motif analysis identifying consensus motifs in DNA captured by NEAT1-TFR1 (399 of top 500 peaks) and by NEAT1-TFR2 (500 of top 500 peaks ranked by peak P-value). Potential Hoogsteen base pairing between motifs and respective TFR sequences are shown; mismatches are marked (*). (E) TDF analysis of the triplex-forming potential of NEAT1-TFR1 and NEAT1-TFR2 RNAs with top 500 TFR-associated and control DNA peaks (ranked by peak P-value) compared to 500 randomized regions (N = 1000, colored grey). P-values were obtained from one-tailed Mann–Whitney test. (F) Scheme presenting antisense oligo (ASO)-based capture of NEAT1-associated DNA. (G) Consensus motif in NEAT1-associated DNA sites (314 of top 500 peaks ranked by peak P-value). (H) TDF analysis predicting the triplex-forming potential of NEAT1 on ASO-captured DNA regions. Significant TFRs along NEAT1 are shown in orange, the number of target sites (DBS) for each TFR in purple. For TFR- and ASO-based capture assays nucleic acids isolated from HeLa S3 chromatin were used.
A triplex-mediated targeting mechanism would explain how NEAT1 and other regulatory RNAs impact expression of specific genes in remote regions. To examine the potential of NEAT1 to bind to genomic DNA, we performed an RNA-based DNA capture assay. To this end, biotinylated RNA oligos corresponding to NEAT1-TFR1 and NEAT1-TFR2 were incubated with sheared genomic DNA to allow triplex formation. After binding to streptavidin beads, associated DNA was eluted and subjected to deep sequencing. Samples incubated with an oligonucleotide that does not contain a potential TFR served as control (Figure 5C). Differential peak calling comparing the read distribution in controls and captured samples revealed 622 putative binding sites for the GA-rich NEAT1-TFR1 and 4423 binding sites for the CU-rich NEAT1-TFR2 (P-value < 10−2; Negative Binomial test; Supplementary Figure S5C and Supplementary Table S6). ‘Control DNA’ represents regions with a higher number of reads in control libraries compared to DNA sequences that are associated with NEAT1-TFRs. MEME motif analysis of the top 500 target regions in DNA associated with NEAT1-TFR1 and NEAT1-TFR2 revealed the prevalence of purine-rich consensus sequences in 399 and 500 regions, respectively, which can engage in triplex structures with respective TFRs (Figure 5D and Supplementary Figure S5D). TDF analysis confirmed that the enriched regions comprise several potential binding sites for NEAT1-TFR1 and NEAT1-TFR2 (Figure 5E). Control DNA regions, on the other hand, did not display significant triplex-forming sequences, supporting that TFR-containing RNAs have the potential to associate with respective captured DNA via Hoogsteen base pairing.
To validate the interaction of NEAT1 with specific genomic sites in vivo, we captured NEAT1-associated DNA from deproteinized chromatin using a biotin-labeled antisense DNA oligonucleotide (ASO) that binds close to the TFR1 and TFR2 of NEAT1. A sense oligo that does not hybridize to NEAT1 served as control (Figure 5F). RT-qPCR analysis revealed that NEAT1 was captured with the NEAT-specific ASO but not with a control sense oligo (Supplementary Figure S5E). Peak calling analysis using DNA libraries from three biological replicates identified 3692 NEAT1-associated DNA regions (P-value < 10−10, Negative Binomial test; Supplementary Figure S5F and Supplementary Table S7). MEME motif analysis revealed a purine-rich consensus sequence in 314 of the top 500 peaks (Figure 5G and Supplementary Figure S5G). Furthermore, TDF analysis showed that NEAT1 has a high potential to bind to the captured DNA and there is at least one canonical triplex-forming sequence (DBS) in 3444 NEAT1-associated DNA regions (Figure 5H). Altogether, these results demonstrate that NEAT1 is a triplex-forming RNA that targets numerous genomic loci via direct binding to DNA.
To examine whether the crosslink-free ASO-mediated capture assay is more advantageous for the identification of genomic DNA:RNA triplexes than previous approaches using crosslinked chromatin, we compared the NEAT1 targets identified by the ASO-mediated capture with CHART-seq data (7). 13% of ASO-mediated capture targets overlapped with 19% of CHART-seq regions (Supplementary Figure S5H, top). While the median length of CHART-seq peaks was 16 511 bp, the median peak length of ASO-captured DNA was 550 bp, which allows a more precise mapping of RNA-associated regions (Supplementary Figure S5H, bottom). Comparison of the signals across co-enriched regions substantiated the higher peak resolution of the ASO-capture approach (Supplementary Figure S5I). Given that CHART-seq does not distinguish between protein- and triplex-mediated interactions, co-enrichment of sequences in CHART-seq and TriplexDNA-seq suggests that a fraction of the CHART-seq data reflects binding of NEAT1 to chromatin via triplex formation. Noteworthy, co-enriched triplex-forming regions did not represent the highest signal-containing part of the CHART-seq peaks (Supplementary Figure S5I), indicating that CHART-seq preferentially identifies protein-mediated interactions. Importantly, most regions identified by ASO-mediated capture were not contained in the CHART-seq data, supporting that removal of protein-DNA interactions facilitates isolation of cellular triplexes.
DISCUSSION
Several studies have indicated that lncRNAs may guide effector proteins to gene regulatory regions via triplex formation (17–22,24). Moreover, genome-wide in silico analyses of lncRNA-mediated DNA:RNA triplex structures have identified a huge number of potential triplex target sequences which are prevalent at promoters, introns, UTRs, super-enhancers and transposable elements (28,45). The presence of putative triplex target sites in regulatory regions suggests that triple helix formation is an important mechanism by which lncRNAs exert their function in target gene recognition and transcriptional control.
Current technological advances, such as CHART (8), ChIRP (6), RAP (9), MARGI (10) and GRID-seq (11) have allowed mechanistic insights into the function of chromatin-associated RNAs by identifying their genomic target sites. The association of RNA with chromatin may be brought about by either DNA-bound proteins, by the formation of R-loops or by DNA:RNA triplexes. All previously used methods included crosslinking to stabilize protein-nucleic acid interactions. Therefore, the majority of target sequences identified by crosslink-based methods may be brought about by binding of RNA to chromatin-associated proteins rather than by direct interaction with DNA. Accordingly, local and cis interactions predominated in GRID-seq data, the majority of chromatin-associated RNA comprising nascent RNA chains that are presumably tethered to chromatin by elongating RNA polymerase. As crosslinking does not stabilize the interactions between RNA and DNA, triplexes may not survive crosslinking and washing unless stabilized by proteins. Therefore, these methods might not be applicable for cataloguing DNA:RNA triplexes. To overcome these limitations, we have established crosslink-free methods making use of conditions which would preserve DNA:RNA triplexes while depleting protein- and R-loop-mediated RNA-DNA interactions. The partial overlap of CHART-seq regions with our ASO-capture data (19%) further supports that the most RNA-DNA interactions identified by CHART-seq are mediated by DNA-bound proteins.
A surprising finding was that a major fraction of DNA-associated RNAs originated from protein-coding genes. Given that 15–20% of isoforms of protein-coding genes do not encode proteins, and a significant fraction of mRNA is retained in the nucleus and exhibits non-coding functions (46–50), triplex-mediated tethering of such RNAs may be fundamental for their function. Moreover, protein-coding genes can give rise to a variety of regulatory RNAs, such as microRNAs, circular RNAs, antisense RNAs, eRNAs and intronic sense RNAs (51). Recent studies have also shown that about half of all annotated enhancers are intragenic. Transcription at intragenic enhancers attenuates expression of the host genes, the extent of attenuation positively correlating with transcription of eRNAs (52–55).
To identify genomic regions that are associated with RNA, we used a ligation-mediated approach to capture DNA:RNA triplexes. ChromHMM chromatin state analysis reinforced that triplex formation occurs at active chromatin domains. This is in accord with studies reporting that the triplex-forming third strand cannot be accommodated in the nucleosome core (56,57) and triplex-formation acts as a nucleosomal barrier (58,59). The combined analysis of TriplexRNA and TriplexDNA data revealed the abundance of trans interactions, which is in agreement with earlier reports showing triplex formation of microRNAs and several lncRNAs with distant genomic loci (20,21,60,61). The finding that super-enhancer sequences were enriched in TriplexRNA but not in TriplexDNA, indicates that RNA originating from super-enhancers does not associate with the super-enhancer itself but with distant regions. Such RNAs might mediate the contact of enhancers with target genes via triplex-formation. Moreover, the presence of long purine-rich sequences in TriplexDNA peaks suggests that several RNAs may either bind simultaneously or form triplexes under specific physiological conditions.
One of the most compelling results was the enrichment of RNA and DNA regions harboring repeat elements. This implies that a regulatory RNA might interact with multiple DNA regions via triplex formation in a nonrandom fashion. Alternatively, multiple RNAs containing similar repeat elements might form triplex structures with specific genomic sites in a temporal and spatial manner, acting as a signal amplifier or a networking tool for a group of co-regulated genes. The importance of repeat-containing transcripts has been supported by several studies showing that many lncRNA comprise repeat sequences which contribute to binding to DNA and tissue-specific gene expression (28,41–43,62). For example, transcription of LINE-1 undergoes dynamic changes during early embryogenesis, reinforcing that RNA-comprising repeat elements have the potential to directly bind to DNA and impact developmental programs (23). Furthermore, sequences enriched in Alu repeats have been shown to promote nuclear localization of RNAs (63). In support of repeat elements having the potential to form DNA:RNA triplex structures, Alu repeats and ERVL LTR sequences were enriched in RNA-associated DNA. LTRs impact gene expression by providing alternative promoter and enhancer sequences (64), Alu repeats affect nucleosome positioning and transcription factor binding (65,66). Moreover, SINE repeats have high densitiy of potential triplex-target sites and are rich in super-enhancers which are targeted by super-lncRNAs (27,45). These results suggest that tethering RNA to DNA via repeat-derived sequences may facilitate the guidance of associated proteins to specific genomic sites.
TriplexRNA harbors pyrimidine (C,U), purine (A,G) and mixed (U,G) motifs. This seems to contradict in vitro studies showing that triplex formation by cytosine residues requires low pH (67,68) and purine-rich RNA sequences do not engage in stable DNA:RNA triplexes (69–71). This suggests that additional parameters, such as chromatin environment, triplex-interacting proteins and the length of RNA and DNA molecules, may create a local environment which either prompts the protonation of cytosines, facilitates Hoogsteen base pairing or stabilizes DNA:RNA triplexes in vivo (72). In support of this notion, triplex-formation by the UC-rich lncRNAs Fendrr and KHPS1 have been shown to occur under physiological pH (18,24) and PARTICLE, MEG3, and HOTAIR engage in DNA:RNA triplexes via purine-rich TFRs (19–21).
Recovery of DNA:RNA triplex structures is influenced by several factors. As pointed out by Britten and Davidson (1969), a single RNA can regulate several genes and different RNAs may activate the same genes (12). Thus the abundance of triplex-forming RNAs and the number of genomic target sites might impact the enrichment of TriplexRNA, that is, higher-copy number transcripts and RNAs that target multiple genes are likely to be more enriched than RNAs that regulate just one or a few genes. Accordingly, NEAT1, which is associated with numerous genomic loci (7), is among the top hits in TriplexRNA-seq. Moreover, if a gene is targeted by several RNAs, potential interacting RNAs may be missed because the most abundant RNAs with the best fitting TFRs will compete for binding of molecules comprising weaker TFRs. Similarly, DNA regions targeted by multiple RNAs might dominate in TriplexDNA-seq data. These possibilities would explain the underrepresentation of local interactions between TriplexRNA and TriplexDNA. The majority of TriplexRNA and TriplexDNA did not overlap with reported R-loop regions, reinforcing the validity of the experimental approach used. However, we cannot rule out that different R-loops might show different sensitivities to RNase H and are not completely depleted. Moreover, there might be noncanonical nucleic acid interactions which we are not aware of yet.
Surely, more work is needed to further optimize our approaches and to functionally analyze the cellular Triplexome. The established protocol and the first analysis of data from DNA-associated RNAs combined with identification of genomic loci that are targeted by triplex-forming RNAs provides information about RNAs that access the genome in a highly discriminating fashion through specific DNA sequences. The most challenging task ahead is to decipher the modes of action by which these RNAs influence their target genes. We are yet to understand how DNA:RNA triplexes form, how triplex formation is regulated and how triplexes impact gene expression.
DATA AVAILABILITY
All raw sequence data and peak predictions have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO accession number GSE120850. ChromHMM and DNase-I hypersensitive site annotations were based on ENCODE data from HeLa S3 cells (accession number ENCSR497SKR and ENCSR959ZXU, respectively). CHART-seq DNA and R-loop regions were obtained from published data (7,40).
Supplementary Material
ACKNOWLEDGEMENTS
We thank David Ibberson and the personnel of the CellNetworks Deep Sequencing Core facility and EMBL GeneCore for Illumina sequencing.
Author contributions: I.G. conceived the study and coordinated the research. N.S.C. established the experimental procedures and conducted almost all wet-lab experiments. N.S.C. and T.R. performed electrophoretic mobility shift experiments. C.K., R.L. and I.C. performed the bioinformatic analyses. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG (GR475/22-2; SFB1036 to I.G.); CellNetworks (EcTop Survey 2014 to I.G.); Baden-Württemberg Stiftung (to I.G.); Aachen Interdisciplinary Center for Clinical Research (IZKF to I.C.); Excellence Initiative of the German federal and state governments (to I.C.); Start program of the RWTH Aachen Medical Faculty (to I.C.). Funding for open access charge: German Cancer Research Center (DKFZ), Molecular Biology of the Cell II.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bonasio R., Shiekhattar R.. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014; 48:433–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Engreitz J.M., Ollikainen N., Guttman M.. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016; 17:756–770. [DOI] [PubMed] [Google Scholar]
- 3. Guttman M., Rinn J.L.. Modular regulatory principles of large non-coding RNAs. Nature. 2012; 482:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Werner M.S., Ruthenburg A.J.. Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep. 2015; 12:1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Werner M.S., Sullivan M.A., Shah R.N., Nadadur R.D., Grzybowski A.T., Galat V., Moskowitz I.P., Ruthenburg A.J.. Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat. Struct. Mol. Biol. 2017; 24:596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y.. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011; 44:667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E.. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014; 55:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simon M.D., Wang C.I., Kharchenko P.V., West J.A., Chapman B.A., Alekseyenko A.A., Borowsky M.L., Kuroda M.I., Kingston R.E.. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20497–20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Engreitz J.M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C., Kadri S., Xing J., Goren A., Lander E.S. et al.. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013; 341:1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sridhar B., Rivas-Astroza M., Nguyen T.C., Chen W., Yan Z., Cao X., Hebert L., Zhong S.. Systematic mapping of RNA-chromatin interactions in vivo. Curr. Biol. 2017; 27:602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li X., Zhou B., Chen L., Gou L.T., Li H., Fu X.D.. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 2017; 35:940–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Britten R.J., Davidson E.H.. Gene regulation for higher cells: a theory. Science. 1969; 165:349–357. [DOI] [PubMed] [Google Scholar]
- 13. Felsenfeld G., Davies D.R., Rich A.. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957; 79:2023–2024. [Google Scholar]
- 14. Hoogsteen K. The structure of crystals containing a hydrogen-bonded complex of 1-methylthymine and 9-methyladenine. Acta Crystallogr. 1959; 12:822–823. [Google Scholar]
- 15. Morgan A.R., Wells R.D.. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J. Mol. Biol. 1968; 37:63–80. [DOI] [PubMed] [Google Scholar]
- 16. Beal P.A., Dervan P.B.. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991; 251:1360–1363. [DOI] [PubMed] [Google Scholar]
- 17. Schmitz K.M., Mayer C., Postepska A., Grummt I.. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010; 24:2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., Macura K., Blass G., Kellis M., Werber M. et al.. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013; 24:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O’Leary V.B., Ovsepian S.V., Carrascosa L.G., Buske F.A., Radulovic V., Niyazi M., Moertl S., Trau M., Atkinson M.J., Anastasov N.. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015; 11:474–485. [DOI] [PubMed] [Google Scholar]
- 20. Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E. et al.. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015; 6:7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kalwa M., Hanzelmann S., Otto S., Kuo C.C., Franzen J., Joussen S., Fernandez-Rebollo E., Rath B., Koch C., Hofmann A. et al.. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016; 44:10631–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao Z., Sentürk N., Song C., Grummt I.. lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 2018; 32:836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fadloun A., Le Gras S., Jost B., Ziegler-Birling C., Takahashi H., Gorab E., Carninci P., Torres-Padilla M.E.. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013; 20:332–338. [DOI] [PubMed] [Google Scholar]
- 24. Postepska-Igielska A., Giwojna A., Gasri-Plotnitsky L., Schmitt N., Dold A., Ginsberg D., Grummt I.. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 2015; 60:626–636. [DOI] [PubMed] [Google Scholar]
- 25. Li Y., Syed J., Sugiyama H.. RNA-DNA triplex formation by long noncoding RNAs. Cell Chem. Biol. 2016; 23:1325–1333. [DOI] [PubMed] [Google Scholar]
- 26. Buske F.A., Bauer D.C., Mattick J.S., Bailey T.L.. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012; 22:1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goni J.R., de la Cruz X., Orozco M.. Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Res. 2004; 32:354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jalali S., Singh A., Maiti S., Scaria V.. Genome-wide computational analysis of potential long noncoding RNA mediated DNA:DNA:RNA triplexes in the human genome. J. Transl. Med. 2017; 15:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu Q., Gaddis S.S., MacLeod M.C., Walborg E.F., Thames H.D., DiGiovanni J., Vasquez K.M.. High-affinity triplex-forming oligonucleotide target sequences in mammalian genomes. Mol. Carcinog. 2007; 46:15–23. [DOI] [PubMed] [Google Scholar]
- 30. Hawley D.K., Roeder R.G.. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J. Biol. Chem. 1985; 260:8163–8172. [PubMed] [Google Scholar]
- 31. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Allhoff M., Sere K., F. Pires J., Zenke M., G. Costa I.. Differential peak calling of ChIP-seq signals with replicates with THOR. Nucleic Acids Res. 2016; 44:e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Durbin R.. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al.. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei Y., Zhang S., Shang S., Zhang B., Li S., Wang X., Wang F., Su J., Wu Q., Liu H. et al.. SEA: a super-enhancer archive. Nucleic Acids Res. 2016; 44:D172–D179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995; 57:289–300. [Google Scholar]
- 40. Sanz L.A., Hartono S.R., Lim Y.W., Steyaert S., Rajpurkar A., Ginno P.A., Xu X., Chedin F.. Prevalent, dynamic, and conserved R-Loop structures associate with specific epigenomic signatures in mammals. Mol. Cell. 2016; 63:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson R., Guigo R.. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. RNA. 2014; 20:959–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kannan S., Chernikova D., Rogozin I.B., Poliakov E., Managadze D., Koonin E.V., Milanesi L.. Transposable element insertions in long intergenic non-coding RNA Genes. Front. Bioeng. Biotechnol. 2015; 3:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kelley D., Rinn J.. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012; 13:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chedin F. Nascent connections: R-loops and chromatin patterning. Trends Genet. 2016; 32:828–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soibam B. Super-lncRNAs: identification of lncRNAs that target super-enhancers via RNA:DNA:DNA triplex formation. RNA. 2017; 23:1729–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bahar Halpern K., Caspi I., Lemze D., Levy M., Landen S., Elinav E., Ulitsky I., Itzkovitz S.. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 2015; 13:2653–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Caudron-Herger M., Muller-Ott K., Mallm J.P., Marth C., Schmidt U., Fejes-Toth K., Rippe K.. Coding RNAs with a non-coding function: maintenance of open chromatin structure. Nucleus. 2011; 2:410–424. [DOI] [PubMed] [Google Scholar]
- 48. Kumari P., Sampath K.. CncRNAs: bi-functional RNAs with protein coding and non-coding functions. Semin. Cell Dev. Biol. 2015; 47–48:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nam J.W., Choi S.W., You B.H.. Incredible RNA: dual functions of coding and noncoding. Mol. Cells. 2016; 39:367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Solnestam B.W., Stranneheim H., Hallman J., Kaller M., Lundberg E., Lundeberg J., Akan P.. Comparison of total and cytoplasmic mRNA reveals global regulation by nuclear retention and miRNAs. BMC Genomics. 2012; 13:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ayupe A.C., Tahira A.C., Camargo L., Beckedorff F.C., Verjovski-Almeida S., Reis E.M.. Global analysis of biogenesis, stability and sub-cellular localization of lncRNAs mapping to intragenic regions of the human genome. RNA Biol. 2015; 12:877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al.. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cinghu S., Yang P., Kosak J.P., Conway A.E., Kumar D., Oldfield A.J., Adelman K., Jothi R.. Intragenic enhancers attenuate host gene expression. Mol. Cell. 2017; 68:104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kowalczyk M.S., Hughes J.R., Garrick D., Lynch M.D., Sharpe J.A., Sloane-Stanley J.A., McGowan S.J., De Gobbi M., Hosseini M., Vernimmen D. et al.. Intragenic enhancers act as alternative promoters. Mol. Cell. 2012; 45:447–458. [DOI] [PubMed] [Google Scholar]
- 55. Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V. et al.. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012; 488:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brown P.M., Fox K.R.. Nucleosome core particles inhibit DNA triple helix formation. Biochem. J. 1996; 319:607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brown P.M., Madden C.A., Fox K.R.. Triple-helix formation at different positions on nucleosomal DNA. Biochemistry. 1998; 37:16139–16151. [DOI] [PubMed] [Google Scholar]
- 58. Espinas M.L., Jimenez-Garcia E., Martinez-Balbas A., Azorin F.. Formation of triple-stranded DNA at d(GA.TC)n sequences prevents nucleosome assembly and is hindered by nucleosomes. J. Biol. Chem. 1996; 271:31807–31812. [DOI] [PubMed] [Google Scholar]
- 59. Westin L., Blomquist P., Milligan J.F., Wrange O.. Triple helix DNA alters nucleosomal histone-DNA interactions and acts as a nucleosome barrier. Nucleic Acids Res. 1995; 23:2184–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. O’Leary V.B., Smida J., Buske F.A., Carrascosa L.G., Azimzadeh O., Maugg D., Hain S., Tapio S., Heidenreich W., Kerr J. et al.. PARTICLE triplexes cluster in the tumor suppressor WWOX and may extend throughout the human genome. Sci. Rep. 2017; 7:7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Paugh S.W., Coss D.R., Bao J., Laudermilk L.T., Grace C.R., Ferreira A.M., Waddell M.B., Ridout G., Naeve D., Leuze M. et al.. MicroRNAs form triplexes with double stranded DNA at sequence-specific binding sites; a eukaryotic mechanism via which microRNAs could directly alter gene expression. PLoS Comput. Biol. 2016; 12:e1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chishima T., Iwakiri J., Hamada M.. Identification of transposable elements contributing to tissue-specific expression of long non-coding RNAs. Genes (Basel). 2018; 9:E23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lubelsky Y., Ulitsky I.. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature. 2018; 555:107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim H.S. Genomic impact, chromosomal distribution and transcriptional regulation of HERV elements. Mol. Cells. 2012; 33:539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Englander E.W., Howard B.H.. Nucleosome positioning by human Alu elements in chromatin. J. Biol. Chem. 1995; 270:10091–10096. [DOI] [PubMed] [Google Scholar]
- 66. Polak P., Domany E.. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006; 7:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. de los Santos C., Rosen M., Patel D.. NMR studies of DNA (R+)n.(Y-)n.(Y+)n triple helices in solution: imino and amino proton markers of T.A.T and C.G.C+ base-triple formation. Biochemistry. 1989; 28:7282–7289. [DOI] [PubMed] [Google Scholar]
- 68. Manzini G., Xodo L.E., Gasparotto D., Quadrifoglio F., van der Marel G.A., van Boom J.H.. Triple helix formation by oligopurine-oligopyrimidine DNA fragments. Electrophoretic and thermodynamic behavior. J. Mol. Biol. 1990; 213:833–843. [DOI] [PubMed] [Google Scholar]
- 69. Escude C., Francois J.C., Sun J.S., Ott G., Sprinzl M., Garestier T., Helene C.. Stability of triple helices containing RNA and DNA strands: experimental and molecular modeling studies. Nucleic Acids Res. 1993; 21:5547–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Semerad C.L., Maher L.J. 3rd. Exclusion of RNA strands from a purine motif triple helix. Nucleic Acids Res. 1994; 22:5321–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Skoog J.U., Maher L.J. 3rd. Repression of bacteriophage promoters by DNA and RNA oligonucleotides. Nucleic Acids Res. 1993; 21:2131–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maldonado R., Filarsky M., Grummt I., Langst G.. Purine- and pyrimidine-triple-helix-forming oligonucleotides recognize qualitatively different target sites at the ribosomal DNA locus. RNA. 2018; 24:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequence data and peak predictions have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO accession number GSE120850. ChromHMM and DNase-I hypersensitive site annotations were based on ENCODE data from HeLa S3 cells (accession number ENCSR497SKR and ENCSR959ZXU, respectively). CHART-seq DNA and R-loop regions were obtained from published data (7,40).