Abstract
Cystic fibrosis is a disease caused by defective function of a chloride channel coupled to a blockade of autophagic flux. It has been proposed to use synthetic chloride transporters as pharmacological agents to compensate insufficient chloride fluxes. Here, we report that such chloride anionophores block autophagic flux in spite of the fact that they activate the pro-autophagic transcription factor EB (TFEB) coupled to the inhibition of the autophagy-suppressive mTORC1 kinase activity. Two synthetic chloride transporters (SQ1 and SQ2) caused a partially TFEB-dependent relocation of the autophagic marker LC3 to the Golgi apparatus. Inhibition of TFEB activation using a calcium chelator or calcineurin inhibitors reduced the formation of LC3 puncta in cells, yet did not affect the cytotoxic action of SQ1 and SQ2 that could be observed after prolonged incubation. In conclusion, the squaramide-based synthetic chloride transporters studied in this work (which can also dissipate pH gradients) are probably not appropriate for the treatment of cystic fibrosis yet might be used for other indications such as cancer.
Introduction
Cystic fibrosis is the most frequent monogenetic lethal disease affecting humans1,2. This pathology is caused by loss-of-function mutation in the cystic fibrosis transmembrane conductance regulator (CFTR), a chloride transporter3,4. Importantly, CFTR mutations do not only compromise chloride flux (resulting in increased chloride concentrations in sweat and alterations in the composition of mucus with subsequent bronchial infections) but also affect autophagic flux in respiratory epithelial cells and macrophages5–10. The perturbation of autophagy may be disease-relevant because autophagy induction by pharmacological inhibitors of E1A-associated protein p300 (where E1A = adenovirus early region 1A), best known as EP300, and transglutaminase-2 can favor the function of certain CFTR mutants (and in particular the del508 mutant protein)11–14 and reduce lung inflammation in patients15–17.
Intrigued by these observations, we wondered whether direct perturbations of chloride homeostasis by means of synthetic chloride transporters (or anionophores) such as the squaramide-based compound SQ1 and the analogous SQ218,19 might affect the autophagic process. SQ1 and SQ2 incorporate into the plasma membrane (and perhaps other cellular membranes), thus causing chloride influx into cells and dissipating chloride gradients in intracellular compartments, and neutralizing lysosomal pH in the process19. Although this ultimately causes cell death (and SQ1 has indeed been developed as a first-in-class anticancer agent), SQ1 and SQ2 can modulate autophagy before cells die19, offering a window of opportunity for investigating their impact on the system.
Here, we report that squaramide-based synthetic chloride transporters dramatically inhibit autophagic flux although they activate a pro-autophagic transcription factor. Moreover, these chloride anionophores trigger the relocation of an autophagic marker (LC3) towards the Golgi apparatus.
Materials and methods
Cell culture and transfection
Human Osteosarcoma U2OS cells were cultured at 37 °C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) supplemented with 100 mM 2-[4-(2-hydroxyethyl) piperazin-1-yl] ethanesulfonic acid (HEPES) buffer, 10% heat-inactivated fetal bovine serum (FBS) (Life Technologies) and 1% penicillin/streptomycin (Life Technologies).
GFP-LC3 stable cell lines were generated by transducing U2OS WT, U2OS ATG5−/−, TFEB−/−, CFBE Delta-F508 with pre-packaged viral particles expressing recombinant GFP-LC3 (LentiBrite GFP-LC3B Lentiviral Biosensor; Millipore, 17–10193). U2OS cells stably expressing GFP-TFEB were transfected with the pEGFP-N1-TFEB plasmid using the FuGENE® HD transfection reagent protocol. The pEGFP-N1-TFEB plasmid was a gift from Shawn Ferguson (Addgene plasmid # 38119). Subsequently, stable expressing cells were selected by means of appropriate selection antibiotics and clones were obtained by single cell sorting using a FACS DIVA (Becton Dickinson, Franklin Lakes, NJ, USA). U2OS GALT-GFP LC3-RFP cells were constructed with the protocol as published in our pervious paper20 with a plasmid coding for GALT1(β1,4-galactosyltransferase 1)-GFP, and then transfected with lentiviral particles coding for RFP-LC3 from Merck Millipore(17–10143). PC12 GFP-Q74 cells are generous gift from David Rubinsztein’s lab from Cambridge21.
Chemicals and antibodies
The squaramide compound SQ1, its analogue SQ2, and the control compound SQ3 (with no chloride transport activity) were previously described18,19. The following chemicals and antibodies were used in this study: Torin 1, bafilomycin A1, BAPTA AM, Cyclosporin A (Tocris Bioscience, Bristol, UK), Cycloheximide, Cyclosporin H, N-Acetyl-L-cysteine, L-Glutathione reduced (Sigma-Aldrich) oleate from Larodan (Solna, Sweden).
Anti-beta actin (Abcam 8226, Cambridge, UK), LC3 (#3868), mTOR (#2983), Phospho-mTOR (Ser2448) (#5536), LAMP1 (#9091), p70 S6 Kinase (#9202), Phospho-p70 S6 Kinase (Thr389) (#9205), SQSTM1/p62 (#7695) antibody were all purchased from Cell Signaling Technology, IF antibody Alexa Fluor® 647 anti-human CD107a (LAMP-1) Antibody from BioLegend.
Cytofluorometry measurement of cell death
Cell viability was evaluated by co-staining the cells, during 30 min at 37 °C, with 40 nM 3,3′dihexiloxalocarbocyanine iodide (DiOC6(3)), Molecular Probes/Invitrogen), a mitochondrial transmembrane potential sensitive dye, and 2 μM DAPI (all from Molecular Probes-Life Technologies, Carlsbad, CA, USA). Cytofluorometric acquisitions were carried out on a Milteny cytofluorometer (MACSQuant® Analyzer 10), and statistical analyses were performed by using the FlowJo software (LLC, Oregon, USA).
Cytofluorometry measurement of ROS generation
The generation of ROS was monitored with hydroethidine (HE) (at a final concentration of 5 μM; stock 10 mM in DMSO; excitation wave length of 488 nM, emission 620 nM; Molecular Probe. Briefly, cells were collected after treatment with the testing compounds followed by co-staining, during 30 min at 37 °C, with HE. Cytofluorometric acquisitions were carried out on a Milteny cytofluorometer (MACSQuant® Analyzer 10), and statistical analyses were performed by using the FlowJo software (LLC, Oregon, USA).
High content image acquisition
Cells were seeded in tissue culture-treated 384-well μClear imaging plates (Greiner BioOne, Frickenhausen, Germany) and incubated under standard tissue culture conditions during 24 h at 37 °C. Then, cells were treated with the indicated compounds and after 6 or 24 h of incubation, cells were fixed with 4% formaldehyde solution containing 1 μM Hoechst 33342 overnight at 4 °C. The fixative was changed to PBS, and the plates were subjected to automated image analysis. For automated fluorescence microscopy, a robot-assisted Molecular Devices IXM XL BioImager (Molecular Devices, Sunnyvale, CA, USA) equipped with Sola light sources (Lumencor, Beaverton, OR, USA), adequate excitation and emission filters (Semrock, Rochester, NY, USA), and a 16-bit monochromes sCMOS PCO.edge 5.5 camera (PCO, Kelheim, Germany) and a ×20 PlanAPO objective (Nikon, Tokyo, Japan) was used to acquire nine view fields/well, followed by image processing with the custom module editor of the MetaXpress software (Molecular Devices). For the latter, the images were segmented and analyzed for GFP, RFP granularity or global fluorescence intensity (depending on the dyes) by comparing the standard deviation of the mean fluorescence intensity of groups of adjacent pixels within the cytoplasm of each cell to the mean fluorescence intensity in the same ROI using the MetaXpress software (Molecular Devices).
Immunoblotting
Immunoblotting was performed following standard procedures. Cells were harvested and the obtained pellet was resuspended in RIPA buffer (89900; Thermo Fisher Scientific) supplemented with phosphatase and protease inhibitors (88669; Thermo Fisher Scientific) followed by sonication and protein content quantification by DCTM Protein Assay kit (5000112; Bio-Rad, Hercules, CA, USA). Then, 10 μg of protein were separated on NuPAGE Novex Bis-Tris 4–12% pre-cast gels (Invitrogen-Life Technologies, Carlsbad, CA, USA) and transferred to Immobilon polyvinylidene difluoride membranes (Merck-Millipore, Darmstadt, Germany). Unspecific binding was reduced by incubating the membranes for 1 h in 0.05% Tween 20 (v/v in TBS) supplemented with 5% w/v bovine serum albumin (Euromedex, Souffelweyersheim, France). Following, proteins were probed with antibodies specific for actin, LC3, mTOR, Phospho-mTOR (Ser2448), LAMP1, p70 S6 Kinase or SQSTM1/p62. Primary antibodies were revealed with species-specific immunoglobulin G conjugated to horseradish peroxidase (Southern Biotech, Birmingham, AL, USA), followed by chemiluminescence analysis with the SuperSignal West Pico reagent by means of an ImageQuant 4000 (GE Healthcare, Little Chalfont, UK).
Statistical analyses
Data are reported as means ± SD of n > 3 replicates and experiments were repeated at least twice yielding similar results. Data were analyzed using Prism (GraphPad Software, Inc., La Jolla, CA, USA), and statistical significance was assessed by means of two-tailed Student’s t-test or ANOVA tests, as appropriate. Unless otherwise specified, data are reported as mean ± SEM. Statistical significance was analyzed using the Student’s-test. Differences in treated and control cells were considered to be significant if *p < 0.05, **p < 0.01, ***p < 0.001.
Results
SQ1 and SQ2 induce autophagic LC3+ puncta in U2OS cells
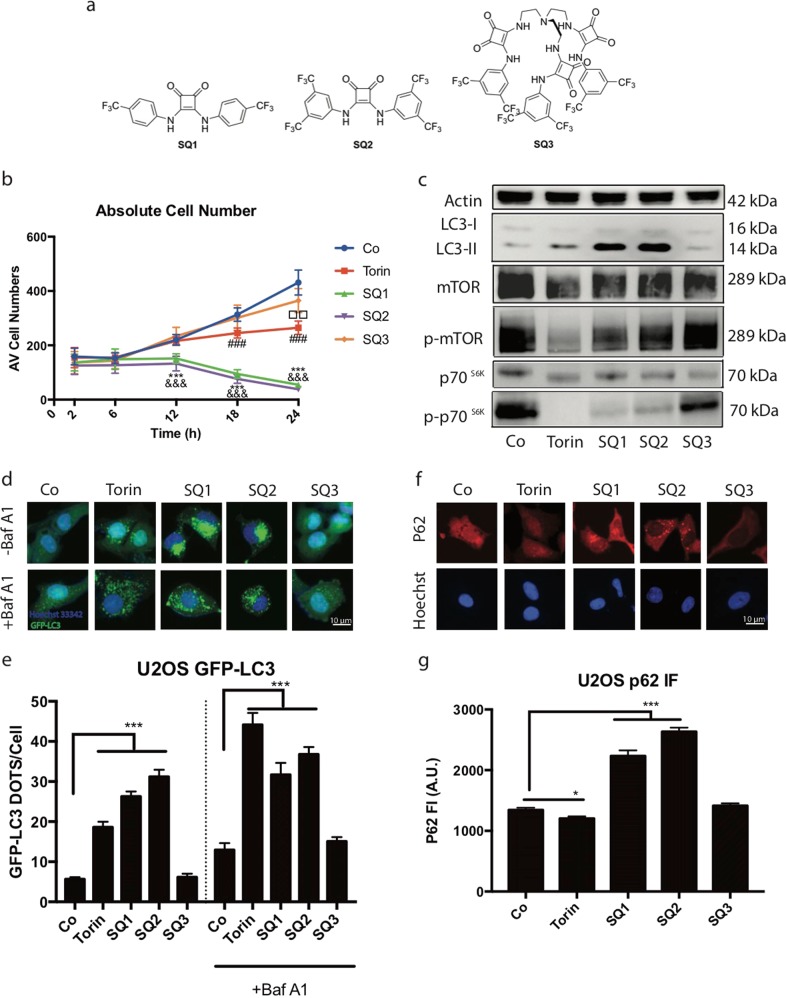
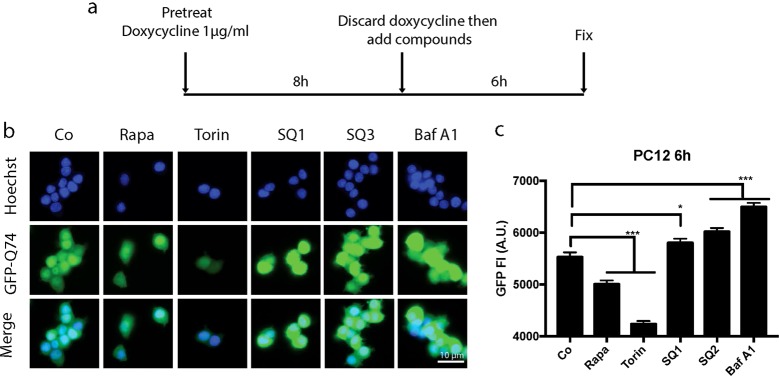
Our laboratory has made extensive use of U2OS biosensor cell lines to measure cellular stress responses including autophagy12,22–28. We first determined the kinetics of potential cytotoxic effects of the synthetic chloride channels SQ1 and SQ2 on these cells to study autophagy in conditions in which U2OS cells fully conserve their viability. These measurements led to the conclusion that the SQ1 and SQ2 have cytostatic effects that become significant at 12 h of incubation and diminishes the number of cells below the control value, indicating cytotoxicity, from 18 h. However, at 6 h neither cytostatic nor cytotoxic effects were detectable (Fig. 1b). Hence, all subsequent experiments were done in this time frame (6 h). SQ1 and SQ2 both induced the lipidation of microtubule-associated proteins 1A/1B light chain 3B (hereafter referred to as LC3 II), as detectable by an increase in the electrophoretic mobility of LC3 (Fig. 1c). Moreover, both synthetic chloride anionophores (but not the control compound SQ3, which shares structural features with SQ2, yet lacks chloride transporter activity19) inhibited the kinase activity of mechanistic target of rapamycin complex-1 (mTORC1), as indicated by a reduced phosphorylation of ribosomal protein S6 kinase beta-1 (S6K1, best known as p70S6K) (Fig. 1c). Commensurate with these effects, SQ1 and SQ2 (but not SQ3) stimulated the aggregation of green fluorescent protein (GFP)-LC3 fusion protein in cytoplasmic dots (Fig. 1d). This effect was found both in the absence and in the presence of bafilomycin A1 (BafA1), an inhibitor of the lysosomal vATPase (Fig. 1e). However, SQ1 and SQ2 did not induce autophagic flux, as indicated by two independent series of experiments. First, in the presence of SQ1 and SQ2, the abundance of the autophagic substrate sequestosome 1 (STQM1, best known as p62) in U2OS cells did not decrease (as this was observed with the positive control, torin, an inhibitor of mTORC1 and mTORC2) but increased (Fig. 1f, g). This increase probably reflects the impairment of lysosomal function due to the decrease of the activity of the pH-sensitive hydrolases. This loss of lysosomal function is due to abnormal pH increase in lysosomes when intracellular chloride concentration is dysregulated. Second, in the neuronal PC12 cell line engineered to express a doxycyclin-inducible autophagic cargo (namely a GFP fused to exon 1 of a pathogenic huntingtin protein variant that contains 74 glutamine repeats, Q74)29, SQ1 and SQ2 increased the level of the cargo, contrasting with the effects of torin or rapamycin that both caused its elimination (Fig. 2).
Fig. 1. Chemical structure of SQ1 and SQ2 and their effect on autophagic parameters.
a Structure of squaramide-based synthetic chloride transporters SQ1, SQ2, and SQ3. b Kinetic studies of Torin 300 nM, SQ1, SQ2, and SQ3 at 10 μM on U2OS human osteosarcoma cells from 2–24 h. Cell numbers were determined by means of fluorescence microscopy upon Hoechst staining. The graph depicts the average cell number for each treatment per site of acquisition in a 384 well plate. c Immunoblot analysis of LC3 lipidation, protein expression level of mTOR, p70S6K and their phosphorylated forms(Thr389) after 6-hour treatments. d, e Representative images and statistical analysis of U2OS GFP-LC3 cells after treatment with Torin 300 nM or SQ1, SQ2, and SQ3 at 10 μM during 6 h having bafilomycin A1(100 nM) presented in the last 2 h of treatment. f, g Representative images and statistical analysis of U2OS submitted to p62 immunofluorescence staining after 6 h of treatment with torin (300 nM) or SQ1, SQ2, and SQ3 (10 μM). The bar chart indicated the global fluorescence intensity of p62 per cell, followed by p62 immunofluorescence staining. Data are expressed as means ± SEM of at least three independent experiments (***/&&&/###p < 0.0001, compared to untreated cells, Co)
Fig. 2. Effects of SQ1 and SQ2 on autophagic flux.
a Scheme of the design of the experiment. b Representative images of PC12 GFP-Q74 cell after treatment with rapamycin (Rapa, 10 μM), torin 300 nM and SQ1, SQ2, SQ3 respectively at 10 μM for 6 h. c Data are expressed as means ± SD of one representative experiment and represent the global GFP fluorescent intensity per cell. (*p < 0.05, ***p < 0.0001, compared to untreated cells, Co)
In sum, SQ1 and SQ2 stimulate LC3 lipidation, its redistribution towards puncta, yet do not stimulate autophagic flux, confirming previous observations made in HeLa cells19.
Golgi localization of LC3 after SQ1/SQ2 treatment
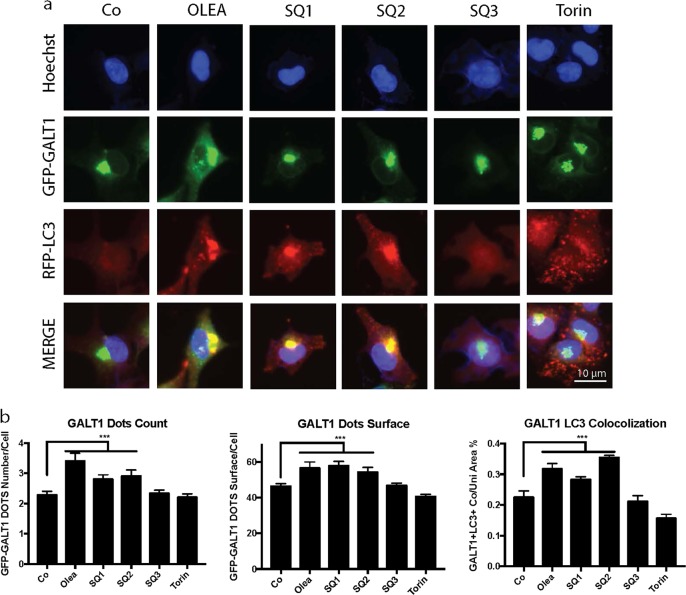
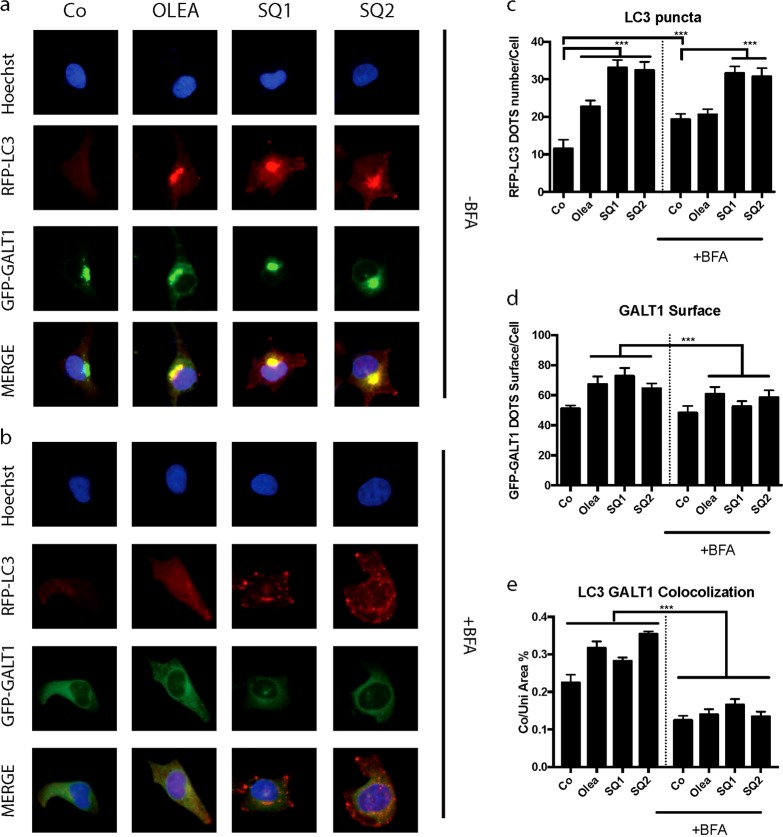
The GFP-LC3 puncta induced by SQ1 or SQ2 were larger than those induced by torin and tended to coalesce at one pole of the nucleus (Fig. 1). We therefore suspected that such puncta might aggregate at or close to the Golgi apparatus. To examine this hypothesis, we took advantage of a U2OS biosensor cell line expressing a red fluorescent protein (RFP)-LC3 fusion protein as a reporter of LC3 localization, as well as a GFP-galactose-1-phosphate uridylyltransferase (GALT) fusion protein as a reporter of Golgi location20,22,24,30,31. After treatment with SQ1 or SQ2, we found a remarkable overlap of both fluorescent signals indicating the translocation of LC3 to the Golgi, a phenotype that closely resembled the one induced by oleate20,25 but differed from that induced by torin (Fig. 3). To further investigate this phenomenon, we pretreated the cells for 1 h with brefeldin A (BFA), an inhibitor of GBF1 (brefeldin A-resistance guanine nucleotide exchange factor 1) that disrupts the Golgi apparatus causing its dispersion into ministacks all over the cytoplasm32–34. Preincubation with BFA reduced the colocalization of GFP-GALT and RFP-LC3, confirming the idea that LC3 coalesces at the Golgi, yet did not prevent the SQ1/SQ2-induced formation of RFP-LC3 dots, suggesting that the Golgi is not required for this phenomenon to occur (Fig. 4).
Fig. 3. SQ1and SQ2 induced Golgi localization of LC3.
Representative images and statistical analysis of U2OS cells expressing GFP-GALT RFP-LC3 cells treated with torin (300 nM), oleate (500 μM), SQ1, SQ2 or SQ3 (10 μM) for 6 h. Quantitative analysis represents the number and area of GALT1+ Golgi structures per cell and the colocalization of LC3+ and GALT1+ structures. Data are expressed as means ± SD of one representative experiment (***p < 0.0001, compared to untreated cells, Co). Representatie images are showin in panel a and quantitative comparisions are detailled in panel b
Fig. 4. Brefeldin A reduced SQ1 and SQ2 induced translocation of LC3 to the Golgi.
a, b Representative images of U2OS GFP-GALT RFP-LC3 cells treated with the compounds for 6 h (oleate 500 μM, SQ1, SQ2 at 10 μM) without (a) or after pretreatment with brefeldinA (10 μg/ml) for 1 h (b). c–e Statistical analysis of RFP-LC3 puncta counts per cell, GFP-GALT marked Golgi surface per cell and the localization of LC3 and GALT (***p < 0.0001, compared to untreated control cells, Co)
Contribution of TFEB to LC3 relocation
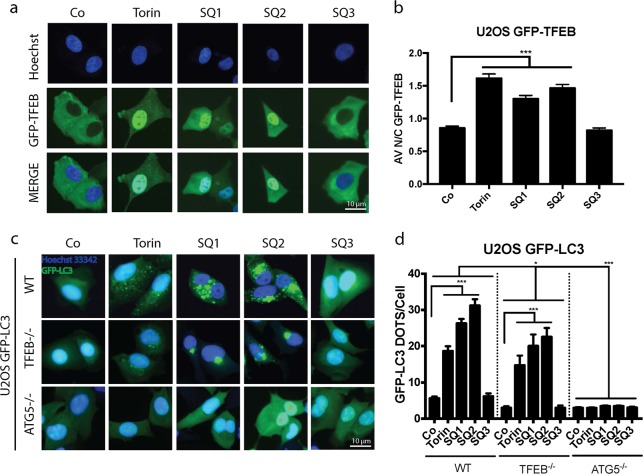
Since SQ1 and SQ2 inhibit mTORC1, and this inhibition has been involved in the activation of the pro-autophagic transcription factor EB (TFEB)35–39, we investigated whether these synthetic chloride transporters would cause the translocation of a GFP-TFEB reporter protein from the cytoplasm to the nucleus. Indeed, SQ1 and SQ2 (but not SQ3) were able to stimulate TFEB activation (Fig. 5a, b). TFEB is known to stimulate lysosomal biogenesis, and both SQ1 and SQ2 caused the overexpression of lysosomal-associated membrane protein 1 (LAMP-1) in U2OS cells (Fig. S1). Moreover, knockout of TFEB partially reduced the formation of LC3B puncta induced by SQ1 or SQ2, an inhibition that was far less prominent than that found for the knockout of ATG5 (Fig. 5c, d). Quantitation of GFP-LC3 puncta that co-stain with an antibody recognizing LAMP1 revealed that SQ1 and SQ2 did not inhibit the fusion between lysosomes and autophagosomes (Fig. S3e). These observations suggest that SQ compounds inhibit autophagic flux at a step downstream of the autophagosome-lysosome fusion, presumably because SQ1 and SQ2 dissipate the (normally acidic) pH gradient on the lysosomal membrane19.
Fig. 5. Contribution of TFEB and ATG5 in LC3 relocation induced by the SQs.
a, b Representative images of U2OS cells stably expressing GFP-TFEB fusion protein, treated with torin 300 nM, SQ1, SQ2, and SQ3 at 10 μM during 6 h. Data are expressed as means ± SEM of at least three independent experiments and demonstrate the average ratio between GFP-TFEB florescence intensity in the nucleus vs. cytoplasm. (*p < 0.05, **p < 0.01, ***p < 0.0001, compared to untreated cells, Co). c, d Representative images and statistics of U2OS GFP-LC3 cells, and the ATG5 KO and TFEB KO counterparts after 6 h of treatment with the mentioned compounds. Bars depicts the absolute number of GFP-LC3+ dots per cell. Data are expressed as means ± SEM of at least three independent experiments (*p < 0.05, ***p < 0.0001, compared to untreated cells, Co)
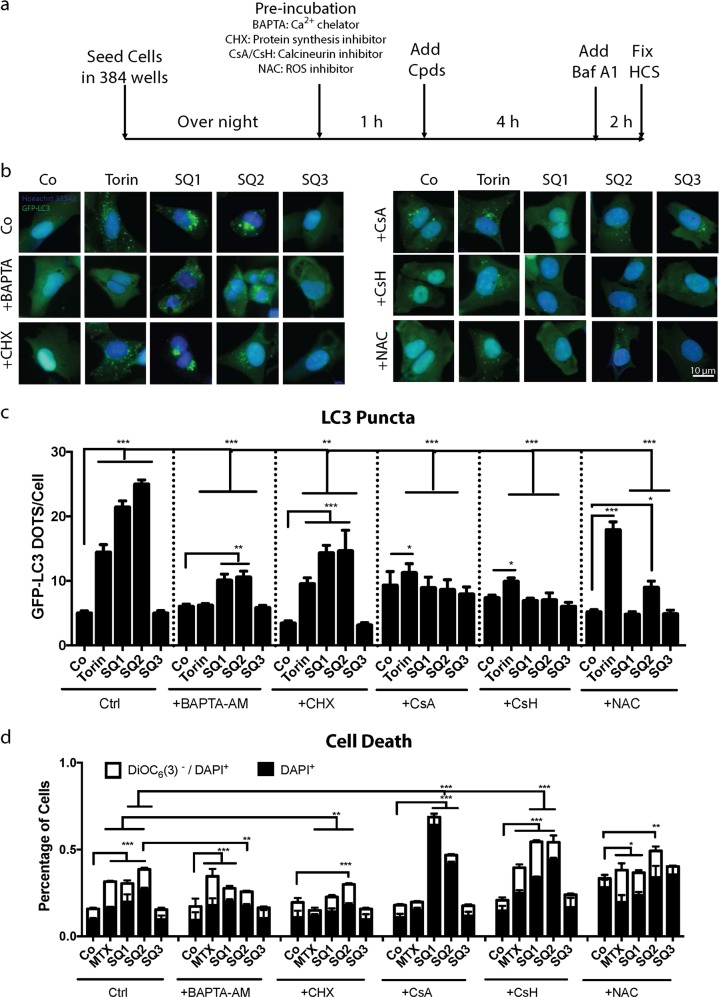
Intrigued by these observations, we decided to investigate the role of TFEB in the induction of LC3B puncta by inhibiting calcineurin, the phosphatase that dephosphorylates TFEB for its activation after mTORC1 inhibition35,40,41. For this, we either used the calcium chelator 2,2′-(ethylenedioxy)dianiline-N,N,N,N-tetraacetic acid (BAPTA) that can be targeted into cells a cell-permeant acetoxymethyl ester derivative (BAPTA-AM) or, more specifically, enzymatic inhibitors of calcineurin such as cyclosporine A (CsA) and its non-immunosuppressive control compound cyclosporin H (CsH). Moreover, we used cycloheximide (CHX) as a general inhibitor of protein translation. Cells were preincubated for 1 h with these reagents and then were treated with SQ1 or SQ2. Of note, BAPTA-AM, CsA, and CsH strongly inhibited the formation of GFP-LC3 puncta (Fig. 6b, c). CsA and CsH failed to inhibit the formation of reactive oxygen species (ROS) induced by SQ1 and SQ2 (as measurable by quantifying the conversion of non-fluorescent hydroethidine into fluorescent ethidium), which however could be quenched by using the antioxidant N-acetyl cysteine (NAC) (Fig. S2). NAC was also capable of inhibiting LC3 puncta formation (Fig. 6b, c). Importantly, none of the inhibitors of GFP-LC3 puncta (i.e., BAPTA-AM, CHX, CsA, CsH, NAC) nor full inhibition of the autophagic process (in ATG5 KO cells) (Fig. S3C) were able to significantly reduce cell killing by SQ1 or SQ2, as measured at 24 h of incubation. Hence, the formation of GFP-LC3 puncta and later cell death can be uncoupled from each other.
Fig. 6. Effects of pharmacological inhibition of TFEB on LC3 relocation and cell death induced by SQ1 and SQ2.
a Experimental design to assess LC3 puncta when the synthetic chloride transporters SQs were combined with different inhibitors. b Representative images of U2OS GFP-LC3 expressing cells when pretreated with the calcium chelator BAPTA-AM, the antioxydant NAC, the calcineurin inhibitor CsA, its analogue CsH, and the protein synthesis inhibitor CHX followed by the co-treatment with the synthetic chloride modulators. c Bars show the average number of GFP-LC3+ dots per cell. Data are expressed as means ± SD of one representative experiment *P < 0.05; **P < 0.01; ***P < 0.001. d Evaluation of cell death by double-staining with DAPI together with DiOC6(3) followed by cytofluorometric detection of dead cells (DAPI+) and dying cells (DAPI+ DiOC6(3)low). Data are expressed as means ± SEM of at least three independent experiments (Co/Ctrl). *P < 0.05; **P < 0.01; ***P < 0.001
Furthermore, to evaluate the possibility that CsA and TFEB act on different steps of the cellular alterations induced by SQ1 and SQ2, we compared the SQ1 and SQ2 effects on the formation of GFP-LC3 dots between U2OS WT and U2OS TFEB KO cells in the presence or absence CsA. The CsA-mediated inhibition SQ1 and SQ2-elicited GFP-LC3 dots was maintained in TFEB KO cells. This argues in favor of the possibility that TFEB and CsA modulate the effects of SQ1 and SQ2 in a differential fashion (Fig. S3a, b).
Finally, we treated a cystic fibrosis bronchial epithelial (CFBE) cell line harboring the most frequent CFTR mutation (delF508) for 24 h with SQ1 or SQ2 in the absence or presence of bafilomycin A1. The results indicate that SQ1 or SQ2 induced the formation of GFP-LC3 dots and that this effect was not further enhanced by bafilomycin A1 (Fig. S3F), confirming that SQ1 or SQ2 inhibit autophagic flux in a disease-relevant cellular model.
Discussion
SQ1 and SQ2 have previously been shown to stimulate the lipidation of LC319. Here, we confirm this finding and link it to the relocation of LC3 to discrete structure in the cytoplasm, which occurs in a strictly ATG5-dependent fashion. We found that SQ1 and SQ2 inhibit mTORC1, a phenomenon that is linked to the activation of TFEB (and other similar transcripition factors)19. Indeed, SQ1 and SQ2 promote the translocation of TFEB from the cytoplasm to the nucleus, and TFEB is partially required for the aggregation of LC3 in discrete cytoplasmic puncta. Although TFEB activation should stimulate autophagic flux, we found that SQ1 and SQ2 were unable to do so. Rather both SQ1 and SQ2 caused the accumulation of autophagic cargo in two different cell types, in human osteosarcoma U2OS cells (in which SQSTM1 became more abundant) and in rat neuronal PC12 cells (in which a mutant Huntingtin Q74 construct accumulated). SQ1 and SQ2 are synthetic chloride transporters that dissipate the lysosomal pH gradient, a phenomenon that previously has been suggested to account for the inhibition of autophagic flux19. Thus, although both agents activate a pro-autophagic transcription factor they ultimately fail to stimulate autophagic flux.
Several strategies designed to inhibit the calcineurin-mediated activation of TFEB, namely intracellular calcium chelation or direct calcineurin inhibition by CsA largely prevented autophagy induction by SQ1 or SQ2, confirming the importance of this pathway for the observed phenotype. However, inhibition of protein neo-synthesis or the calcineurin pathway was unable to prevent SQ1 and SQ2-induced cell death in conditions in which such an inhibition fully suppressed the formation of LC3 puncta. We conclude from these results that the generation of such puncta apparently is not required for the cytotoxic action of synthetic chloride transporters, which must rely on other damage pathways to kill cells.
Surprisingly, SQ1 and SQ2 caused the relocation of LC3 into discrete perinuclear structures, usually presenting as one single cap in immediate vicinity of the nucleus. These structures turned out to co-localize with the Golgi apparatus. We previously found a similar Golgi-specific coalescence of LC3 in cells treated with unsaturated fatty acids (such as oleate)20,42, the photosensitizer redaporfin22,43, and the cytolytic peptide derivative LTX-401, which itself has a strong tropism for the Golgi25. Thus, structurally rather distinct compounds can induce a similar coalescence of LC3 in or at the Golgi apparatus. For all these compounds (SQ1, SQ2, oleate, redaporfin, LTX401), the Golgi-disrupting agent BFA prevented the relocalization of LC3 to perinuclear caps. The mechanisms explaining this organelle-specific pattern of LC3 redistribution remain to be elucidated.
Synthetic chloride transporters have been developed as hypothetical remedies against cystic fibrosis44–49. Indeed, at least theoretically, such ionophores might be used as substitutes to compensate for the defective function of CFTR44,50. As shown in this paper, consistent with precedent reports19,51, chloride ionophores that can dissipate pH gradients, such as the compounds studied here that have been shown to be capable of transporting both chloride and protons across lipid bilayers, block autophagic flux in a peculiar manner, arguing against their therapeutic utility. Indeed, it appears to be advantageous to restore autophagic turnover in cystic fibrosis12,15,17, implying that autophagy inhibition would ultimately worsen the disease. These types of transporter should be exploited for therapeutic use against other diseases, for instance cancer, in which their cytotoxic potential might be advantageous. The development of chloride transporters which do not dissipate pH gradients is ongoing in Sydney52.
Supplementary information
Acknowledgements
S.Z., Y.W., W.X. are supported by the China Scholarship Council. G.K. is supported by the Ligue contre le Cancer (équipes labelisées); Agence National de la Recherche (ANR)— Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de- France; Institut National du Cancer (INCa); Fondation Bettencourt-Schueller; Fondation de France; Fondation pour la Recherche Médicale (FRM); the European Commission (ArtForce); the LabEx Immuno-Oncology; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); and the SIRIC Cancer Research and Personalized Medicine (CARPEM). P.A.G. is supported by the Australian Research Council (DP180100612).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Piacentini
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shaoyi Zhang, Yan Wang, Wei Xie
These authors jointly supervised this work: Naoufal Zamzami, Guido Kroemer
Change history
4/3/2019
In the version of this article originally submitted, it was stated that the first three authors (Shaoyi_ Than, Yan Wang, Wei Xie) had contributed equally. However, in the published version this information was missing.
Contributor Information
Naoufal Zamzami, Email: zamzami66@gmail.com.
Guido Kroemer, Email: kroemer@orange.fr.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-1474-8).
References
- 1.Southern KW, et al. A survey of newborn screening for cystic fibrosis in Europe. J. Cyst. Fibros. 2007;6:57–65. doi: 10.1016/j.jcf.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Buzzetti R, et al. An overview of international literature from cystic fibrosis registries: 1. Mortality and survival studies in cystic fibrosis. J. Cyst. Fibros. 2009;8:229–237. doi: 10.1016/j.jcf.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Stoltz DA, Meyerholz DK, Welsh MJ. Origins of cystic fibrosis lung disease. New Engl. J. Med. 2015;372:351–362. doi: 10.1056/NEJMra1300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riordan JR, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 5.Fahy JV, Dickey BF. Airway mucus function and dysfunction. New Engl. J. Med. 2010;363:2233–2247. doi: 10.1056/NEJMra0910061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoegger MJ, et al. Impaired mucus detachment disrupts mucociliary transport in a piglet model of cystic fibrosis. Science. 2014;345:818–822. doi: 10.1126/science.1255825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quinton PM. Cystic fibrosis: impaired bicarbonate secretion and mucoviscidosis. Lancet. 2008;372:415–417. doi: 10.1016/S0140-6736(08)61162-9. [DOI] [PubMed] [Google Scholar]
- 8.Bruscia EM, et al. Macrophages directly contribute to the exaggerated inflammatory response in cystic fibrosis transmembrane conductance regulator−/− mice. Am. J. Respir. Cell Mol. Biol. 2009;40:295–304. doi: 10.1165/rcmb.2008-0170OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villella, V. R. et al. A pathogenic role for cystic fibrosis transmembrane conductance regulator in celiac disease. EMBO J., 38, pii: e100101 (2019). [DOI] [PMC free article] [PubMed]
- 10.Maiuri L, Kroemer G. Autophagy delays progression of the two most frequent human monogenetic lethal diseases: cystic fibrosis and Wilson disease. Aging. 2018;10:3657–3661. doi: 10.18632/aging.101736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mariño G, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol. Cell. 2014;53:710–725. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, et al. Evaluation of autophagy inducers in epithelial cells carrying the ΔF508 mutation of the cystic fibrosis transmembrane conductance regulator CFTR. Cell Death Dis. 2018;9:191. doi: 10.1038/s41419-017-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izzo V, et al. Metabolic interactions between cysteamine and epigallocatechin gallate. Cell Cycle. 2017;16:271–279. doi: 10.1080/15384101.2016.1249550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luciani A, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat. Cell Biol. 2010;12:863. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- 15.Stefano DD, et al. Restoration of CFTR function in patients with cystic fibrosis carrying the F508del-CFTR mutation. Autophagy. 2014;10:2053–2074. doi: 10.4161/15548627.2014.973737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luciani A, et al. Targeting autophagy as a novel strategy for facilitating the therapeutic action of potentiators on ΔF508 cystic fibrosis transmembrane conductance regulator. Autophagy. 2012;8:1657–1672. doi: 10.4161/auto.21483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tosco A, et al. A novel treatment of cystic fibrosis acting on-target: cysteamine plus epigallocatechin gallate for the autophagy-dependent rescue of class II-mutated CFTR. Cell Death Differ. 2016;23:1380. doi: 10.1038/cdd.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marques I, et al. Full elucidation of the transmembrane anion transport mechanism of squaramides using in silico investigations. Phys. Chem. Chem. Phys. 2018;20:20796–20811. doi: 10.1039/C8CP02576B. [DOI] [PubMed] [Google Scholar]
- 19.Busschaert N, et al. A synthetic ion transporter that disrupts autophagy and induces apoptosis by perturbing cellular chloride concentrations. Nat. Chem. 2017;9:667. doi: 10.1038/nchem.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niso‐Santano M, et al. Unsaturated fatty acids induce non‐canonical autophagy. EMBO J. 2015;34:1025–1041. doi: 10.15252/embj.201489363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyttenbach A, et al. Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington’s disease. Hum. Mol. Genet. 2001;10:1829–1845. doi: 10.1093/hmg/10.17.1829. [DOI] [PubMed] [Google Scholar]
- 22.Gomes‐da‐Silva LC, et al. Photodynamic therapy with redaporfin targets the endoplasmic reticulum and Golgi apparatus. EMBO J. 2018;37:e98354. doi: 10.15252/embj.201798354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietrocola F, et al. Aspirin recapitulates features of caloric restriction. Cell Rep. 2018;22:2395–2407. doi: 10.1016/j.celrep.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauvat A, et al. Trans-fats inhibit autophagy induced by saturated fatty acids. EBioMedicine. 2018;30:261–272. doi: 10.1016/j.ebiom.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H, et al. The oncolytic compound LTX-401 targets the Golgi apparatus. Cell Death Differ. 2018;25:227. doi: 10.1038/cdd.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P, et al. Identification of pharmacological agents that induce HMGB1 release. Sci. Rep. 2017;7:14915. doi: 10.1038/s41598-017-14848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morselli E, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 2011;192:615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen S, et al. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol. Cell. 2012;48:667–680. doi: 10.1016/j.molcel.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 30.Zhou H, et al. The oncolytic compound LTX-401 targets the Golgi apparatus. Cell Death Differ. 2016;23:2031. doi: 10.1038/cdd.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes-da-Silva, L. C. et al. Recruitment of LC3 to damaged Golgi apparatus. Cell Death Differ. (2018) [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 32.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donaldson JG, Finazzi D, Klausner RD. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 34.Helms JB, Rothman JE. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- 35.Medina DL, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015;17:288. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Settembre C, et al. A lysosome‐to‐nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roczniak-Ferguson A, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 2012;5:ra42–ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong Y, Song F. Intracellular calcium signaling regulates autophagy via calcineurin-mediated TFEB dephosphorylation. Autophagy. 2015;11:1192–1195. doi: 10.1080/15548627.2015.1054594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao Z, Klionsky DJ. The symphony of autophagy and calcium signaling. Autophagy. 2015;11:973. doi: 10.1080/15548627.2015.1058475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grose C, Klionsky D. Alternative autophagy, brefeldin A and viral trafficking pathways. Autophagy. 2016;12:1429. doi: 10.1080/15548627.2016.1203489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomes-da-Silva LC, Zhao L, Arnaut LG, Kroemer G, Kepp O. Redaporfin induces immunogenic cell death by selective destruction of the endoplasmic reticulum and the Golgi apparatus. Oncotarget. 2018;9:31169. doi: 10.18632/oncotarget.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. Efficient, non-toxic anion transport by synthetic carriers in cells and epithelia. Nat. Chem. 2016;8:24–32. doi: 10.1038/nchem.2384. [DOI] [PubMed] [Google Scholar]
- 45.Shen B, Li X, Wang F, Yao X, Yang D. A synthetic chloride channel restores chloride conductance in human cystic fibrosis epithelial cells. PLoS ONE. 2012;7:e34694. doi: 10.1371/journal.pone.0034694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mall MA, Galietta LJ. Targeting ion channels in cystic fibrosis. J. Cyst. Fibros. 2015;14:561–570. doi: 10.1016/j.jcf.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Jiang C, et al. Partial correction of defective Cl− secretion in cystic fibrosis epithelial cells by an analog of squalamine. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2001;281:L1164–L1172. doi: 10.1152/ajplung.2001.281.5.L1164. [DOI] [PubMed] [Google Scholar]
- 48.Seganish, J. L. & Davis, J. T. Prodigiosin is a chloride carrier that can function as an anion exchanger. Chem. Commun., 14, 5781–5783 (2005). [DOI] [PubMed]
- 49.Saha T, Hossain MS, Saha D, Lahiri M, Talukdar P. Chloride-mediated apoptosis-inducing activity of bis (sulfonamide) anionophores. J. Am. Chem. Soc. 2016;138:7558–7567. doi: 10.1021/jacs.6b01723. [DOI] [PubMed] [Google Scholar]
- 50.Hernando E, et al. Small molecule anionophores promote transmembrane anion permeation matching CFTR activity. Sci. Rep. 2018;8:2608. doi: 10.1038/s41598-018-20708-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodilla AM, et al. Synthetic tambjamine analogues induce mitochondrial swelling and lysosomal dysfunction leading to autophagy blockade and necrotic cell death in lung cancer. Biochem. Pharmacol. 2017;126:23–33. doi: 10.1016/j.bcp.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 52.Wu X, et al. Nonprotonophoric electrogenic Cl− transport mediated by valinomycin-like carriers. Chem. 2016;1:127–146. doi: 10.1016/j.chempr.2016.04.002. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.