Abstract
Metagenomes from uncultured microorganisms are rich resources for novel enzyme genes. The methods used to screen the metagenomic libraries fall into two categories, which are based on sequence or function of the enzymes. The sequence-based approaches rely on the known sequences of the target gene families. In contrast, the function-based approaches do not involve the incorporation of metagenomic sequencing data and, therefore, may lead to the discovery of novel gene sequences with desired functions. In this review, we discuss the function-based screening strategies that have been used in the identification of enzymes from metagenomes. Because of its simplicity, agar plate screening is most commonly used in the identification of novel enzymes with diverse functions. Other screening methods with higher sensitivity are also employed, such as microtiter plate screening. Furthermore, several ultra-high-throughput methods were developed to deal with large metagenomic libraries. Among these are the FACS-based screening, droplet-based screening, and the in vivo reporter-based screening methods. The application of these novel screening strategies has increased the chance for the discovery of novel enzyme genes.
Keywords: Metagenomics, Function-based screening, Agar plate screening, Microfluidics, FACS-based screening
Introduction
The total number of microbial cells on Earth is estimated to be around 1030 [1]. The largest proportion of the microbial population is made up of prokaryotes that comprise 106–108 separate species [2]. It is widely accepted that prokaryotes possess unique microbial diversity and represent a largely-unexplored biological and genetic reservoir that can be exploited for novel enzymes with unique metabolic capabilities [3], [4]. However, conventional techniques used in the laboratory to culture bacteria are often inefficient and limited as most of bacteria taken from environmental samples are currently unculturable [4], [5]. This could be attributed to a number of different factors such as alterations in atmospheric oxygen levels, osmotic conditions, specific nutrients required for survival, as well as pH and temperature conditions [6]. Hence the routine culture environments provided in the laboratory impose a selective pressure that would prevent the majority of microbes from growing [5], [6]. Therefore, a suite of culture-independent techniques is essential to understand the population structure, ecological roles, evolution, and genetic diversity of unculturable microbes found in natural environments [5], [6], [7].
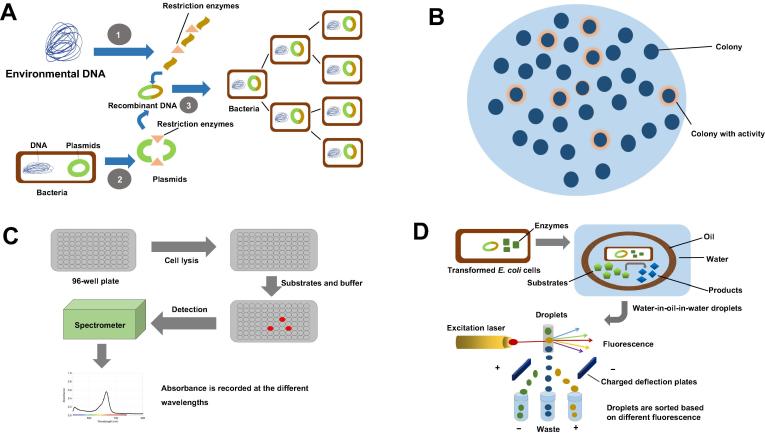
Metagenomics holds great promise for tapping the rich genetic resources in the uncultured microorganisms, by skipping culturing microorganisms and isolating genomic DNA directly from an environmental sample [8]. The rich genetic resource in the resulting metagenomic libraries can be explored in two ways: sequence-driven and function-driven screening [3], [9]. The sequence-based screening is used more frequently than the function-based approaches, due to the easy access to the metagenomics sequencing data and many software available for data analysis. However, sequence-based approaches have inherent drawbacks, with their effectiveness largely affected by the accuracy of genome annotation and the completeness of the data available [10]. These approaches reply on the algorithms available and information present in databases to infer the functions of newly-discovered genes. Thus they may not work well if the sequence similarity does not correspond to a functional relationship, or if the novel gene has only a weak similarity to any genes whose products have been examined biochemically, or if a particular gene is able to carry out numerous functions in the cell [10], [11]. Therefore, function-driven screening is the preferred approach when it comes to discovering genes with novel functions or exploring the sequence diversity of protein families with certain functions [5]. The basic procedure of the functional metagenomic screening, which includes the construction of metagenomic libraries and the different assay techniques employed to identify novel genes with desired functions, is illustrated in Figure 1.
Figure 1.
An overview of metagenomic screening approaches
A. Steps involved in the construction of a library from environmental metagenome. B. Agar plate activity screening. C. Microtiter plate screening. D. Microfluidics coupled with FACS. FACS, fluorescence-activated cell sorting.
This review is directed toward a concise synopsis of the different function-driven metagenomics screening techniques that have recently been used. A variety of novel genes encoding novel enzymes have been discovered in this way. Nonetheless, some inherent limitations of conventional function-driven screening strategies call for attention, such as high time consumption, low hit rate, and labor-intensive operations [3], [8]. To cope with these problems, several high-throughput screening strategies have been developed during the past two decades, such as microfluidics-based screening and reporter-based screening [10], [12]. The metagenomic studies and corresponding functional screening strategies discussed in this review are summarized in Table 1.
Table 1.
Recent examples of functional screening strategies employed to obtain metagenome-derived biocatalysts
| Screening approach | Target gene | Detection method | Inducer | Source | Host, vector | Ref. |
|---|---|---|---|---|---|---|
| Agar plate screening | Proteases | Phenotypical detection | 1% skim-milk | Goat skin | E. coli, plasmid | [13] |
| Agar plate screening | Proteases | Phenotypical detection | 1% skim-milk | Desert sands | E. coli, plasmid | [14] |
| Agar plate screening | Proteases | Phenotypical detection | AZCL-casein | Deciduous forest soil | E. coli, plasmid | [15] |
| Agar plate screening | Esterases | Phenotypical detection | 1% Tributyrin | Marine mud | E. coli, plasmid | [16] |
| Agar plate screening | Esterases | Phenotypical detection | X-caprylate | Vegetable soil | E. coli, plasmid | [17] |
| Agar plate screening | Esterases | Phenotypical detection | α-Naphtyl-acetate, fast blue RR | Salted shrimp | E. coli, plasmid | [18] |
| Agar plate screening | Esterases | Phenotypical detection | 1% Tributyrin | Alluvial soil | E. coli, plasmid | [19] |
| Agar plate screening | Esterases | Phenotypical detection | Dimethyl phthalate | Biofilm waste treatment plant | E. coli, fosmid | [20] |
| Agar plate screening | Lipase | Phenotypical detection | 1% Tributyrin | Cymbastela concentrica, etc. | E. coli, fosmid | [21] |
| Agar plate screening | Lipase | Phenotypical detection | 1% Tributyrin | Marine sponge (Haliclona simulans) | E. coli, fosmid | [22] |
| Agar plate screening | Lipase | Phenotypical detection | 1% Tributyrin | Marine sponge (Ircinia sp.) | E. coli, plasmid | [23] |
| Agar plate screening | Lipase | Phenotypical detection | 1% Tributyrin | Biomass | E. coli, cosmid | [24] |
| Agar plate screening | Cellulase | Phenotypical detection | Carboxymethyl cellulose | Coral (Siderastrea stellata) | E. coli, fosmid | [25] |
| Agar plate screening | Cellulase | Phenotypical detection | AZCL-HE-cellulase | Brown alga (Ascophyllum nodosum) | Yeast, plasmid | [26] |
| Agar plate screening | Starch hydrolyzing enzyme | Phenotypical detection | Starch, Lugol solution | Acid mine drainage | E. coli, plasmid | [27] |
| Agar plate screening | β-Glycosidases | Phenotypical detection | X-gal | Baltic Sea water | E. coli, plasmid | [28] |
| Agar plate screening | β-Glycosidases | Phenotypical detection | 4-Nitorphenyl-β-d-glucoside | Alkaline polluted soil (Guangxi) | E. coli, plasmid | [29] |
| Agar plate screening | β-Glycosidases | Phenotypical detection | AZCL-xylan, xyloglucan | Cow dung | E. coli, phage | [30] |
| Agar plate screening | Pectinases | Phenotypical detection | Pectin | Forest soil (Western Ghats, India) | E. coli, plasmid | [31] |
| Agar plate screening | Tannase | Phenotypical detection | X-caprylate | Cotton field soil | E. coli, plasmid | [32] |
| Agar plate screening | Nuclease | Heterologous complementation | Hydrogen peroxide | Soil (Joao, Brazil) | E. coli, plasmid | [33] |
| Agar plate screening | RNase H | Heterologous complementation | Leaf and branch compost | E. coli, plasmid | [34] | |
| Agar plate screening | Dioxygenases | Phenotypical detection | Indole | Aromatic compounds polluted soil | P. putida, phage | [35] |
| Agar plate screening | Dioxygenases | Phenotypical detection | Catechol | Activated sludge | E. coli, plasmid | [36] |
| Agar plate screening | Di-chlorophenol hydroxylase | Phenotypical detection | 3,5-Dichlorocatehol | Polychlorinated biphenyl contaminated soil | E. coli, plasmid | [37] |
| Agar plate screening | Polyhydroxyalkanoate synthase | Heterologous complementation | Sandy loam surface soil | S. meliloti, cosmid | [38] | |
| Agar plate screening | Laccase | Phenotypical detection | 2,6-DMP, etc. | Mangrove soil | E. coli, plasmid | [39] |
| Agar plate screening | DNA polymerase | Heterologous complementation | Cold sensitive mutant strain | Glacial ice | E. coli, fosmid | [40] |
| Agar plate screening | Hydrogenase | Heterologous complementation | Freshwater enrichment | Not stated | E. coli, plasmid | [41] |
| Agar plate screening | Genes resistant to toxic elements | Phenotypical detection | pH 1.8 | Acidic water (Tinto river) | E. coli, plasmid | [42] |
| Agar plate screening | Genes resistant to toxic elements | Phenotypical detection | Different number of antibiotics | Human fecal | E. coli, fosmid | [43] |
| Agar plate screening | Genes resistant to toxic elements | Phenotypical detection | Different number of antibiotics | Soil | E. coli, plasmid | [44] |
| Agar plate screening | Genes resistant to toxic elements | Phenotypical detection | Several antibiotics | Dairy cow manure | E. coli, fosmid | [45] |
| Agar plate screening | Genes resistant to toxic elements | Phenotypical detection | Several antibiotics | Cheese food matrix | E. coli, fosmid | [46] |
| Agar plate screening | Genes resistant to toxic elements | Phenotypical detection | Chloride salts | Surface water (Mississippi River) | E. coli, fosmid | [47] |
| Agar plate screening | Genes resistant to toxic elements | Heterologous complementation | Acrylate | Wastewater treatment plant | E. coli, cosmid | [48] |
| Microtiter plate screening | Cellulase | Absorbance measurement | Dinitrophenol-cellobioside | Soil, Buffalo rumen, etc. | E. coli, fosmid | [49] |
| Microtiter plate screening | Esterase | Absorbance measurement | Nitrophenyl acetate | Oil reservoir | Not stated, fosmid | [50] |
| Microtiter plate screening | Plant polymer decomposing enzymes | Absorbance measurement | Multiple substrates | Leaf compost | E. coli, fosmid | [51] |
| Microtiter plate screening | Laccase, exochitinase, etc. | Absorbance measurement | ABTS, methylumbelliferone | Forest soil (Morvan, France) | Not stated | [52] |
| SIGEX-FACS | Aromatic hydrocarbon catabolic operons | Fluorescence | Benzoate, naphthalene | Ground water contaminated with crude oil | E. coli, plasmid | [53] |
| SIGEX-FACS | Transcriptional regulators | Fluorescence | Salicylate, etc. | Ground water contaminated with crude oil | E. coli, plasmid | [54] |
| SIGEX-FACS | Aromatic hydrocarbon induced genes | Fluorescence | Salicylate Benzoate, etc. |
Soil contaminated with polyaromatic hydrocarbon | E. coli, plasmid | [55] |
| GESS-FACS | Phenol generating enzymes | Fluorescence | p-Nitrophenyl | Sea tidal flat sediments | E. coli, plasmid | [56] |
| GESS-FACS | Lipase, cellulase, etc. | Fluorescence | p-Nitrophenyl acetate, etc. | Ocean tidal flat sediment | E. coli, fosmid | [57] |
| GESS-FACS | Phosphatase | Fluorescence | p-Nitrophenyl phosphate | Ocean tidal flat sediments | E. coli, fosmid | [58] |
| GMD | Lipolytic enzymes | Fluorescence | Fluorescein dicaprylate | Soil (Quercus serrata forest) | E. coli, plasmid | [59] |
| GMD, FACS | Screening for antibiotics | Fluorescence | S. aureus | 3 strains of Staphylococcus obtained from an ARS culture collection |
E. coli S. cerevisiae, plasmid |
[60] |
| Microfluidics (water in oil droplets), FACS | Hydrolases | Fluorescence | Sulfate monoester Phosphate triester |
Variety of sources (soil, degraded plant material, cow rumen) | E. coli, plasmid | [61] |
Note: AZCL, azurine-cross-linked; AZCL-HE, azurine-cross-linked hydroxyl-ethylcellulose; DMP, dimethoxyphenol; ABTS, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonate); GESS, genetic enzyme screening system; GMD, gel micro-droplet; FACS, fluorescence-activated cell sorting; SIGEX, substrate induced gene expression; ARS, agriculture research service.
Agar plate screening
Agar plate screening for hydrolytic enzymes
Functional screening of metagenomic libraries using agar plates provides a simple and straightforward approach to identify novel enzymes that function under diverse conditions. Numerous novel hydrolytic enzymes, including lipases, esterases, cellulases, proteases, laccases, glycosylases, nitrilases, and dehalogenases, have been identified using this method [62]. Assays used to discover these enzymes are based on the production of a chromophore or fluorophore in colonies incubated with a chromogenic or fluorogenic substrate. The throughput of agar plate based assays is usually 105–106 clones per day [63]. Despite the low throughput, agar plate screening has led to the successful isolation of a large number of unique enzymes from various environments. In the following section, we describe the approaches used to identify novel hydrolytic enzymes.
Proteases
The catabolism of proteins is catalyzed by proteases via cleaving the peptide bonds that link amino acids within proteins [64], [65], [66]. Screening of protease activity involves incubating 1% skim-milk on LB agar plates and a positive signal for protease activity is shown as halos due to the degradation of milk proteins [13], [14], [65]. A lot of proteases with desirable properties have been identified using this simple technique [64], [65], [66], [67]. For instance, a metagenomic library containing DNA from bacteria dwelling on goat skin was functionally screened for protease activity, which leads to the discovery of a serine protease enzyme with a high alkalinity tolerance [13]. A metagenomic library containing clones derived from a forest soil sample was functionally screened for protease activity on yeast extract and tryptone agar plates supplemented with azurine-cross-linked casein. The screening result in the discovery of a novel alkaline serine protease with high oxidant stability [15]. Similarly, activity screening of metagenome libraries of soil samples from Death Valley and Gobi deserts revealed two serine proteases with distinctly thermophilic profiles [14].
Esterases and lipases
Ester bonds can be synthesized and hydrolyzed by enzymes in carboxylic ester hydrolase class, which contains lipases and esterases [16], [68], [69]. Agar plate screening has been used to identify novel esterases [15], [19], [65], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78] and lipases [67], [79], [80]. Detection of lipase or esterase activity is usually performed using tributyrin since hydrolysis of emulsified tributyrin leads to the formation of halos around positive colonies. To specifically detect lipase activity, LB agar media containing olive oil is used [68], [81], [82], with the addition of a fluorescent dye Rhodamine B. The orange fluorescence, from a complex of fatty acids and Rhodamine B, serves as a good indicator of lipase activity [62], [68].
A novel pyrethroid-hydrolyzing esterase enzyme, which has potential applications in insecticide production, was identified from a metagenomic library of a soil sample at vegetable garden [17]. A similar screening approach resulted in the identification of a highly organic solvent-tolerant and thermostable esterase [16]. When using fast blue RR staining to identify clones that exhibited esterase activity from a salted shrimp derived metagenomic library, a unique salt-tolerant esterase was identified [18]. Agar plate activity screening of an alluvial soil metagenomic library revealed the identity of two unique esterases that exhibited chloramphenicol reactivating activity [83], whereas two esterase with distinctly cold adapted activity were identified when activity-based screening was carried out with an arctic soil derived metagenomic library under low temperature [84]. Additionally, a substrate set, composed of di-butyl phthalate, di-propyl phthalate, and di-pentyl phthalate, was utilized to carry out activity-based screening of a wastewater-derived metagenomic library [20]. As a result, three novel phthalate esters hydrolase enzymes were discovered, which exhibited an adaptation to cold activity.
To identify lipases, metagenomic libraries derived from microbe societies associated with green alga (Ulva australis) and with a temperate marine sponge (Cymbastela concentrica) were functionally screened. This leads to the identification of three unique lipases that exhibited potential antibacterial properties [21]. Additionally, functional screening of a marine sponge (Haliclona simulans) derived metagenomic library revealed a lipase that adapted to high salinity concentrations [22]. Similarly, a lipase displaying alkalophilic activity was identified from the metagenomic library constructed from marine sponge Ircinia sp [23]. Following functional screening of metagenomic libraries derived from a cell culture enriched at hot and high pH condition, a thermo-stable alkaline lipase was discovered [24].
Cellulases
Cellulases are a group of enzymes which hydrolyze cellulose. Agar plate activity screening for cellulases and glycosyl hydrolases often involves the use of carboxymethyl cellulose as a substrate and Congo red dye as an indicator [85], with unstained haloes representing positive clones. Such method has been used to discover many novel cellulases from different environments, some of which are from unexpected locations with no selective pressure for cellulases [65], [67], [86], [87], [88], [89]. For instance, cellulases are found in a metagenomic library from coral Siderastrea stellata [25]. The coral is not a place you would expect to find cellulases, because it contains very little cellulosic substance. Other cellulases are found to have properties matching their surrounding environments. For example, a halo-tolerant and cold-active cellulase was identified in a metagenomic library constructed with microbial DNA from a cold-active marine brown alga Ascophyllum nodosum [26].
Xylanases
Xylanases are hydrolytic enzymes that degrade the linear polysaccharide beta-1,4 xylan into xylose, thus breaking down hemicellulose. Detection of xylanase activity is done using birchwood xylan as substrate and Congo Red as indicator dye, with positive clones identified by unstained halos. Identification of novel xylanases from metagenomics has been reported in a number of studies [90], [91], [92], [93], [94].
Starch hydrolyzing enzymes
Screening for starch hydrolyzing activity in the metagenomic library uses starch as substrate and Lugol solution (iodine) as the indicator dye, with positive clones identified as clear halo zones around the colonies [27], [95]. Agar plate activity screening for novel starch hydrolyzing enzymes, such as amylases, pullulanases, and glucoamylases, has been reported in many studies [65], [67], [95], [96]. These starch hydrolyzing enzymes show special characteristics adapted to the surrounding environmental elements, such as coldness and high pH. Some amylases with no sequence homolog to known proteins were found, which expands the sequence diversity of the amylase enzyme family. For example, function-based screening of an acid mine drainage-derived metagenomic library led to the identification of two amylases containing no known amylolytic domain [27]. It is noteworthy that these amylases do not share any sequence similarity with known amylases or glycoside hydrolase either. Therefore, they represent a new subgroup of the amylase enzyme family.
β-Glycosidases
β-Glycosidases are enzymes that cleave beta-glycosidic bonds [97]. Agar plate activity screening for β-glucosidase activity is carried out using ammonium ferric citrate-esculin, while p-nitrophenyl-β-d-xyloside (pNPX) is used to identify β-xylosidase [97], [98]. Using agar plate activity screening, novel β-glycosidase enzymes had been reported in several studies [29], [97], [98], [99], [100], [101]. A novel cold-active glycosidase that exhibited beta-glucosidase, beta-fucosidase, and beta-galactosidase activities was detected from of a metagenomic library derived from a marine sample [28]. Another novel β-glucosidase with both glycosidase and lipolytic activity was identified from clones of metagenomic library derived from an alkaline polluted soil sample [102]. Similar screening was also used to explore the microbiota in animal stomach samples. A screening of genes encoding fibrolytic enzymes resulted in the discovery of four protozoan glycoside hydrolases from a bovine ruminal protozoan-enriched metagenomic library [30].
Pectinases
Screening for pectinolytic activity is based on growing clones on pectin-containing LB plates using Ruthenium red solution as indicator, with positive clones shown as clear zones. Activity-driven analysis of clones from a metagenomic library obtained from a tropical forest soil sample led to the identification of a pectinase active at broad pH and temperature [31], indicating that metagenomes could be a useful source for pectinase discovery.
Tannases
Tannases catalyze the hydrolysis of digallate, which is found in many bacteria, such as those dwelling in rumen. Functional screening of tannases involves the use of isopropyl-β-d-thiogalactopyranoside (IPTG) as the inducer and X-caprylate as the indicator dye, with positive clones showing a distinctive blue color. Function-driven analysis of clones from a metagenomic library constructed with a cotton field soil sample resulted in the detection of a gene encoding tannase [32]. This tannase shows excellent resistance to salt concentration as high as 4 M NaCl. It also has a broad spectrum of substrates, making it a potential biocatalyst with potential industrial application.
Nucleases
Functional screening was also used to identify novel enzymes responsible for DNA repair that are vital in preserving the integrity of DNA [33]. Clones from the metagenomic library were analyzed through complementation assay using Escherichia coli strain DH10B, which is sensitive to hydrogen peroxide due to the lack of DNA repair gene encoding recA. A positive clone was identified based on the increased resistance to hydrogen peroxide treatment. The resulting gene encodes a novel salt-tolerant exo-nuclease with no significant similarity to any known nuclease.
Similarly, complementary genetic screening was employed to identify unique RNase H genes from a metagenomic library constructed from a leaf and branch compost [34]. Twelve unique genes encoding type 1 RNases H were identified, of which eleven genes showed 40%–72% protein sequence identities to those found in the National Centre for Biotechnology Information (NCBI) database. Interestingly, another enzyme identified in this study lacks a conserved DEDD/E active site motif, but still exhibited RNases activity, indicating distinctive catalytic mechanism.
Agar plate activity screening for non-hydrolytic enzymes
Typically, the targets for metagenomics screening are various hydrolytic enzymes, due to the easy detection of the phenotypes, such as the formation of halos around the colonies. Occasionally, metagenomics screenings are used to screen non-hydrolytic enzymes as well.
Dioxygenases
Aimed to identify enzymes that are able to degrade aromatic compounds, Nagayama et al. performed an indigo-forming activity analysis on clones from a metagenomic library derived from an aromaticallypolluted soil sample. In this study, a gene encoding a multicomponent hydroxylase was found to be responsible for the phenol degradation [35]. A similar analysis was also performed to identify the novel enzymes able to carry out microbial degradation of aromatic compounds from sludge. Consequently, 38 new genes coding for extradiol dioxygenases were identified, forming a new subfamily of extradiol dioxygenases [36].
Dichlorophenol hydroxylases
Lu and colleagues collected an environmental sample from an area contaminated by polychlorinated bi-phenyl compounds for more than two decades and used it for the construction of a metagenomic library [37]. They used 3,5-dichlorocatechol to screen for chlorophenol hydroxylase genes and identified a 2.4-dichlorophenol hydroxylase with a Km of 5 µM for 2,4-dichlorophenol and 6 µM for NADPH. This was the first report of identifying a unique TfdB gene using the functional metagenomic approach.
Polyhydroxyalkanoate synthases
The phenotypic complementation was employed when screening for novel genes encoding polyhydroxyalkanoate synthases in a metagenomic library from soil. Several genes with low identity to known genes that are involved in polyhydroxyalkanoate synthesis were isolated, which expands the diversity of this group of genes [38].
Laccases
Based on a metagenomic library constructed from a mangrove soil sample, function-driven screening resulted in the identification of a multi-copper oxidase with laccase activity [39]. This laccase shows good alkaline activity and good solubility, which set it apart from other laccases.
DNA polymerases
Complementation assay using E. coli polA mutant was employed in the screening of glacial ice derived metagenomic libraries. This mutant strain cannot survive at the temperature below 20 °C due to a mutation in the DNA polymerase gene. Therefore, during the screening process, only the cells carrying the metagenomic clones conferring DNA polymerase activity can survive at 18 °C. Such screening resulted in the detection of nine genes encoding DNA polymerases [40].
Hydrogenases
In an activity-based approach to identify an acid-tolerant hydrogenase from metagenomic libraries, the clones were transferred via tri-parental mating into a hydrogenase-deleted mutant Shewanella oneidensis and grown on freshwater medium [41]. The clones that exhibited hydrogenase activity were distinguished by the change in color of the media from yellow to colorless.
Screening for the genes responsible for the resistance to toxic elements
The agar-plate activity screening method is often used to screen for the genes responsible for the resistance to the toxic elements such as antibiotic, extreme salt concentration, extreme pH, and heavy metals. Function-driven analysis of metagenomic libraries derived from rhizosphere and planktonic microbial communities revealed 15 novel genes with the encoded proteins conferring acid resistance [42]. Most products of these genes are putative or unknown proteins, which implies the unknown mechanism for acid resistance. Guazzaroni et al. also identified nine genes that were expressed at high levels in P. putida and Bacillus subtilis, which enhanced host cells’ ability to withstand extreme acidic conditions [42].
In a search for antibiotic resistance genes, seven different antibiotics were used to screen a fosmid library constructed from gut microbiota of four healthy humans, which resulted in the discovery of eight new antibiotic resistance genes [43]. Similarly, functional screening of clones from a soil metagenomic library revealed 41 novel genes that encode antibiotic resistance proteins across eight protein families [44]. In another study, metagenomic libraries derived from a cow manure sample were functionally screened for resistance to a number of antibiotics, resulting in the detection of 80 unique antibiotic enzymes together with a novel clade of chloramphenicol acetyl-transferases [45]. Similar screening was also performed with metagenomic libraries derived from food fermenting microbiota in the presence of a wide range of antibiotics. Novel kanamycin and ampicillin resistant clones originating from Lactobacillus helveticus and Streptococcus salivarius were reported [46]. Like the antibiotic resistant genes’ (ARGs) screening, metal resistance activity screening on metagenomic libraries derived from the Upper Mississippi River showed the highest frequency of clones with resistance to Mn2+ but no clone with resistance to Cu2+ [47]. In the quest to find unique genes that confer bacterial resistance to acrylate, activity screening of clones from a metagenomic library derived from a sewage treatment plant revealed three enzyme classes that conferred an acrylate-resistant (AcrR) phenotype [48].
Microtiter plate screening
Agar plate screening approaches are simple to perform and provide an easy way to identify active clones. However, they have major setbacks, with low dynamic range and weak visualization of differences in catalytic rates of enzyme variants, and, therefore, difficulty in quantification [9], [12]. In order to improve sensitivity, a number of different approaches are applied. The most commonly applied screening strategy is based on microtiter plates, which involves incubation of bacterial culture with enzyme substrate in the micro-wells [9], [103]. Usually, a single colony or diluted cell culture containing several colonies is distributed into each individual well manually or through a liquid handling station. The cells are grown in microtiter plates and assessed in a second plate after cell lysis, with the original plates stored as back-up [9], [104]. With the occurrence of substrate conversion, a visual signal emerges, such as color or fluorescence, which is used to identify colonies expressing an enzyme with desirable properties. The throughput of microtiter plates has been enhanced through the incorporation of robotic technologies [104]. Also, fluorescence-activated cell sorting (FACS) has been used to distribute single cells into microtiter plates [105]. However, these kind of experiments require a strong prior knowledge on the best convenient substrates and also the chemistry on how it works. This presents a bottleneck and consequently, microtiter plate screening is not applicable in many occasions [104].
In a study, chromogenic dinitrophenol (DNP)-cellobioside was used as the substrate to identify cellulase activity [49]. The 384-well microplates allow for quantitative analysis since the plate reader can be used to measure the absorbance. Moreover, fosmid library was constructed to improve the efficiency and reliability of the activity-based screening. It was noticed that adding the inducer L-arabinose in the culturing media resulted in an increase in the copy number of the fosmid library by up to 100 folds in the E. coli cells while a single copy of the fosmid library was maintained during the growth period for stability [9].
In a metagenomic analysis of an oil reservoir metagenome sample, E. coli cells containing fosmid library were grown in microtiter plates. The cells in the duplicate plates were lysed, and the resulting supernatants were transferred to 384-well microtiter plates. The enzyme assays for short chain and long chain esterase activity were performed by adding the crude cell extracts to microtiter plates containing nitrophenyl acetate and nitrophenyl palmitate as substrate, respectively. One enzyme showing the highest activity on short chain ester substrate was identified as a novel esterase exhibiting high thermo-stability and high tolerance to metal ions and solvents [50].
A high-throughput screening technique was also developed for the identification of enzymes involved in starch, hemicellulose, cellulose, chitin, lignin decomposition, protein hydrolysis, and phosphate mineralization, using the multiple substrate approach to allow for concurrent identification of diverse activities essential during the various stages of biomass depolymerization [51]. Enzyme assays were performed on microtiter plates and resulted in the identification of phosphatases, carbohydrate hydrolase, and protease activity. It is noticeable that such a method requires the extensive use of sophisticated equipment, such as liquid handling system and automatic colony picker. This may limit its use in the labs without such setups.
To investigate the functions of microbial communities at different soil layers, special microtiter plates, Biolog Ecoplates, were used to study the metabolic profiles of the soil microbiome [52]. Uroz et al. analyzed metagenomic libraries derived from two acidic forest soil samples that were incubated with different carbon sources on Ecoplate microplates. They detected positive results by measuring color development at 590 nm on a microplate reader. In addition, functional screening for laccase, β-glucosidase, xylosidase, cellobiohydrolase and exochitinase activity were also performed by inoculating with relevant substrates on regular microtiter plates. It was found that the substrates that were readily metabolized differed between the two soil samples, suggesting that the microbial communities had a specialization in response to nutritional conditions [52].
FACS-based screening
There are a number of challenges associated with the conventional function-driven screening approaches in the analysis of metagenomic libraries, such as low throughput, low hit frequency for positive clones, and increased evidence of catalytic promiscuity [3]. Moreover, these approaches are often labor intensive and time-consuming [7]. One solution to such problems is to use FACS to screen the libraries. FACS enables the selection of cells based on cell size, shape, and fluorescence [12], [59], [106], [107], [108]. FACS have many advantages: (1) it deposits single events into a variety of vessels quickly and accurately; (2) the laminar flow fluidics of FACS prevents disruption of cells during sorting; and (3) the contamination is limited because of the small volume of each droplet [105], [109], [110]. Due to its powerful sorting abilities, FACS is easily coupled to a number of different high-throughput screening methods such as droplet sorting and reporter-based screening. The reporter-based screening method depends on the expression of the reporter genes, such as GFP genes [12], [111]. In these systems, the metagenomic library is transformed into the host cells which harbor the reporter genes. Then the gene products of the metagenomic library activate the expression of reporter genes through transcriptional regulation or post-translational modifications [12].
A good example of reporter-based screening system is the substrate induced gene expression (SIGEX) system which is coupled with FACS to sort GFP-expressing E. coli cells that respond to the presence of aromatic compounds [53]. Using SIGEX, Uchiyama et al. identified numerous transcriptional regulators which turn on the report gene expression in response to aromatic compounds, such as salicylate, 3-methyl catechol, 4-chlorocatechol, and chlorohydroquinone [54]. In another study, several aromatic hydrocarbon (AH)-degrading genes were discovered using SIGEX in a metagenomic library derived from AH-contaminated soil sample [53]. Subsequently, the library was sequenced for contig assembly. The AH-degrading genes were mapped to the contigs, resulting in the identification of several complete operons involved in the AH metabolism [55]. This approach combines the SIGEX technique and the next-generation sequencing (NGS) technology, which is very instrumental in studying the operons or clusters in the unculturable microorganisms.
Similarly, a genetic circuit termed genetic enzyme screening system (GESS) was developed for the high-throughput function-driven analysis of unique phenol-generating enzymes from metagenomic libraries [56]. Through a phenol-binding transcriptional activator, phenol generated by these enzymes can trigger the expression of GFP reporter gene. Three such enzymes were identified, including alkaline phosphatase, lipase, and cellulose, with the help of phenolic substrates (phenyl phosphate, p-nitrophenyl acetate, and p-nitrophenyl-β-d-cellobioside) respectively [57]. Also, p-nitrophenyl phosphate (pNPP)-GESS was used to identify a novel psychrophilic alkaline phosphatase, together with the phenol-recognizing dimethylphenol regulator (DmpR) as the transcriptional activator [58].
Microfluidics-based screening
In addition to the conventional FACS-based screening, a more advanced high-throughput screening strategy was developed which use microfluidic chips to generate monodispersed microdroplets [12], [112]. Microdroplets are produced in large numbers at speeds of thousands of droplets per second and a single droplet functions as a reaction chamber. Cells, enzyme variants, substrates and products are confined in the picoliter volume of the droplets, where reactions take place [9]. Subsequently, the droplets are sorted according to fluorescence or color of the product. The coupling of microfluidics with FACS results in the ultrahigh-throughput screening of metagenomic libraries. However this technique has its own limitation that only fluorogenic substrates not capable of crossing the oil phase barrier may be used [9]. Sometimes, agarose is included in the droplets to increase the strength of structure, which forms the microfluidic gel microdroplets (GMD) [59].
The GMD technique was employed to co-encapsulate a fluorogenic substrate together with clones from a metagenomic library to screen for unique genes with lipolytic activity. Using this method, a lipolytic enzyme EstT was identified [59]. Additionally, GMD-FACS was also used as an ultrahigh-throughput screening in the detection of novel antibiotics [60]. Clones of metagenomic library constructed from three strains of Staphylococcus were co-encapsulated in GMD with target pathogen Staphylococcus aureus. The GMD was treated with fluorescent dye SyTox Orange and the fluorescence from the DNA-bound SyTox Orange indicates the antibiotic activity. The screening resulted in the discovery of a lytic hydrolase specific for S. aureus.
In order to overcome the limitations associated with microfluidics coupled to FACS, efforts were made to improve the microfluidic devices. To bypass the expensive FACS operation, an ultrahigh-throughput technique based on the water-in-oil droplets was developed [61]. The biomimetic compartments within the droplets act as a genotype phenotype linkage in analogy to cells. Such a method was used to screen for novel hydrolase genes from a large metagenomic library derived from a wide range of sources, with sulfate monoester and phosphate tri-ester as fluorescence substrates. The resulting hydrolases span three protein super-families, most of which have very low sequence homolog with known proteins.
In another study, expression of alkaline phosphatase was carried out by encapsulating single E. coli cells in approximately 800-picoliter droplets. The reaction in the droplets was monitored by connecting a photomultiplier to a microscope to detect the presence of the fluorescent product. The catalytic turnover of the substrate was measured at several locations along the microfluidic channel. The enzyme activities at different time points were used to provide time-resolved kinetic measurements [113].
It is noticeable that almost all ultrahigh-throughput micro-droplet techniques rely on the detection of a fluorescent product. In the cases lacking this type of readout, droplet screening becomes impossible. In order to deal with this unsatisfactory situation, a microfluidic absorbance activated droplet sorter (AADS) was developed [114]. AADS is based on the NAD+ dependent deamination of amino acids catalyzed by phenylalanine dehydrogenase (PheDH). The reaction does not produce fluorescence. Instead, it is coupled with a reaction producing an absorbing dye, which triggers the optical sensor next to the microfluidic chip and diverge the flow of droplets. Such method achieved the enrichment of active variants of PheDH up to 2800 folds. However, due to the low sensitivity of absorbance detection, the sorting speed of this method (100 Hz) is significant lower than those reported for the fluorescent-based method (2000 Hz) [61].
Conclusion and future perspectives
Metagenomes derived from unculturable microorganisms is a great reservoir for the genes with unknown functions. A variety of function-based metagenomic screening methods have been developed to explore these rich genetic resources, resulting in the identification of large number of novel enzymes with unique metabolic activities. However, there are a number of challenges that hamper the discovery of genes with novel functions, such as poor expression of target genes in the host cells and inefficient activity assays for the gene products. The traditional agar plate screening method suffers from the low sensitivity and low throughput. To overcome such problems, high-throughput methods such as FACS-driven screening, and microfluidics-driven screening have been developed. These methods have greatly expanded toolkits for exploring the vast sequence diversity in the metagenomes. In particular, the microfluidics-based screening has shown its potential to screen over 107 variants per day. When coupled with FACS, the microfluidic devices can greatly expand the scope of high-throughput metagenomic screening. The major bottleneck of such technique is the detection method, which is mostly limited to fluorescent signal. In the future, other detection methods, such as mass spectrometry, nuclear magnetic resonance (NMR) and colorimetric assay, may be combined with microfluidic devices to accelerate the discovery of novel biocatalysts or other genes with important functions in the microbiota. On the other hand, most function-based approaches for metagenomic screening are hindered by the biased and insufficient expression in E. coli. There is an urgent need to develop a greater range of alternative hosts with good expression of foreign genes of metagenomic origins. To this end, it is essential to develop efficient shuttle vectors that have extended host ranges so that the metagenomic library carried by these vectors can be expressed in various hosts. Alternatively, the translation profiles in E. coli may be altered to accommodate the foreign genes by engineering the ribosome proteins or manipulating the factors involved in transcriptional/translational control.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This work is financially supported by the National Natural Science Foundation of China (Grant No. 31670793).
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Turnbaugh P.J., Gordon J.I. An invitation to the marriage of metagenomics and metabolomics. Cell. 2008;134:708–713. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Sleator R.D., Shortall C., Hill C. Metagenomics. Lett Appl Microbiol. 2008;47:361–366. doi: 10.1111/j.1472-765X.2008.02444.x. [DOI] [PubMed] [Google Scholar]
- 3.Simon C., Daniel R. Metagenomic analyses: past and future trends. Appl Environ Microbiol. 2011;77:1153–1161. doi: 10.1128/AEM.02345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennedy J., Marchesi J.R., Dobson A.D. Marine metagenomics: strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb Cell Fact. 2008;7:27. doi: 10.1186/1475-2859-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh J., Behal A., Singla N., Joshi A., Birbian N., Singh S. Metagenomics: concept, methodology, ecological inference and recent advances. Biotechnol J. 2009;4:480–494. doi: 10.1002/biot.200800201. [DOI] [PubMed] [Google Scholar]
- 6.Vartoukian S.R., Palmer R.M., Wade W.G. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol Lett. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- 7.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon C., Daniel R. Achievements and new knowledge unraveled by metagenomic approaches. Appl Microbiol Biotechnol. 2009;85:265–276. doi: 10.1007/s00253-009-2233-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leemhuis H., Kelly R.M., Dijkhuizen L. Directed evolution of enzymes: library screening strategies. IUBMB Life. 2009;61:222–228. doi: 10.1002/iub.165. [DOI] [PubMed] [Google Scholar]
- 10.Ferrer M., Beloqui A., Vieites J.M., Guazzaroni M.E., Berger I., Aharoni A. Interplay of metagenomics and in vitro compartmentalization. Microb Biotechnol. 2009;2:31–39. doi: 10.1111/j.1751-7915.2008.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallin P.F., Binnewies T.T., Ussery D.W. The genome BLASTatlas-a GeneWiz extension for visualization of whole-genome homology. Mol Biosyst. 2008;4:363–371. doi: 10.1039/b717118h. [DOI] [PubMed] [Google Scholar]
- 12.van Rossum T., Kengen S.W., van der Oost J. Reporter-based screening and selection of enzymes. FEBS J. 2013;280:2979–2996. doi: 10.1111/febs.12281. [DOI] [PubMed] [Google Scholar]
- 13.Pushpam P.L., Rajesh T., Gunasekaran P. Identification and characterization of alkaline serine protease from goat skin surface metagenome. AMB Express. 2011;1:3. doi: 10.1186/2191-0855-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neveu J., Regeard C., DuBow M.S. Isolation and characterization of two serine proteases from metagenomic libraries of the Gobi and Death Valley deserts. Appl Microbiol Biotechnol. 2011;91:635–644. doi: 10.1007/s00253-011-3256-9. [DOI] [PubMed] [Google Scholar]
- 15.Biver S., Portetelle D., Vandenbol M. Characterization of a new oxidant-stable serine protease isolated by functional metagenomics. Springerplus. 2013;2:410. doi: 10.1186/2193-1801-2-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao W., Wu K., Chen L., Fan H., Zhao Z., Gao B. A novel esterase from a marine mud metagenomic library for biocatalytic synthesis of short-chain flavor esters. Microb Cell Fact. 2016;15:41. doi: 10.1186/s12934-016-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G., Wang K., Liu Y.H. Molecular cloning and characterization of a novel pyrethroid-hydrolyzing esterase originating from the Metagenome. Microb Cell Fact. 2008;7:38. doi: 10.1186/1475-2859-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S.Y., Shin H.J., Kim G.J. Screening and identification of a novel esterase EstPE from a metagenomic DNA library. J Microbiol. 2011;49:7–14. doi: 10.1007/s12275-011-0201-7. [DOI] [PubMed] [Google Scholar]
- 19.Tao W., Lee M.H., Wu J., Kim N.H., Lee S.W. Isolation and characterization of a family VII esterase derived from alluvial soil metagenomic library. J Microbiol. 2011;49:178–185. doi: 10.1007/s12275-011-1102-5. [DOI] [PubMed] [Google Scholar]
- 20.Jiao Y., Chen X., Wang X., Liao X., Xiao L., Miao A. Identification and characterization of a cold-active phthalate esters hydrolase by screening a metagenomic library derived from biofilms of a wastewater treatment plant. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yung P.Y., Burke C., Lewis M., Kjelleberg S., Thomas T. Novel antibacterial proteins from the microbial communities associated with the sponge Cymbastela concentrica and the green alga Ulva australis. Appl Environ Microbiol. 2011;77:1512–1515. doi: 10.1128/AEM.02038-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selvin J., Kennedy J., Lejon D.P., Kiran G.S., Dobson A.D. Isolation identification and biochemical characterization of a novel halo-tolerant lipase from the metagenome of the marine sponge Haliclona simulans. Microb Cell Fact. 2012;11:72. doi: 10.1186/1475-2859-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su J., Zhang F., Sun W., Karuppiah V., Zhang G., Li Z. A new alkaline lipase obtained from the metagenome of marine sponge Ircinia sp. World J Microbiol Biotechnol. 2015;31:1093–1102. doi: 10.1007/s11274-015-1859-5. [DOI] [PubMed] [Google Scholar]
- 24.Meilleur C., Hupe J.F., Juteau P., Shareck F. Isolation and characterization of a new alkali-thermostable lipase cloned from a metagenomic library. J Ind Microbiol Biotechnol. 2009;36:853–861. doi: 10.1007/s10295-009-0562-7. [DOI] [PubMed] [Google Scholar]
- 25.Sousa F.M., Moura S.R., Quinto C.A., Dias J.C., Pirovani C.P., Rezende R.P. Functional screening for cellulolytic activity in a metagenomic fosmid library of microorganisms associated with coral. Genet Mol Res. 2016;15:235–237. doi: 10.4238/gmr.15048770. [DOI] [PubMed] [Google Scholar]
- 26.Martin M., Biver S., Steels S., Barbeyron T., Jam M., Portetelle D. Identification and characterization of a halotolerant, cold-active marine endo-beta-1,4-glucanase by using functional metagenomics of seaweed-associated microbiota. Appl Environ Microbiol. 2014;80:4958–4967. doi: 10.1128/AEM.01194-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delavat F., Phalip V., Forster A., Plewniak F., Lett M.C., Lievremont D. Amylases without known homologues discovered in an acid mine drainage: significance and impact. Sci Rep. 2012;2:354. doi: 10.1038/srep00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wierzbicka-Wos A., Bartasun P., Cieslinski H., Kur J. Cloning and characterization of a novel cold-active glycoside hydrolase family 1 enzyme with beta-glucosidase, beta-fucosidase and beta-galactosidase activities. BMC Biotechnol. 2013;13:22. doi: 10.1186/1472-6750-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang C., Li S.X., Luo F.F., Jin K., Wang Q., Hao Z.Y. Biochemical characterization of two novel beta-glucosidase genes by metagenome expression cloning. Bioresour Technol. 2011;102:3272–3278. doi: 10.1016/j.biortech.2010.09.114. [DOI] [PubMed] [Google Scholar]
- 30.Findley S.D., Mormile M.R., Sommer-Hurley A., Zhang X.C., Tipton P., Arnett K. Activity-based metagenomic screening and biochemical characterization of bovine ruminal protozoan glycoside hydrolases. Appl Environ Microbiol. 2011;77:8106–8113. doi: 10.1128/AEM.05925-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sathya T.A., Jacob A.M., Khan M. Cloning and molecular modelling of pectin degrading glycosyl hydrolase of family 28 from soil metagenomic library. Mol Biol Rep. 2014;41:2645–2656. doi: 10.1007/s11033-014-3123-8. [DOI] [PubMed] [Google Scholar]
- 32.Yao J., Fan X.J., Lu Y., Liu Y.H. Isolation and characterization of a novel tannase from a metagenomic library. J Agric Food Chem. 2011;59:3812–3818. doi: 10.1021/jf104394m. [DOI] [PubMed] [Google Scholar]
- 33.Silva-Portela R.C., Carvalho F.M., Pereira C.P., de Souza-Pinto N.C., Modesti M., Fuchs R.P. ExoMeg1: a new exonuclease from metagenomic library. Sci Rep. 2016;6:19712. doi: 10.1038/srep19712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanaya E., Sakabe T., Nguyen N.T., Koikeda S., Koga Y., Takano K. Cloning of the RNase H genes from a metagenomic DNA library: identification of a new type 1 RNase H without a typical active-site motif. J Appl Microbiol. 2010;109:974–983. doi: 10.1111/j.1365-2672.2010.04724.x. [DOI] [PubMed] [Google Scholar]
- 35.Nagayama H., Sugawara T., Endo R., Ono A., Kato H., Ohtsubo Y. Isolation of oxygenase genes for indigo-forming activity from an artificially polluted soil metagenome by functional screening using Pseudomonas putida strains as hosts. Appl Microbiol Biotechnol. 2015;99:4453–4470. doi: 10.1007/s00253-014-6322-2. [DOI] [PubMed] [Google Scholar]
- 36.Suenaga H., Ohnuki T., Miyazaki K. Functional screening of a metagenomic library for genes involved in microbial degradation of aromatic compounds. Environ Microbiol. 2007;9:2289–2297. doi: 10.1111/j.1462-2920.2007.01342.x. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y., Yu Y., Zhou R., Sun W., Dai C., Wan P. Cloning and characterisation of a novel 2,4-dichlorophenol hydroxylase from a metagenomic library derived from polychlorinated biphenyl-contaminated soil. Biotechnol Lett. 2011;33:1159–1167. doi: 10.1007/s10529-011-0549-0. [DOI] [PubMed] [Google Scholar]
- 38.Schallmey M., Ly A., Wang C., Meglei G., Voget S., Streit W.R. Harvesting of novel polyhydroxyalkanaote (PHA) synthase encoding genes from a soil metagenome library using phenotypic screening. FEMS Microbiol Lett. 2011;321:150–156. doi: 10.1111/j.1574-6968.2011.02324.x. [DOI] [PubMed] [Google Scholar]
- 39.Ye M., Li G., Liang W.Q., Liu Y.H. Molecular cloning and characterization of a novel metagenome-derived multicopper oxidase with alkaline laccase activity and highly soluble expression. Appl Microbiol Biotechnol. 2010;87:1023–1031. doi: 10.1007/s00253-010-2507-5. [DOI] [PubMed] [Google Scholar]
- 40.Simon C., Herath J., Rockstroh S., Daniel R. Rapid identification of genes encoding DNA polymerases by function-based screening of metagenomic libraries derived from glacial ice. Appl Environ Microbiol. 2009;75:2964–2968. doi: 10.1128/AEM.02644-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adam N., Perner M. Activity-based screening of metagenomic libraries for hydrogenase enzymes. Methods Mol Biol. 2017;1539:261–270. doi: 10.1007/978-1-4939-6691-2_17. [DOI] [PubMed] [Google Scholar]
- 42.Guazzaroni M.E., Morgante V., Mirete S., Gonzalez-Pastor J.E. Novel acid resistance genes from the metagenome of the Tinto River, an extremely acidic environment. Environ Microbiol. 2013;15:1088–1102. doi: 10.1111/1462-2920.12021. [DOI] [PubMed] [Google Scholar]
- 43.Cheng G., Hu Y., Yin Y., Yang X., Xiang C., Wang B. Functional screening of antibiotic resistance genes from human gut microbiota reveals a novel gene fusion. FEMS Microbiol Lett. 2012;336:11–16. doi: 10.1111/j.1574-6968.2012.02647.x. [DOI] [PubMed] [Google Scholar]
- 44.McGarvey K.M., Queitsch K., Fields S. Wide variation in antibiotic resistance proteins identified by functional metagenomic screening of a soil DNA library. Appl Environ Microbiol. 2012;78:1708–1714. doi: 10.1128/AEM.06759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wichmann F., Udikovic-Kolic N., Andrew S., Handelsman J. Diverse antibiotic resistance genes in dairy cow manure. MBio. 2014;5 doi: 10.1128/mBio.01017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devirgiliis C., Zinno P. Functional screening of antibiotic resistance genes from a representative metagenomic library of food fermenting microbiota. BioMed Res Int. 2014;1 doi: 10.1155/2014/290967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staley C., Johnson D., Gould T.J., Wang P., Phillips J., Cotner J.B. Frequencies of heavy metal resistance are associated with land cover type in the Upper Mississippi River. Sci Total Environ. 2015;511:461–468. doi: 10.1016/j.scitotenv.2014.12.069. [DOI] [PubMed] [Google Scholar]
- 48.Curson A.R., Burns O.J., Voget S., Daniel R., Todd J.D., McInnis K. Screening of metagenomic and genomic libraries reveals three classes of bacterial enzymes that overcome the toxicity of acrylate. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mewis K., Taupp M., Hallam S.J. A high throughput screen for biomining cellulase activity from metagenomic libraries. J Vis Exp. 2011 doi: 10.3791/2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewin A., Strand T.A., Haugen T., Klinkenberg G., Kotlar H.K., Valla S. Discovery and characterization of a thermostable esterase from an oil reservoir metagenome. Advances in Enzyme Research. 2016;4:68–86. [Google Scholar]
- 51.Nyyssonen M., Tran H.M., Karaoz U., Weihe C., Hadi M.Z., Martiny J.B. Coupled high-throughput functional screening and next generation sequencing for identification of plant polymer decomposing enzymes in metagenomic libraries. Front Microbiol. 2013;4:282. doi: 10.3389/fmicb.2013.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uroz S., Ioannidis P., Lengelle J., Cébron A., Morin E., Buée M. Functional assays and metagenomic analyses reveals differences between the microbial communities inhabiting the soil horizons of a Norway spruce plantation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0055929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uchiyama T., Abe T., Ikemura T., Watanabe K. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat Biotechnol. 2005;23:88–93. doi: 10.1038/nbt1048. [DOI] [PubMed] [Google Scholar]
- 54.Uchiyama T., Miyazaki K. Metagenomic screening for aromatic compound-responsive transcriptional regulators. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meier M.J., Paterson E.S., Lambert I.B. Use of substrate-induced gene expression in metagenomic analysis of an aromatic hydrocarbon-contaminated soil. Appl Environ Microbiol. 2015;82:897–909. doi: 10.1128/AEM.03306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi S.L., Rha E., Lee S.J., Kim H., Kwon K., Jeong Y.S. Toward a generalized and high-throughput enzyme screening system based on artificial genetic circuits. ACS Synth Biol. 2014;3:163–171. doi: 10.1021/sb400112u. [DOI] [PubMed] [Google Scholar]
- 57.Kim H., Kwon K.K., Seong W., Lee S.G. Multi-enzyme screening using a high-throughput genetic enzyme screening system. J Vis Exp. 2016 doi: 10.3791/54059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee D.H., Choi S.L., Rha E., Kim S.J., Yeom S.J., Moon J.H. A novel psychrophilic alkaline phosphatase from the metagenome of tidal flat sediments. BMC Biotechnol. 2015;15:1. doi: 10.1186/s12896-015-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hosokawa M., Hoshino Y., Nishikawa Y., Hirose T., Yoon D.H., Mori T. Droplet-based microfluidics for high-throughput screening of a metagenomic library for isolation of microbial enzymes. Biosens Bioelectron. 2015;67:379–385. doi: 10.1016/j.bios.2014.08.059. [DOI] [PubMed] [Google Scholar]
- 60.Scanlon T.C., Dostal S.M., Griswold K.E. A high-throughput screen for antibiotic drug discovery. Biotechnol Bioeng. 2014;111:232–243. doi: 10.1002/bit.25019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colin P.Y., Kintses B., Gielen F., Miton C.M., Fischer G., Mohamed M.F. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat Commun. 2015;6:10008. doi: 10.1038/ncomms10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Popovic A., Tchigvintsev A., Tran H., Chernikova T.N., Golyshina O.V., Yakimov M.M. Metagenomics as a tool for enzyme discovery: hydrolytic enzymes from marine-related metagenomes. Adv Exp Med Biol. 2015;883:1–20. doi: 10.1007/978-3-319-23603-2_1. [DOI] [PubMed] [Google Scholar]
- 63.Turner N.J. Directed evolution of enzymes for applied biocatalysis. Trends Biotechnol. 2003;21:474–478. doi: 10.1016/j.tibtech.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 64.Waschkowitz T., Rockstroh S., Daniel R. Isolation and characterization of metalloproteases with a novel domain structure by construction and screening of metagenomic libraries. Appl Environ Microbiol. 2009;75:2506–2516. doi: 10.1128/AEM.02136-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goncalves A.C., dos Santos A.C., dos Santos T.F., Pessoa T.B., Dias J.C., Rezende R.P. High yield of functional metagenomic library from mangroves constructed in fosmid vector. Genet Mol Res. 2015;14:11841–11847. doi: 10.4238/2015.October.2.17. [DOI] [PubMed] [Google Scholar]
- 66.Apolinar-Hernandez M.M., Pena-Ramirez Y.J., Perez-Rueda E., Canto-Canche B.B., De Los Santos-Briones C., O'Connor-Sanchez A. Identification and in silico characterization of two novel genes encoding peptidases S8 found by functional screening in a metagenomic library of Yucatan underground water. Gene. 2016;593:154–161. doi: 10.1016/j.gene.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 67.Berlemont R., Pipers D., Delsaute M., Angiono F., Feller G., Galleni M. Exploring the Antarctic soil metagenome as a source of novel cold-adapted enzymes and genetic mobile elements. Rev Argent Microbiol. 2011;43:94–103. doi: 10.1590/S0325-75412011000200005. [DOI] [PubMed] [Google Scholar]
- 68.Kouker G., Jaeger K.E. Specific and sensitive plate assay for bacterial lipases. Appl Environ Microbiol. 1987;53:211–213. doi: 10.1128/aem.53.1.211-213.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim Y.J., Choi G.S., Kim S.B., Yoon G.S., Kim Y.S., Ryu Y.W. Screening and characterization of a novel esterase from a metagenomic library. Protein Expr Purif. 2006;45:315–323. doi: 10.1016/j.pep.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 70.Chu X., He H., Guo C., Sun B. Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl Microbiol Biotechnol. 2008;80:615–625. doi: 10.1007/s00253-008-1566-3. [DOI] [PubMed] [Google Scholar]
- 71.Martin M., Barbeyron T., Michel G., Daniel P., Vandenbol M. Functional screening of a metagenomic library from algal biofilms. Commun Agric Appl Biol Sci. 2013;78:37–41. [PubMed] [Google Scholar]
- 72.Tirawongsaroj P., Sriprang R., Harnpicharnchai P., Thongaram T., Champreda V., Tanapongpipat S. Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J Biotechnol. 2008;133:42–49. doi: 10.1016/j.jbiotec.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 73.Sulaiman S., Yamato S., Kanaya E., Kim J.J., Koga Y., Takano K. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl Environ Microbiol. 2012;78:1556–1562. doi: 10.1128/AEM.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu X.P., Heath C., Taylor M.P., Tuffin M., Cowan D. A novel, extremely alkaliphilic and cold-active esterase from Antarctic desert soil. Extremophiles. 2012;16:79–86. doi: 10.1007/s00792-011-0407-y. [DOI] [PubMed] [Google Scholar]
- 75.Jeon J.H., Lee H.S., Kim J.T., Kim S.J., Choi S.H., Kang S.G. Identification of a new subfamily of salt-tolerant esterases from a metagenomic library of tidal flat sediment. Appl Microbiol Biotechnol. 2012;93:623–631. doi: 10.1007/s00253-011-3433-x. [DOI] [PubMed] [Google Scholar]
- 76.Heath C., Hu X.P., Cary S.C., Cowan D. Identification of a novel alkaliphilic esterase active at low temperatures by screening a metagenomic library from antarctic desert soil. Appl Environ Microbiol. 2009;75:4657–4659. doi: 10.1128/AEM.02597-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Navarro-Fernandez J., Nechitaylo T.Y., Guerrero J.A., Golyshina O.V., Garcia-Carmona F., Sanchez-Ferrer A. A novel platelet-activating factor acetylhydrolase discovered in a metagenome from the earthworm-associated microbial community. Environ Microbiol. 2011;13:3036–3046. doi: 10.1111/j.1462-2920.2011.02581.x. [DOI] [PubMed] [Google Scholar]
- 78.Fan X., Liu X., Huang R., Liu Y. Identification and characterization of a novel thermostable pyrethroid-hydrolyzing enzyme isolated through metagenomic approach. Microb Cell Fact. 2012;11:33. doi: 10.1186/1475-2859-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jeon J.H., Kim S.J., Lee H.S., Cha S.S., Lee J.H., Yoon S.H. Novel metagenome-derived carboxylesterase that hydrolyzes beta-lactam antibiotics. Appl Environ Microbiol. 2011;77:7830–7836. doi: 10.1128/AEM.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng J., Liu C., Liu L., Jin Q. Characterisation of a thermo-alkali-stable lipase from oil-contaminated soil using a metagenomic approach. Syst Appl Microbiol. 2013;36:197–204. doi: 10.1016/j.syapm.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 81.Kok R.G., Christoffels V.M., Vosman B., Hellingwerf K.J. Growth-phase-dependent expression of the lipolytic system of Acinetobacter calcoaceticus BD413: cloning of a gene encoding one of the esterases. J Gen Microbiol. 1993;139:2329–2342. doi: 10.1099/00221287-139-10-2329. [DOI] [PubMed] [Google Scholar]
- 82.De Santi C., Altermark B., Pierechod M.M., Ambrosino L., de Pascale D., Willassen N.P. Characterization of a cold-active and salt tolerant esterase identified by functional screening of Arctic metagenomic libraries. BMC Biochem. 2016;17:1. doi: 10.1186/s12858-016-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tao W., Lee M.H., Yoon M.Y., Kim J.C., Malhotra S., Wu J. Characterization of two metagenome-derived esterases that reactivate chloramphenicol by counteracting chloramphenicol acetyltransferase. J Microbiol Biotechnol. 2011;21:1203–1210. doi: 10.4014/jmb.1107.07034. [DOI] [PubMed] [Google Scholar]
- 84.Yu E.Y., Kwon M.A., Lee M., Oh J.Y., Choi J.E., Lee J.Y. Isolation and characterization of cold-active family VIII esterases from an arctic soil metagenome. Appl Microbiol Biotechnol. 2011;90:573–581. doi: 10.1007/s00253-011-3132-7. [DOI] [PubMed] [Google Scholar]
- 85.Teather R.M., Wood P.J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang W., Archbold T., Kimber M.S., Li J., Lam J.S., Fan M.Z. The porcine gut microbial metagenomic library for mining novel cellulases established from growing pigs fed cellulose-supplemented high-fat diets. J Anim Sci. 2012;90:400–402. doi: 10.2527/jas.53942. [DOI] [PubMed] [Google Scholar]
- 87.Mewis K., Armstrong Z., Song Y.C., Baldwin S.A., Withers S.G., Hallam S.J. Biomining active cellulases from a mining bioremediation system. J Biotechnol. 2013;167:462–471. doi: 10.1016/j.jbiotec.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 88.Zhang L., Fan Y., Zheng H., Du F., Zhang K.Q., Huang X. Isolation and characterization of a novel endoglucanase from a Bursaphelenchus xylophilus metagenomic library. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mai Z., Su H., Yang J., Huang S., Zhang S. Cloning and characterization of a novel GH44 family endoglucanase from mangrove soil metagenomic library. Biotechnol Lett. 2014;36:1701–1709. doi: 10.1007/s10529-014-1531-4. [DOI] [PubMed] [Google Scholar]
- 90.Nimchua T., Thongaram T., Uengwetwanit T., Pongpattanakitshote S., Eurwilaichitr L. Metagenomic analysis of novel lignocellulose-degrading enzymes from higher termite guts inhabiting microbes. J Microbiol Biotechnol. 2012;22:462–469. doi: 10.4014/jmb.1108.08037. [DOI] [PubMed] [Google Scholar]
- 91.Nacke H., Engelhaupt M., Brady S., Fischer C., Tautzt J., Daniel R. Identification and characterization of novel cellulolytic and hemicellulolytic genes and enzymes derived from German grassland soil metagenomes. Biotechnol Lett. 2012;34:663–675. doi: 10.1007/s10529-011-0830-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bastien G., Arnal G., Bozonnet S., Laguerre S., Ferreira F., Faure R. Mining for hemicellulases in the fungus-growing termite Pseudacanthotermes militaris using functional metagenomics. Biotechnol Biofuels. 2013;6:78. doi: 10.1186/1754-6834-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matsuzawa T., Kaneko S., Yaoi K. Screening, identification, and characterization of a GH43 family beta-xylosidase/alpha-arabinofuranosidase from a compost microbial metagenome. Appl Microbiol Biotechnol. 2015;99:8943–8954. doi: 10.1007/s00253-015-6647-5. [DOI] [PubMed] [Google Scholar]
- 94.Gong X., Gruniniger R.J., Forster R.J., Teather R.M., McAllister T.A. Biochemical analysis of a highly specific, pH stable xylanase gene identified from a bovine rumen-derived metagenomic library. Appl Microbiol Biotechnol. 2013;97:2423–2431. doi: 10.1007/s00253-012-4088-y. [DOI] [PubMed] [Google Scholar]
- 95.Lee Y.S., Seo S.H., Yoon S.H., Kim S.Y., Hahn B.S., Sim J.S. Identification of a novel alkaline amylopullulanase from a gut metagenome of Hermetia illucens. Int J Biol Macromol. 2016;82:514–521. doi: 10.1016/j.ijbiomac.2015.10.067. [DOI] [PubMed] [Google Scholar]
- 96.Mai Z., Su H., Zhang S. Isolation and characterization of a glycosyl hydrolase family 16 beta-agarase from a mangrove soil metagenomic library. Int J Mol Sci. 2016;17:1360. doi: 10.3390/ijms17081360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang M., Lai G.L., Nie Y., Geng S., Liu L., Zhu B. Synergistic function of four novel thermostable glycoside hydrolases from a long-term enriched thermophilic methanogenic digester. Front Microbiol. 2015;6:509. doi: 10.3389/fmicb.2015.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Biver S., Stroobants A., Portetelle D., Vandenbol M. Two promising alkaline beta-glucosidases isolated by functional metagenomics from agricultural soil, including one showing high tolerance towards harsh detergents, oxidants and glucose. J Ind Microbiol Biotechnol. 2014;41:479–488. doi: 10.1007/s10295-014-1400-0. [DOI] [PubMed] [Google Scholar]
- 99.Uchiyama T., Miyazaki K., Yaoi K. Characterization of a novel beta-glucosidase from a compost microbial metagenome with strong transglycosylation activity. J Biol Chem. 2013;288:18325–18334. doi: 10.1074/jbc.M113.471342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jiang C., Ma G., Li S., Hu T., Che Z., Shen P. Characterization of a novel beta-glucosidase-like activity from a soil metagenome. J Microbiol. 2009;47:542–548. doi: 10.1007/s12275-009-0024-y. [DOI] [PubMed] [Google Scholar]
- 101.Fang Z., Fang W., Liu J., Hong Y., Peng H., Zhang X. Cloning and characterization of a beta-glucosidase from marine microbial metagenome with excellent glucose tolerance. J Microbiol Biotechnol. 2010;20:1351–1358. doi: 10.4014/jmb.1003.03011. [DOI] [PubMed] [Google Scholar]
- 102.Jiang C.J., Chen G., Huang J., Huang Q., Jin K., Shen P.H. A novel beta-glucosidase with lipolytic activity from a soil metagenome. Folia Microbiol (Praha) 2011;56:563–570. doi: 10.1007/s12223-011-0083-4. [DOI] [PubMed] [Google Scholar]
- 103.Uchiyama T., Miyazaki K. Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr Opin Biotechnol. 2009;20:616–622. doi: 10.1016/j.copbio.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 104.Xiao H., Bao Z., Zhao H. High throughput screening and selection methods for directed enzyme evolution. Ind Eng Chem Res. 2015;54:4011–4020. doi: 10.1021/ie503060a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kodzius R., Gojobori T. Single-cell technologies in environmental omics. Gene. 2016;576:701–707. doi: 10.1016/j.gene.2015.10.031. [DOI] [PubMed] [Google Scholar]
- 106.Kintses B., Hein C., Mohamed M.F., Fischlechner M., Courtois F., Laine C. Picoliter cell lysate assays in microfluidic droplet compartments for directed enzyme evolution. Chem Biol. 2012;19:1001–1009. doi: 10.1016/j.chembiol.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 107.Uchiyama T., Miyazaki K. Substrate-induced gene expression screening: a method for high-throughput screening of metagenome libraries. Methods Mol Biol. 2010;668:153–168. doi: 10.1007/978-1-60761-823-2_10. [DOI] [PubMed] [Google Scholar]
- 108.Yun J., Ryu S. Screening for novel enzymes from metagenome and SIGEX, as a way to improve it. Microb Cell Fact. 2005;4:8. doi: 10.1186/1475-2859-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stepanauskas R., Sieracki M.E. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc Natl Acad Sci U S A. 2007;104:9052–9057. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ibrahim S.F., van den Engh G. Flow cytometry and cell sorting. In: Kumar A., Galaev I.Y., Mattiasson B., editors. Cell separation: fundamentals, analytical and preparative methods. Springer, Berlin Heidelberg; Berlin, Heidelberg: 2007. pp. 19–39. [Google Scholar]
- 111.Dietrich J., McKee A., Keasling J. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu Rev Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- 112.Shah R.K., Shum H.C., Rowat A.C., Lee D., Agresti J.J., Utada A.S. Designer emulsions using microfluidics. Mater Today. 2008;11:18–27. [Google Scholar]
- 113.Huebner A., Olguin L.F., Bratton D., Whyte G., Huck W.T.S., de Mello A.J. Development of quantitative cell-based enzyme assays in microdroplets. Anal Chem. 2008;80:3890–3896. doi: 10.1021/ac800338z. [DOI] [PubMed] [Google Scholar]
- 114.Gielen F., Hours R., Emond S., Fischlechner M., Schell U., Hollfelder F. Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS) Proc Natl Acad Sci U S A. 2016;113:E7383–E7389. doi: 10.1073/pnas.1606927113. [DOI] [PMC free article] [PubMed] [Google Scholar]